Abstract
The environment and events that we are exposed to in utero, during birth and in early childhood influence our future physical and mental health. The underlying mechanisms that lead to these outcomes are unclear, but long-term changes in epigenetic marks, such as DNA methylation, could act as a mediating factor or biomarker. DNA methylation data were assayed at 713 522 CpG sites from 9537 participants of the Generation Scotland: Scottish Family Health Study, a family-based cohort with extensive genetic, medical, family history and lifestyle information. Methylome-wide association studies of eight early life environment phenotypes and two adult mental health phenotypes (major depressive disorder and brief resilience scale) were conducted using DNA methylation data collected from adult whole blood samples. Two genes involved with different developmental pathways (PRICKLE2, Prickle Planar Cell Polarity Protein 2 and ABI1, Abl-Interactor-1) were annotated to CpG sites associated with preterm birth (P < 1.27 × 10−9). A further two genes important to the development of sensory pathways (SOBP, Sine Oculis Binding Protein Homolog and RPGRIP1, Retinitis Pigmentosa GTPase Regulator Interacting Protein) were annotated to sites associated with low birth weight (P < 4.35 × 10−8). The examination of methylation profile scores and genes and gene-sets annotated from associated CpGs sites found no evidence of overlap between the early life environment and mental health conditions. Birth date was associated with a significant difference in estimated lymphocyte and neutrophil counts. Previous studies have shown that early life environments influence the risk of developing mental health disorders later in life; however, this study found no evidence that this is mediated by stable changes to the methylome detectable in peripheral blood.
Introduction
The diathesis-stress model posits that behaviours and psychological disorders are the result of underlying biological factors (diatheses) plus exposure to stressful events or environments. Childhood adversity increases the risk of poorer physical and mental health outcomes in later life (1,2), with neglect, sexual and emotional abuse, and violence providing greater risk (3–6). However, multiple studies have reported additional perinatal and early life stressors and environments that are also associated with adult mental health. Individuals born preterm or with a low birth weight are more likely to experience problems with attentiveness and hyperactivity, as well as elevated levels of anxiety and depression (7–9). Seasonality of birth has also been associated with psychiatric outcomes in later life (10–12), with those born in January having higher risk of schizophrenia and bipolar and those born in June and July having greater risk of depression. Parental factors, including the age of the parents at birth (13) or having an absent parent (14), are reported to increase the risk of depression. Where we live has also been shown to be detrimental to mental health (15) and is likely due to a variety of environmental and social factors (16,17).
Adverse environments in the gestational and postnatal periods are known to cause long-term alterations to DNA methylation across the genome (18). These modifications are reported to mediate resilience and responses to stress-related disorders throughout the life course (19). Epigenomic variation has also been implicated in a range of psychiatric conditions (20), including bipolar disorder (21), schizophrenia (22) and major depressive disorder [MDD (23)]. Much of the epigenetic research seeking to link early life environments and MDD has been conducted using small cohort samples sizes (typically <1000) and focussed on candidate regions (24). However, this approach is likely to be suboptimal as demonstrated by the lack of reproducible results from candidate gene association studies for MDD (25).
The principal aim of the current research was to investigate whether alterations to DNA methylation have the potential to mediate the stress component in the diathesis-stress model. This was achieved by conducting a methylome-wide association study (MWAS) of early life environments and later mental health outcomes in a single study cohort of over 9500 adults from the Generation Scotland: Scottish Family Health Study cohort (26). Linked electronic health records, responses at interview and questionnaire data were used to ascertain the early life environments, MDD status and psychological resilience measured using the brief resilience scale (BRS). MDD and BRS were examined due to their reported association with early life environments (27,28). The associated CpG sites identified by the MWAS of these phenotypes were then annotated to genes and gene-sets. The gene and gene-set overlap between the early life environments and mental health outcomes were then examined. In addition, methylation profile scores were used to assess the broader methylome-wide overlap between the early life environment and mental health traits.
Results
We conducted analyses of eight early life environments (preterm birth, low birth weight, birth month, birth date, having a young parent, having a lone parent, urban environment, and population density) and two measures of adult mental health (MDD and BRS) using the Generation Scotland: Scottish Family Health Study cohort (n = 24 080). First, we used regression to assess the association between the early life environments and MDD and BRS using phenotypic data. Second, we conducted a MWAS to identify CpG sites associated with each phenotype. Finally, we used the summary statistics from the MWAS to determine the extent of any overlap between the early life environments and the mental health phenotypes using genes and gene-set analysis and methylation profile scores.
Regression of adult mental health on early life environments
Generalized linear mixed models were used to assess each early life environment in turn and its association with either MDD or BRS (Supplementary Material, Table 1), after adjusting for sex and relatedness between individuals. There were no significant associations between the early life environments and the adult mental health phenotypes after adjusting for multiple testing (P > 6.25 × 10−3). All early life environments, except for preterm birth, marginally increased the risk of developing MDD in adulthood. All early life environments, except for having a lone parent, marginally lowered BRS suggesting a negative effect on psychological resilience. The associations between birth date and birth month with BRS were nominally significant (P < 0.05) with those born across the summer months scoring lower on the BRS; however, these associations did not remain after correction for multiple testing.
MWAS of early life environments and adult mental health
To identify CpG sites associated with each of the early life environments and the mental health phenotypes a MWAS was conducted. Normalized M-values for 713 522 CpG sites profiled from combined blood samples of 9537 GS:SFHS individuals (5087 in Set 1 and 4450 in Set 2) remained after quality control procedures. The associations between these M-values and the early life environments and mental health phenotypes were estimated using two association study methods (MWAS 1 and MWAS 2). MWAS 1 fits a linear regression model to the data with the M-values as the dependent variable, whereas MWAS 2 fits the phenotype as the dependent variable while accounting for methylome-wide correlational structure and with an additional correction for predicted blood cell type composition.
The significant CpG sites (P < 7.01 × 10−8) from MWAS 1 and MWAS 2 are in Table 1, except those associated with birth month and with birth date which are in Supplementary Material, Tables 2 and 3. In Table 1, the six significant CpG sites identified in MWAS 2 were also significant in MWAS 1 for the same phenotypes. In MWAS 1, there were 93 significant sites for birth month and 637 significant sites for birth date. However, no CpG sites were significant in MWAS 2 for either birth month or birth date; further analysis examining this discrepancy is covered in the Supplementary Information.
Table 1.
CpG sites associated with phenotypes in either MWAS 1 or MWAS 2
| Phenotype | CpG site | Chr | BP position | Annotated Gene | MWAS 1 | MWAS 2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect size | Standard error | P-value | Effect size | Standard error | P-value | |||||
| Preterm birth | cg00725333 | 3 | 64 189 256 | PRICKLE2 | −0.2747 | 0.0376 | 6.24 × 10 −13 | −0.0364 | 0.0060 | 1.15 × 10 −9 |
| cg17668848 | 3 | 108 029 973 | HHLA2 | −0.2871 | 0.0396 | 9.13 × 10 −13 | −0.0356 | 0.0058 | 8.21 × 10 −10 | |
| cg24329141 | 10 | 27 095 369 | ABI1 | −0.2368 | 0.0352 | 2.98 × 10 −11 | −0.0345 | 0.0057 | 1.27 × 10 −9 | |
| Low birth weight | cg15582176 | 1 | 1 183 528 | C1QTNF12 | −0.3604 | 0.0661 | 6.53 × 10 −8 | −0.0254 | 0.0054 | 2.49 × 10−6 |
| cg19909717 | 6 | 107 924 415 | SOBP | −0.2225 | 0.0403 | 4.35 × 10 −8 | −0.0329 | 0.0055 | 1.59 × 10 −9 | |
| cg21803443 | 7 | 126 547 786 | GRM8 | −0.3656 | 0.0618 | 4.62 × 10 −9 | −0.0278 | 0.0056 | 7.22 × 10−7 | |
| cg12090821 | 14 | 21 755 658 | RPGRIP1 | −0.0857 | 0.0152 | 2.43 × 10 −8 | −0.0329 | 0.0058 | 1.47 × 10 −8 | |
| cg05905731 | 16 | 85 485 785 | −0.1695 | 0.0300 | 2.23 × 10 −8 | −0.0294 | 0.0055 | 7.50 × 10−8 | ||
| Young parent | cg00528572 | 11 | 92 703 433 | MTNR1B | −0.1369 | 0.0200 | 9.04 × 10 −12 | −0.0145 | 0.0025 | 4.47 × 10 −9 |
| cg02427109 | 18 | 77 917 459 | PARD6G, PARD6G-AS1 | −0.1163 | 0.0213 | 4.96 × 10 −8 | −0.0101 | 0.0025 | 4.12 × 10−5 | |
| Population density | cg06759845 | 1 | 156 460 474 | MEF2D | 5.20 × 10−6 | 9.40 × 10−7 | 3.40 × 10 −8 | 90.39 | 17.02 | 1.10 × 10−7 |
| cg12433043 | 6 | 100 619 893 | −6.65 × 10−6 | 1.20 × 10−6 | 3.37 × 10 −8 | −88.86 | 16.75 | 1.13 × 10−7 | ||
| cg03623878 | 13 | 113 655 560 | MCF2L | 9.10 × 10−6 | 1.58 × 10−6 | 9.56 × 10 −9 | 81.18 | 16.88 | 1.51 × 10−6 | |
| cg08036492 | 17 | 13 976 536 | COX10 | 3.09 × 10−6 | 5.71 × 10−7 | 6.74 × 10 −8 | 87.75 | 18.29 | 1.61 × 10−6 | |
| MDD | cg02280719 | 1 | 6 802 222 | 0.0400 | 0.0064 | 5.99 × 10 −10 | 0.0174 | 0.0043 | 5.32 × 10−5 | |
| cg08548783 | 6 | 57 903 690 | 0.0378 | 0.0067 | 1.37 × 10 −8 | 0.0142 | 0.0043 | 9.79 × 10−4 | ||
Note: MWAS 1 results are from a methylome-wide association analysis using linear regression and MWAS 2 results are from a methylome-wide association analysis using OSCA. Bold P-values indicate an association after correction for multiple testing (P < 7.01 × 10−8). Sites are ordered by phenotype and then genomic position. Chromosome number (Chr) and base pair (BP) position are based on genome assembly GRCh37 (hg19). Annotation of genes is provided by missMethyl.
