Abstract
In this study we explored the use of super-resolution track density imaging (TDI) for neuroanatomical characterization of the adult zebrafish brain. We compared the quality of image contrast and resolution obtained with T2* magnetic resonance imaging (MRI), diffusion tensor based imaging (DTI), TDI, and histology. The anatomical structures visualized in 5 μm TDI maps corresponded with histology. Moreover the super-resolution property and the local-directional information provided by directionally-encoded color TDI facilitated delineation of a larger number of brain regions, commissures and small white matter tracks when compared to conventional MRI and DTI. In total, we were able to visualize 17 structures that were previously unidentifiable using MR microimaging, such as the four layers of the optic tectum. This study demonstrates the use of TDI for characterization of the adult zebrafish brain as a pivotal tool for future phenotypic examination of transgenic models of neurological diseases.
Keywords: zebrafish, brain, magnetic resonance, diffusion-weighted imaging, track-density imaging, probabilistic tractography
Introduction
Zebrafish are an established experimental model for the investigation of neurodegenerative disorders. Large clutch sizes, transparent larval stages, and the ability to perform forward or reverse genetic analyses have particularly facilitated the use of embryonic and larval zebrafish (Lieschke and Currie 2007). Mature zebrafish, which possess well-developed endocrine, sensory, and motor systems (Bally-Cuif and Vernier 2010), a range of complex behaviours such as social interactions (Saverino and Gerlai 2008), learning and memory (Gomez-Laplaza and Gerlai 2010; Norton and Bally-Cuif 2010; Al-Imari and Gerlai 2008) and a physiology similar to mammals (Bally-Cuif and Vernier 2010), have been recognized as beneficial to investigations of neurological diseases (Alfaro et al. 2011; Siebel et al. 2011; Wong et al. 2010; Keller and Murtha 2004; Mathur and Guo 2010; Ramirez et al. 2012; Haud et al. 2011; Williams et al. 2011). As adult zebrafish continue to be used as an experimental model, new techniques are being developed to better characterize their behaviour (Cachat et al. 2011), neuronal activity (Pineda et al. 2011), neurochemistry (Kabli et al. 2009) and neuromorphology (Rao et al. 2009; Ullmann et al. 2010b; Ullmann et al. 2010a; Kabli et al. 2006).
Magnetic resonance imaging (MRI) is an important technique for investigating the morphology, connectivity, and function of the brain. Although initially developed for imaging humans, MRI has become a key platform for imaging small animals and is now routinely performed on mice (Richards et al. 2011; Ullmann et al. 2012), rats (Petiet et al. 2007), and zebrafish (Ullmann et al. 2010b; Kabli et al. 2009; Ullmann et al. 2010a). However, MRI becomes increasingly challenging as the size of the animal decreases. To delineate ever-smaller brain regions, higher resolution and contrast are required. Tradeoffs between image acquisition parameters must carefully be considered to accommodate these requirements. For example, the adult zebrafish brain has a relatively small size (5mm3), with a volume approximately 64 times smaller than that of an adult mouse brain. Consequently, an increase of at least four times image resolution in each plane is necessary to resolve the zebrafish brain microstructures. This, however, would result in a reduction in the signal to noise ratio by a factor of eight. In this case, ultra-high resolution imaging is undertaken on fixed brains to enable longer scan times and an increased number of averages.
A new method called track density imaging (TDI) has recently been developed, which offers both high resolution and novel contrast (Calamante et al. 2010) that cannot be obtained with conventional MRI sequences. During post-processing, TDI incorporates data from whole-brain fiber tracking to yield image maps at a higher resolution than the acquired data, i.e. it achieves super-resolution (Calamante et al. 2011). These TDI maps can also encode information on fiber directionality, which can be displayed in directionally-encoded colour (DEC) (Calamante et al. 2010). In particular, the technique of DEC short-tracks TDI (DEC stTDI) is a modified variant specifically designed to provide high contrast fiber-direction information, while avoiding intensity saturation from dense white matter structures (Calamante et al. 2012b). The high resolution and high contrast properties of these maps make this technique ideal for the study of the fine structures in the zebrafish brain. In this paper, we therefore explore the use of super-resolution track density imaging (TDI) as a contrast mechanism for enhanced characterization of the zebrafish brain, and demonstrate its potential role in phenotypic examination of animal models of neurodegenerative disorders.
Materials and Methods
Sample Preparation
All animal experiments were performed according to procedures approved by the University of Queensland Animal Ethics Committee (AEC No SBMS/074/08). Four adult wildtype zebrafish (AB strain) were euthanized with an overdose of tricaine methane sulfonate (MS222, 1:10,000 in water). The dorsal cranium was removed from the fish and the head was immersion fixed and stained in a solution of 4% paraformaldehyde in PBS and 0.5% Magnevist (Bayer) for 12 hours. The brain and ventral cranium, were mounted onto a small piece of plastic, and then placed in a 5 mm NMR tube and filled with Fomblin (perfluoropolyether, Ausimont, Morristown, NJ, USA) for imaging. The hydrophobic nature of Fomblin, an inert perfluoropolyether oil solution, prevents leaching of Magnevist out from the sample to the medium.
MR Data Acquisition
Diffusion-weighted magnetic resonance images were acquired using a Bruker (Ettlingen, Germnay) 16.4 Tesla vertical magnet with a bore diameter of 89 mm, a Bruker micro 2.5 imaging gradient, and a custom-made horizontal 5 × 4 mm solenoid radiofrequency (rf) coil. Sample temperature was maintained at 22°C using a circulating water gradient cooling system. Three-dimensional diffusion-weighted spin-echo images were acquired with the following parameters: TR/TE 450/21.83 ms, FOV 6.9 × 2.6 × 2.6 mm3, acquisition matrix 144 × 54 × 54, NEX 4, resulting in an isotropic resolution of 48 μm. Each dataset was composed of two low diffusion weighted images (b= 0 s/mm2) and thirty high diffusion weighted (b=5000 s/mm2) images with encoding gradient (δ/Δ = 2.5/14 ms) vectors uniformly distributed using an electrostatic approach (Jones et al. 1999). Total imaging time was 46 hours and 40 minutes. The 3D data set was symmetrically zero-filled to 288 × 108 × 108 matrix resulting in an isotropic resolution of 24 μm.
For each sample, two high-resolution anatomical scans were also acquired with a 3D FLASH (Frahm et al. 1986) gradient echo T2*-weighted image sequence. The first scan had a 48 μm isotropic image resolution and the following imaging parameters: TR/TE = 50/8.3 ms; matrix 128 × 54 × 54; NEX = 16, and 39 min imaging time; while the second scan was obtained at a 10 μm isotropic resolution TR/TE = 50/8.3 ms; matrix 592 × 260 × 260; NEX = 16, and a 15 hour imaging time.
Super-resolution TDI Map Reconstruction
To correct for possible drift during the long scan time, a 3D rigid body 3-parameter model registration with correlation ratio cost function was performed using the first b0 image in each dataset as a template using the program FSL FLIRT 4.1.9 (http://fsl.fmrib.ox.ac.uk/fsl/fsl-4.1.9). Fiber-tracks were then generated using the program MRtrix 0.2.9 (brain.org.au/software/mrtrix) (Tournier et al. 2012), which uses constrained spherical deconvolution (CSD) to resolve crossing-fibers by estimating the fiber orientation distribution (FOD) at each voxel (Tournier et al. 2007). Probabilistic fiber-tracking (Behrens et al., 2003) was performed using the iFOD1 algorithm (Tournier et al. 2010), with 0.024 mm step-size, maximum angle between steps = 45°, lmax = 6. Fiber-tracks were terminated when they exited the brain or when the FOD < 0.02. A whole brain CSD probabilistic-tractography dataset of 60 million short fiber tracks was calculated, with a minimum track length of 2-voxel size and a maximum length of 10 voxel size (Calamante et al. 2012b). This whole-brain tractography dataset was then used to generate DEC stTDI maps, using a 5 μm isotropic (super-resolution) voxel size. In these maps, the colour in each voxel is determined by averaging the colours of all track segments contained within the voxel. Therefore, the colour-coding indicates the local fiber orientation (red: rostral-caudal, green: medial-lateral, blue: dorsal-ventral).
For the purpose of comparison, the same diffusion weighted imaging (DWI) data set was also analysed using the diffusion tensor model (Basser 1995), and the results displayed as DEC fractional anisotropy (FA) maps (Pajevic and Pierpaoli 1999). These maps have the same resolution as the acquired DWI data, and the colour encodes the direction of the principal eigenvector of the diffusion tensor.
