Abstract
The Mucorales fungal genus Rhizopus is used for the industrial production of organic acids, enzymes and fermented foods. The metabolic engineering efficiency of Rhizopus could be improved using gene manipulation; however, exogenous DNA rarely integrates into the host genome. Consequently, a genetic tool for Mucorales fungi needs to be developed. Recently, programmable nucleases that generate DNA double-strand breaks (DSBs) at specific genomic loci have been used for genome editing in various organisms. In this study, we examined gene disruption in Rhizopus oryzae using transcription activator-like effector nucleases (TALENs), with and without exonuclease overexpression. TALENs with an overexpressing exonuclease induced DSBs, followed by target site deletions. Although DSBs are repaired mainly by nonhomologous end joining in most organisms, our results suggested that in R. oryzae microhomology-mediated end joining was the major DSB repair system. Our gene manipulation method using TALENs coupled with exonuclease overexpression contributes to basic scientific knowledge and the metabolic engineering of Rhizopus.
Keywords: TALEN, genome editing, Mucorales fungi, Rhizopus oryzae, exonuclease, metabolic engineering
Gene disruption was achieved in Rhizopus oryzae using Platinum TALEN (transcription activator-like effector nuclease) with newly identified exonuclease that stimulated microhomology-mediated end joining.
Introduction
The filamentous fungi of the genus Rhizopus are important microorganisms that have positive applications but negative health effects on humans. Some Rhizopus strains, such as Rhizopus oryzae, are used in the industrial production of organic acids, enzymes and fermented foods (Nahas 1988, Hachmeister and Fung 1993, Zhang et al. 2007). However, R. oryzae is also the main causative microorganism of mucormycosis, which results in a severe infection with a high mortality rate (Ibrahim et al. 2012). Additionally, the plant pathogen Rhizopus microsporus causes a severe crop disease (Partida-Martinez and Hertweck 2005). Therefore, the genetic modification of Rhizopus increases metabolite production rates and provides significant information on pathogenic mechanism-associated gene functions.
Currently, there are several genetic tools applicable to Rhizopus species, including Rhizopus niveus (Yanai et al. 1990, Liou et al. 1992, Takaya et al. 1994), Rhizopus delemar (Horiuchi et al. 1995) and R. oryzae (Skory 2002, Michielse et al. 2004). For example, vectors that complement the auxotrophy of some mutants have been developed to screen transformants. Additionally, an Agrobacterium-mediated transformation system has been developed for efficient DNA integration into R. oryzae. However, the only example of targeted gene disruption in Rhizopus strains facilitated by spontaneous homologous recombination (HR) is that of the high-affinity iron permease gene (ftr1) in R. oryzae (Ibrahim et al. 2010). Targeted gene manipulation by HR is a useful strategy for gene replacement, including the use of constructs that result in gene disruption and modification. However, spontaneous HR rarely occurs in most filamentous fungi (Weld et al. 2006). Moreover, exogenous DNA rarely integrates into the host genomes of Mucorales (e.g. Rhizopus, Mucor, Absidia, Phycomyces and Rhizomucor) compared with other filamentous fungi, because their exogenous DNA connects extrachromosomally (Skory 2004). In addition, the autonomous replication of exogenous DNA may occur without an origin of replication (Revuelta and Jayaram 1986, Yanai et al. 1990, Benito et al. 1995, Appel et al. 2004, Skory 2004). Therefore, targeted gene manipulation by spontaneous HR using exogenous donor DNA is very rare and unreliable in Mucorales.
Recently, programmable nucleases have enabled the introduction of DNA double-strand breaks (DSBs) at specific genomic loci. Programmable nucleases are customizable site-specific nucleases that trigger genetic engineering (Sakuma and Woltjen 2014). A variety of programmable nuclease-mediated genome manipulations, such as gene knockout, gene knock-in and chromosomal rearrangement, utilizing endogenous DSB repair pathways have been performed in various organisms. Among such genetic modifications, gene disruptions resulting from short insertions and deletions that are introduced by mutagenic end-joining repair pathways, such as nonhomologous end joining (NHEJ) and microhomology-mediated end joining (MMEJ), are particularly easy to apply. The reason for the ease of application is that these repair mechanisms are independent of HR and do not require the targeted integration of exogenous donor DNA (Kim and Kim 2014).
Transcription activator-like effector (TALE) nuclease (TALEN) is a programmable nuclease that consists of a DNA-binding domain and a DNA cleavage domain. The DNA-binding and cleavage domains originated from TALE and FokI endonucleases, respectively (Joung and Sander 2013). The efficiency of TALEN-mediated genome editing may be enhanced by several improvements. First, TALENs with variable repeats, known as Platinum TALENs, have higher DNA cleavage activity levels than conventional TALENs with constant repeats (Sakuma et al. 2013). Second, the introduction of TALENs along with exonuclease 1 has enhanced mutagenic efficiencies in rat fibroblasts and zygotes (Mashimo et al. 2013), supposedly because deletion mutations could efficiently be introduced by the resection of DSB ends by exonucleases. Another major programmable nuclease is part of the CRISPR (clustered regularly interspaced short palindromic repeats)–Cas system (Cong et al. 2013, Mali et al. 2013), which includes a Cas protein and single-guide RNA. In Mucorales, gene manipulation has been performed using the CRISPR–Cas system as the programmable nuclease (Nagy et al. 2017, Bruni et al. 2019). Although the usefulness of CRISPR–Cas system in basic research is indisputable, the rate of the off-target effects of TALENs is reportedly lower than that of the CRISPR–Cas system (Kim and Kim 2014). In addition, genome-editing efficiency of TALENs in heterochromatin target sites was reportedly higher than that of CRISPR–Cas system (Jain et al. 2021); therefore, gene manipulation with TALENs appears to result in safer and more versatile gene editing.