Miami plots for preterm birth, low birth weight, having a young parent, population density, and MDD are in Figures 1–5, respectively. The remaining Miami plots are in Supplementary Material, Figures 1–5 and QQ-plots and 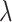 for all phenotypes are in Supplementary Material, Figures 6–15.
for all phenotypes are in Supplementary Material, Figures 6–15.
Figure 1.
Miami plot of the observed P-values of each CpG site for an association with preterm birth. Log10P-values are shown for MWAS 1 and −log10P-values are shown for MWAS 2. The dotted lines indicate methylome-wide significance (P = 7.01 × 10−8). The annotation of genes for significant sites is reported by missMethyl for MWAS 1 and by OSCA for MWAS 2.
Figure 5.
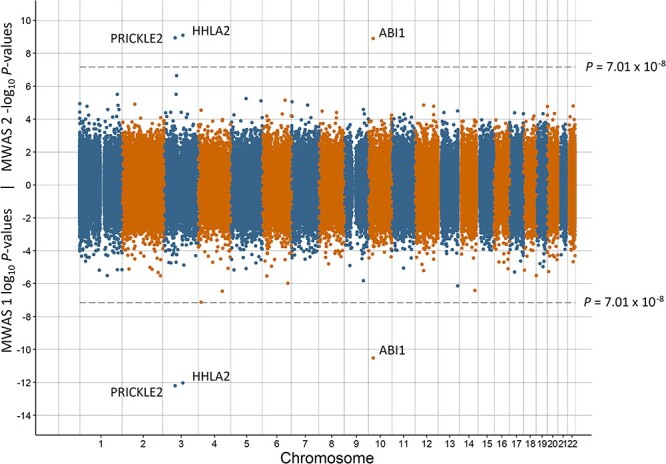
Miami plot of the observed P-values of each CpG site for an association with MDD. Log10P-values are shown for MWAS 1 and −log10P-values are shown for MWAS 2. The dotted lines indicate methylome-wide significance (P = 7.01 × 10−8). Significant sites did not annotate to any genes.
Figure 2.
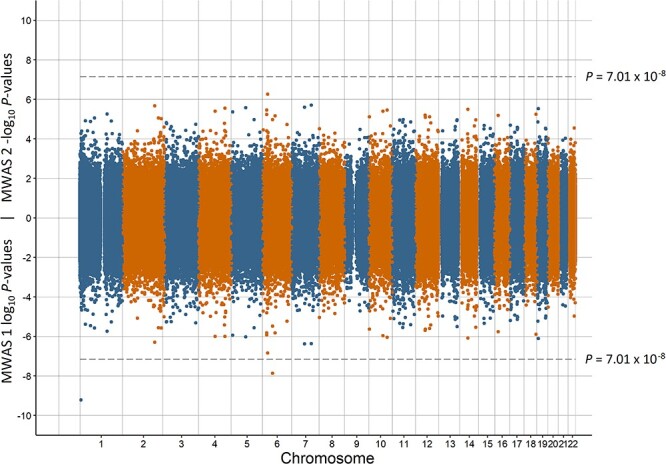
Miami plot of the observed P-values of each CpG site for an association with low birth weight. Log10P-values are shown for MWAS 1 and −log10P-values are shown for MWAS 2. The dotted lines indicate methylome-wide significance (P = 7.01 × 10−8). The annotation of genes for significant sites is reported by missMethyl for MWAS 1 and by OSCA for MWAS 2.
Figure 3.
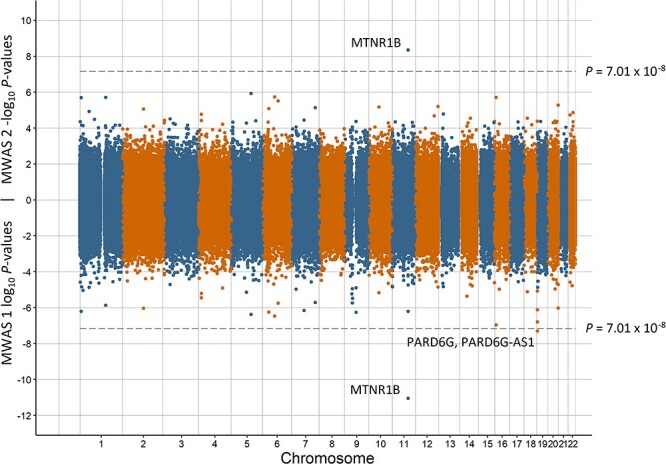
Miami plot of the observed P-values of each CpG site for an association with having a young parent. Log10P-values are shown for MWAS 1 and −log10P-values are shown for MWAS 2. The dotted lines indicate methylome-wide significance (P = 7.01 × 10−8). The annotation of genes for significant sites is reported by missMethyl for MWAS 1 and by OSCA for MWAS 2.
Figure 4.
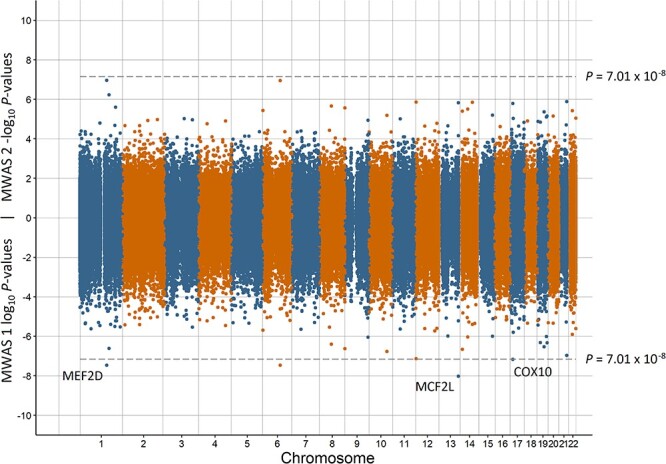
Miami plot of the observed P-values of each CpG site for an association with population density. Log10 P-values are shown for MWAS 1 and −log10 P-values are shown for MWAS 2. The dotted lines indicate methylome-wide significance (P = 7.01 × 10−8). The annotation of genes for significant sites is reported by missMethyl for MWAS 1.
Gene and gene-set analysis
The genes annotated to associated CpG sites from MWAS 1 and MWAS 2 were examined for overlap between the early life environment and adult mental health phenotypes. For the significant sites in MWAS 1, there were three annotated genes for preterm birth, having a young parent and population density, four annotated genes for low birth weight, 20 annotated genes for birth month and 167 annotated genes for birth date. There were no annotated genes for the mental health phenotypes and therefore no overlap was observed with the early life environment phenotypes.
For significant sites in MWAS 2, there were three annotated genes for preterm birth, two annotated genes for low birth weight and one annotated gene for having a young parent. As there were no annotated genes for the mental health phenotypes, there was no overlap of annotated genes with the early life environment phenotypes.
There were no enriched Gene Ontology or the KEGG gene-sets (P > 0.05) for any of the phenotypes from either MWAS 1 or MWAS 2.
Mediation analysis
cg21803443 was associated with low birth weight in MWAS 1 and was annotated to the GRM8 (Glutamate Metabotropic Receptor 8) protein-coding gene. Küpers et al. (29) also reported a CpG site (cg15908975) annotated to GRM8 associated with birth weight. GRM8 has also been associated with depression (30). Therefore, a mediation analysis with low birth weight as the independent variable, either cg21803443 or cg15908975 as the mediator variable, and MDD as the dependent variable was conducted. There were no significant direct or indirect effects (P > 0.05; Supplementary Material, Figs 16 and 17) when fitting either CpG site as the mediator variable.