Bootstrap method was not performed to test the general stTDI map reproducibility due to high demand on computational resources. To calculate the mean and deviations of stTDI map dispersion values using bootstrap method (e.g. as described in Jeurissen et al., 2011), this would require at least 50 repetitions of 60 million streamlines of stTDI runs or generation of 3billion streamlines. Instead, to characterise in more detail the CNR behaviour with increasing number of tracks and how this affect visualisation of structures in the zebrafish brain, stTDI maps were generated using 1, 5, 10, 20, 40 and 60 millions tracks. Excellent brain structure identification, contrast and colour saturation were observed in stTDI generated using > 20 million tracks. 60 million stTDI for all dataset was chosen at the end to ensure superior reproduction of stTDI maps (see Supplementary Fig. 1). Each stTDI calculation was completed in 6 hours using 6-core Xeon workstation using 70 GB RAM.
Targeted Probabilistic Fiber-Tracking of White Matter Structures
Due to the rich contrast present in the DEC stTDI maps, they can be used to delineate the extent of white matter structures. Specific regions of interest (ROIs) were identified and then combined with target ROIs to assess structural connectivity using targeted tractography (Mori and van Zijl 2002). Approximately 5,000 streamlines were generated from each seed ROI using similar probabilistic fiber-tracking parameters as above, except that full-length streamlines (in contrast to short tracks) were generated to visualise the extent of the structural connection. In particular, targeted tractography was carried out for seed ROIs located in the forebrain and midbrain.
Histology
Upon completion of diffusion and anatomical scans two scanned brains were dissected from their respective craniums and embedded in Paraplast Plus tissue embedding medium (Oxford Labware), sectioned at 10 μm, stained with luxol fast blue for myelin and counterstained with cresyl violet for neurons. Briefly, the sections were deparaffinised in xylene, rehydrated in ethanol and water and then incubated in a 1% luxol fast blue stain for 4 hours. Subsequently the sections differentiated in 0.05% Lithium carbonate and 70% ethanol solutions. Slides were then incubated in a 0.25% cresyl violet solution for 3 min before dehydrating, clearing, and mounting. Histological sections closest to the TDI slices were visually identified and digitally scanned for comparison using a Zeiss Axio Imager Z2 microscope fitted with a motorised stage and a Metafer Slide Scanning Platform (MetaSytems, Germany). A Zeiss 20× 0.8NA PlanApochromatic objective and Metasystems CoolCube camera were used for image acquisition. All images were processed using Adobe Photoshop CS5 and figures compiled with Adobe Illustrator CS5.
Results
Figure 1 demonstrates the results of whole-brain DEC stTDI processing at 5μm resolution produced from 48μm 30-directions HARDI data acquired at 16.4T for one of the brain samples studied; similar results were obtained for the other samples (data not shown). Directionally encoded color (DEC) stTDI maps enable exceptional visualization of fine white matter tracks, characterizing not only the spatial extent of numerous brain structures, but also fiber directionality.
Figure 1:
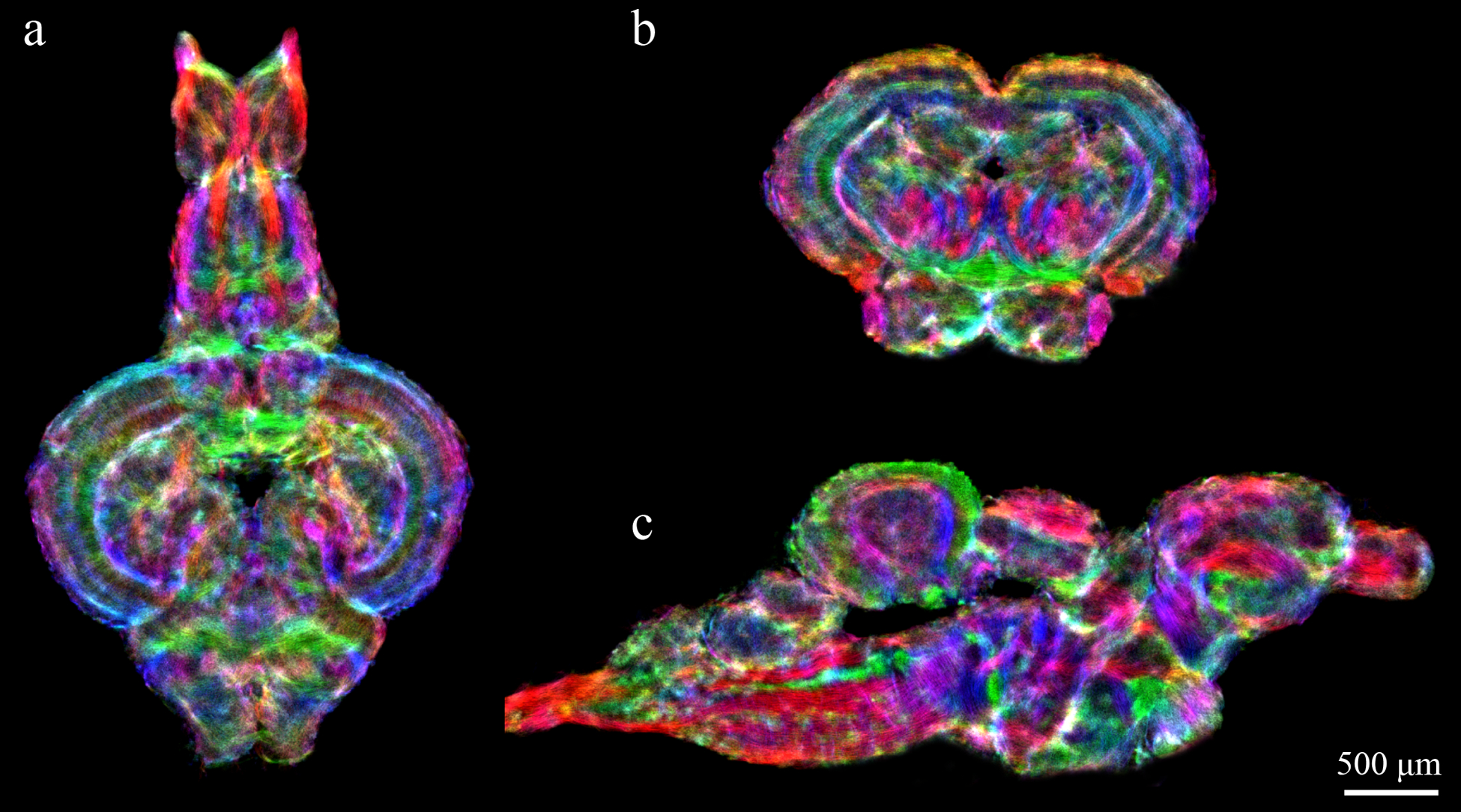
Sample DEC stTDI maps (5μm isotropic resolution) calculated from whole-brain fiber-tracking results, which were generated from DWI data acquired on a 16.4T scanner (at 48μm isotropic resolution). Horizontal section (a), axial section (b), and sagittal section (c) of whole brain fiber-tracking results. Color-coding indicates the local fiber orientation (with red: rostral-caudal, green: medial-lateral, and blue: dorsal-ventral.
Figure 2 shows a comparison of T2*-weighted images, DEC FAmaps, and DEC-stTDI maps of a series of slices through the zebrafish brain. While T2* images are of high-resolution (10μm3), limited contrast between tissue types, particularly noticeable in the telencephalon (Fig. 2a,b), restricts detailed delineation of substructures. In the DEC FA maps contrast is enhanced by local directional information but low-resolution impedes detailed visualization. In contrast, the high resolution and high contrast in DEC-stTDI maps results in vastly improved definition of fiber tracks, cellular layers and nuclei.