In this study, we developed a gene disruption method in R. oryzae using Platinum TALENs coupled with exonuclease overexpression, RoKem1 and RoXrn1, which were newly identified. We attempted to disrupt the trpC gene, one of the genes involved in the tryptophan biosynthesis, in which deficient mutants could be obtained by positive selection using 5-fluoroanthranilic acid (5-FAA). We found that the overexpression of one of the exonucleases, RoKem1, along with the introduction of Platinum TALENs was required for gene disruption in R. oryzae. This is the first report of TALEN-mediated gene disruption in Mucorales, and thus, the results contribute to our basic scientific knowledge, as well as metabolic engineering in Rhizopus strains.
Materials and methods
Strains, media and culture conditions
Rhizopus oryzae AM002, an adenine auxotrophic mutant, was used in this study (Fig. S1, Supporting Information). The strain was generated from R. oryzae NBRC5384 (NBRC, National Institute of Technology and Evaluation, Japan) by UV mutagenesis and is an adeA-deficient mutant. Three types of media were used in this study: (i) potato dextrose agar (BD Biosciences) was used as the complete medium containing adenine; (ii) minimum (M) medium without adenine was used for the selection of transformants; and (iii) M medium supplemented with cellobiose (CM) without adenine was used to induce expression from the amyA promoter. Rhizopus oryzae AM002 was grown on a plate containing potato dextrose agar or cultivation medium prepared in accordance with a previous report (Zhou et al. 1999) with some modifications. The M medium (20 g/L glucose, 1 g/L (NH4)2SO4, 0.6 g/L KH2PO4, 0.25 g/L MgSO4·7H2O, 0.09 g/L ZnSO4·7H2O and 15 g/L agar) and CM medium (glucose was replaced with cellobiose in the M medium) were prepared with and without 0.006 g/L tryptophan. Rhizopus oryzae AM002 was cultured at 30°C for 7 days after plating.
Sample preparation for the genome and transcriptome analyses
The fungal sample cultured on a plate containing potato dextrose agar for 7 days was used for genome sequencing. The sample for the RNA-seq analysis was prepared as described later. The germination culture was prepared by inoculating R. oryzae NBRC5384 spores (at a final concentration of 103 spores/mL) into a 500-mL Erlenmeyer flask with baffles containing 200 mL of potato dextrose broth (BD Biosciences) at 27°C and 170 rpm for 3 days. After broth filtration, the supernatant was removed. The growth culture was prepared by inoculating half the cells into a 500-mL Erlenmeyer flask containing 100 mL of agar-free M medium supplemented with 5 g/L CaCO3 at 27°C and 220 rpm for 40 h. After cell collection, the production culture was prepared by inoculating 6 g cells into a 200-mL Erlenmeyer flask containing 40 mL of agar-free M medium supplemented with 5 g/L CaCO3 at 27°C and 170 rpm for 8 h.
Genome sequencing and RNA-seq analyses
The fungal hyphae were frozen in liquid nitrogen and fractured with metal corn using a Multi-Beads Shocker (Yasui Kikai) at 4°C. Subsequently, the whole genome was extracted using Dr. GenTLE (from Yeast) High Recovery (TaKaRa), and total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen), in accordance with the manufacturers’ protocols. The draft genome sequencing using the HiSeq 2000 System (Illumina) was performed by Hokkaido System Science. The mRNA sequencing libraries were prepared using Illumina TruSeq RNA Sample Prep Kit v2 (Illumina) and sequenced using the MiSeq System (Illumina) with MiSeq Reagent Kit (300 cycles, Illumina). Sequencing data produced by the MiSeq System were analyzed using CLC Genomics Workbench (CLC bio).
Development of R. oryzae expression plasmids
For exogenous gene expression in R. oryzae, the following two vectors were constructed: pAmy_L_empty and pAmy_R_empty, which contained an empty gene cassette harboring the amyA promoter and pdcA terminator, with and without the adeA selection marker, respectively. The primers used and isolated promoter sequences are listed in Table S1 and Supplementary Sequences (Supporting Information), respectively.
pAde1, the adeA-expressing plasmid, was constructed by fusing the following two fragments using an In-Fusion HD Cloning Kit (TaKaRa): (i) the vector fragment from pPTR I DNA (TaKaRa) amplified with pPTR1-ptrProup-F and pPTR1-Nae1up-R primers and (ii) the insert adeA-gene fragment from the R. oryzae genome DNA amplified with ade1-up6 and ade1-down6 primers.
The pAmy_L_empty was constructed by fusing the following three fragments using an In-Fusion HD Cloning Kit: (i) the vector fragment from pAde1 amplified with pPTR1-Pst1-F and pPTR1-Pst1-R primers; (ii) the insert amyA-promoter fragment from the R. oryzae genome amplified with pPTR1-amyApro-F and amyApro-pdcAter-R primers; and (iii) the insert pdcA-terminator fragment from the R. oryzae genome amplified with amyApro-pdcAter-F and pdcAter-pPTR1-R.