Methylation profile scores
The MWAS 2 effect size estimates from Set 1 results for the early life environment phenotypes were used to calculate profile scores for individuals in Set 2. The utility of these profile scores for predicting MDD, BRS or the same phenotype (Table 2) in Set 2 was assessed. There was significant prediction of birth month using birth month profile scores (R2 = 0.002, P = 5.19 × 10−3) and of having a young parent using the profile scores from young parent (R2 = 0.013, P = 3.46 × 10−5). There was nominal prediction of MDD from preterm birth (R2 = 0.002, P = 0.034) and birth month (R2 = 0.002, P = 0.043), and nominal prediction of BRS from birth month (R2 = 0.003, P = 0.016). However, none of the profile scores predicting mental health phenotypes were significant after correction for multiple testing (P > 6.25 × 10−3). The MWAS results from a continuous measure of gestation length conducted by Merid et al. (31) and from a continuous measure of birth weight conducted by Küpers et al. (29) were used to construct profile scores in GS:SFHS; however, these score was not associated with either MDD or BRS (P > 0.05).
Table 2.
Prediction of MDD, brief resilience scale and the same early life environment in Set 2 using methylation profile scores
| MDD | Brief resilience scale | Same early life environment | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Early life environment | Threshold | R 2 | P-value | Threshold | R 2 | P-value | Threshold | R 2 | P-value |
| Preterm birth | 1 × 10−4 | 0.0023 | 0.034 | 1 × 10−7 | 0.0001 | 0.74 | 1 × 10−7 | 0.0575 | 0.11 |
| Low birth weight | 1 × 10−5 | 0.0005 | 0.32 | 1 × 10−6 | 0.0009 | 0.21 | 1 × 10−7 | 0.0177 | 0.26 |
| Birth month | 1 × 10−6 | 0.0021 | 0.043 | 1 × 10−6 | 0.0032 | 0.016 | 1 × 10−4 | 0.0024 | 5.19 × 10 −3 |
| Birth date | 1 × 10−5 | 0.0013 | 0.10 | 1 × 10−5 | 0.0016 | 0.09 | 1 × 10−4 | 0.0009 | 0.042 |
| Young parent | 1 × 10−3 | 0.0006 | 0.28 | 1 × 10−5 | 0.0010 | 0.19 | 1 × 10−5 | 0.0128 | 3.46 × 10 −5 |
| Lone parent | 1 × 10−5 | 0.0005 | 0.34 | 1 × 10−4 | 0.0001 | 0.66 | 1 × 10−3 | 0.0004 | 0.49 |
| Urban environment | 1 × 10−5 | 0.0004 | 0.40 | 1 × 10−2 | 0.0008 | 0.25 | 1 × 10−5 | 0.0006 | 0.21 |
| Population density | 1 × 10−4 | 0.0015 | 0.08 | 1 × 10−6 | 0.0010 | 0.19 | 1 × 10−5 | 0.0013 | 0.037 |
Notes: Scores were calculated using the CpG site effect sizes from MWAS 2 of early life environment phenotypes in Set 1. The effect sizes in Set 1 were obtained from methylome-wide association studies using OSCA (MWAS 2). Multiple thresholds based on CpG site P-values were examined, with the threshold explaining the greatest phenotypic variance reported. An adjusted R2 from a simple linear regression was used for the prediction of quantitative phenotypes, else Nagelkerke’s R2 was used to calculate the proportion of phenotypic variance explained on the liability scale. Bold P-values indicate an association after correction for multiple testing (P < 6.25 × 10−3).
Discussion
The impact of early life environments on later life is of critical importance across multiple clinical and research domains. We sought to examine eight early life environments experienced around the time of birth and quantify: (i) their association with mental health in adulthood; (ii) whether they were associated with detectable changes to the methylome in to adulthood; and (iii) whether there was any shared association on the methylome between early life environments and adult mental health.
Previous studies have reported an effect of extreme preterm birth (gestation length <28 weeks) on mental health (8,32). The population-based GS:SFHS cohort had four participants (out of 3134) born at <28 weeks gestation length and therefore a 37-week threshold for a preterm birth was used. Three CpG sites were associated with preterm birth in both the MWAS analyses: cg00725333, cg17668848 and cg24329141. These three sites mapped to PRICKLE2 (Prickle Planar Cell Polarity Protein 2), HHLA2 (Human Endogenous Retrovirus-H Long Terminal Repeat-Associating Protein 2) and ABI1 (Abl-Interactor-1) protein coding genes, respectively. In humans, PRICKLE2 has been associated with myelomeningocele (33), a severe form of spina bifida. Myelomeningocele compromises the development of the spine and spinal cord in the womb and has been associated with preterm birth (34). HHLA2 regulates T-cell function (35), however its role in gestation length is unclear. Knockouts of ABI1 in mice have demonstrated it to be essential for embryonic development, survival (36) and placental development (37). PRICKLE2 and ABI1 have plausible roles in gestation length with altered methylation of proximal CpG sites into adulthood. A MWAS conducted using preterm infants (38) revealed a different set of associated CpG sites to those observed here in adulthood.
Similar to preterm birth, published studies of birth weight and mental health report that lower birth weight thresholds identify stronger associations with poorer mental health (39,40). In the current study, a sex and gestational-age adjusted measure of birth weight was used increasing the distinction with the preterm birth phenotype. Five CpG sites were associated with birth weight in MWAS 1 of which two were also significant in MWAS 2. The two sites that were significant in both MWAS annotated to genes important for the development of the sensory systems: SOBP (Sine Oculis Binding Protein Homolog) and RPGRIP1 (Retinitis Pigmentosa GTPase Regulator Interacting Protein). SOBP has been implicated in the embryonic development of the mouse cochlea (41). In a meta-analysis of DNA methylation data in neonates for low birth weight, a CpG site annotated to the SOBP gene was also significant after correcting for false discovery rate [but not after Bonferroni correction; (29)]. The critical role of RPGRIP1 in the remodelling of rod photoreceptors has been demonstrated in humans (42). The association between low birth weight and the long-term expression of CpG sites requires further investigation, certainly as low birth weight has been associated with both hearing impairment (43) and ophthalmic deficits (44). cg21803443 on chromosome 7 was significant in MWAS 1 and annotated to the GRM8 protein-coding gene. Küpers et al. (29) also identified a CpG site (cg15908975, P = 4.52 × 10−7 in a European meta-analysis) close to GRM8 associated with birth weight. GRM8 is involved in the inhibition of the cyclic AMP cascade influencing glutamatergic neurotransmission and had an association with depression (P = 1.80 × 10−12) in a genome-wide association meta-analysis (30). Mediation analysis of two CpG sites close to GRM8 (cg21803443 and cg15908975), fitting birth weight as the independent variable and MDD as the dependent variable did not reveal a significant direct or indirect effect. There was no predictive ability of a methylation profile score for low birth weight to predict either MDD or BRS. A previous analysis of birth weight as a continuous trait in GS:SFHS identified one significant CpG site [cg00966482; (45)], however this site was not significant in either MWAS 1 (P = 0.22) or MWAS 2 (P = 0.53).
Schnittker (12) suggests that the role of seasonality of birth on mental health was more prevalent in the early part of the 20th century and was partly attributable to poorer prenatal nutrition across the winter months. However, Disanto et al. (10) analysed post-1950 data from England and reported an effect of seasonality of birth on schizophrenia, bipolar disorder and to a lesser extent recurrent MDD. In the current study, there was nominal evidence for an effect of birth date and birth month on BRS and nominal prediction of both MDD and BRS using a profile score for birth month calculated from DNA methylation data. The greatest difference between the MWAS 1 and MWAS 2 results was for birth date and birth month. Additional prediction into the Lothian Birth Cohorts of 1921 and 1936 [LBC (46); see Supplementary Information], using methylation profile scores, demonstrated that this divergence is likely due to differing blood cell type composition (for neutrophils and lymphocytes) between those born in the winter and those born in the summer. Altered gene expression due to seasonality has been reported previously (47), with seasonality of birth also influencing neonatal immune development (48) and thymic output (49). The current work provides further substantial evidence that birth date is associated with blood cell type composition. This association is detectable throughout the life course and needs to be accounted for in future research on traits influenced by seasonality of birth.
In the present study, there was no effect of having a young parent on mental health. However, a much larger study (2.9 M individuals) on Danish participants found an increased risk of mood disorders (International Classification of Disease codes F30–39) for those with teenage mothers [incidence risk ratio = 1.35 (95% CI = 1.30–1.40)] and those with teenage fathers [incidence risk ratio = 1.20 (95% CI = 1.13–1.27); (13)]. The methylation profile for having a young parent from Set 1 had significant prediction of having a young parent in Set 2, suggesting replication across Sets.
Two correlated early life phenotypes, urban environment and population density, were studied to measure the effect of geographical environment at birth. There was no effect of either early life phenotype on MDD or BRS and there were no associated CpG sites. Urbanicity has been associated with depression (50,51), although no association has been observed in low- and middle-income countries (52) and the United States (53). The contradictory findings and the lack of observable effects here may be due to the multiple factors incorporated in these phenotypes, such as pollution (54) and socio-economic status (55), but may be offset by access to mental health services.