Figure 2:
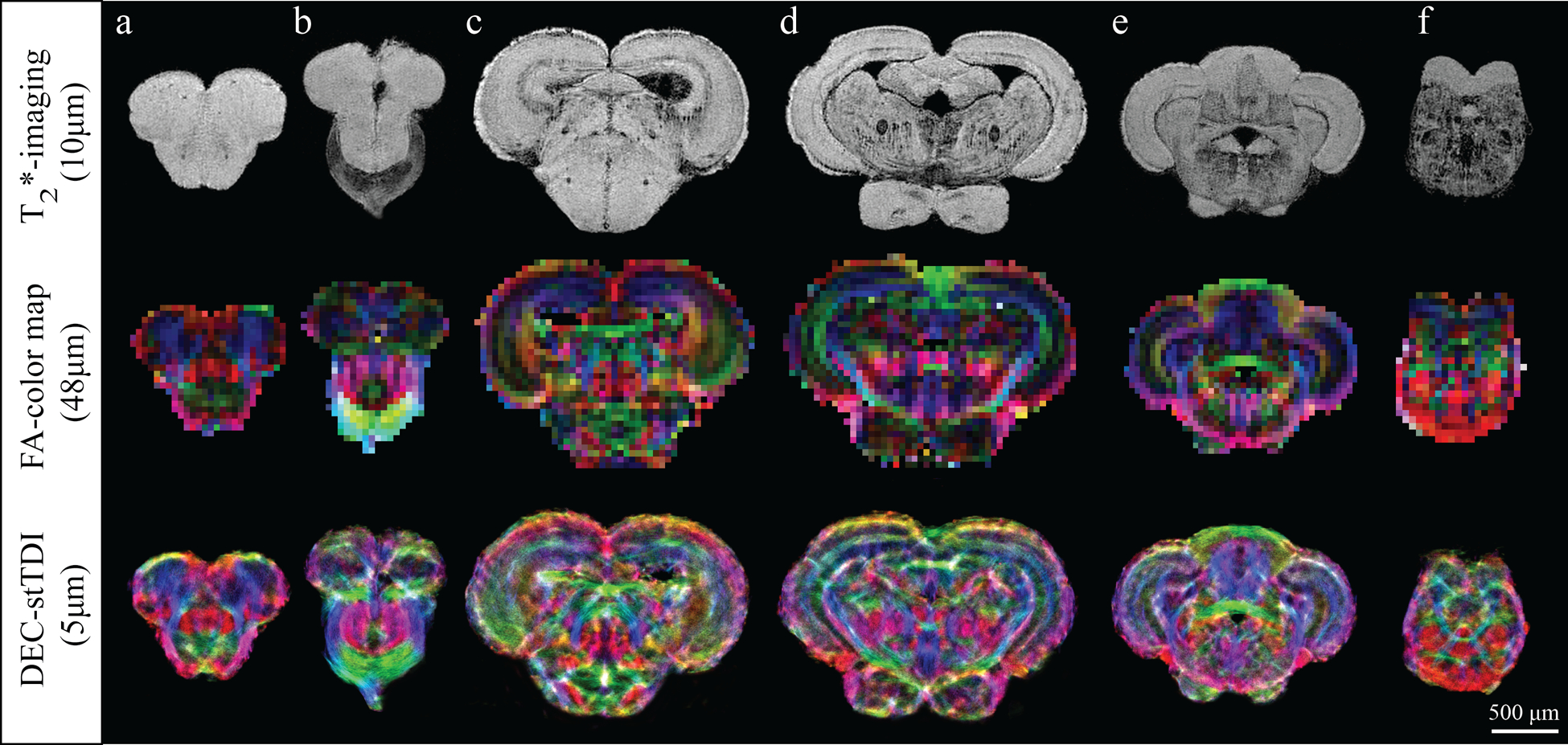
Comparison of T2*-weighted images, DEC FA maps, and DEC-stTDI maps in six different axial slices of the zebrafish brain. Top row: T2*-images with a 10μm isotropic resolution. Middle row: corresponding DEC FA maps with a 48μm isotropic resolution. Bottom row: corresponding super-resolution DEC-stTDI maps with 5μm isotropic resolution. DEC-stTDI maps were generated from the original 48μm HARDI using a 5μm super-resolution grid and 60 million tracks. The color-coding indicates the primary local orientation (red: rostral-caudal, green: medial-lateral, blue: dorsal-ventral).
Figures 3 and 4 demonstrate the anatomical consistency between super-resolution track density imaging and conventional histology of the zebrafish brain. Three selected regions were shown at higher magnification, through the telencephalon and optic tectum with corresponding histological sections at approximately the same position. In Figure 3 numerous structural subdivisions could be visualized based upon color changes in the directionally encoded color TDI maps. In our maps, the color green indicates a medial-lateral fiber orientation, red to a rostral-caudal fiber orientation, and blue a dorsal-ventral fiber orientation. Color variations indicate fiber tracks with orientations in between the major directions listed above. In Figure 3a,b the subdivisions of the dorsal telencephalon (D) can be delineated as follows: the medial zone (Dm) appears bright yellow, the ventral surface of the dorsal zone (Dd) appears green, the lateral zone (Dl) appears bright pink at the surface and dark more medially, the posterior zone (Dp) characterized by the hyperintense white color; and the remaining central area the central zone (Dc). In the ventral telencephalon, the anterior part of the parvicellular preoptic nucleus (PPa) can also be identified as bright pink region. DEC stTDI also enabled characterization of numerous fiber tracks and commissures, such as the lateral forebrain bundle (LFB), which is visible in blue as it projects dorso-ventrally through Dc to the level of the anterior commissure, where it then extends caudally into the diencephalon (Fig. 3a,b); the anterior commissure which can be distinguished as it divides into its dorsal (CantD) and ventral (CantV) components (Fig. 1c,d); and the caudal end of the medial olfactory tract (MOT), which is shown in red, lateral to dorsal anterior commissure (Fig. 3c,d).
Figure 3:
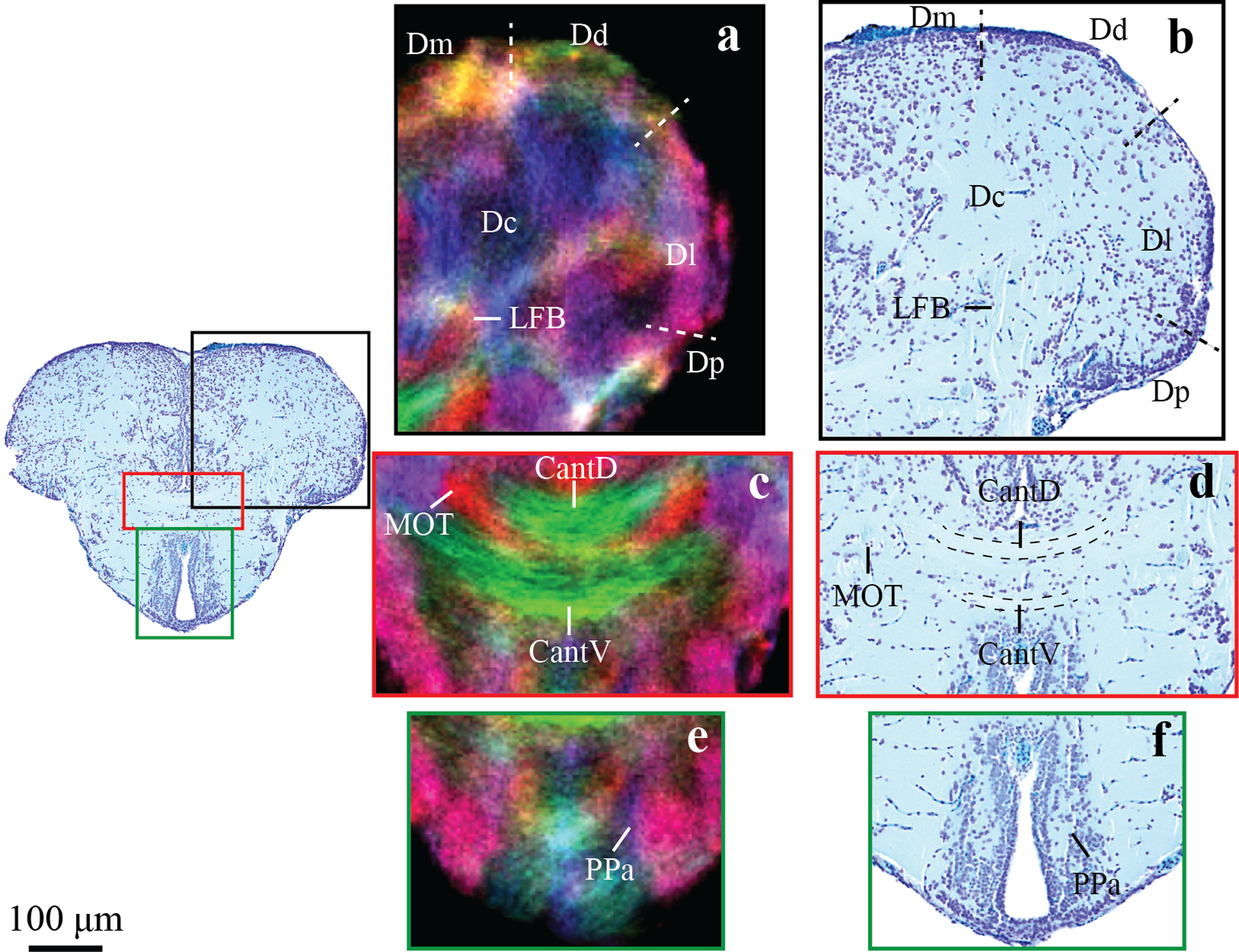
Zoomed in regions of an axial image through the telencephalon in DEC stTDI maps (a,c,e) and histological sections (b,d,f). Identified structures include the dorsal anterior commissure, CantD; ventral anterior commissure, CantV; central zone of the dorsal telencephalon, Dc; dorsal zone of the dorsal telencephalon, Dd; lateral zone of the dorsal telencephalon, Dl; medial zone of the dorsal telencephalon, Dm; posterior zone of the dorsal telencephalon, Dp; lateral forebrain bundle, LFB; anterior part of the parvocellular preoptic nucleus, PPa. The DEC stTDI maps have 5μm isotropic resolution, and the color-coding indicates the local orientation (red: rostral-caudal, green: medial-lateral, blue: dorsal-ventral).