The pAmy_R_empty was constructed by fusing the following three fragments using an In-Fusion HD Cloning kit: (i) the vector fragment from pUC18 amplified with pPTR1-sal1-F and pPTR1-sal1-R primers; (ii) the insert amyA-promoter fragment from the R. oryzae genome amplified with sal1-amyApro-F and amyApro-pdcAter-R primers; and (iii) the insert pdcA-terminator fragment from the R. oryzae genome amplified with amyApro-pdcAter-F and pdcAter-sal1-R primers.
After the cloning, BamHI and Esp3I sites in the adeA gene and pdcA terminator, respectively, were removed from pAmy_L_empty. Two and one Esp3I sites in the pUC18 vector and pdcA terminator, respectively, were removed from pAmy_R_empty.
Construction of TALEN destination vectors for R. oryzae
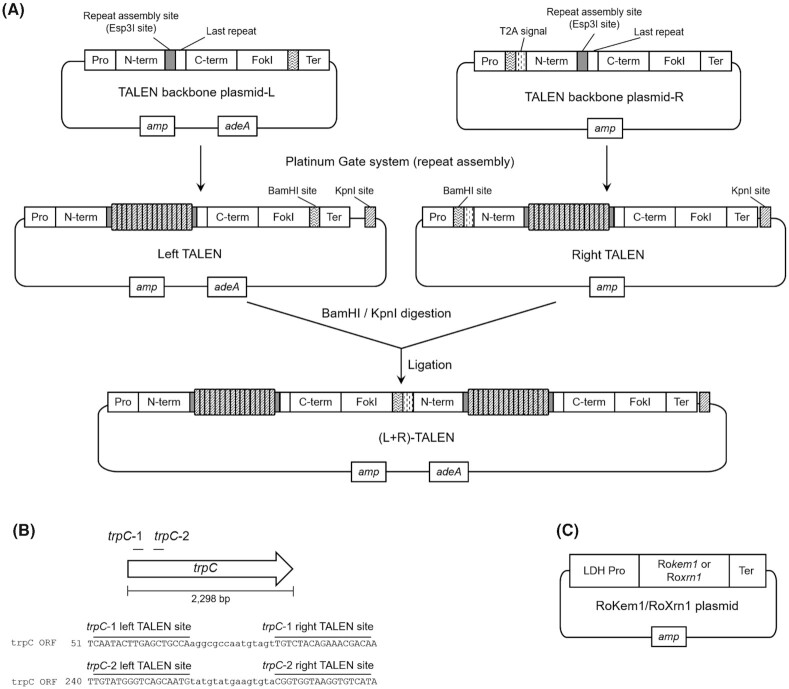
To construct the TALEN destination vectors for R. oryzae, we cloned the Platinum TALEN scaffold into R. oryzae expression vectors, with coding sequence modifications based on the previously established fast unification of separate endonucleases (FUSE) system (Tokumasu et al. 2014). Briefly, modified Platinum TALEN scaffolds for the Golden Gate assembly containing an NN or NI module as the last repeat were cloned into the pAmy_L_empty and pAmy_R_empty vectors to create pAmy_L and pAmy_R, respectively. For the left TALEN, a BamHI site was added just upstream of the stop codon. For the right TALEN, a T2A-coding sequence (Daniels et al. 2014) containing a BamHI site was added just downstream of the start codon (Fig. 1A). pLdh_L and pLdh_R, as well as pAdh_L and pAdh_R, were constructed by replacing the amyA promoter in pAmy_L and pAmy_R with the ldhA and adh promoters, respectively.
Figure 1.
Schematic illustrations of plasmids and TALEN target sites. (A) The constructed TALEN system in R. oryzae with Platinum Gate system; (B)trpC gene and TALEN target sites; and (C) exonuclease expression plasmids. Pro, promoter; N-term, N-terminal domain of TALE; C-term, C-terminal domain of TALE; Ter, terminator; amp, ampicillin resistance gene. trpC-1 and trpC-2 indicate TALEN target sites.
TALEN target site design, DNA-binding repeat assembly and TALEN cDNA unification using the FUSE method
Two TALEN target sites in the trpC locus, trpC-1 and trpC-2, were designed using the TALEN Targeter web tool (Doyle et al. 2012) (https://tale-nt.cac.cornell.edu/node/add/talen). The important residues involved in the activity of TrpC in Escherichia coli are K114, E163 and N184 (Kos et al. 1988). According to a homology search, these residues were equivalent to K356, E405 and N426, respectively, of TrpC in R. oryzae. The TALEN target sites were designed upstream of K356. The left and right target sequences of trpC-1 were 5′-caatacttgagctgcca-3′ and 5′-tgtcgtttctgtagaca-3′, respectively, and those of trpC-2 were 5′-tgtatgggtcagcaatg-3′ and 5′-atgacaccttaccaccg-3′, respectively (Fig. 1B). The DNA-binding repeats of the TALENs were assembled as previously described (Sakuma et al. 2013). Subsequently, to obtain the all-in-one TALEN vector, TALEN-T2A-TALEN, the left and right TALEN expression cassettes were unified by restriction digestion with BamHI and KpnI, followed by ligation (Fig. 1A).