Published DNA methylation analyses of MDD have typically been conducted on relatively small samples. The two CpG sites associated with MDD in the current study did not annotate to known protein coding genes. These two sites were outside of the ten differentially methylated regions for major depression identified by Roberson-Nay et al. (56) (39 major depression cases and 111 controls) and located away from the locations identified for MDD by Oh et al. (57) (103 cases and 97 controls) and Starnawska et al. (58) (724 individuals assessed for depression symptomatology score). A longitudinal MWAS of MDD (199 cases and 382 in remittance) identified six plausible CpG sites based on function (59), although none of these sites remained significant after applying correction for multiple testing. A further study of profile scores for MDD in GS:SFHS found that prediction of MDD in an independent subset was possible, but was reliant on capturing lifestyle factors associated with MDD (60). An association meta-analysis (7948 individuals) identified 20 CpG sites with a suggestive association (P < 1 × 10−5) with depressive symptoms (23), of which one was nominally significant (P = 0.048) in a replication cohort of 3308 individuals. None of these 20 suggestive sites were close to either of the associated sites observed in the current study. To reach replicable findings for CpG sites associated with MDD it is likely that larger sample sizes will be needed as has been demonstrated in genome-wide associations studies (30).
The results reported here are based on a single European population and their applicability to other countries and ancestries is unknown. GS:SFHS is a family-based sample drawn from the general population. Therefore, the more extreme phenotypes that have been analysed in the published literature would not have provided adequate power in the studied cohort. Ascertainment bias may also be present in GS:SFHS; for example, preterm birth can compromise normal neurodevelopment and increase an individual’s risk of chronic disease in later life (61), which in turn may limit participation in the study. There was also no measure of childhood abuse or neglect collected in GS:SFHS and those phenotypes may provide additional avenues for investigation using a similar methodology to that used here. Studies with larger sample sizes have reported phenotypic associations between the early life variables and mental health; however, in GS:SFHS many of these associations were not significant. This suggests that the current study was underpowered, there are differences in the variables examined, other studies were false positives or a combination of those factors. Finally, the DNA methylation data analysed was obtained from blood and the analysis of other tissue samples may reveal additional associations with the phenotypes examined.
In conclusion, there were plausible CpG sites associated with preterm birth, low birth weight and having a young parent in both MWAS 1 and MWAS 2. Further, one of the more interesting findings was the association between birth date and blood cell type composition for neutrophils and lymphocytes. It was not possible to predict either MDD or BRS from methylation profile scores calculated from early life phenotypes. Although, there was significant predictive ability of the methylation profile scores across the two sets of GS:SFHS data for birth month and having a young parent for their respective phenotypes.
Materials and Methods
Generation Scotland: Scottish Family Health Study (GS:SFHS)
GS:SFHS (26) is a family-based cohort study of 24 080 participants (14 154 female and 9926 male) aged between 18 and 100 (mean = 47.6 years, standard deviation [SD] = 15.4 years). Baseline data were collected between 2006 and 2011 and covered medical, behaviour and lifestyle factors with a subset recontacted (N = 9618) in 2015 and 2016 with additional phenotypes collected (62). Clinical information from linked electronic hospital records across the life course was also available. At the baseline appointment, a blood draw was taken from each participant, which has so far been used to obtain DNA methylation data for 9773 individuals.
Phenotypes
Multiple phenotypes were generated from the baseline and recontact data focused on either early life environments or adult mental health. The early life environments were broadly categorized as either biological (preterm birth, low birth weight, birth date and birth month) or sociodemographic (having a lone parent, having a young parent, urbanicity and population density). The adult mental health measures were MDD and psychological resilience (measured using the BRS). The demographics of GS:SFHS are provided in Table 3.
Table 3.
Demographic information for Generation Scotland: Scottish Family Health Study
| Whole sample | Methylation sample | |||||
|---|---|---|---|---|---|---|
| Item | N | Present/absent (%) | Mean, SD | N | Present/absent (%) | Mean, SD |
| Female | 24 080 | 14 154/9926 (58.8) | 9537 | 5638/3899 (59.1) | ||
| Age | 24 080 | 47.64, 15.41 | 9537 | 49.81, 13.70 | ||
| Preterm birth | 3134 | 109/3025 (3.5) | 950 | 31/919 (3.3) | ||
| Low birth weight | 3129 | 103/3026 (3.3) | 949 | 30/919 (3.2) | ||
| Birth month | 24 080 | 12 842/11 238 (53.3) | 9516 | 5459/4057 (57.4) | ||
| Birth date | 22 656 | 0.01, 0.71 | 9509 | 0.01, 0.71 | ||
| Young parent | 23 546 | 1241/22 305 (5.3) | 9398 | 534/8864 (5.7) | ||
| Lone parent | 22 543 | 1333/21 210 (5.9) | 9086 | 580/8506 (6.4) | ||
| Urban environment | 19 428 | 11 739/7689 (60.4) | 8017 | 5468/2549 (68.2) | ||
| Population density | 19 426 | 1949.72, 1611.32 | 8017 | 2019.44, 1489.35 | ||
| Major depressive disorder | 21 340 | 2766/18 574 (13.0) | 9481 | 1624/7857 (17.1) | ||
| Brief resilience scale | 9354 | 0.00, 3.80 | 4839 | −0.19, 3.90 | ||
Notes: Number of individuals (N) available for each item. The present N, absent N and prevalence (%) are reported for binary items with mean and SD reported for continuous items.
Age was reported in years, a preterm birth was recorded as present for those born < 37 weeks gestation length, a low birth weight was recorded as present for individuals born in the 3rd centile for birth weight adjusted for sex and gestation length, birth month compares those born between April and October inclusive (present) with those born the rest of the year (absent), birth date is a continuous measure of when an individual was born during the year with a maximum value for those born 21st June and a minimum value for those born 21st December, a young parent was present for individuals when either parent was <21 years old when the individual was born, a lone parent was recorded as present for individuals who were not living with both parents when the individual was born, an urban environment was present for individuals recorded as living in Edinburgh, Glasgow, Aberdeen or Dundee when they were born, population density was the number of individuals per square kilometre in their region when an individual was born, MDD status was based on a structured clinical interview and brief resilience scale was a continuous measure of an individual’s psychological resilience.
Preterm births were categorized as a recorded gestation of period of <37 weeks [based on World Health Organization guidance (63)] using the SMR02—Maternity Inpatient and Day Case linked electronic health records. These records were available for individuals born after 1992. Birth weight for GS:SFHS participants was also obtained from SMR02 electronic health records. The threshold for low birth weight was based on the sex and gestation length adjusted 3rd centile for birth weight reported using a Scottish sample and SMR02 records by Bonellie et al. (64). Preterm birth and low birth weight were assessed as binary traits following Nosarti et al. (9) and Colman et al. (65).
Seasonality of birth was assessed using two phenotypes. First, a binary phenotype for birth month was generated with those born between April and October inclusive compared with those born during the remaining months. These months were selected based on the review of birth month and depression by Schnittker (12), with increased risk of depression reported for those born April through October. Second, the birth date during the year was assessed as a continuous phenotype (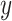 ) and calculated as:
) and calculated as:
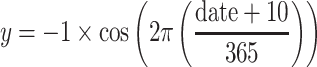 |
where 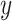 ranged from −1 for those born on the winter solstice (21st December) to +1 for those born on the summer solstice (21st June).
ranged from −1 for those born on the winter solstice (21st December) to +1 for those born on the summer solstice (21st June). 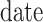 was the day of birth during the year for each participant (January 1st = 1, January 2nd = 2, etc.). For those born on the 29th February,
was the day of birth during the year for each participant (January 1st = 1, January 2nd = 2, etc.). For those born on the 29th February, 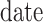 = 59.5.
= 59.5.
Using the self-reported age of an individual’s parents at the time of birth, an individual was classified as having a young parent if either parent was under the age of 21. Where an individual was recorded as having a lone parent (see below), the age of the parent that the individual lived with was used.
An individual was recorded as having a lone parent if either their mother or father was recorded as living in a different country or region to the individual and the other parent at the time of birth of the individual. The regions reflect the 32 council areas of Scotland. Individuals with missing information, or where an individual was reported not to live with either parent, were excluded.
The region that an individual was living in at the time of their birth was also used to define an urban environment phenotype. Individuals living in Edinburgh, Glasgow, Aberdeen or Dundee were classified as being urban and those living in other regions as non-urban. The year of an individual’s birth and the region they were living in were also used to obtain a measure of population density based on population estimates from the National Records of Scotland (66) and was recorded as the number of individuals per square kilometre.
To determine MDD status, the initial screening questions from the Structured Clinical Interview for DSM-IV (SCID) Non-Patient Version (67) were used to identify those individuals that would subsequently complete the mood sections of the SCID. The SCID was administered by nurses trained in its application and further information on the MDD criteria used in GS:SFHS is reported in Fernandez-Pujals et al. (68). Participants who met the criteria for at least one MDD episode in the mood sections of the SCID were classified as cases and those who did not meet this criterion or did not report MDD symptoms in the initial screening were classified as controls.
The BRS (69) was used to determine a continuous measure of an individual’s psychological resilience and ability to ‘bounce back’ from stressful events. BRS was obtained for a subset (N = 9505) of the cohort during a recontact of all GS:SFHS participants in 2015 and 2016. BRS was ascertained from the response to six questions using a 5-point Likert scale with further details on the assessment provided by Navrady et al. (62). Individuals that responded to five or more questions were retained (N = 9354) with 55 missing values imputed using the missForest package (70) applying 500 trees per forest. A principal component analysis using the Psych package (71) was applied to the data to extract the first unrotated principal component which was then scaled to create a continuous measure of BRS.