Figure 4:
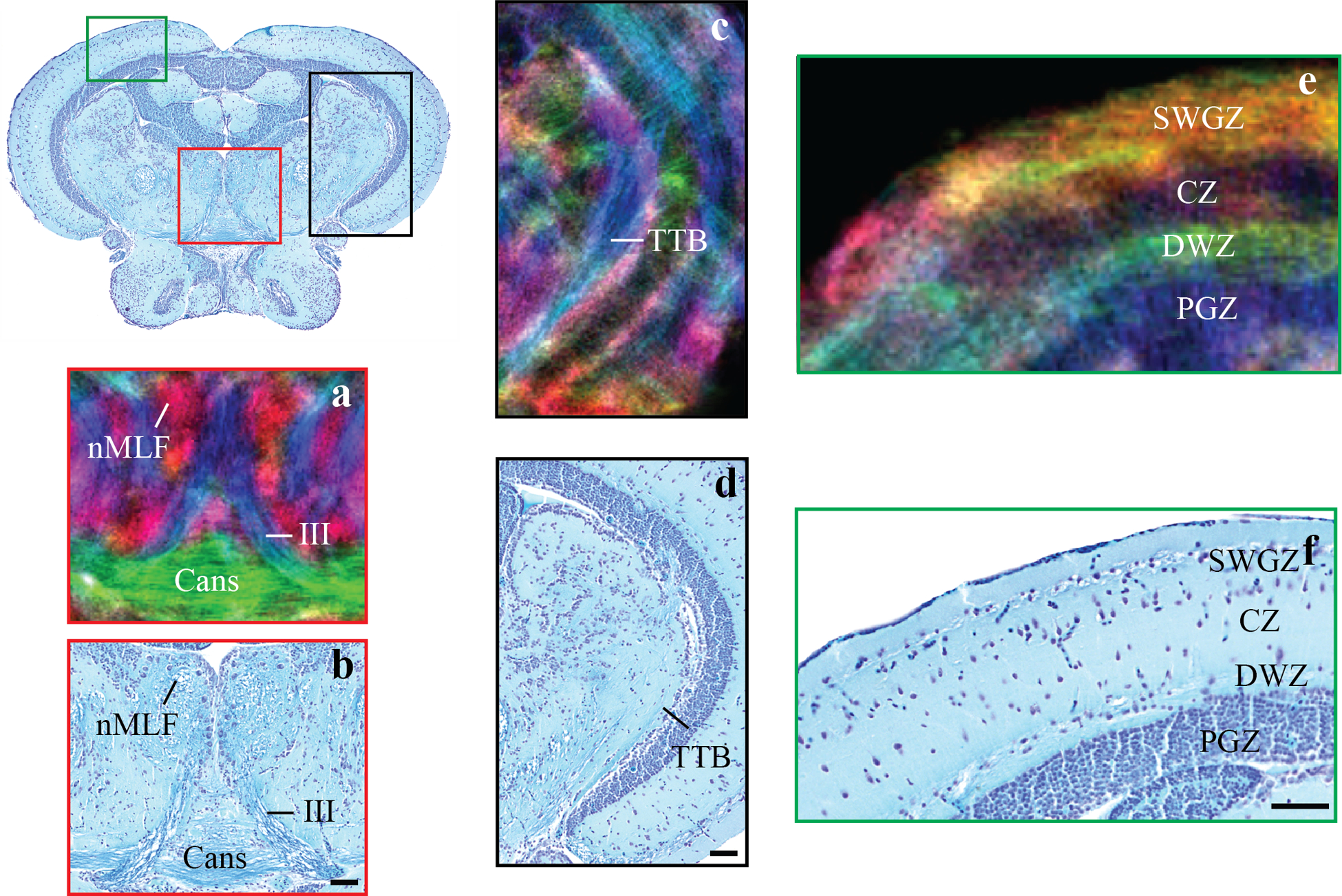
Zoomed in regions of an axial image through the mesencephalon in DEC stTDI maps (a, c, e) and histological sections (b,d,f). Identified structures include: the oculomotor nerves, III; the commissura ansulata, Cans; the central zone of the optic tectum, CZ; the deep white zone of the optic tectum, DWZ, the nucleus of the medial longitudinal fascicle, nMLF; the periventricular grey zone of the optic tectum, PGZ; and the superficial grey and white zone of the optic tectum, SWGZ. The DEC stTDI maps have 5μm isotropic resolution, and the color-coding indicates the local orientation (red: rostral-caudal, green: medial-lateral, blue: dorsal-ventral).
A similar correspondence is also found when examining the optic tectum. DEC stTDI maps clearly reveal nuclei, white matter tracks and commissures including the nucleus of the medial longitudinal fascicle (nMLF, Fig 4a,b), the oculomotor nerves (III, Fig 4a,b), the fiber-tracks that make up the commissura ansulata (Cans, Fig 4a, b), and the tractus tectobulbaris (TTB, Fig 4c,d). The distinct lamination pattern of the teleost optic tectum can also be visualized with DEC stTDI. The findings from DEC stTDI maps are consistent with previous studies (Kishida 1979; Laufer and Vanegas 1974; Corbo et al. 2012) which have shown the optic tectum to contain four primary cellular layers, the superficial grey and white zone (SWGZ), the central zone (CZ), the deep white zone (DWZ) and the periventricular grey zone (PGZ) (Northcutt 1983)
Figure 5 demonstrates the enhanced ability to identify neuroanatomical structures in the zebrafish brain. Many of these structures could not be delineated in the MRI-based zebrafish brain atlas (Ullmann et al. 2010b). In particular, a greater number of fiber tracks and commissures could be delineated and a complete list can be found in Table 1. TDI also facilitated the visualization of a number of additional brain regions not visible on T2* imaging, including the caudal lobe of the cerebellum (LCa), caudal octavolateralis nucleus (CON), and the different cellular layers in the olfactory bulb.