Cloning of R. oryzae exonucleases
The coding sequences of two candidate R. oryzae exonuclease genes, Rokem1 (Accession No. LC638492) and Roxrn1 (Accession No. LC638493), were amplified using the R. oryzae NBRC5384 genome as a template with the primer sets ldhApro-en1-F/en1-pdcAter-R and ldhApro-en2-F/en2-pdcAter-R, respectively. The vector fragment was amplified with the primers pdcAter-F and ldhApro-R, using pLdh_R as a template. The Rokem1 and Roxrn1 gene fragments were cloned independently into the vector fragment using an In-Fusion HD Cloning Kit (Fig. 1C).
Transformation
The transformation of R. oryzae AM002 was performed using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories) as previously described (Skory 2002) with slight modifications. To linearize the plasmids, the pAmy-, pAdh- and pLdh-based TALEN vectors were digested with ScaI, ScaI and NdeI, respectively. pLdh-RoKem1 and pLdh-RoXrn1 were digested with PstI and SphI, respectively. The linearized plasmid weight ratios for the transformation were as follows: for the samples without exonuclease, the left TALEN to right TALEN ratio was ∼1:3, whereas for the samples with exonuclease, the left TALEN:right TALEN:exonuclease ratio was ∼1:3:3 and the (L + R)-TALEN to exonuclease ratio was ∼1:4. The amount of total DNA was 1–2 μg per bombardment. The preparation of DNA-coated tungsten particles was performed as previously described (Herzog et al. 1996) with slight modifications. Briefly, tungsten particles were coated with the following mixture: 12.5 μL of tungsten particle solution, 1 μL DNA solution (1–2 mg/mL), 12.5 μL of 2 M CaCl2 and 5 μL of 0.1 M spermidine. Spores of AM002 on filter paper were transformed.
Screening of 5-FAA-resistant mutants and tryptophan auxotroph identification
Spores transformed with TALEN plasmids were grown on CM or M medium (CM for pAmy and M for pAdh and pLdh) supplemented with tryptophan at 30°C for 7 days. After sporulation-stimulated segregation, mutant spores were screened in 5-FAA medium (10 g/L glucose, 3.36 g/L yeast nitrogen without amino acids, 5 g/L 5-FAA, 0.006 g/L tryptophan and 15 g/L agar) adjusted to pH 6 with 1 N NaOH as the first screening. In the second screening, the colonies were grown again in the 5-FAA medium. Tryptophan auxotrophy among the grown colonies in the second screening was identified using M medium with or without tryptophan.
Analysis of trpC mutations
For genotyping, genomic PCR products amplified region around the target site were sequenced directly or after bacterial cloning into the pUC18 vector. KOD FX Neo (TOYOBO) or PrimeSTAR MAX DNA polymerase (TaKaRa) was used to amplify the insert and cloning vector. The hyphae of tryptophan auxotrophic mutants were suspended in PCR buffer [100 mM Tris–HCl (pH 9.5), 1 M KCl and 10 mM EDTA], heated at 95°C for 10 min, vortexed and used as templates for genomic PCR. The genomic DNA fragments containing the two TALEN target sites, trpC-1 and trpC-2, were amplified with trpC-up-F2/trpC-down-R2 primers and trpC-up-F4/trpC-down-R3 primer pairs, respectively. The vector fragment was amplified with the primer set trpC-trpCR3-F/pUC18-trpCF4-R. The sequences of the primers are shown in Table S1 (Supporting Information). The PCR products amplified with trpC-up-F4 and trpC-down-R3 were cloned into the vector fragment prepared as described earlier using an In-Fusion HD Cloning Kit, and then, the cloned plasmids were sequenced. The PCR products amplified with trpC-up-F2 and trpC-down-R2 were sequenced directly.
Results
Vector system for the constitutive or inducible expression of TALENs in R. oryzae
To evaluate promoters in R. oryzae NBRC5384, an RNA-seq analysis was performed using total RNA collected from a fungal sample grown on M medium. Using the read counts of the RNA-seq analysis, we identified the ldhA and adh promoters as being stronger than the pdcA promoter, which had been previously characterized for exogenous expression (Mertens et al. 2006) (Fig. 2). Additionally, the amyA promoter was chosen as an inducible promoter (Mertens et al. 2006). In our system, the left and right TALENs were expressed either using independent plasmids or a single plasmid containing an integrated coding sequence divided by the T2A peptide driven by a single promoter (Fig. 1A).
Figure 2.
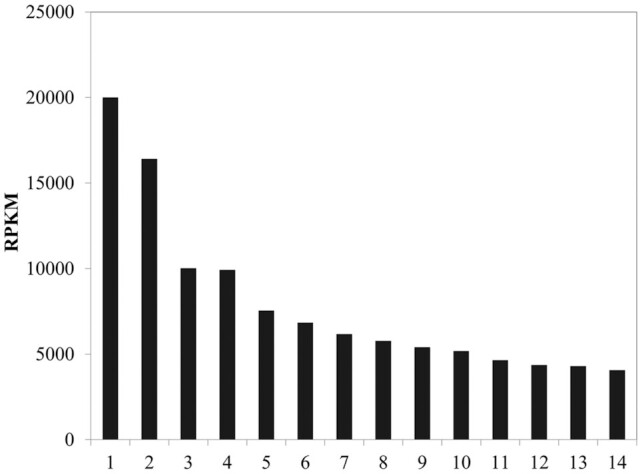
Identification of highly expressed R. oryzae genes by RNA-seq analysis. Reads per kilobase of exon per million mapped reads (RPKM) of the top 14 highly expressed genes are shown. 1, unknown; 2, l-lactate dehydrogenase (ldhA); 3, glyceraldehyde-3-phosphate dehydrogenase; 4, alcohol dehydrogenase (adh); 5, glucoamylase; 6, elongation factor 1-alpha; 7, enolase; 8, bZIP transcription factor; 9, concanamycin-induced protein C (cipC); 10, fructose-bisphosphate aldolase; 11, LysM domain-containing protein; 12, NADH-ubiquinone oxidoreductase MLRQ subunit; 13, calcium-dependent lipid-binding domain-containing protein; 14, pyruvate decarboxylase (pdcA).