Regression of adult mental health on early life environments
To determine whether there was an association between early life stressors and the measures of adult mental health a generalized linear mixed model was used from the lme4qtl package (72). MDD and BRS were assessed separately as the dependent variable for an association with each early life factor in turn as the independent variable. The total genetic value derived from pedigree data [with the variance/covariance structure defined by a kinship matrix using the kinship2 package (73)] was fitted as a random effect, jointly with sex fitted as a fixed effect. Binary traits were fitted as factors and continuous traits were centred and scaled to have a mean of 0 and a SD of 1. A binomial regression with a logit function was used to assess associations with MDD. For a significant association between a mental health trait and an early life environment a Bonferroni correction was applied to the P-values within each mental health trait, P < 6.25 × 10−3 (α = 0.05/8).
Methylation data
The Infinium MethylationEPIC BeadChip (Illumina Inc., USA) was used to profile DNA methylation data at 853 307 CpG sites generated in two sets of individuals. There were 5190 individuals in Set 1 and 4583 unrelated individuals in Set 2, with no related individuals between the sets. Quality control was then applied to both Sets and full details are provided by Barbu et al. (60) and McCartney et al. (74). In summary, individuals were excluded if they were outliers based on multidimensional scaling, the predicted sex from the methylation data mismatched the recorded sex, or where ≥1% of CpG sites had a detection P-value > 0.05. CpG sites were excluded where (i) the beadcount was <3 in >5% of individuals or (ii) where sites in which ≥0.5% of individuals had a detection P-value > 0.05. The sites identified as non-specific binding and/or polymorphisms at the target site by McCartney et al. (75) were removed. The log2 ratio of the intensities of methylated probe versus unmethylated probe data was used to generate methylation M-values separately in each Set (76). In total there were 713 522 CpG sites remaining with 5087 individuals in Set 1 and 4450 individuals in Set 2.
After combining the Sets (9537 individuals), correction was applied for:
(1) Technical variation, where M-values were included as dependent variables in a mixed linear model adjusting for the plate used to profile the DNA methylation data and the date of the individual’s blood draw as random effects, jointly with plate position, Set, clinic, year of appointment, day of week of appointment and the first 10 principal components (from the EPIC array control sites) as fixed effects; and
(2) Biological variation by fitting variables of (1) as dependent variables in a second mixed linear model adjusting for genetic and common family shared environmental contributions [classed as G: common genetic; K: kinship; F: nuclear family; C: couple and S: sibling, see Xia et al. (77) and Zeng et al. (78) for further information] as random effects, jointly with sex, age and estimated blood cell type composition [CD8T, CD4T, NK, Bcell, Mono, Gran; obtained using the Houseman algorithm (79) within the Meffil package (80)] as fixed effects.
MWAS of early life environments and adult mental health
Two methods (MWAS 1 and MWAS 2) were used to conduct the MWAS generating two sets of summary statistics for each phenotype. MWAS 1 was conducted using the eBayes function within the limma package (81), which applies a linear mixed model to the data. The M-values, corrected for technical and biological variation, were the dependent variable with each phenotype included separately as the independent variable. Set, smoking status (ever or never), smoking pack years, and the first 20 principal components created from the M-values using the FactoMineR package (82), were fitted as fixed effects.
MWAS 2 was conducted using the software tool OmicS-data-based Complex trait Analysis (OSCA) (83). First, to account for methylome-wide correlational structure, this tool uses a linear regression analysis to identify groups of lead CpG sites based on the association test statistics. Each phenotype in turn was the dependent variable with the corrected M-values as the independent variable adjusted for the same covariates used in MWAS 1 (Set, smoking status, smoking pack years, and the first 20 principal components). Second, each group of lead sites were fitted as random effects in a Multi-component MLM-based association excluding the target (MOMENT) analysis to assess the effect of each target probe in turn, with the respective phenotype as the dependent variable and the M-values as the independent variable. MOMENT also fits predicted blood cell type composition [basophil, eosinophil, lymphocyte, monocyte and neutrophil; based on haematological analysis of the LBC (84)] as a fixed effect.
A methylome-wide significance threshold was determined using a Bonferroni correction based on the number of CpG sites analysed for each analysis: P < 7.01 × 10−8 (α = 0.05/713 522). To visualise the MWAS output, Miami plots were created using ggplot2 (85) with the P-values for MWAS 1 on the log10 scale and for MWAS 2 on the -log10 scale. QQ-plots were created using Haplin (86) with the shaded error representing the 95% confidence interval. Genomic inflation (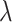 ) was calculated for each output as the median of the observed chi-squared distribution of P-values divided by the median of the expected chi-squared distribution.
) was calculated for each output as the median of the observed chi-squared distribution of P-values divided by the median of the expected chi-squared distribution.
Gene and gene-set analysis
The significant CpG sites (P < 7.01 × 10−8) for each phenotype and each MWAS were annotated to genes based on location. For MWAS 1, the missMethyl package (87) was used to annotate genes according to location. For MWAS 2, the annotations reported by the OSCA package were used. The gometh function within missMethyl was used to analyse the data for enrichment of Gene Ontology and KEGG gene-sets. The overlap of the annotated genes and enriched gene-sets between the early life environment and adult mental health phenotypes were examined.
Mediation analysis
During the revision of the manuscript a reviewer suggested including a mediation analysis of two CpG sites associated with birthweight and located close to GRM8: cg21803443 identified in the present study and cg15908975 identified by Küpers et al. (29). GRM8 has previously been associated with depression (30). A mediation analysis (88) fitting birth weight as the independent variable, MDD as the dependent variable, and either cg21803443 or cg15908975 as the mediator variable was conducted.
Methylation profile scores
MWAS 2 was used to calculate summary statistics for each early life environment phenotype using the individuals in Set 1 (N = 5190). Multiple P-value thresholds (< 10−7, < 10−6, < 10−5, < 10−4, < 10−3, < 10−2 and < 10−1) were used to identify those sites for inclusion. For each individual in Set 2, an aggregated methylation profile score was calculated by multiplying the CpG effect sizes from Set 1 by their respective M-values in Set 2. There were no related individuals between Set 1 and Set 2. The ability of these profile scores to predict the same phenotype, MDD or BRS, was assessed after adjusting for sex, age, and the first 20 principal components (derived from single nucleotide polymorphism data) as fixed effects. For prediction of binary phenotypes, a coefficient of determination was calculated using Nagelkerke’s R2 (89) using a population prevalence of that equal to the sample. For quantitative phenotypes, a simple linear regression was used with the adjusted R2 reported. A Bonferroni correction was used to identify significant prediction based on the number of early life phenotypes examined in each case: P < 6.25 × 10−3 (α = 0.05/8).
During the revision of the manuscript a reviewer suggested examining whether methylation profile scores calculated from continuous measures of preterm birth or birth weight predicted MDD or BRS. The most significant CpG site from each of the 2375 differentially methylated region identified for gestational age from Merid et al. (31) and the 914 CpG sites prioritized (P < 1.06 × 10−7 and I2 ≤ 50%) by Küpers et al. (29) for birth weight were used to calculate the respective profile score for prediction of MDD or BRS in Set 2. The same fixed effects and method for calculating R2 as described in the previous paragraph were used.
Supplementary Material
Acknowledgements
This investigation represents independent research part-funded by the National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
This research was funded in whole, or in part, by the Wellcome Trust (Reference 213674/Z/18/Z). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR03006) and is currently supported by the Wellcome Trust (216767/Z/19/Z). Genotyping of the GS:SFHS samples was carried out by the Genetics Core Laboratory at the Edinburgh Clinical Research Facility, University of Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award ‘STratifying Resilience and Depression Longitudinally’ (STRADL) Reference 104036/Z/14/Z). We are grateful to all the families who took part, the general practitioners and the Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, including interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, healthcare assistants and nurses. Ethics approval for the study was given by the NHS Tayside committee on research ethics (reference 05/S1401/8). We would like to thank Simon Cox for his contribution to running analyses on the Lothian Birth Cohorts of 1921 and 1936.
LBC1921 funding has been received from the UK’s Biotechnology and Biological Sciences Research Council (BBSRC) (15/SAG09977, wave 1), a Royal Society-Wolfson Research Merit Award to IJD (wave 2), the Chief Scientist Office (CSO) of the Scottish Government’s Health Directorates (CZG/3/2/79, post-wave 1 questionnaire study; CZB/4/505, wave 3; ETM/55, wave 4) and the UK’s Medical Research Council (MRC) Centenary Early Career Award to Dr Tom Booth (wave 5). Funding for LBC1936 has been received from Research Into Ageing (Programme grant 251; wave 1), and Age UK (Disconnected Mind Programme grant) and the UK’s Medical Research Council (G0701120, wave 2; G1001245, wave 3; MR/M013111/1, wave 4). The Alzheimer Scotland Dementia Research Centre funded LBC1936 dementia ascertainment. BBSRC funded whole-genome sequencing of both cohorts. Wellcome, the University of Edinburgh, the University of Queensland and Age UK funded DNA methylation analysis in both cohorts.
Conflict of interest statement. A.M.McI. has received research support from Eli Lilly and Company, Janssen and the Sackler Trust and speaker fees from Illumina and Janssen. C.M.L is a member of the Myriad Neuroscience SAB.