Figure 5:
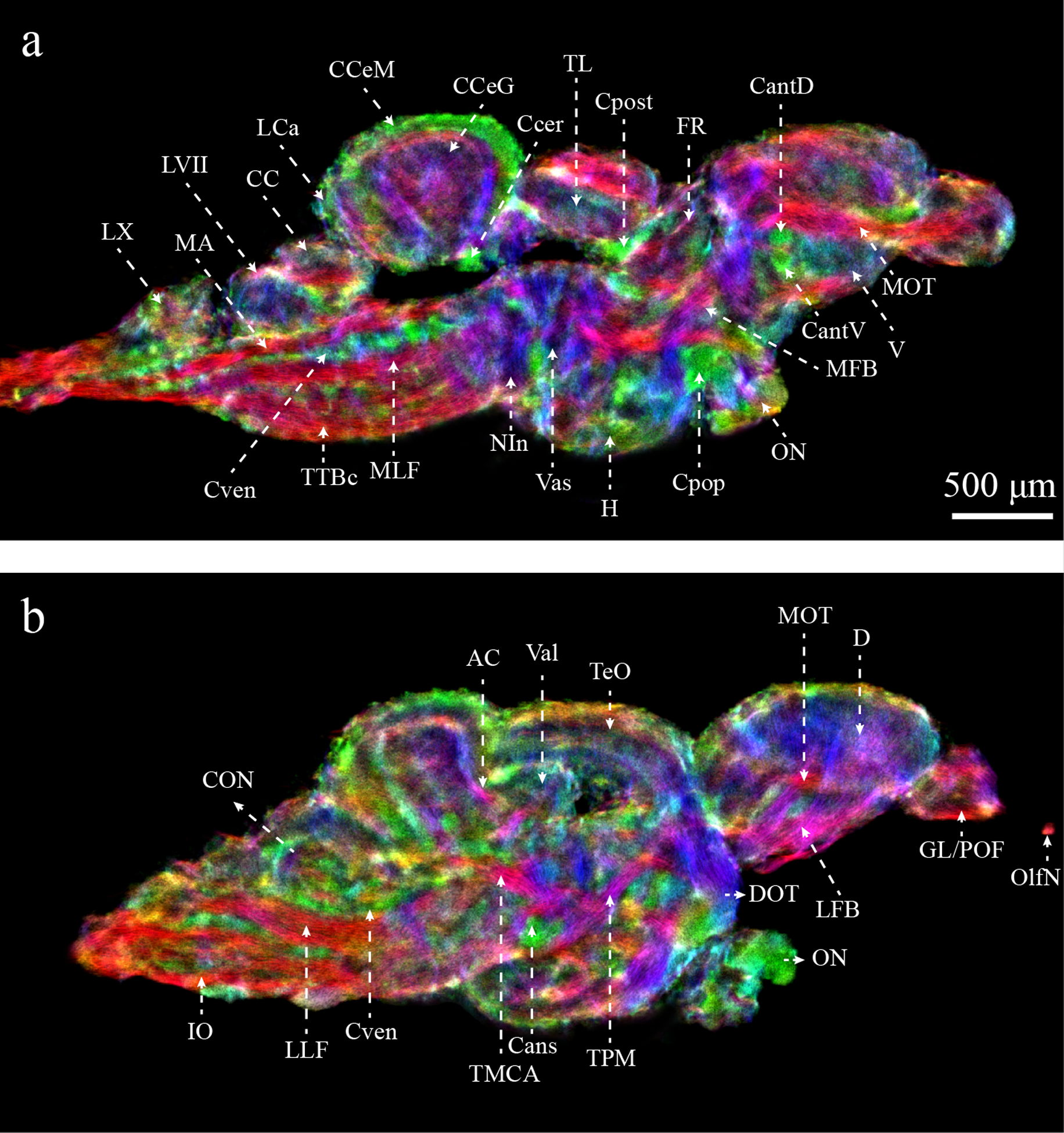
Enhanced characterization of the zebrafish brain from DEC-stTDI images (5μm isotropic resolution). Identified structures include: AC, anterior cerebellar tract; Cans, ansulate commissure; CantD, anterior commissure, dorsal part; CantV, anterior commissure, ventral part; CC, cerebellar crest; CCeG, cerebellar corpus, granular layer; CCeM, cerebellar corpus, molecular layer; Ccer, cerebellar commissure; CON, caudal octavolateralis nucleus; Cpop, postoptic commissure; Cpost, posterior commissure; Cven, ventral rhombencephalic commissure; DOT, dorsomedial optic tract; FR, habenula-interpeduncular tract; GL/POF, glomerular/primary olfactory layers; IO, inferior olive; LCa, caudal lobe of the cerebellum; LFB, lateral forebrain bundle; LLF, lateral longitudinal fascicle; LVII, facial lobe; LX, vagal lobe; MA, Mauthner axon; MFB, medial forebrain bundle; MLF, medial longitudinal fascicle; MOT, medial olfactory tract; NIn, interpeduncular nucleus; OlfN, olfactory nerve; ON, optic nerve; TeO, optic tectum; TL, longitudinal torus; TMCA, anterior mesencephalon-cerebellar tract; TPM, pretecto-mammillary tract; TTBc, crossed tecto-bulbar tract; Val, lateral division of the valvula cerebelli; Vas, vascular lacuna of area postrema. Color-coding indicates the local orientation (red: rostral-caudal, green: medial-lateral, blue: dorsal-ventral).
Table 1.
List of fiber tracks visible with DEC stTDI.
| Commissures | Fiber Tracts | Nerves |
|---|---|---|
| Ansulate commissure | Medial olfactory tract | Oculomotor nerve |
| Dorsal anterior commissure* | Lateral forebrain bundle* | Trochlear nerve* |
| Ventral anterior commissure* | Medial forebrain bundle* | Octaval nerve* |
| Habenular commissure | Pretecto-mammillary tract | Anterior lateral line* |
| Horizontal commissure* | Lateral longitudinal fascicle | Posterior lateral line* |
| Posterior commissure* | Medial longitudinal fascicle | |
| Optic chiasma | Crossed tecto-bulbar tract* | |
| Supraoptic commissure* | Uncrossed tecto-bulbar tract | |
| Secondary gustatory commissure* | Anterior mesencephalo-cerebellar tract | |
| Ventral rhombencephalic commissure* | Anterior cerebellar tract* | |
| Posterior cerebellar tract | ||
| Secondary gustatory tract | ||
| Inferior olive* | ||
| Optic tract | ||
| Mauthner axon* |
Indicates fiber tracts previously unidentifiable with magnetic resonance histology.
Figure 6 shows the result of targeted tractography that was generated from specific seeding regions of interest (ROIs) identified on the DEC stTDI maps. The following white matter fiber-track profiles were generated: (1) The medial (Fig 6e) and lateral (Fig 6c) olfactory tracts, which are shown in red/pink and yellow/orange, respectively. (2) The dorsal and ventral anterior commissures, which are shown in blue and green, respectively (Fig. 6b). (3) The oculomotor nerve which is shown in pastel green and pink (Fig. 6g). (4) The crossed tecto-bulbar tract shown in purple (Fig. 6g). Fiber tracking throughout the brain confirms established zebrafish neuroanatomy. Fig 6. visualizes nicely the medial olfactory tract coursing more ventrally than the lateral olfactory tract and the oculomotor nerves projecting from between the tectum and hypothalamus dorsally to the oculmotor nerve that is located below the tectal ventricle.
Figure 6:
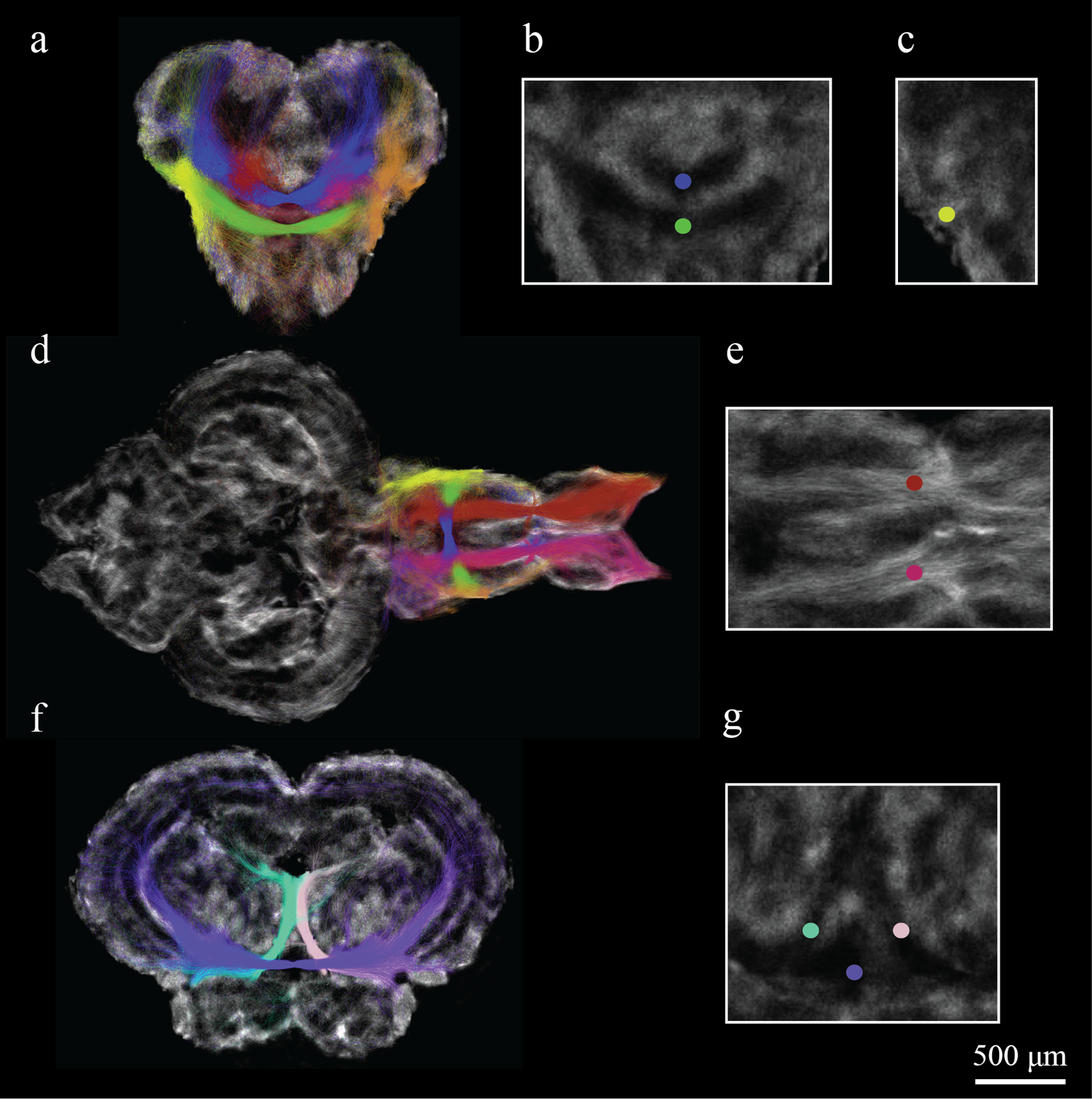
Targeted tractography for seeds defined in the forebrain and midbrain of the zebrafish brain. Seed ROIs and fiber tracks are labeled with different colors including: (a-e) red/pink for the medial and lateral olfactory tracts, blue/green for the dorsal and ventral anterior commissure; (f, g) pastel green/pink for the oculomotor nerves and purple for the crossed tecto-bulbar tract. Fiber tracking illustrates nicely the connections that the olfactory tracts make between the olfactory bulbs and the ventral telencephalon with a few connections projecting into the diencephalon.