Targeted gene disruption using TALENs
To examine the gene disruption activity of TALENs in R. oryzae, we targeted the trpC gene, which produces deficient mutants that can be positively selected using 5-FAA (Toyn et al. 2000) (Fig. S2, Supporting Information). Two kinds of target sequences were designed at the trpC locus using the TALEN Targeter web tool (Fig. 1B). Both separate and unified TALEN expression plasmids for R. oryzae were constructed using three promoters as described earlier. Using various transformant conditions with each TALEN pair, we obtained 5-FAA-resistant clones; however, they were not tryptophan auxotrophic mutants, because they grew in the M medium without tryptophan. These results indicated that it was difficult to disrupt genes effectively in R. oryzae using TALENs alone.
Identification of exonuclease orthologs in R. oryzae
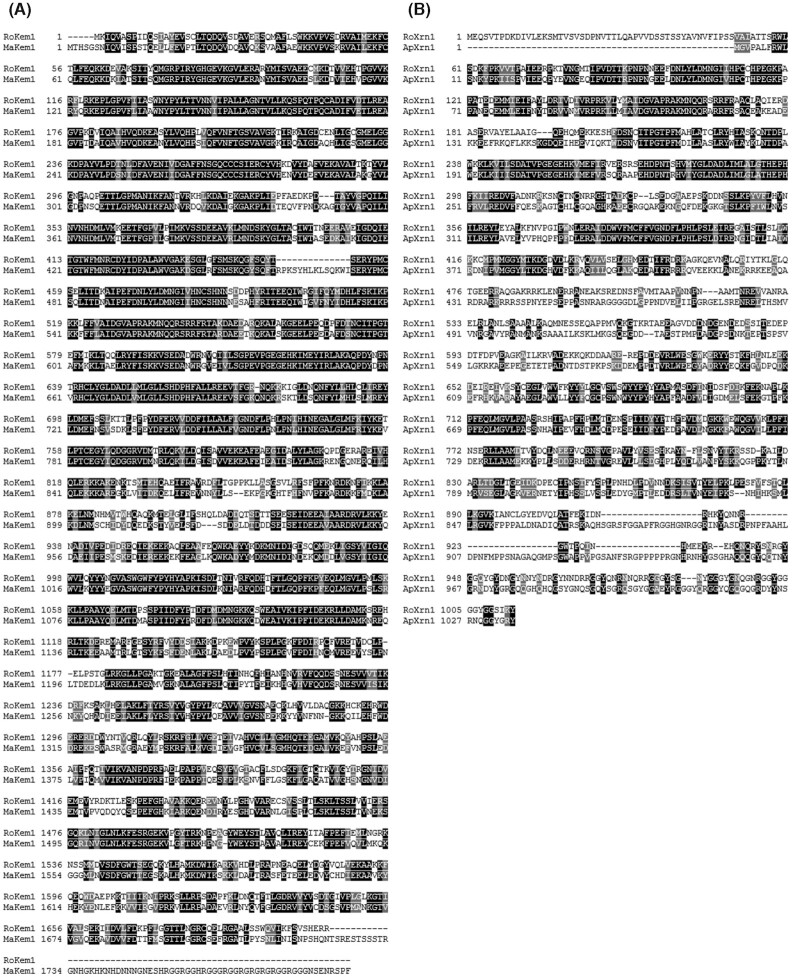
The overexpression of exonuclease 1 increases the gene disruption efficiency of TALENs in rat zygotes (Mashimo et al. 2013). Consequently, we searched for exonuclease orthologs in R. oryzae to facilitate TALEN-mediated genome editing. This search was performed using the in-house genome sequence data. Although typical exonucleases were not annotated, multifunctional exoribonucleases that have exonuclease activities (Heyer et al. 1995) were annotated. RoKem1, consisting of 1704 aa, shares a 68% identity with KEM1 in Mucor ambiguus, and RoXrn1, consisting of 1013 aa, shares a 43% identity with XRN1-like protein in Aspergillus parasiticus.
Targeted gene disruption using TALENs with exonucleases
Two exonuclease orthologous genes were cloned from R. oryzae, and their expression vectors, pLdh-RoKem1 and pLdh-RoXrn1, were constructed (Figs 1C and 3A and B). Each pair of TALEN and exonuclease plasmids was co-transformed, and the transformants were screened using 5-FAA plates. Many 5-FAA-resistant clones were obtained, similar to the results without the exonuclease; however, here, five tryptophan auxotrophic mutants were contained (Table 1).
Figure 3.
Amino acid alignments of potential exonuclease orthologs in R. oryzae and known exonucleases. Alignments of (A) the newly identified RoKem1 and Kem1 from M. ambiguus (MaKem1) and (B) the newly identified RoXrn1 and Xrn1-like protein from A. parasiticus (ApXrn1). The Xrn 5ʹ–3ʹ exonuclease N-terminal domains of MaKem1 and ApXrn1 are located at 471–1429 and 1–869 aa, respectively.
Table 1.