Contributor Information
David M Howard, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London SE5 8AF, UK; Division of Psychiatry, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh EH10 5HF, UK.
Oliver Pain, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London SE5 8AF, UK.
Ryan Arathimos, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London SE5 8AF, UK; NIHR Maudsley Biomedical Research Centre, South London and Maudsley NHS Trust, London SE5 8AF, UK.
Miruna C Barbu, Division of Psychiatry, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh EH10 5HF, UK.
Carmen Amador, MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh EH4 2XU, UK.
Rosie M Walker, Centre for Genomic and Experimental Medicine, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH4 2XU, UK; Centre for Clinical Brain Sciences, Chancellor’s Building, 49 Little France Crescent, University of Edinburgh, Edinburgh EH16 4SB, UK.
Bradley Jermy, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London SE5 8AF, UK; NIHR Maudsley Biomedical Research Centre, South London and Maudsley NHS Trust, London SE5 8AF, UK.
Mark J Adams, Division of Psychiatry, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh EH10 5HF, UK.
Ian J Deary, Lothian Birth Cohorts, Department of Psychology, University of Edinburgh, Edinburgh EH8 9JZ, UK.
David Porteous, Centre for Genomic and Experimental Medicine, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH4 2XU, UK.
Archie Campbell, Centre for Genomic and Experimental Medicine, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH4 2XU, UK; Usher Institute for Population Health Sciences and Informatics, University of Edinburgh, Edinburgh EH16 4UX, UK.
Patrick F Sullivan, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm 171 77, Sweden; Department of Genetics, University of North Carolina, Chapel Hill, NC 27599, USA; Department of Psychiatry, University of North Carolina, Chapel Hill, NC 27514, USA.
Kathryn L Evans, Centre for Genomic and Experimental Medicine, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH4 2XU, UK.
Louise Arseneault, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London SE5 8AF, UK.
Naomi R Wray, Queensland Brain Institute, University of Queensland, Brisbane, Queensland 4072, Australia.
Michael Meaney, Douglas Hospital Research Centre, Douglas Mental Health University Institute, McGill University, Montréal, QC H4H 1R3, Canada; Ludmer Centre for Neuroinformatics and Mental Health, McGill University, Montréal, QC H3T 1E2, Canada; Singapore Institute for Clinical Sciences, Singapore 117609, Singapore.
Andrew M McIntosh, Division of Psychiatry, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh EH10 5HF, UK; Lothian Birth Cohorts, Department of Psychology, University of Edinburgh, Edinburgh EH8 9JZ, UK.
Cathryn M Lewis, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London SE5 8AF, UK; NIHR Maudsley Biomedical Research Centre, South London and Maudsley NHS Trust, London SE5 8AF, UK.
Data availability
According to Wellcome Trust’s Policy on data, software and materials management and sharing, all data supporting this study will be openly available at https://doi.org/10.7488/ds/3126.
Funding
Sir Henry Wellcome Postdoctoral Fellowship (Reference 213674/Z/18/Z to D.M.H.); the Brain & Behavior Research Foundation (2018 NARSAD Young Investigator Grant; Ref: 27404 to D.M.H. and Ref: 21956 to K.L.E); Wellcome Trust (104036/Z/14/Z and 216767/Z/19/Z to A.M.McI); UKRI MRC funding (MC PC 17209 and MR/S035818/1 to A.M.McI and MR/N015746/1 to C.M.L); the Medical Research Council (MRC) UK (grants MC_PC_U127592696 and MC_PC_U127561128 to C.A.); Medical Research Council and the Biotechnology and Biological Sciences Research Council (MR/K026992/1 to I.J.D.); Alzheimer’s Research UK major project grant (ARUK-PG2017B-10 to R.E.M.); NHMRC (grants 1078901 and 1087889 to N.R.W.). C.M.L. acknowledges MRC grant MR/N015746/1. K.L.E is supported by a NARSAD Independent Investigator Award (Ref: 21956).
References
- 1. Kessler, R.C., Davis, C.G. and Kendler, K.S. (1997) Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychol. Med., 27, 1101–1119. [DOI] [PubMed] [Google Scholar]
- 2. Bellis, M.A., Lowey, H., Leckenby, N., Hughes, K. and Harrison, D. (2013) Adverse childhood experiences: retrospective study to determine their impact on adult health behaviours and health outcomes in a UK population. J. Public Health, 36, 81–91. [DOI] [PubMed] [Google Scholar]
- 3. Horwitz, A.V., Widom, C.S., McLaughlin, J. and White, H.R. (2001) The impact of childhood abuse and neglect on adult mental health: a prospective study. J. Health Soc. Behav., 42, 184–201. [PubMed] [Google Scholar]
- 4. Molnar, B.E., Berkman, L.F. and Buka, S.L. (2001) Psychopathology, childhood sexual abuse and other childhood adversities: relative links to subsequent suicidal behaviour in the US. Psychol. Med., 31, 965–977. [DOI] [PubMed] [Google Scholar]
- 5. Springer, K.W., Sheridan, J., Kuo, D. and Carnes, M. (2007) Long-term physical and mental health consequences of childhood physical abuse: results from a large population-based sample of men and women. Child Abuse Negl., 31, 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morgan, C., Gayer-Anderson, C., Beards, S., Hubbard, K., Mondelli, V., Di Forti, M., Murray, R.M., Pariante, C., Dazzan, P., Craig, T.J.et al. (2020) Threat, hostility and violence in childhood and later psychotic disorder: population-based case–control study. Br. J. Psychiatry, 217, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mathewson, K.J., Chow, C.H.T., Dobson, K.G., Pope, E.I., Schmidt, L.A. and Van Lieshout, R.J. (2017) Mental health of extremely low birth weight survivors: a systematic review and meta-analysis. Psychol. Bull., 143, 347–383. [DOI] [PubMed] [Google Scholar]
- 8. Johnson, S. and Marlow, N. (2014) Growing up after extremely preterm birth: lifespan mental health outcomes. Semin. Fetal Neonatal Med., 19, 97–104. [DOI] [PubMed] [Google Scholar]
- 9. Nosarti, C., Reichenberg, A., Murray, R.M., Cnattingius, S., Lambe, M.P., Yin, L., MacCabe, J., Rifkin, L. and Hultman, C.M. (2012) Preterm birth and psychiatric disorders in young adult life. Arch. Gen. Psychiatry, 69, 610–617. [DOI] [PubMed] [Google Scholar]
- 10. Disanto, G., Morahan, J.M., Lacey, M.V., DeLuca, G.C., Giovannoni, G., Ebers, G.C. and Ramagopalan, S.V. (2012) Seasonal distribution of psychiatric births in England. PLoS One, 7, e34866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joiner, T.E., Pfaff, J.J., Acres, J.G. and Johnson, F. (2002) Birth month and suicidal and depressive symptoms in Australians born in the Southern vs. the Northern hemisphere. Psychiatry Res., 112, 89–92. [DOI] [PubMed] [Google Scholar]
- 12. Schnittker, J. (2018) Season of birth and depression in adulthood: revisiting historical forerunner evidence for in-utero effects. SSM-Population Health, 4, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGrath, J.J., Petersen, L., Agerbo, E., Mors, O., Mortensen, P.B. and Pedersen, C.B. (2014) A comprehensive assessment of parental age and psychiatric disorders. JAMA Psychiat., 71, 301–309. [DOI] [PubMed] [Google Scholar]
- 14. Mäkikyrö, T., Sauvola, A., Moring, J., Veijola, J., Nieminen, P., Järvelin, M.R. and Isohanni, M. (1998) Hospital-treated psychiatric disorders in adults with a single-parent and two-parent family background: a 28-year follow-up of the 1966 Northern Finland Birth Cohort. Fam. Process, 37, 335–344. [DOI] [PubMed] [Google Scholar]
- 15. Tost, H., Champagne, F.A. and Meyer-Lindenberg, A. (2015) Environmental influence in the brain, human welfare and mental health. Nat. Neurosci., 18, 1421–1431. [DOI] [PubMed] [Google Scholar]
- 16. Gruebner, O., Rapp, M.A., Adli, M., Kluge, U., Galea, S. and Heinz, A. (2017) Cities and mental health. Dtsch. Arztebl. Int., 114, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okkels, N., Kristiansen, C.B., Munk-Jørgensen, P. and Sartorius, N. (2018) Urban mental health: challenges and perspectives. Curr. Opin. Psychiatry, 31, 258–264. [DOI] [PubMed] [Google Scholar]
- 18. Feil, R. and Fraga, M.F. (2012) Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet., 13, 97–109. [DOI] [PubMed] [Google Scholar]
- 19. Klengel, T., Pape, J., Binder, E.B. and Mehta, D. (2014) The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology, 80, 115–132. [DOI] [PubMed] [Google Scholar]
- 20. Tsankova, N., Renthal, W., Kumar, A. and Nestler, E.J. (2007) Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci., 8, 355–367. [DOI] [PubMed] [Google Scholar]