Discussion
This study demonstrates the role of super-resolution track density imaging (stTDI) for high contrast whole-brain imaging and for guiding target probabilistic tractography to reveal specific white matter structures in the adult zebrafish. We compared DEC stTDI maps with conventional T2* images, diffusion tensor based images (DEC FA), and histological sections to demonstrate the efficacy of DEC stTDI maps for neuroanatomical characterization of the zebrafish brain. The structures visualized in the DEC stTDI maps were consistent with our histological analysis, supporting the veracity of the anatomical contrast in these maps, which is in agreement with previous TDI studies in the mouse (Calamante et al. 2012b) and human (Calamante et al. 2012a) brains. Moreover the super-resolution property and the local-directional information provided by DEC stTDI, combined with the high signal to noise ratio provided by ultra-high MRI at 16.4T, enabled enhanced examination of the zebrafish brain.
Detailed descriptions of adult zebrafish brain anatomy and connections have already been established with histology (Wullimann et al. 1996; Rink and Wullimann 2004; Mueller et al. 2011) and conventional MRI (Ullmann et al. 2010b), yet both methods possess shortcomings, which limit their use. In conventional histology, a large library of markers exists facilitating the identification of many neuroanatomical structures. However, the low resolution along the slice orientation and the difficulty of obtaining a perfect series of sections in a single brain limits whole brain analyses (Yang et al. 2012). MRI lacks a library of cellular markers but permits acquisition of isotropic images at a lower resolution than microscopy. As a result, researchers rely on a variety of pulse sequences to exploit variations in either proton density and tissue relaxation properties (Johnson et al. 2010; McLaren et al. 2009; Johnson et al. 2012), and/or a limited number of contrast agents. To date the majority of studies have utilized gadolinium-based contrast agents, such as Magnevist® and Pro-Hance®, however Manganese, a functional contrast agent (Koretsky and Silva 2004) (Leergaard et al. 2003), and more recently potassium dichromate (Rowland 2011; Zhang et al. 2010) and myelin-specific (Frullano et al. 2012) contrast agents have also been used.
In contrast to T1/T2* MRI sequences, TDI is based on the information contained in a whole-brain tractogram generated from DWI data, and therefore relies on the innate structure and connectivity of the tissue to provide contrast. Furthermore, the super-resolution properties of the TDI technique (Calamante et al. 2011) provide a major boost to the spatial resolution achievable by this MRI method. In this study, we were able to delineate a greater number of structures using DEC stTDI than when compared to T2* maps. For example, the clear visualization of the anterior commissure facilitated the demarcation of a border between the dorsal telencephalon and the ventral telencephalon (Fig. 5a), which corroborates previous immunohistochemical and molecular results (Mueller et al. 2011; Ganz et al. 2012). Similarly, some fiber tracks such as the lateral and medial forebrain bundles (LFB and MFB), which are very small in diameter (45μm) and therefore only appear as faint lines in the 10μm T2*-weighted image and as coarse red pixels in the 48μm DEC FA maps, could clearly be distinguished as discrete fiber bundles in the 5μm DEC stTDI maps. As a result, DEC stTDI, with its very high spatial resolution and the high anatomical contrast provided by the orientation information, enabled us to delineate 30 fiber bundles, including commissures, fiber tracks and nerves. Table 1 lists all of the tracts, commissures and nerves that were successfully identified, which includes 17 structures that were previously unidentifiable using MR histology.
The very high resolution and contrast achievable using DEC stTDI maps could play an important role in creating more detailed multi-modal brain atlases. Previous studies have demonstrated the utility of DEC stTDI for visualization of small brain structures, such as the thalamic nuclei (Calamante et al. 2012a), the cerebellar peduncles (Calamante et al. 2010), and other structures in the human brain (Cho et al. 2013). By registering the TDI data to other MR images and histology, more detailed high-resolution atlases could be created, due to the complementary information of all these imaging modalities.
DEC stTDI can also assist in creating more detailed connectivity maps. The enhanced resolution facilitates the placement of very small regions of interest enabling more specific targeted tractography. These improvements would be particularly useful for brains such as the zebrafish, which have a volume over 100 times smaller than the human brain. In this study, we found this to be the case for the targeted tractography analyses we performed as tractography results confirm previous retrograde and anterograde labeling experiments (von Bartheld et al. 1984; Rink and Wullimann 2004; Wullimann et al. 1996).
While this study and previous studies have found that DEC stTDI maps replicate histological data, these comparisons have been limited to qualitative analyses because of the different contrast mechanisms. For example, our histological sections demonstrate the location of neurons and fiber tracts but do not contain directional information. However, a new tractography technique, three-dimensional polarized light imaging (3D-PLI) has been recently developed that can visualize the orientation of nerve fibers based upon their birefringence (Axer et al. 2011; Palm et al. 2010). Birefringence, is the product of the arrangement of proteins and lipids in myelin, and has an optical anisotropy that reflects the spatial fiber architecture of nerve fibers. By carefully collecting a series of sections and calculating fiber orientation maps, whole brain fiber tracking can be performed and compared with DWI data. Furthermore, as birefringence data is collected from histological data, the acquired resolution is very high with voxel sizes as small as 100μm3 in the human brain (Palm et al. 2010). This voxel size is directly comparable to that achieved in human TDI data (Calamante et al. 2010), and could potentially be used to validate the DEC stTDI technique. However, it should be noted that 3D-PLI has a number of technical difficulties when used for whole-brain fiber-tracking, including accurate registration of the large number of PLI sections into a 3D volume, vector reorientation, and partial volume effects in voxels containing more than one fiber population (akin to the limitations of the diffusion tensor model).
The quality of DEC stTDI is dependent, among other factors (see references Calamante et al. 2010 and Calamante et al. 2012b for a more detailed discussion), on the spatial resolution and angular resolution of the acquired DWI data. This is necessary to obtain a reliable fiber orientation distribution (FOD) for modeling of fiber crossing, which in turn has a direct influence on the quality of whole-brain tractography. For examining the zebrafish brain with a volume of only (3mm)3 and containing substructures that are in the order of 10–50μm in diameter, imaging with a high spatial resolution is paramount. Three-dimensional gradient echo (3D GE) MRI sequences at ultra-high field strength are capable of acquiring 10μm isotropic resolution images but a large number of scan averages (NEX=16) are necessary to achieve a good signal to noise ratio (SNR). While it is possible to acquire a data with a longer scan time, this will mostly contribute to an increase SNR rather than additional tissue contrast. Theoretically, to obtain 3D GE image at 5 μm isotropic resolution and maintaining SNR level as in the 10μm images, NEX needs to be increased by 23, which is increasing the scan time from 15h to 120h (considerable time cost). In contrast, the combination of such spatial resolution and number of scans is not practical for HARDI acquisition. For example, it took 55h to collect 48μm isotropic resolution data with 30 diffusion directions, 2 b0 images and 4 NEX at 16.4T in the current study. For reducing the DWI scan to 15h, we would have only been able to collect data with 6 diffusion encoding directions and 2b0, which is suitable for DTI processing only. Therefore, super-resolution 5um DEC stTDI provides additional value by enabling nuclei and fiber-tracks not previously seen in the 3D GE image to be visualised at even higher spatial resolution, although at the expense of longer acquisition times than those required for standard DTI.
