Abstract
Androgen receptor (AR) inhibitor therapy is a developing treatment for AR-positive breast cancer (BC) with ongoing clinical trials. AR splicing variant-7 (AR-V7) is a truncated variant of AR that leads to AR inhibitor therapy resistance in prostate cancer; recent studies have identified AR-V7 in BC and theorized that AR-V7 can have a similar impact. This study assessed the prevalence and clinicopathologic features associated with AR-V7 in a large BC cohort. BC samples were evaluated by MSK-Fusion targeted RNAseq for AR-V7 detection and MSK-IMPACT targeted DNAseq, including triple-negative tumors with no driver alteration and estrogen receptor-positive/ESR1 wildtype tumors progressing on therapy. Among 196 primary and metastatic/recurrent cases (196 RNAseq, 194DNAseq), 9.7% (19/196) were AR-V7 positive and 90.3% (177/196) AR-V7 negative. All AR-V7 positive BC were AR-positive by immunohistochemistry (19/19). The prevalence of AR-V7 by receptor subtype (N=189) was: 18% (12/67) in ER-/PgR-/HER2-negative BC, 3.7% (4/109) in ER-positive/HER2-negative BC, and 15.4% (2/13) in HER2-positive BC; AR-V7 was detected in one ER-positive/HER2-unknown BC. Apocrine morphology was observed in 42.1% (8/19) of AR-V7 positive BC and 3.4% (6/177) AR-V7 negative BC (P < 0.00001). Notably, AR-V7 was detected in 2 primary BC and 7 metastatic/recurrent BC patients with no prior endocrine therapy. We conclude that positive AR IHC and apocrine morphology are pathologic features that may indicate testing for AR-V7 is warranted in both primary and metastatic BC in the appropriate clinical context. The study findings further encourage the assessment of AR-V7 as a predictive biomarker for AR antagonist benefit in ongoing clinical BC trials.
Keywords: Androgen receptor, splice variant, breast cancer, RNA sequencing
INTRODUCTION
Androgen receptor (AR), encoded by the AR gene on chromosome Xq11–12, is a member of the ligand-activated steroid-hormone family of receptors, which also includes the estrogen receptor (ER) and progesterone receptor (PgR). AR is an emerging biomarker in breast cancer (BC) and is expressed in 70–90% of invasive carcinomas 1–6. Although the prevalence of AR expression is higher in ER-positive tumors (80–90%), it is also positive in approximately 60% of human epidermal growth factor receptor 2 (HER2)-positive BC, and up to 35% of triple-negative breast cancers (ER, PgR, and HER2-negative) (TNBC) 1–9, depending upon the definition of AR positivity used. The role of AR inhibitor therapy in BC, including AR antagonists bicalutamide and enzalutamide, is an area of active research and still evolving, with initial clinical trials showing promising results, particularly in TNBC 10,11.
The AR gene (NM_000044) consists of eight canonical exons 12,13 which encode four regions: N-terminal domain (NTD), DNA binding domain (DBD), hinge region and ligand binding domain (LBD). Exons 2–3 form the DBD while exons 5–8 form the LBD. Several cryptic exons (CE), defined as being present in some but not all transcripts, as well as exon 9 are involved in the formation of truncated AR splice variants that lack the LBD 14. One alternative splicing variant, AR splicing variant 7 (AR-V7), is generated through splicing of exon 3 to downstream CE3 (Figure 1). This results in an RNA transcript and subsequent protein sequence that excludes exons 4 – 8 and lacks the LBD.
Figure 1:
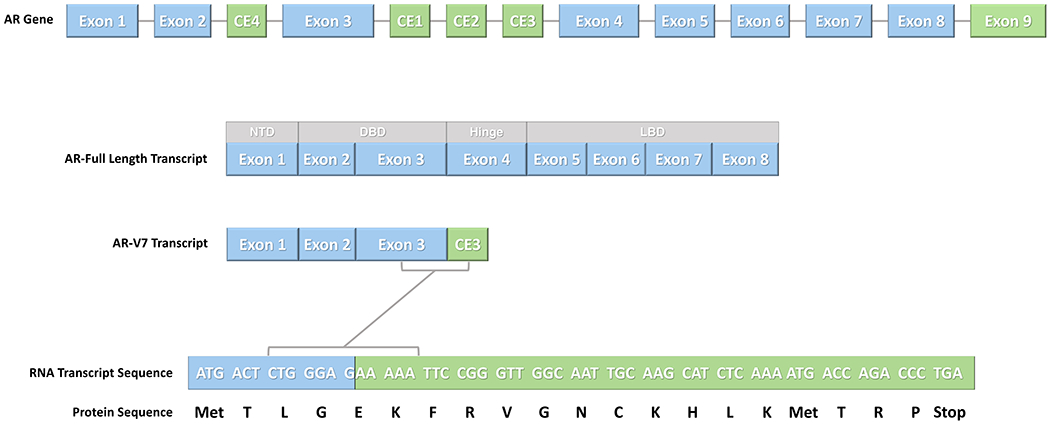
The AR gene consists of eight canonical exons (NM_000044). Several cryptic exons (CE) as well as exon 9 are involved in the formation of several truncated AR splice variants which lack the ligand binding domain (LBD). AR-V7 is generated through splicing of exon 3 to downstream CE3. This results in a RNA transcript sequence and subsequent protein sequence that excludes exons 4 – 8 and lacks the LBD. NTD, N-terminal domain; DBD, DNA binding domain.