Evaluation of trpC gene disruption by TALEN with exonucleases.
| Target site | Promoter | Joint or severalty | Exonuclease | First screening of 5-FAA | Second screening of 5-FAA | Tryptophan auxotrophic mutants |
|---|---|---|---|---|---|---|
| trpC-1 | amy | J | RoKem1 | 70 | 47 | 0 |
| S | RoKem1 | 28 | 9 | 0 | ||
| adh | J | RoKem1 | 28 | 3 | 0 | |
| S | RoKem1 | 56 | 14 | 5 | ||
| ldh | J | RoKem1 | 28 | 13 | 0 | |
| S | RoKem1 | 21 | 7 | 0 | ||
| adh | J | RoXrn1 | 28 | 6 | 0 | |
| S | RoXrn1 | 28 | 8 | 0 | ||
| ldh | J | RoXrn1 | 21 | 7 | 0 | |
| S | RoXrn1 | 21 | 6 | 0 | ||
| trpC-2 | amy | J | RoKem1 | 76 | 51 | 0 |
| S | RoKem1 | 21 | 5 | 0 | ||
| adh | J | RoKem1 | 40 | 8 | 0 | |
| S | RoKem1 | 21 | 6 | 0 | ||
| ldh | J | RoKem1 | 21 | 1 | 0 | |
| S | RoKem1 | 22 | 1 | 0 | ||
| adh | J | RoXrn1 | N.T. | N.T. | N.T. | |
| S | RoXrn1 | 21 | 7 | 0 | ||
| ldh | J | RoXrn1 | 21 | 5 | 0 | |
| S | RoXrn1 | 49 | 28 | 0 |
Promoter, promoter for the expression of TALENs; joint or severalty, TALENs in a single vector (joint; J) or in multiple vectors (severalty; S); exonuclease, kinds of exonuclease expressed with TALENs; first screening of 5-FAA, number of colonies grown on the initial 5-FAA medium; second screening of 5-FAA, number of colonies grown on the second 5-FAA medium; tryptophan auxotrophic mutants, number of mutants that did not grow in the absence of tryptophan from colonies identified in the second 5-FAA screening; N.T., not tested.
Sequencing analysis of trpC-deficient mutants
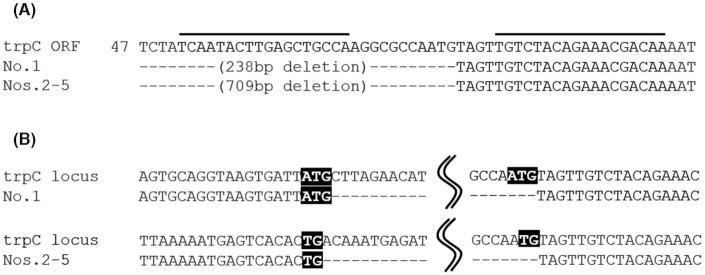
A sequencing analysis was performed at the trpC locus of the five tryptophan auxotrophic mutants described earlier, as well as the loci of 14 5-FAA-resistant clones that were not identified as tryptophan auxotrophic mutants. As expected, the clones without tryptophan auxotrophy did not have trpC mutations. However, one of the five tryptophan auxotrophic mutants (No. 1) harbored a 238-bp deletion, and the others (Nos 2–5) harbored 709-bp deletions in the upstream region starting from the TALEN cleavage site (Fig. 4A). Although the main DSB repair pathway is NHEJ in fungi, like many eukaryotic organisms (Ninomiya et al. 2004), these deletions are the results of MMEJ repair, because there were 2–3-bp direct repeats at both end-joining sites (Fig. 4B).
Figure 4.
Sequence analysis of the TALEN cleavage site in the tryptophan auxotrophic mutants. (A)TALEN target site in trpC-1 and positions of base deletions in the tryptophan auxotrophic mutants 1–5. Black bars indicate TALEN-binding sites. Gaps generated by deletion are shown as dashes. (B) DSB repair junctions in the tryptophan auxotrophic mutants 1–5. Shaded backgrounds indicate microhomologies of the 3′ and 5′ termini in each junction. Gaps generated by deletion are shown as dashes.
Discussion
We attempted to develop a gene disruption method in R. oryzae using programmable nucleases because there have been only two examples of targeted gene disruption in Rhizopus strains (Ibrahim et al. 2010, Bruni et al. 2019). Although genome editing technology has been applied to various fungal organisms (Arazoe et al. 2015, Katayama et al. 2016, Liu et al. 2017), only one successful example of genome editing using the CRISPR–Cas system in Rhizopus strains has been reported (Bruni et al. 2019). This is the first report of a TALEN-mediated gene disruption in Mucorales.
We attempted to disrupt the target gene using a template-free deletion induced by a TALEN in R. oryzae because exogenous DNA is rarely integrated into the host genomes of Rhizopus strains compared with other organisms. A gene deletion at the specific target genomic loci was not observed in R. oryzae using a TALEN only, even though we used the highly active Platinum TALEN. During the mutant screening process, we determined that most of the 5-FAA-resistant clones were not actual mutants. This may result from the transcriptional repression of the trpC gene by the temporal binding of TALEN molecules at the target locus. Therefore, we performed a three-step selection (two screenings of growth in 5-FAA and tryptophan auxotrophic selection) process.