- 21. Fries, G.R., Li, Q., McAlpin, B., Rein, T., Walss-Bass, C., Soares, J.C. and Quevedo, J. (2016) The role of DNA methylation in the pathophysiology and treatment of bipolar disorder. Neurosci. Biobehav. Rev., 68, 474–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smigielski, L., Jagannath, V., Rössler, W., Walitza, S. and Grünblatt, E. (2020) Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings. Mol. Psychiatry, 25, 1718–1748. [DOI] [PubMed] [Google Scholar]
- 23. Story Jovanova, O., Nedeljkovic, I., Spieler, D., Walker, R.M., Liu, C., Luciano, M., Bressler, J., Brody, J., Drake, A.J., Evans, K.L.et al. (2018) DNA methylation signatures of depressive symptoms in middle-aged and elderly persons: meta-analysis of multiethnic epigenome-wide studies. JAMA Psychiat., 75, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park, C., Rosenblat, J.D., Brietzke, E., Pan, Z., Lee, Y., Cao, B., Zuckerman, H., Kalantarova, A. and McIntyre, R.S. (2019) Stress, epigenetics and depression: a systematic review. Neurosci. Biobehav. Rev., 102, 139–152. [DOI] [PubMed] [Google Scholar]
- 25. Border, R., Johnson, E.C., Evans, L.M., Smolen, A., Berley, N., Sullivan, P.F. and Keller, M.C. (2019) No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am. J. Psychiatry, 176, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith, B.H., Campbell, A., Linksted, P., Fitzpatrick, B., Jackson, C., Kerr, S.M., Deary, I.J., MacIntyre, D.J., Campbell, H., McGilchrist, M.et al. (2013) Cohort profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. Int. J. Epidemiol., 42, 689–700. [DOI] [PubMed] [Google Scholar]
- 27. Navrady, L., Zeng, Y., Clarke, T., Adams, M., Howard, D., Deary, I. and McIntosh, A. (2018) Genetic and environmental contributions to psychological resilience and coping [version 1; peer review: 2 approved]. Wellcome Open Research, 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heim, C. and Binder, E.B. (2012) Current research trends in early life stress and depression: review of human studies on sensitive periods, gene–environment interactions, and epigenetics. Exp. Neurol., 233, 102–111. [DOI] [PubMed] [Google Scholar]
- 29. Küpers, L.K., Monnereau, C., Sharp, G.C., Yousefi, P., Salas, L.A., Ghantous, A., Page, C.M., Reese, S.E., Wilcox, A.J., Czamara, D.et al. (2019) Meta-analysis of epigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat. Commun., 10, 1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howard, D.M., Adams, M.J., Clarke, T.-K., Hafferty, J.D., Gibson, J., Shirali, M., Coleman, J.R.I., Hagenaars, S.P., Ward, J., Wigmore, E.M.et al. (2019) Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci., 22, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merid, S.K., Novoloaca, A., Sharp, G.C., Küpers, L.K., Kho, A.T., Roy, R., Gao, L., Annesi-Maesano, I., Jain, P., Plusquin, M.et al. (2020) Epigenome-wide meta-analysis of blood DNA methylation in newborns and children identifies numerous loci related to gestational age. Genome Med., 12, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Lieshout, R.J., Ferro, M.A., Schmidt, L.A., Boyle, M.H., Saigal, S., Morrison, K.M. and Mathewson, K.J. (2018) Trajectories of psychopathology in extremely low birth weight survivors from early adolescence to adulthood: a 20-year longitudinal study. J. Child Psychol. Psychiatry, 59, 1192–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tarui, T., Kim, A., Flake, A., McClain, L., Stratigis, J.D., Fried, I., Newman, R., Slonim, D.K. and Bianchi, D.W. (2017) Amniotic fluid transcriptomics reflects novel disease mechanisms in fetuses with myelomeningocele. Am. J. Obstet. Gynecol., 217, 217, 587.e211–587.e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Purisch, S.E., DeFranco, E.A., Muglia, L.J., Odibo, A.O. and Stamilio, D.M. (2008) Preterm birth in pregnancies complicated by major congenital malformations: a population-based study. Am. J. Obstet. Gynecol., 199, 287.e281–287.e288. [DOI] [PubMed] [Google Scholar]
- 35. Zhao, R., Chinai, J.M., Buhl, S., Scandiuzzi, L., Ray, A., Jeon, H., Ohaegbulam, K.C., Ghosh, K., Zhao, A., Scharff, M.D.et al. (2013) HHLA2 is a member of the B7 family and inhibits human CD4 and CD8 T-cell function. Proc. Natl. Acad. Sci., 110, 9879–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dubielecka, P.M., Ladwein, K.I., Xiong, X., Migeotte, I., Chorzalska, A., Anderson, K.V., Sawicki, J.A., Rottner, K., Stradal, T.E. and Kotula, L. (2011) Essential role for ABI1 in embryonic survival and WAVE2 complex integrity. Proc. Natl. Acad. Sci., 108, 7022–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ring, C., Ginsberg, M.H., Haling, J. and Pendergast, A.M. (2011) Abl-interactor-1 (ABI1) has a role in cardiovascular and placental development and is a binding partner of the α4 integrin. Proc. Natl. Acad. Sci., 108, 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sparrow, S., Manning, J.R., Cartier, J., Anblagan, D., Bastin, M.E., Piyasena, C., Pataky, R., Moore, E.J., Semple, S.I., Wilkinson, A.G.et al. (2016) Epigenomic profiling of preterm infants reveals DNA methylation differences at sites associated with neural function. Transl. Psychiatry, 6, e716–e716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lund, L.K., Vik, T., Lydersen, S., Løhaugen, G.C.C., Skranes, J., Brubakk, A.-M. and Indredavik, M.S. (2012) Mental health, quality of life and social relations in young adults born with low birth weight. Health Qual. Life Outcomes, 10, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Husby, I.M., Stray, K.M.-T., Olsen, A., Lydersen, S., Indredavik, M.S., Brubakk, A.-M., Skranes, J. and Evensen, K.A.I. (2016) Long-term follow-up of mental health, health-related quality of life and associations with motor skills in young adults born preterm with very low birth weight. Health Qual. Life Outcomes, 14, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen, Z., Montcouquiol, M., Calderon, R., Jenkins, N.A., Copeland, N.G., Kelley, M.W. and Noben-Trauth, K. (2008) JXC1/SOBP, encoding a nuclear zinc finger protein, is critical for cochlear growth, cell fate, and patterning of the organ of corti. J. Neurosci., 28, 6633–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roepman, R., Bernoud-Hubac, N., Schick, D.E., Maugeri, A., Berger, W., Ropers, H.-H., Cremers, F.P.M. and Ferreira, P.A. (2000) The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum. Mol. Genet., 9, 2095–2105. [DOI] [PubMed] [Google Scholar]
- 43. Cristobal, R. and Oghalai, J.S. (2008) Hearing loss in children with very low birth weight: current review of epidemiology and pathophysiology. Arch. Dis. Child. Fetal Neonatal Ed., 93, F462–F468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Connor, A.R., Wilson, C.M. and Fielder, A.R. (2007) Ophthalmological problems associated with preterm birth. Eye, 21, 1254–1260. [DOI] [PubMed] [Google Scholar]
- 45. Madden, R.A., McCartney, D.L., Walker, R.M., Hillary, R.F., Bermingham, M.L., Rawlik, K., Morris, S.W., Campbell, A., Porteous, D.J., Deary, I.J.et al. (2020) Birth weight associations with DNA methylation differences in an adult population. Epigenetics, 16, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor, A.M., Pattie, A. and Deary, I.J. (2018) Cohort profile update: the Lothian birth cohorts of 1921 and 1936. Int. J. Epidemiol., 47, 1042–1042r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dopico, X.C., Evangelou, M., Ferreira, R.C., Guo, H., Pekalski, M.L., Smyth, D.J., Cooper, N., Burren, O.S., Fulford, A.J., Hennig, B.J.et al. (2015) Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun., 6, 7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thysen, A.H., Rasmussen, M.A., Kreiner-Møller, E., Larsen, J.M., Følsgaard, N.V., Bønnelykke, K., Stokholm, J., Bisgaard, H. and Brix, S. (2016) Season of birth shapes neonatal immune function. J. Allergy Clin. Immunol., 137, 1238–1246.e1213. [DOI] [PubMed] [Google Scholar]
- 49. Disanto, G., Watson, C.T., Meier, U.C., Ebers, G.C., Giovannoni, G. and Ramagopalan, S.V. (2013) Month of birth and thymic output. JAMA Neurol., 70, 527–528. [DOI] [PubMed] [Google Scholar]
- 50. Sundquist, K., Frank, G. and Sundquist, J. (2004) Urbanisation and incidence of psychosis and depression: follow-up study of 4.4 million women and men in Sweden. Br. J. Psychiatry, 184, 293–298. [DOI] [PubMed] [Google Scholar]
- 51. Hoare, E., Jacka, F. and Berk, M. (2019) The impact of urbanization on mood disorders: an update of recent evidence. Curr. Opin. Psychiatry, 32, 198–203. [DOI] [PubMed] [Google Scholar]
- 52. DeVylder, J.E., Kelleher, I., Lalane, M., Oh, H., Link, B.G. and Koyanagi, A. (2018) Association of urbanicity with psychosis in low- and middle-income countries. JAMA Psychiat., 75, 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kessler, R.C., Berglund, P., Demler, O., Jin, R., Koretz, D., Merikangas, K.R., Rush, A.J., Walters, E.E. and Wang, P.S. (2003) The epidemiology of major depressive disorder: results from the National Comorbidity Survey replication (NCS-R). JAMA, 289, 3095–3105. [DOI] [PubMed] [Google Scholar]