AR-V7 is one mechanism of resistance to androgen deprivation therapy (ADT) in prostate cancer, as the lack of the LBD results in a constitutively-active and androgen-independent receptor 14–16. In a study by Hickey et al examining the expression of AR splice variants in primary BC, transcripts of AR-V7 were identified in approximately half of the BCs tested with highest expression in ER-negative cases that were HER2-enriched 8, which corresponds to the frequency of expression in the TCGA dataset.
Resistance to targeted therapy in BC remains a challenge. With ongoing trials targeting AR in BC, the exploration of resistance mechanisms and susceptible populations is a necessary area of investigation. In this study, we examined the frequency of AR-V7 in BC detected by RNA sequencing and examined the clinical and histopathologic features of primary and metastatic tumors with this alteration.
MATERIALS AND METHODS
Case Selection and Molecular Testing
This study was conducted under institutional review board approval. BCs profiled by MSK-Fusion, an RNA-based targeted assay that utilizes the Anchored multiplex PCR technology and next-generation sequencing (NGS) for the identification of gene fusions and oncogenic isoforms 17,18, between September 25, 2018 to July 3, 2019 were assessed for AR-V7 transcripts. Gene specific primers were designed towards the 3’ end of exon 3 and in the 3’ direction to specifically capture an intron 3 sequence corresponding to CE3 (Figure 1). To confirm AR-V7 positivity, transcript sequences are manually translated and compared to previously published AR-V7 protein sequence 14. Concurrent DNA-based NGS results using MSK-IMPACT 19 on the same specimen was also performed. Breast tumors submitted for DNA sequencing include predominantly patients with advanced or metastatic breast cancer. After DNA sequencing, BCs reflexed to RNA sequencing during the period studied included triple-negative tumors with no driver alteration and ER-positive/ESR1 wildtype tumors progressing on therapy with no clear mechanism of resistance. All BC samples with and without AR-V7 identified by RNA sequencing tested during this time period were included.
Clinicopathologic Data Collection and Receptor Testing
Clinical information, including prior BC history and treatment, for all patients was retrieved from the electronic medical records. Tumor grade and histologic subtype of the primary BC were extracted from the pathology reports; representative slides of the primary tumor were re-reviewed (DSR) when material was available to assess for apocrine morphology. Apocrine morphology was characterized by abundant eosinophilic granular cytoplasm and enlarged nuclei with prominent nucleoli (Figure 2A). A diagnosis of apocrine carcinoma was rendered when apocrine morphology was present in >90% of invasive tumor cells 20 and invasive carcinoma with apocrine features was rendered if apocrine morphology was present in a subset of tumor cells. Immunohistochemistry (IHC) for ER and PgR and IHC and/or fluorescence in situ hybridization (FISH) for HER2 were performed at the time of diagnosis on the specimen submitted for molecular testing and reported according to the ASCO/CAP guideline recommendations 21,22 using a Food and Drug Administration (FDA)-approved method either at MSKCC or an outside institution. AR IHC results performed in-house on the specimen submitted for molecular testing were recorded if available. AR IHC results were also documented if performed on alternate specimens or performed at an outside institution; 97% (83/86) of cases with AR IHC results were stained at MSKCC. In accordance with the standard practice at our institution, AR IHC (clone SP107, 1:125, retrieval ER2 30’, Ventana, Tucson, AZ) is performed on TNBC at the time of diagnosis if material is available at MSKCC and on select ER-positive or HER2-positive BC if clinically requested. The percent nuclear staining and staining intensity for AR IHC was retrieved from pathology reports, with a positive AR IHC result considered with at least 1% nuclear staining (Figure 2B).