Despite the imperfect selection procedure, we successfully obtained genome-edited clones resulting from exonuclease overexpression along with TALEN introduction. The genomic cleavage by the TALEN, followed by the exonuclease-driven resectioning, might result in relatively long deletions spanning hundreds of bases. In addition, importantly, we found traces of MMEJ repair in these deletion events, although NHEJ is generally dominant in the DSB repair of fungi.
This may be explained by the low expression levels of genes involved in DSB repair pathways other than MMEJ. In fact, the RNA-seq data suggested that the expression level of ku70, a major player in NHEJ repair, was very low in R. oryzae. The expression level of replication protein A (rpa) was also low. RPA is a single-strand DNA-binding protein that plays an essential role in DNA replication, recombination and repair (especially important for HR repair). Another possible reason is the low functionality of the molecules, such as RPA, which suppresses MMEJ, as indicated by the increase in MMEJ activity in RPA mutants compared with wild-type strains (Deng et al. 2014, McVey 2014). Although the RPA gene in R. oryzae has a conserved D228, which, if mutated, increases the frequency of MMEJ (Deng et al. 2014), an ∼200-aa C-terminal part of RPA was lost in R. oryzae in comparison with other organisms (Fig. S3, Supporting Information). Further investigation is needed to clarify the detailed mechanisms of the DSB repair pathway in R. oryzae. For genetic engineering, the gene insertion strategy based on MMEJ, called PITCh (Nakade et al. 2014), will help advance gene manipulations in R. oryzae.
To our best knowledge, this study is the first report of the employment of TALEN/exonuclease approach in fungal strains; however, we believe that our approach can be applied in a wide range of fungal strains because our approach is not affected by the HR efficiency of hosts, which is a major limitation to apply genome editing in various organisms. In addition, TALEN activity in heterochromatin target sites is reportedly higher than the activity of CRISPR–Cas system (Jain et al. 2021); therefore, the combination with TALENs and exonuclease might be a broadly accessible strategy of genome editing even in a silenced gene locus. Since the mutants created by genome editing can stably be maintained, our approach will contribute to the implementation of the collection of fungal mutant resources.
In summary, our study demonstrated that genome editing using TALENs with exonuclease overexpression was applicable in R. oryzae. Our method contributes to basic scientific knowledge and will aid in the metabolic engineering of Rhizopus strains.
Acknowledgments
We thank Edanz (https://jp.edanz.com/ac) for editing this manuscript.
Supplementary Material
Contributor Information
Yuichi Tsuboi, Biological Science Laboratories, KAO Corporation, 1334 Minato, Wakayama, Wakayama 640-8580, Japan; Graduate School of Integrated Sciences for Life, Hiroshima University, 1-3-1 Kagamiyama, Higashi-Hiroshima, Hiroshima 739-8526, Japan.
Tetsushi Sakuma, Graduate School of Integrated Sciences for Life, Hiroshima University, 1-3-1 Kagamiyama, Higashi-Hiroshima, Hiroshima 739-8526, Japan.
Takashi Yamamoto, Graduate School of Integrated Sciences for Life, Hiroshima University, 1-3-1 Kagamiyama, Higashi-Hiroshima, Hiroshima 739-8526, Japan.
Hiroyuki Horiuchi, Department of Biotechnology, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan; Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan.
Fumikazu Takahashi, Biological Science Laboratories, KAO Corporation, 1334 Minato, Wakayama, Wakayama 640-8580, Japan.
Kazuaki Igarashi, Biological Science Laboratories, KAO Corporation, 1334 Minato, Wakayama, Wakayama 640-8580, Japan.
Hiroshi Hagihara, Biological Science Laboratories, KAO Corporation, 1334 Minato, Wakayama, Wakayama 640-8580, Japan.
Yasushi Takimura, Biological Science Laboratories, KAO Corporation, 1334 Minato, Wakayama, Wakayama 640-8580, Japan.
Conflict of interest
None declared.
References
- Appel KF, Wolff AM, Arnau J. A multicopy vector system for genetic studies in Mucorcircinelloides and other zygomycetes. Mol Genet Genomics. 2004;271:595–602. [DOI] [PubMed] [Google Scholar]
- Arazoe T, Ogawa T, Miyoshi Ket al. Tailor-made TALEN system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng. 2015;112:1335–42. [DOI] [PubMed] [Google Scholar]
- Benito EP, Campuzano V, Lopez-Matas MAet al. Isolation, characterization and transformation, by autonomous replication, of Mucorcircinelloides OMPdecase-deficient mutants. Mol Gen Genet. 1995;248:126–35. [DOI] [PubMed] [Google Scholar]
- Bruni GO, Zhong K, Lee SCet al. CRISPR–Cas9 induces point mutation in the mucormycosis fungus Rhizopusdelemar. Fungal Genet Biol. 2019;124:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox Det al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Rossano AJ, Macleod GTet al. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS One. 2014;9:e100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng SK, Gibb B, de Almeida MJet al. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DSet al. TAL effector-nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmeister KA, Fung DY. Tempeh: a mold-modified indigenous fermented food made from soybeans and/or cereal grains. Crit Rev Microbiol. 1993;19:137–88. [DOI] [PubMed] [Google Scholar]