- 54. Bakolis, I., Hammoud, R., Stewart, R., Beevers, S., Dajnak, D., MacCrimmon, S., Broadbent, M., Pritchard, M., Shiode, N., Fecht, D.et al. (2020) Mental health consequences of urban air pollution: prospective population-based longitudinal survey. Soc. Psychiatry Psychiatr. Epidemiol., 56, 1587–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lorant, V., Deliège, D., Eaton, W., Robert, A., Philippot, P. and Ansseau, M. (2003) Socioeconomic inequalities in depression: a meta-analysis. Am. J. Epidemiol., 157, 98–112. [DOI] [PubMed] [Google Scholar]
- 56. Roberson-Nay, R., Lapato, D.M., Wolen, A.R., Lancaster, E.E., Webb, B.T., Verhulst, B., Hettema, J.M. and York, T.P. (2020) An epigenome-wide association study of early-onset major depression in monozygotic twins. Transl. Psychiatry, 10, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oh, G., Wang, S.-C., Pal, M., Chen, Z.F., Khare, T., Tochigi, M., Ng, C., Yang, Y.A., Kwan, A., Kaminsky, Z.A.et al. (2015) DNA modification study of major depressive disorder: beyond locus-by-locus comparisons. Biol. Psychiatry, 77, 246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Starnawska, A., Tan, Q., Soerensen, M., McGue, M., Mors, O., Børglum, A.D., Christensen, K., Nyegaard, M. and Christiansen, L. (2019) Epigenome-wide association study of depression symptomatology in elderly monozygotic twins. Transl. Psychiatry, 9, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Clark, S.L., Hattab, M.W., Chan, R.F., Shabalin, A.A., Han, L.K.M., Zhao, M., Smit, J.H., Jansen, R., Milaneschi, Y., Xie, L.Y.et al. (2020) A methylation study of long-term depression risk. Mol. Psychiatry, 25, 1334–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barbu, M.C., Shen, X., Walker, R.M., Howard, D.M., Evans, K.L., Whalley, H.C., Porteous, D.J., Morris, S.W., Deary, I.J., Zeng, Y.et al. (2020) Epigenetic prediction of major depressive disorder. Mol. Psychiatry, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mwaniki, M.K., Atieno, M., Lawn, J.E. and Newton, C.R.J.C. (2012) Long-term neurodevelopmental outcomes after intrauterine and neonatal insults: a systematic review. Lancet, 379, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Navrady, L.B., Wolters, M.K., MacIntyre, D.J., Clarke, T.K., Campbell, A.I., Murray, A.D., Evans, K.L., Seckl, J., Haley, C., Milburn, K.et al. (2018) Cohort profile: stratifying resilience and depression longitudinally (STRADL): a questionnaire follow-up of Generation Scotland: Scottish Family Health Study (GS:SFHS). Int. J. Epidemiol., 47, 13–14g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. World Health Organization . (2018), https://www.who.int/news-room/fact-sheets/detail/preterm-birth.
- 64. Bonellie, S., Chalmers, J., Gray, R., Greer, I., Jarvis, S. and Williams, C. (2008) Centile charts for birthweight for gestational age for Scottish singleton births. BMC Pregnancy Childbirth, 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Colman, I., Ataullahjan, A., Naicker, K. and Van Lieshout, R.J. (2012) Birth weight, stress, and symptoms of depression in adolescence: evidence of Fetal programming in a National Canadian Cohort. Can. J. Psychiatr., 57, 422–428. [DOI] [PubMed] [Google Scholar]
- 66. National Records of Scotland . (2019), https://www.nrscotland.gov.uk/statistics-and-data/statistics/statistics-by-theme/population/population-estimates.
- 67. First, M.B., Spitzer, R.L., Miriam Gibbon and Williams, J.B.W. (2002). Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition. (SCID-I/P). Biometrics Research, New York State Psychiatric Institute, New York.
- 68. Fernandez-Pujals, A.M., Adams, M.J., Thomson, P., McKechanie, A.G., Blackwood, D.H.R., Smith, B.H., Dominiczak, A.F., Morris, A.D., Matthews, K., Campbell, A.et al. (2015) Epidemiology and heritability of major depressive disorder, stratified by age of onset, sex, and illness course in Generation Scotland: Scottish Family Health Study (GS:SFHS). PLoS One, 10, e0142197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith, B.W., Dalen, J., Wiggins, K., Tooley, E., Christopher, P. and Bernard, J. (2008) The brief resilience scale: assessing the ability to bounce back. Int. J. Behav. Med., 15, 194–200. [DOI] [PubMed] [Google Scholar]
- 70. Stekhoven, D.J. and Bühlmann, P. (2011) MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics, 28, 112–118. [DOI] [PubMed] [Google Scholar]
- 71. Revelle, W. (2020) Procedures for psychological, psychometric, and personality research. Northwestern University, Evanston, Illinois. https://www.scholars.northwestern.edu/en/publications/psych-procedures-for-personality-and-psychological-research.
- 72. Ziyatdinov, A., Vázquez-Santiago, M., Brunel, H., Martinez-Perez, A., Aschard, H. and Soria, J.M. (2018) lme4qtl: linear mixed models with flexible covariance structure for genetic studies of related individuals. BMC Bioinformatics, 19, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sinnwell, J.P., Therneau, T.M. and Schaid, D.J. (2014) The kinship2 R package for pedigree data. Hum. Hered., 78, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McCartney, D.L., Hillary, R.F., Stevenson, A.J., Ritchie, S.J., Walker, R.M., Zhang, Q., Morris, S.W., Bermingham, M.L., Campbell, A., Murray, A.D.et al. (2018) Epigenetic prediction of complex traits and death. Genome Biol., 19, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McCartney, D.L., Walker, R.M., Morris, S.W., McIntosh, A.M., Porteous, D.J. and Evans, K.L. (2016) Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data, 9, 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Du, P., Zhang, X., Huang, C.-C., Jafari, N., Kibbe, W.A., Hou, L. and Lin, S.M. (2010) Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics, 11, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xia, C., Amador, C., Huffman, J., Trochet, H., Campbell, A., Porteous, D., Generation, S., Hastie, N.D., Hayward, C., Vitart, V.et al. (2016) Pedigree- and SNP-associated genetics and recent environment are the major contributors to anthropometric and cardiometabolic trait variation. PLoS Genet., 12, e1005804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zeng, Y., Amador, C., Xia, C., Marioni, R., Sproul, D., Walker, R.M., Morris, S.W., Bretherick, A., Canela-Xandri, O., Boutin, T.S.et al. (2019) Parent of origin genetic effects on methylation in humans are common and influence complex trait variation. Nat. Commun., 10, 1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Houseman, E.A., Accomando, W.P., Koestler, D.C., Christensen, B.C., Marsit, C.J., Nelson, H.H., Wiencke, J.K. and Kelsey, K.T. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Min, J.L., Hemani, G., Davey Smith, G., Relton, C. and Suderman, M. (2018) Meffil: efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics (Oxford, England), 34, 3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smyth, G.K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol., 3, 1. [DOI] [PubMed] [Google Scholar]
- 82. Lê, S., Josse, J. and Husson, F. (2008) FactoMineR: an R package for multivariate analysis. 2008, 25, 18. [Google Scholar]
- 83. Zhang, F., Chen, W., Zhu, Z., Zhang, Q., Nabais, M.F., Qi, T., Deary, I.J., Wray, N.R., Visscher, P.M., McRae, A.F.et al. (2019) OSCA: a tool for omic-data-based complex trait analysis. Genome Biol., 20, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Starr, J.M. and Deary, I.J. (2011) Sex differences in blood cell counts in the Lothian birth cohort 1921 between 79 and 87 years. Maturitas, 69, 373–376. [DOI] [PubMed] [Google Scholar]
- 85. Wickham, H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- 86. Gjessing, H.K. and Lie, R.T. (2006) Case-parent triads: estimating single- and double-dose effects of fetal and maternal disease gene haplotypes. Ann. Hum. Genet., 70, 382–396. [DOI] [PubMed] [Google Scholar]
- 87. Phipson, B., Maksimovic, J. and Oshlack, A. (2015) missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics, 32, 286–288. [DOI] [PubMed] [Google Scholar]
- 88. Tingley, D., Yamamoto, T., Hirose, K., Keele, L. and Imai, K. (2014) Mediation: R package for causal mediation analysis. (2014) J. Stat. Softw., 59, 38. [Google Scholar]
- 89. Lee, S.H., Goddard, M.E., Wray, N.R. and Visscher, P.M. (2012) A better coefficient of determination for genetic profile analysis. Genet. Epidemiol., 36, 214–224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
According to Wellcome Trust’s Policy on data, software and materials management and sharing, all data supporting this study will be openly available at https://doi.org/10.7488/ds/3126.