Figure 2:
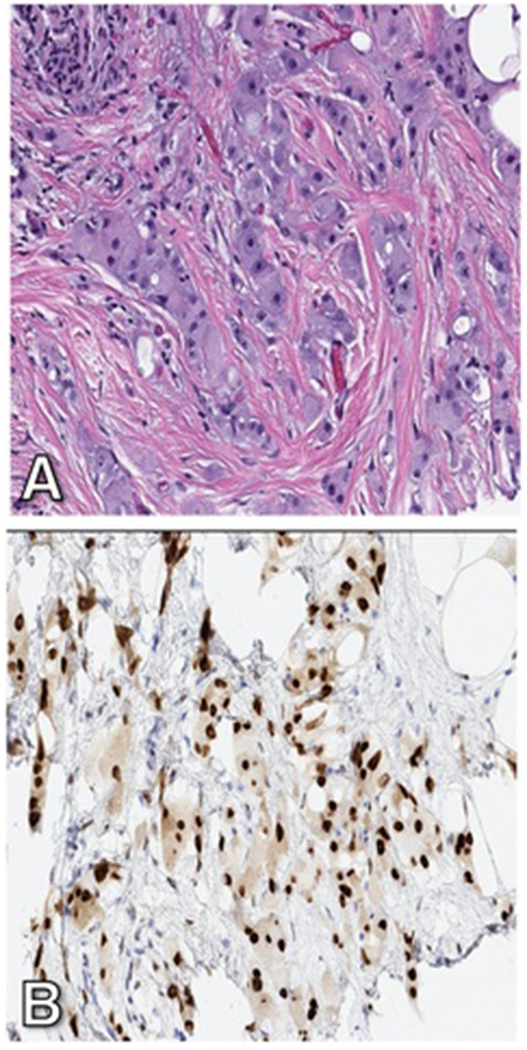
Representative invasive carcinoma displaying apocrine morphology on hematoxylin and eosin stained section (A), characterized by abundant eosinophilic granular cytoplasm and enlarged nuclei with prominent nucleoli, and strong nuclear staining with AR immunohistochemistry (B). Original magnification, 200x (A, B).
MSK-IMPACT, MSK-Fusion, IHC staining for ER, PgR, HER2 and AR and HER2 FISH are NYSDOH clinically validated assays that were performed in CLIA-accredited laboratories.
Association between clinicopathological parameters and AR-V7 were analyzed using Fischer exact, with P < 0.05 considered significant.
RESULTS
Study Cohort
Of 196 BC samples analyzed by MSK-Fusion during the indicated time period, the truncated AR-V7 transcript was detected in 9.7% (19/196) of cases (Figure 3). The number and percentage of transcript reads supporting AR-V7 for these 19 cases is listed in Table 1. Only reads spanning the breakpoint are considered to support the isoform. The clinicopathologic features of the AR-V7 positive and AR-V7 negative cases are summarized in Table 2. BC samples were from the primary tumor in 28.6% (56/196) of cases, a locoregional recurrence in 12.7% (25/196) of cases, and distant metastatic site in 58.7% (115/196) of cases. The mean age at diagnosis was 50.9 years (interquartile range, 40.8–60.6 years) for 190 patients in which the date of diagnosis was available.
Figure 3:
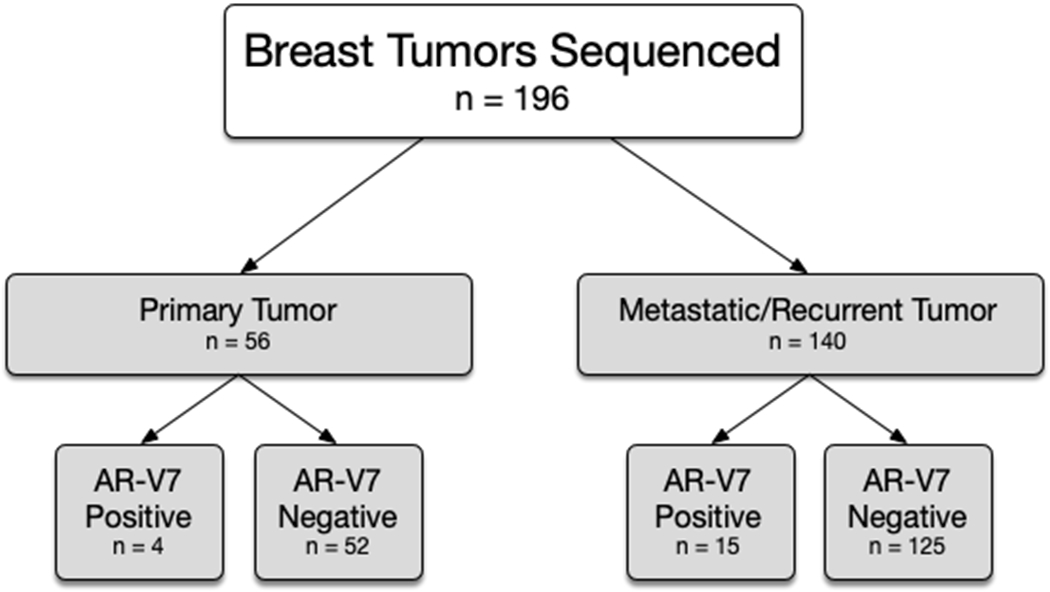
Breast tumors sequenced.
Table 1.