It should be noted that TDI contrast is the reverse of that in a GE image. In general, myelinated white matter tracks appear hyperintense in TDI, hypointense in GE, and blue after luxol fast blue staining. This could make comparisons confusing and potentially misleading. More importantly, hypointense areas in TDI maps do not necessarily represent nuclei but could instead represent areas with a low number of streamlines. For example, throughout the telencephalon there are numerous hypointense areas, such as in the medial zone of the dorsal telencephalon (see sagittal section in Fig. 1) or the dorsolateral zone in the dorsal telencephalon (Dl in Fig. 3a). As seen in histological sections, these areas do not correspond to large nuclei but to areas with a reduced number of fibers.
Conclusion
This study demonstrates super-resolution TDI provides enhanced characterization of the zebrafish brain. DEC stTDI maps corresponded to those provided using histology, and allowed delineation of a greater number of brain regions, commissures, and fiber tracks when compared to T2* imaging. With the increased use of transgenic adult zebrafish for examination of neurological diseases, we expect this methodology to be an important additional imaging tool for phenotypic examination.
Supplementary Material
Supplementary Figure 1: TDI bootstrapping to test the optimal number of tracks to be used for zebrafish brain stTDI. TDI maps were generated with an incremental number of tracks including a) 1 million tracks; b) 5 million tracks; c) 20 million tracks; d) 40 million tracks; e) 60 million tracks. Scale bar = 500μm.
Acknowledgement
We are grateful to the National Health and Medical Research Council (NHMRC) of Australia, and the Australian Research Council (ARC LE100100074 and FT110100726) for their support. We would also like to thank the National Imaging Facility (NIF) and the Queensland NMR Network (QNN) for access to the 16.4T scanner and technical support. The Florey Institute of Neuroscience and Mental Health is supported by Victorian State Government infrastructure funds. We would also like to thank Jane Ellis for assistance with the histology.
Abbreviations:
- 3D GE
Three-dimensional gradient echo
- 3D-PLI
Three-dimensional polarized light imaging
- AC
Anterior cerebellar tract
- Cans
Ansulate commissure
- CantD
Anterior commissure, dorsal part
- CantV
Anterior commissure, ventral part
- CC
Cerebellar crest
- CCeG
Cerebellar corpus, granular layer
- CCeM
Cerebellar corpus, molecular layer
- Ccer
Cerebellar commissure
- CON
Caudal octavolateralis nucleus
- Cpop
Postoptic commissure
- Cpost
Posterior commissure
- CSD
Constrained spherical deconvolution
- Cven
Ventral rhombencephalic commissure
- CZ
Central zone of the optic tectum
- D
Dorsal telencephalon
- Dc
Central zone of the dorsal telencephalon
- Dd
Dorsal zone of the dorsal telencephalon
- DEC
Directionally encoded color
- Dl
Lateral zone of the dorsal telencephalon
- Dm
Medial zone of the dorsal telencephalon
- DOT
Dorsomedial optic tract
- Dp
Posterior zone of the dorsal telencephalon
- DTI
Diffusion tensor imaging
- DWI
Diffusion weighted imaging
- DWZ
Deep white zone of the optic tectum
- FA
Fractional anisotropy
- FOD
Fiber orientation distribution
- FOV
Field of view
- FR
Habenula-interpeduncular tract
- GL
Glomerular layer
- HARDI
High angular-resolution diffusion-weighted imaging
- III
Oculomotor nerve
- IO
Inferior olive
- LCa
Caudal lobe of the cerebellum
- LFB
Lateral forebrain bundle
- LLF
Lateral longitudinal fascicle
- LOT
Lateral olfactory tract
- LVII
Facial lobe
- LX
Vagal lobe
- MA
Mauthner axon
- MFB
Medial forebrain bundle
- MLF
Medial longitudinal fascicle
- MOT
Medial olfactory tract
- MRI
Magnetic resonance imaging
- NEX
Number of averages
- NIN
Interpeduncular nucleus
- nMLF
Nucleus of the medial longitudinal fascicle
- OlfN
Olfactory nerve
- ON
Optic nerve
- PGZ
Periventricular grey zone of the optic tectum
- POF
Primary olfactory layer
- PPa
Parvocellular preoptic nucleus
- RF
Radio frequency
- SNR
Signal to noise ratio
- SWGZ
Superficial grey and white zone of the optic tectum
- TDI
Track density imaging
- TE
Echo time
- TeO
Optic tectum
- TL
Longitudinal torus
- TMCA
Anterior mesencephalon-cerebellar tract
- TPM
Pretecto-mammillary tract
- TR
Repetition time
- TTB
Tractus tectobulbaris
- TTBc
Crossed tecto-bulbar tract
- Val
Lateral division of the valvula cerebelli
- Vas
Vascular lacuna of area postrema
References
- Al-Imari L, Gerlai R (2008) Sight of conspecifics as reward in associative learning in zebrafish (Danio rerio). Behav Brain Res 189:216–219. [DOI] [PubMed] [Google Scholar]
- Alfaro JM, Ripoll-Gomez J, Burgos JS (2011) Kainate administered to adult zebrafish causes seizures similar to those in rodent models. European Journal of Neuroscience 33 (7):1252–1255. [DOI] [PubMed] [Google Scholar]
- Axer M, Amunts K, Grassel D, Palm C, Dammers J, Axer H, Pietrzyk U, Zilles K (2011) A novel approach to the human connectome: ultra-high resolution mapping of fiber tracts in the brain. NeuroImage 54 (2):1091–1101. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Vernier P (2010) Organization and physiology of the zebrafsih nervous system. In: Zebrafish, vol 29. Fish Physiology. Elsevier Inc., [Google Scholar]
- Basser PJ (1995) Inferring microstructural features and the physiological state of tissues from diffusion-weighted images. NMR Biomed 8 (7–8):333–344. [DOI] [PubMed] [Google Scholar]
- Best JD, Alderton WK (2008) Zebrafish: An in vivo model for the study of neurological diseases. Neuropsychiatric Disease and Treatment 4 (3):567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat J, Stewart A, Utterback E, Hart P, Gaikwad S, Wong K, Kyzar E, Wu N, Kalueff AV (2011) Three-dimensional neurophenotyping of adult zebrafish behavior. Plos One 6 (3):e17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F, Tournier J-D, Jackson GD, Connelly A (2010) Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. NeuroImage 53 (4):1233–1243. [DOI] [PubMed] [Google Scholar]
- Calamante F, Tournier JD, Heidemann RM, Anwander A, Jackson GD, Connelly A (2011) Track density imaging (TDI): Validation of super resolution property. Neuroimage 56 (3):1259–1266. [DOI] [PubMed] [Google Scholar]
- Calamante F, Oh SH, Tournier JD, Park SY, Son YD, Chung JY, Chi JG, Jackson GD, Park CW, Kim YB, Connelly A, Cho ZH (2012a) Super-resolution track-density imaging of thalamic substructures: Comparison with high-resolution anatomical magnetic resonance imaging at 7.0T. Hum Brain Mapp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F, Tournier JD, Kurniawan ND, Yang Z, Gyengesi E, Galloway GJ, Reutens DC, Connelly A (2012b) Super-resolution track-density imaging studies of mouse brain: Comparison to histology. NeuroImage 59 (1):286–296. [DOI] [PubMed] [Google Scholar]
- Cho ZH, Calamante F, Chi JG (2013) 7.0 Tesla MRI Brain White Matter Atlas In. Panmun Co, Ltd, Seoul, South Korea, [Google Scholar]
- Corbo CP, Othman NA, Gutkin MC, Alonso Adel C, Fulop ZL (2012) Use of different morphological techniques to analyze the cellular composition of the adult zebrafish optic tectum. Microsc Res Techniq 75 (3):325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm J, Haase A, Matthaei D (1986) Rapid three-dimensional MR imaging using the FLASH technique. Journal of Comp Assist Tomography 10 (2):363–368. [DOI] [PubMed] [Google Scholar]
- Frullano L, Zhu J, Wang C, Wu C, Miller RH, Wang Y (2012) Myelin imaging compound (MIC) enhanced magnetic resonance imaging of myelination. J Med Chem 55 (1):94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz J, Kaslin J, Freudenreich D, Machate A, Geffarth M, Brand M (2012) Subdivisions of the adult zebrafish subpallium by molecular marker analysis. J Comp Neurol 520 (3):633–655. [DOI] [PubMed] [Google Scholar]