- Herzog RW, Daniell H, Singh NKet al. A comparative study on the transformation of Aspergillus nidulans by microprojectile bombardment of conidia and a more conventional procedure using protoplasts treated with polyethyleneglycol. Appl Microbiol Biotechnol. 1996;59:143–52. [Google Scholar]
- Heyer WD, Johnson AW, Reinhart Uet al. Regulation and intracellular localization of Saccharomycescerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol Cell Biol. 1995;15:2728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H, Takaya N, Yanai Ket al. Cloning of the Rhizopusniveus pyr4 gene and its use for the transformation of Rhizopusdelemar. Curr Genet. 1995;27:472–8. [DOI] [PubMed] [Google Scholar]
- Ibrahim AS, Gebremariam T, Lin Let al. The high affinity iron permease is a key virulence factor required for Rhizopusoryzae pathogenesis. Mol Microbiol. 2010;77:587–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim AS, Spellberg B, Walsh TJet al. Pathogenesis of mucormycosis. Clin Infect Dis. 2012;54:S16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Shukla S, Yang Cet al. TALEN outperforms Cas9 in editing heterochromatin target sites. Nat Commun. 2021;12:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Tanaka Y, Okabe Tet al. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillusoryzae. Biotechnol Lett. 2016;38:637–42. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–34. [DOI] [PubMed] [Google Scholar]
- Kos T, Kuijvenhoven A, Hessing HGet al. Nucleotide sequence of the Aspergillusniger trpC gene: structural relationship with analogous genes of other organisms. Curr Genet. 1988;13:137–44. [DOI] [PubMed] [Google Scholar]
- Liou CM, Yanai K, Horiuchi Het al. Transformation of a Leu− mutant of Rhizopusniveus with the leuA gene of Mucorcircinelloides. Biosci Biotechnol Biochem. 1992;56:1503–4. [DOI] [PubMed] [Google Scholar]
- Liu P, Wang W, Wei D. Use of transcription activator-like effector for efficient gene modification and transcription in the filamentous fungus Trichodermareesei. J Ind Microbiol Biotechnol. 2017;44:1367–73. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KMet al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Kaneko T, Sakuma Tet al. Efficient gene targeting by TAL effector nucleases coinjected with exonucleases in zygotes. Sci Rep. 2013;3:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M. RPA puts the brakes on MMEJ. Nat Struct Mol Biol. 2014;21:348–9. [DOI] [PubMed] [Google Scholar]
- Mertens JA, Skory CD, Ibrahim AS. Plasmids for expression of heterologous proteins in Rhizopusoryzae. Arch Microbiol. 2006;186:41–50. [DOI] [PubMed] [Google Scholar]
- Michielse CB, Salim K, Ragas Pet al. Development of a system for integrative and stable transformation of the zygomycete Rhizopusoryzae by Agrobacterium-mediated DNA transfer. Mol Genet Genomics. 2004;271:499–510. [DOI] [PubMed] [Google Scholar]
- Nagy G, Szebenyi C, Csernetics Aet al. Development of a plasmid free CRISPR–Cas9 system for the genetic modification of Mucorcircinelloides. Sci Rep. 2017;7:16800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahas E. Control of lipase production by Rhizopusoligosporus under various growth conditions. J Gen Microbiol. 1988;134:227–33. [Google Scholar]
- Nakade S, Tsubota T, Sakane Yet al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii Cet al. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA. 2004;101:12248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–8. [DOI] [PubMed] [Google Scholar]
- Revuelta JL, Jayaram M. Transformation of Phycomycesblakesleeanus to G-418 resistance by an autonomously replicating plasmid. Proc Natl Acad Sci USA. 1986;83:7344–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Ochiai H, Kaneko Tet al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep. 2013;3:3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Woltjen K. Nuclease-mediated genome editing: at the front-line of functional genomics technology. Dev Growth Differ. 2014;56:2–13. [DOI] [PubMed] [Google Scholar]
- Skory CD. Homologous recombination and double-strand break repair in the transformation of Rhizopusoryzae. Mol Genet Genomics. 2002;268:397–406. [DOI] [PubMed] [Google Scholar]
- Skory CD. Repair of plasmid DNA used for transformation of Rhizopusoryzae by gene conversion. Curr Genet. 2004;45:302–10. [DOI] [PubMed] [Google Scholar]
- Takaya N, Yanai K, Horiuchi Het al. Cloning and characterization of two 3-phosphoglycerate kinase genes of Rhizopusniveus and heterologous gene expression using their promoters. Curr Genet. 1994;25:524–30. [DOI] [PubMed] [Google Scholar]
- Tokumasu D, Sakuma T, Hayashi Yet al. FAST-id system for enrichment of cells with TALEN-induced mutations and large deletions. Genes Cells. 2014;19:419–31. [DOI] [PubMed] [Google Scholar]
- Toyn JH, Gunyuzlu PL, White WHet al. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast. 2000;16:553–60. [DOI] [PubMed] [Google Scholar]
- Weld RJ, Plummer KM, Carpenter MAet al. Approaches to functional genomics in filamentous fungi. Cell Res. 2006;16:31–44. [DOI] [PubMed] [Google Scholar]
- Yanai K, Horiuchi H, Takagi Met al. Preparation of protoplasts of Rhizopusniveus and their transformation with plasmid DNA. Agric Biol Chem. 1990;54:2689–96. [Google Scholar]
- Zhang ZY, Jin B, Kelly JM. Production of lactic acid from renewable materials by Rhizopus fungi. Biochem Eng J. 2007;35:251–63. [Google Scholar]
- Zhou Y, Dominguez JM, Cao Net al. Optimization of l-lactic acid production from glucose by Rhizopusoryzae ATCC 52311. Appl Biochem Biotechnol. 1999;78:401–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.