Number and percentage of transcript reads supporting AR-V7
| Case Number | AR-V7 Transcript Read Numbers | AR-V7 Transcript Percentage* |
|---|---|---|
| 1 | 8 | 100 |
| 2 | 44 | 62.9 |
| 3 | 298 | 9.3 |
| 4 | 17 | 1.5 |
| 5 | 5 | 0.5 |
| 6 | 429 | 23.4 |
| 7 | 5 | 100 |
| 8 | 154 | 74 |
| 9 | 6 | 3.5 |
| 10 | 421 | 28.3 |
| 11 | 8 | 100 |
| 12 | 8 | 0.5 |
| 13 | 33 | 52.4 |
| 14 | 17 | 100 |
| 15 | 5 | 100 |
| 16 | 9 | 100 |
| 17 | 9 | 0.9 |
| 18 | 211 | 43.9 |
| 19 | 5 | 100 |
The percentage is calculated in reference to the total number of reads covering this target, including wild-type transcripts.
Table 2.
Clinicopathologic features of AR-V7 positive and AR-V7 negative cases
| Total (n=196) | AR-V7 positive (n=19) | AR-V7 negative (n=177) | P value |
|---|---|---|---|
|
| |||
| Age at BC Diagnosis, Median (IQR), Years (n = 190) | 54.4 (44.9 – 66.5) | 50.5 (39.1 – 60.0) | |
|
| |||
| Sample Type (n=196) | |||
| Primary (n=56) | |||
| Post ET1 | 10.5% (2/19) | 2.3% (4/177) | |
| No ET | 10.5% (2/19) | 27.1% (48/177) | |
| Metastasis/Recurrence (n=140) | |||
| Post ET | 42.1% (8/19) | 52.0% (92/177) | |
| No ET | 36.9% (7/19) | 16.9% (30/177) | |
| Treatment N/A | 0% (0/19) | 1.7% (3/177) | |
|
| |||
| Histologic Subtype (n = 196) 2 | |||
| Ductal Carcinoma, NOS | 52.6% (10/19) | 85.3% (151/177) | |
| Apocrine features | 42.1% (8/19) | 3.4% (6/177) | <0.00001 |
| Apocrine Carcinoma | 10.5% (2/19) | 0.0% (0/177) | |
| Ductal Carcinoma with Apocrine Features | 26.3% (5/19) | 2.8% (5/177) | |
| Lobular Carcinoma with Apocrine Features | 5.3% (1/19) | 0.6% (1/177) | |
| Other3 | 5.3% (1/19) | 11.3% (20/177) | |
|
| |||
| Grade (n = 184) 2 | |||
| Well Differentiated | 0.0% (0/18) | 1.2% (2/166) | |
| Well to Moderately Differentiated | 0.0% (0/18) | 0.6% (1/166) | |
| Moderately Differentiated | 22.2% (4/18) | 42.2% (70/166) | |
| Moderately to Poorly Differentiated | 22.2% (4/18) | 4.2% (7/166) | |
| Poorly Differentiated | 55.6% (10/18) | 51.8% (86/166) | |
|
| |||
| Receptor Status | |||
| Estrogen receptor (ER) positive (n = 193) | 36.8% (7/19) | 64.4% (112/174) | |
| Progesterone receptor positive (n = 190) | 11.1% (2/18) | 32.6% (56/172) | |
| HER2 positive (n = 190) | 11.1% (2/18) | 6.4% (11/172) | |
| Triple negative (TNBC) (n = 189) | 66.7% (12/18) | 32.2% (55/171) | 0.008 |
| Androgen receptor (AR) positive (n = 86) | 100.0% (19/19) | 70.2% (47/67) | 0.005 |
| AR positive, ER positive | 36.8% (7/19) | 57.4% (27/47) | |
| AR positive, TNBC | 63.2% (12/19) | 38.3% (18/47) | |
ET Endocrine therapy, including selective estrogen receptor modulators, selective estrogen receptor degraders, and aromatase inhibitors
Subtype and grade based on primary tumor
Other category encompasses cases of invasive ductal carcinoma with special histologic features, including micropapillary, mucinous, metaplastic, neuroendocrine and papillary, invasive lobular carcinoma, small cell carcinoma and cystic hypersecretory carcinoma.
BC breast cancer, IQR interquartile range, N/A not available, HER2 human epidermal growth factor receptor 2
Histologic Subtype and Tumor Grade
The histologic subtype and tumor grade are listed in Table 2. Histologic subtype was widely variable in both the AR-V7 positive and AR-V7 negative groups (Table 2, Figure 4). Of the AR-V7 positive cases, 42.1% (8/19) had apocrine features compared to 3.4% (6/177) of AR-V7 negative cases (P < 0.00001). The relationship with apocrine morphology also held true when controlling for the AR IHC status. Of the 66 AR IHC positive cases, 42.1% (8/19) of the AR-V7 positive / AR IHC positive cases and 0% (0/47) of AR-V7 negative / AR IHC positive cases exhibited apocrine morphology (P < 0.0001). There was no significant difference between the two groups for any other histologic feature.