- Gomez-Laplaza LM, Gerlai R (2010) Latent learning in zebrafish (Danio rerio). Behav Brain Res 208 (2):509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haud N, Kara F, Diekmann S, Henneke M, Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gartner J, Alia A, Hurlstone AF (2011) rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci U S A 108 (3):1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Badea A, Brandenburg J, Cofer G, Fubara B, Liu S, Nissanov J (2010) Waxholm space: An image-based reference for coordinating mouse brain research. NeuroImage 53 (2):365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Calabrese E, Badea A, Paxinos G, Watson C (2012) A multidimensional magnetic resonance histology atlas of the Wistar rat brain. NeuroImage 62 (3):1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Simmons A, Williams SCR, Horsfield MA (1999) Non-invasive assessment of axonal fiber connectivity in the human brain via diffusion tensor MRI. Magnetic Resonance in Medicine 42 (1):37–41. [DOI] [PubMed] [Google Scholar]
- Kabli S, Alia A, Spaink HP, Verbeek FJ, de Groot HJM (2006) Magnetic resonance microscopy of adult zebrafish. Zebrafish 3 (4):431–439. [DOI] [PubMed] [Google Scholar]
- Kabli S, Spaink HP, De Groot HJM, Alia A (2009) In vivo metabolite profile of adult zebrafish brain obtained by high-resolution localized magnetic resonance spectroscopy. Journal of Magnetic Resonance Imaging 29 (2):275–281. [DOI] [PubMed] [Google Scholar]
- Keller ET, Murtha JM (2004) The use of mature zebrafish (Danio rerio) as a model for human aging and disease. Comparative Biochemistry and Physiology, Part C 138:335–341. [DOI] [PubMed] [Google Scholar]
- Kishida R (1979) Comparative study on the teleostean optic tectum. Lamination and cytoarthitecture. Journal fur Hirnforschung 20:57–67. [PubMed] [Google Scholar]
- Koretsky AP, Silva AC (2004) Manganese-enhanced magnetic resonance imaging (MEMRI). NMR Biomed 17 (8):527–531. [DOI] [PubMed] [Google Scholar]
- Laufer M, Vanegas H (1974) The optic tectum of a perciform teleost. II. Fine structure. J Comp Neurol 154:61–96. [DOI] [PubMed] [Google Scholar]
- Leergaard TB, Bjaalie JG, Devor A, Wald LL, Dale AM (2003) In vivo tracing of major rat brain pathways using manganese-enhanced magnetic resonance imaging and three-dimensional digital atlasing. NeuroImage 20 (3):1591–1600. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD (2007) Animal models of human disease: zebrafish swim into view. Nature Reviews 8:353–367. [DOI] [PubMed] [Google Scholar]
- Mathur P, Guo S (2010) Use of zebrafish as a model to understand mechanisms of addiction and complex neurobehavioral phenotypes. Neurobiol Dis 40 (1):66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren DG, Kosmatka KJ, Oakes TR, Kroenke CD, Kohama SG, Matochik JA, Ingram DK, Johnson SC (2009) A population-average MRI-based atlas collection of the rhesus macaque. NeuroImage 45 (1):52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC (2002) Fiber tracking: principles and strategies - a technical review. NMR Biomed 15 (7–8):468–480. [DOI] [PubMed] [Google Scholar]
- Mori S, Zhang J (2006) Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51 (5):527–539. [DOI] [PubMed] [Google Scholar]
- Mueller T, Dong ZQ, Berberoglu MA, Guo S (2011) The dorsal pallium in zebrafish, Danio rerio (Cyprinidae, Teleostei). Brain Res 1381:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG (1983) Evolution of the optic tectum in ray-finned fishes. In: Davis RE, Northcutt RG (eds) Fish Neurobiology, Vol 2. Higher brain areas and functions, vol 2. University of Michigan Press, Ann Arbor, pp 1–42 [Google Scholar]
- Norton W, Bally-Cuif L (2010) Adult zebrafish as a model organism for behavioural genetics. BMC Neuroscience 11 (1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C (1999) Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med 42 (3):526–540. [PubMed] [Google Scholar]
- Palm C, Axer M, Grassel D, Dammers J, Lindemeyer J, Zilles K, Pietrzyk U, Amunts K (2010) Towards ultra-high resolution fibre tract mapping of the human brain - registration of polarised light images and reorientation of fibre vectors. Front Hum Neurosci 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiet A, Hedlund L, Johnson GA (2007) Staining methods for magnetic resonance microscopy of the rat fetus. Journal of Magnetic Resonance Imaging 25:1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda R, Beattie CE, Hall CW (2011) Recording the adult zebrafish cerebral field potential during pentylenetetrazole seizures. J Neurosci Meth 200 (1):20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez IB, Pietka G, Jones DR, Divecha N, Alia A, Baraban SC, Hurlstone AF, Lowe M (2012) Impaired neural development in a zebrafish model for Lowe syndrome. Hum Mol Genet 21 (8):1744–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KD, Alex A, Verma Y, Thampi S, Gupta PK (2009) Real-time in vivo imaging of adult zebrafish brain using optical coherence tomography. Journal of Biophotonics:1–4. [DOI] [PubMed] [Google Scholar]
- Richards K, Watson C, Buckley RF, Kurniawan ND, Yang Z, Keller MD, Beare R, Bartlett PF, Egan GF, Galloway GJ, Paxinos G, Petrou S, Reutens DC (2011) Segmentation of the mouse hippocampal formation in magnetic resonance images. NeuroImage 58 (3):732–740. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF (2004) Connections of the ventral telencephalon (subpallium) in the zebrafish (Danio rerio). Brain Res 1011 (2):206–220. [DOI] [PubMed] [Google Scholar]
- Rowland IJ (2011) Dichromate as a Stain for Mr Microscopy. I S Biomed Imaging:742–745. [Google Scholar]
- Saverino C, Gerlai R (2008) The social zebrafish: behavioral responses to conspecific, heterospecific, and computer animated fish. Behav Brain Res 191:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel AM, Piato AL, Capiotti KM, Seibt KJ, Bogo MR, Bonan CD (2011) PTZ-induced seizures inhibit adenosine deamination in adult zebrafish brain membranes. Brain Res Bull 86 (5–6):385–389. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A (2007) Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. NeuroImage 35 (4):1459–1472. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. In: International Society for Magnetic Resonance in Medicine, Stockholm, Sweden, 2010. p 1670 [Google Scholar]
- Tournier JD, Calamante F, Connelly A (2012) MRtrix: Diffusion tractography in crossing fiber regions. Int J Imag Syst Tech 22 (1):53–66. [Google Scholar]
- Ullmann JF, Keller MD, Watson C, Janke AL, Kurniawan ND, Yang Z, Richards K, Paxinos G, Egan GF, Petrou S, Bartlett P, Galloway GJ, Reutens DC (2012) Segmentation of the C57BL/6J mouse cerebellum in magnetic resonance images. NeuroImage 62 (3):1408–1414. [DOI] [PubMed] [Google Scholar]
- Ullmann JFP, Cowin G, Kurniawan ND, Collin SP (2010a) Magnetic resonance histology of the adult zebrafish brain: optimization of fixation and gadolinium contrast enhancement. NMR Biomed 23 (4):341–346. [DOI] [PubMed] [Google Scholar]
- Ullmann JFP, Cowin G, Kurniawan ND, Collin SP (2010b) A three-dimensional digital atlas of the zebrafish brain NeuroImage 51 (1):76–82. [DOI] [PubMed] [Google Scholar]
- von Bartheld CS, Meyer DL, Fiebig E, Ebbesson SOE (1984) Central connections of the olfactory bulb in the goldfish, Carassius auratus. Cell and Tissue Research 238 (3):475–487. [DOI] [PubMed] [Google Scholar]
- Williams LR, Wong K, Stewart A, Suciu C, Gaikwad S, Wu N, Dileo J, Grossman L, Cachat J, Hart P, Kalueff AV (2011) Behavioral and physiological effects of RDX on adult zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. [DOI] [PubMed] [Google Scholar]
- Wong K, Stewart A, Gilder T, Wu N, Frank K, Gaikwad S, Suciu C, DiLeo J, Utterback E, Chang K, Grossman L, Cachat J, Kalueff AV (2010) Modeling seizure-related behavioral and endocrine phenotypes in adult zebrafish. Brain Res 1348:209–215. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Rupp B, Reichert H (1996) Neuroanatomy of the zebrafish brain: a topological atlas. Birkhäuser Verlag, Basel [Google Scholar]
- Yang Z, Richards K, Kurniawan ND, Petrou S, Reutens DC (2012) MRI-guided volume reconstruction of mouse brain from histological sections. J Neurosci Meth 211 (2):210–217. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bearer EL, Perles-Barbacaru AT, Jacobs RE (2010) Increased anatomical detail by in vitro MR microscopy with a modified Golgi impregnation method. Magn Reson Med 63 (5):1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: TDI bootstrapping to test the optimal number of tracks to be used for zebrafish brain stTDI. TDI maps were generated with an incremental number of tracks including a) 1 million tracks; b) 5 million tracks; c) 20 million tracks; d) 40 million tracks; e) 60 million tracks. Scale bar = 500μm.