Figure 4:
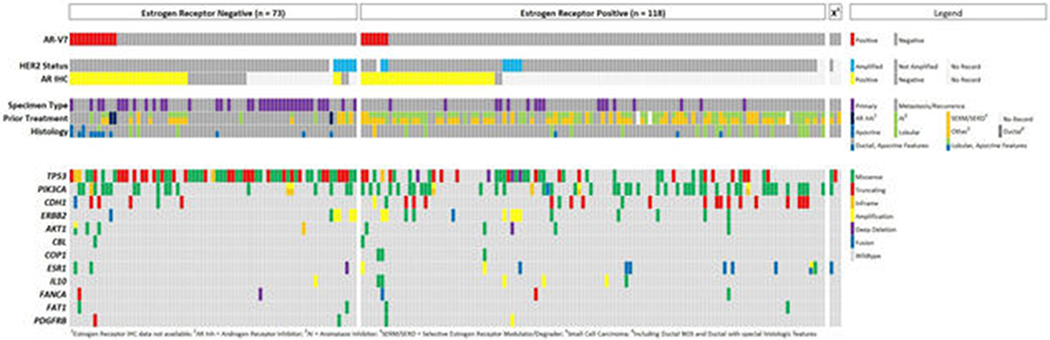
The oncoprint displays the most common somatic alterations in the data set for cases with concurrent DNA and RNA Sequencing (n=194). Each column is a sample and each row is a gene. Alterations are represented with various colors. The specimen type, receptor status, history of treatment and histology subtype are also listed.
Receptor Status
All ARV-7 positive cases were positive for AR IHC (100%, 19/19), with 84.2% (16/19) exhibiting staining in >90% of nuclei. AR-V7 positive cases were more likely than AR-V7 negative cases to be positive for AR IHC (100% versus 70.2%, P = 0.005). Of the 19 AR-V7 positive cases, they were positive for ER in 36.8% (7/19), PgR in 11.1% (2/18) and HER2 in 11.1% (2/18) of cases (Table 2, Figure 4) (the PgR and HER2 status for one case was unknown). The prevalence of AR-V7 by ER, PR and HER2 subtype (N=189 for available ER, PR and HER2 status) was: 18% (12/67) in TNBC, 3.7% (4/109) in ER-positive/HER2-negative BC, and 15.4% (2/13) in HER2-positive BC (AR-V7 was detected in one ER-positive/HER2-unknown BC). The majority of AR-V7 positive cases were TNBC (66.7%, 12/18) which was significantly higher than AR-V7 negative cases (P = 0.008). Of the 14 tumors that had apocrine features on histologic review, 71.4% (10/14) were triple negative.
Treatment Prior to RNA Sequencing
The 19 AR-V7 positive cases included 4 primary and 15 metastatic/recurrent specimens; 16 specimens were analyzed by RNA sequencing status post any treatment and 3 were treatment naïve. Ten cases were from patients who were exposed to endocrine therapy (ET) (selective estrogen receptor modulators [SERM], selective estrogen receptor degraders [SERD], and/or aromatase inhibitors [AI]) prior to testing, including 2 primary tumors in which the patients received ET for a prior diagnosis of breast cancer (Table 2, Figure 4). Four specimens were from patients who received chemotherapy only (triple negative / AR positive) prior to analysis and 2 with prior exposure to an AR inhibitor and chemotherapy. In total, AR-V7 was detected in 2 primary tumors and 7 metastatic/recurrent specimens in patients with no prior ET in particular, although 28.6% (2/7) of the metastatic/recurrent specimens were from patients who received prior AR inhibitor therapy. Prior exposure to AI therapy did not increase the likelihood of detecting AR-V7 (P = 1.0).
Androgen Receptor Inhibitor Therapy
Six patients, all with AR IHC positive BC, received AR inhibitor therapy (bicalutamide, n=3; enzalutamide, n=3), including 3 AR-V7 positive BC and 3 AR-V7 negative BC. The mean AR inhibitor therapy duration was 483 days (range, 141–903 days) for the AR-V7 positive BC patients and 142 days (range, 49–315 days) for the AR-V7 negative BC patients. Enzalutamide was given as monotherapy in 3/3 patients and bicalutamide was given in combination with the CDK 4/6 inhibitor palbociclib in 2/3 patients. All 3 of the AR-V7 positive samples (1 recurrence, 2 metastases) were AR-positive/TNBC; one patient was confirmed to have the variant transcript before receiving AR inhibitor therapy and the other 2 patients received an AR inhibitor before testing. The receptor status for the 3 AR-V7 negative samples (3 metastases) varied: 2 were ER-low positive (1–10% nuclear staining) and HER2-negative, 1 ER-negative and HER2 status not available. All 3 AR-V7 negative BC were AR IHC positive on either the same sample submitted for molecular testing or a prior specimen. Two patients were confirmed to lack the variant transcript before receiving AR inhibitor therapy and the other patient received an AR inhibitor before testing. AR inhibitor therapy was stopped in all six patients due to progression of disease.
Of the 3 patients who received AR inhibitor therapy and had AR-V7 identified in a metastatic lesion, two had primary tumors available for testing. MSK-fusion testing was available for both the pre-AR inhibitor treatment primary tumor and post-AR inhibitor treatment metastasis for one patient who received enzalutamide. As shown in Supplemental Figure 1A, the primary and metastatic lesions are clonally related. AR-V7 was not identified in the pre-treatment primary tumor but was identified in the post-treatment bone metastasis.
An additional one metastasis which showed AR-V7 prior to AR inhibitor treatment, also had a pre-treatment breast primary sample for MSK-fusion testing. Both samples showed an ATG7-RAF1 in-frame fusion but AR-V7 was only identified in the metastatic tumor (Supplemental Figure 1B).
Concurrent Molecular Profile by DNA and RNA Sequencing
DNA sequencing was performed on the same tumor block for 99.0% (194/196) of samples. AR-V7 positive BCs were more likely to have mutations in AKT1 (P = 0.004), CBL (P = 0.009), COP1 (P = 0.03), ESR1 (P = 0.03), IL10 (P = 0.03), PDGFRB (P = 0.03), FAT1 (P = 0.05) and PIK3R1 (P = 0.05) (Figure 4). However, if adjusting for hormone receptor status by only including TNBC cancers that are AR IHC positive (12 AR-V7 positive samples, 18 AR-V7 negative samples) there is no significant difference in mutational profile or amplification status, except for a slightly increased frequency of MYC amplification in AR-V7 negative cases (P = 0.04). Only one AR-V7 positive case had a concurrent fusion (ATG-RAF1).
DISCUSSION
AR inhibition is evolving as a potential treatment option for patients with AR IHC positive TNBC, for which the current standard of care offers limited therapeutic options besides traditional cytotoxic chemotherapy agents 10,11. The oncogenic role of AR in TNBC is a relatively new area of study compared to prostate cancer, where AR is considered a key oncogenic driver and has been treated with ADT for several decades 23,24. Resistance to AR inhibition after prolonged exposure is a known challenge in prostate cancer and the outcomes and limitations of this targeted therapy in TNBC are still being ascertained. The identification of resistance mechanisms, including AR splicing variants, in BC may therefore be necessary when developing treatment protocols. This study investigated the characteristics of AR-V7 positive BC by analyzing the clinical and histopathologic features of a large cohort of primary and metastatic BC cases.
AR-positive TNBC belongs to a molecular apocrine BC subgroup which is often represented by the MDA-MB-453 cell line in translational studies 9. Studies using this cell line have shown that AR binds to the same sites in molecular apocrine BC cells as it does in prostate cancer cell lines, which suggests that AR may have similar behavior and effects in AR positive TNBC as it does in prostate cancer 25,26. Accordingly, the known mechanisms of therapeutic resistance in prostate cancer should be further investigated as to its applicability to AR-positive TNBC.
A well-established limitation to ADT in prostate cancer is that despite an initial positive response, men will frequently develop incurable castration-resistant prostate cancer 24. Increased transcription of AR-V7 is a mechanism for resistance to ADT, as the lack of the LBD results in constitutively-active, androgen-independent transcription factor activity 14–16. Contrary to the upregulation of AR-V7 in prostate cancer as a means of resistance to AR inhibitors, the current study shows that AR-V7 transcripts are found in both primary and metastatic BCs, and that prior treatment (SERM, SERD, AI or AR inhibition) does not appear to affect its frequency. These findings are in concordance with previous studies that showed AR-V7 is not only present in metastatic tumors, but also in primary tumors and breast cancer cell lines 8,9. Altogether, these findings confirm that prior exposure to ADT is not necessary for the expression of AR‐V7 in BC.
Although the time during the disease course at which AR-V7 presents is different between prostate and breast tumors, early studies indicate that when AR-V7 is identified in BC, the resistance to AR inhibition found in prostate cancer already exists. This theory was explored in a study by Hickey et al 8 utilizing the MDA-MB-453 BC cell line which demonstrated that the normally functioning full length AR transcript only underwent nuclear translocation upon androgen stimulation and its activity was significantly diminished in the presence of AR inhibition. Conversely, AR-V7 was present in the nucleus regardless of androgen stimulation and its activity was not altered by treatment with AR inhibition. Although not yet proven in randomized prospective clinical trials, these findings indicate a strong possibility that the existence of AR-V7 diminishes the benefit of therapeutic AR inhibition and deserves exploration as a predictive biomarker for AR antagonist benefit in ongoing studies.
In addition to analyzing the clinical features of AR-V7 positive tumors, this study assessed the histologic features and looked for commonalities within this group. It was previously demonstrated that AR IHC positive tumors, especially those that are ER-negative, are more likely to have apocrine morphology. In fact, the subgroup of ER-negative and AR-positive tumors was re-defined by Farmer et al 27 as the “molecular apocrine” subtype by genome wide expression analysis. This subtype was shown to cluster more closely with luminal-type ER positive breast tumors despite lacking ER-positivity and was significantly enriched for apocrine morphology and HER2 overexpression 5,26,28,29. Our study finds that AR-V7 positive tumors are more likely to have apocrine morphology and that the enrichment of apocrine morphology within the AR-V7 positive cohort holds true even when limiting the comparison to known AR IHC positive tumors. Therefore, apocrine morphology may be a useful screening mechanism for selecting cases to be analyzed for the presence of AR-V7 if AR inhibitor therapy is being considered.
Of note, the AR antibody clone SP107 recognizes the NTD and does not bind the LBD. This is consistent with our findings that all AR-V7 positive BC showed positive nuclear staining by AR IHC and no loss of staining was appreciated.
The primary benefit of this study is that it confirms the presence of AR-V7 in primary BCs prior to treatment. Accordingly, informative testing for AR-V7 can be performed at any point after the initial diagnosis. This study also has limitations. First, the cases included in this study had certain clinicopathologic features that first resulted in DNA sequencing and then subsequent RNA sequencing based on reflex guidelines established at our institution during the study period. As these features may alter the prevalence of AR-V7 cases within this cohort, the frequency at which AR-V7 occurs here is not necessarily reflective of true biologic proportions. The TCGA and the cohort tested by Hickey collectively indicated that AR-V7 is expressed in about 50% of primary breast tumors in particular 8. The present study included 28.6% primary tumors (versus 12.7% locoregional recurrences and 58.7% metastases) and it is possible that this and other clinicopathologic features leading to advanced disease and NGS testing are explanations for the discordance in AR-V7 prevalence rates. A second limitation to this study is that data on AR IHC status was not available for all AR-V7 negative tumors, as there are currently no standard testing or reporting guidelines. However, given the limited information regarding AR-V7 in BC at this time, it was deemed appropriate to include any case that was evaluated by RNA sequencing in order to gather a larger sample size for other observations. Lastly, to determine how AR-V7 affects the outcome of BC patients on AR inhibitor therapy, randomized prospective studies comparing AR-V7 positive and negative treatment groups are necessary. Although this study does provide insight into the varied types of cases that may harbor AR-V7 transcripts, it does not prove that the presence of AR-V7 is an accurate predictor of response to AR inhibition in BC patients. As an AR-V7 specific antibody is available for IHC 8, a future area of study for this dataset also includes correlating the IHC and RNA sequencing findings as a potential screening tool.
At this time, clinical outcomes for AR inhibition in BC trials have not taken into account the presence of AR-V7. This study demonstrates that AR-V7 can be present in primary BC without a history of anti-estrogen therapy or AR inhibition. In BC patients being considered for AR inhibitor therapy, apocrine morphology and positive AR IHC are pathologic features that may indicate further molecular testing for AR-V7 is warranted. These findings further encourage the assessment of AR-V7 as a predictive biomarker for AR antagonist benefit in ongoing clinical BC trials.
Supplementary Material
ACKNOWLEDGEMENTS
This work could not have been possible without the help of the highly specialized technicians in the Diagnostic Molecular Pathology Laboratory at MSKCC, including Purvil Sukhadia and Mike Zaidinski.
FUNDING
This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748 and supported by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Disclosures:
Tiffany A. Traina has received honoraria for ad hoc consulting for Genentech/Roche, Pfizer, AstraZeneca, Merck, Puma, Celgene, Athenex, Innocrin, BMS, Daiichi Sankyo, Seattle Genetics, Exact Sciences, Immunomedics/Gilead; research support to the institution from Daiichi Sankyo, Astellas, Genentech, AstraZeneca, Immunomedics/Gilead.
Ayca Gucalp has received honoraria for ad hoc consulting for Innocrin Pharma, Pfizer; research support to the institution from BioAtla, Bristol-Myers Squibb, Innocrin Pharma, Merck, Novartis, Oncotherapy, Pfizer, Roche, Zenith Epigenetics.
Sarat Chandarlapaty has received honoraria for ad hoc consulting for Eli Lilly, Sermonix, BMS, Paige AI, Context Therapeutics, Revolution Medicine, and Novartis; research support to the institution from Daiichi Sankyo, Novartis, Eli Lilly, Genentech, and Sanofi.
Marc Ladanyi has received honoraria for advisory board participation from Merck, Astra-Zeneca, Bristol Myers Squibb, Takeda, Lilly Oncology, Bayer, ADC Therapeutics, and Paige AI, and research support from LOXO Oncology, Merus, and Helsinn Therapeutics.
Maria Arcila received consulting and speaker fees from Raindance Technologies, Bristol-Myers Squibb, Clinical Care Options, PVI Peerview, Janssen Global Services LLC, Biocartis US Inc., Physicians’ Education Resource LLC, Astra Zeneca, Invivoscribe.
Ryma Benayed has received a grant and travel credit from ArcherDx, honoraria for advisory board participation from Loxo oncology and Roche Dx, and speaking fees from Illumina.
All other authors declare no potential conflict of interest.
Footnotes
Ethics approval/consent to participate: This study was conducted under institutional review board approval (17-287).
DATA AVAILABILITY
The data in this manuscript are available from the corresponding author upon reasonable request.
References
- 1.Kensler KH, Regan MM, Heng YJ, Baker GM, Pyle ME, Schnitt SJ et al. Prognostic and predictive value of androgen receptor expression in postmenopausal women with estrogen receptor-positive breast cancer: results from the Breast International Group Trial 1–98. Breast Cancer Res. 21, 30 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Mod Pathol. 24, 924–931 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez LO, Corte MD, Vazquez J, Junquera S, Sanchez R, Alvarez AC et al. Androgen receptor expresion in breast cancer: relationship with clinicopathological characteristics of the tumors, prognosis, and expression of metalloproteases and their inhibitors. BMC Cancer. 8, 149 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemeier LA, Dabbs DJ, Beriwal S, Striebel JM, Bhargava R. Androgen receptor in breast cancer: expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol. 23, 205–212 (2010) [DOI] [PubMed] [Google Scholar]
- 5.Agoff SN, Swanson PE, Linden H, Hawes SE, Lawton TJ. Androgen receptor expression in estrogen receptor-negative breast cancer. Immunohistochemical, clinical, and prognostic associations. Am J Clin Pathol. 120, 725–731 (2003) [DOI] [PubMed] [Google Scholar]
- 6.Park S, Koo J, Park HS, Kim JH, Choi SY, Lee JH et al. Expression of androgen receptors in primary breast cancer. Ann Oncol. 21, 488–492 (2010) [DOI] [PubMed] [Google Scholar]
- 7.Moinfar F, Okcu M, Tsybrovskyy O, Regitnig P, Lax SF, Weybora W et al. Androgen receptors frequently are expressed in breast carcinomas: potential relevance to new therapeutic strategies. Cancer. 98, 703–711 (2003) [DOI] [PubMed] [Google Scholar]
- 8.Hickey TE, Irvine CM, Dvinge H, Tarulli GA, Hanson AR, Ryan NK et al. Expression of androgen receptor splice variants in clinical breast cancers. Oncotarget. 6, 44728–44744 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickey TE, Robinson JL, Carroll JS, Tilley WD. Minireview: The androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol. 26, 1252–1267 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res. 19, 5505–5512 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traina TA, Miller K, Yardley DA, Eakle J, Schwartzberg LS, O’Shaughnessy J et al. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J Clin Oncol. 36, 884–890 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubahn DB, Joseph DR, Sullivan PM, Willard HF, French FS, Wilson EM. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 240, 327–330 (1988) [DOI] [PubMed] [Google Scholar]
- 13.Tilley WD, Marcelli M, Wilson JD, McPhaul MJ. Characterization and expression of a cDNA encoding the human androgen receptor. Proc Natl Acad Sci U S A. 86, 327–331 (1989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 69, 16–22 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 68, 5469–5477 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 69, 2305–2313 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benayed R, Offin M, Mullaney K, Sukhadia P, Rios K, Desmeules P et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res. 25, 4712–4722 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD et al. Anchored multiplex PCR for targeted next-generation sequencing. Nature medicine. 20, 1479–1484 (2014) [DOI] [PubMed] [Google Scholar]
- 19.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 17, 251–264 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO Classification of Tumours Editorial Board. WHO Classification of Tumours. Breast Tumours, 5th ed. IARC: Lyon, 2019. [Google Scholar]
- 21.Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J Clin Oncol. 38, 1346–1366 (2020) [DOI] [PubMed] [Google Scholar]
- 22.Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol. 36, 2105–2122 (2018) [DOI] [PubMed] [Google Scholar]
- 23.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 20, 3001–3015 (2002) [DOI] [PubMed] [Google Scholar]
- 24.Scher HI, Buchanan G, Gerald W, Butler LM, Tilley WD. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocr Relat Cancer. 11, 459–476 (2004) [DOI] [PubMed] [Google Scholar]
- 25.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 20, 119–131 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C et al. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 25, 3994–4008 (2006) [DOI] [PubMed] [Google Scholar]
- 27.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 24, 4660–4671 (2005) [DOI] [PubMed] [Google Scholar]
- 28.He J, Peng R, Yuan Z, Wang S, Peng J, Lin G et al. Prognostic value of androgen receptor expression in operable triple-negative breast cancer: a retrospective analysis based on a tissue microarray. Med Oncol. 29, 406–410 (2012) [DOI] [PubMed] [Google Scholar]
- 29.Micello D, Marando A, Sahnane N, Riva C, Capella C, Sessa F. Androgen receptor is frequently expressed in HER2-positive, ER/PR-negative breast cancers. Virchows Arch 457, 467–476 (2010) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data in this manuscript are available from the corresponding author upon reasonable request.


