Abstract
Systolic and diastolic blood pressure (S/DBP) are highly correlated modifiable risk factors for cardiovascular disease (CVD). We report here a bidirectional Mendelian Randomization (MR) and horizontal pleiotropy analysis of S/DBP summary statistics from the UK Biobank (UKB)‐International Consortium for Blood Pressure (ICBP) (UKB‐ICBP) BP genome‐wide association study and construct a composite genetic risk score (GRS) by including pleiotropic variants. The composite GRS captures greater (1.11–3.26 fold) heritability for BP traits and increases (1.09‐ and 2.01‐fold) Nagelkerke's R 2 for hypertension and CVD. We replicated 118 novel BP horizontal pleiotropic variants including 18 novel BP loci using summary statistics from the Million Veteran Program (MVP) study. An additional 219 novel BP signals and 40 novel loci were identified after a meta‐analysis of the UKB‐ICBP and MVP summary statistics but without further independent replication. Our study provides further insight into BP regulation and provides a novel way to construct a GRS by including pleiotropic variants for other complex diseases.
Keywords: gene–age interaction, genetic risk score, Mendelian Randomization, pleiotropy
1. INTRODUCTION
Poorly controlled blood pressure (BP) accounts for a large portion of the risk for cardiovascular disease (CVD), stroke, and heart failure (Rapsomaniki et al., 2014). Understanding biological mechanisms for BP regulation could thus potentially help improve BP control and lead to a reduction in the burden of CVD. BP, characterized by systolic and diastolic blood pressure (SBP/DBP), are long‐standing risk predictors for CVD. To date, genome‐wide association studies (GWAS) have been performed on BP traits by focusing on main effects and in studies that included subjects of diverse ancestry over 1000 BP‐associated loci have been identified (Ehret et al., 2016; Evangelou et al., 2018; Franceschini et al., 2013; Giri et al., 2019; Hoffmann et al., 2018; International Consortium for Blood Pressure Genome‐Wide Association et al., 2011; Liang et al., 2017; C. Liu et al., 2016; Sung et al., 2018, 2019; Surendran et al., 2016; Warren et al., 2017; Zhu et al., 2015). Genome‐wide search of gene–environment interactions on BP traits have also been recently conducted, however, only a few new gene–environment interactions have been identified, in part owing to low statistical power (Sung et al., 2018, 2019). Although many GWAS variants are shared between SBP and DBP, both of which are correlated, some seem associated with SBP or DBP differently or even in opposite directions, suggesting evidence of different biological mechanisms. It has been reported that joint analysis of SBP and DBP leads to the identification of BP variants missed by analyzing SBP or DBP separately (Zhu et al., 2015). However prior studies have not addressed the mechanisms underlying the SBP–DBP relationship, reflecting arterial stiffness or arterial compliance (Schillaci & Pucci, 2010). Dissecting the causal relationships of SBP and DBP variants, in particular, whether they affect SBP and DBP through the same (mediation) or different (horizontal pleiotropic) paths, and how many horizontal pleiotropic variants contribute jointly to these highly correlated traits, is thus important for understanding the biology of BP regulation.
Genetic risk scores (GRS) are constructed as weighted linear combinations of individual variant effects estimated from GWAS to predict the individual‐level risk of a common disease. An overall GRS is the average of SBP‐ and DBP‐specific GRS (Evangelou et al., 2018; International Consortium for Blood Pressure Genome‐Wide Association et al., 2011). However, published BP GRS's have explained ~6% of the heritability of SBP and DBP, and have limited predictive power for hypertension (HTN) and cardiovascular disease (CVD). GWAS of gene–age interaction analysis has also identified genetic variants with age‐dependent effect sizes, including for BP (Shi et al., 2009; Simino et al., 2014), lipid levels (Dumitrescu et al., 2011), and body mass index (BMI) (Lasky‐Su et al., 2008). A recent study based on a proportional hazards model reported age‐varying risk profiles in nine diseases, including HTN (Jiang et al., 2021). However, these studies were underpowered because the interactive contribution by variant and age is often weak.
In this study, we address the mechanistic relationship between SBP and DBP by performing a bidirectional Mendelian Randomization (MR) (Smith & Ebrahim, 2003) and GWAS horizontal pleiotropy analysis using summary statistics from >750,000 subjects of European ancestry from the UK Biobank (UKB) and International Consortium for Blood Pressure (ICBP) consortium (Evangelou et al., 2018), followed by summary statistics of 318,891 multiethnic subjects from the Million Veteran Program (MVP) (Giri et al., 2019). We searched for novel BP variants with horizontal pleiotropic effects and constructed a composite GRS using variants with and without horizontal pleiotropic effects and studied the age‐varying effects of GRS for prediction of BP, HTN, and CVD in European, African, and Asian descent individuals.
2. MATERIAL AND METHODS
2.1. Summary statistics of UKB and ICBP
UKB and the ICBP consist of data on 458,577 UK and 299,024 European descent subjects. GWAS of SBP and DBP were conducted in UKB and ICBP separately and the results were meta‐analyzed (Evangelou et al., 2018; International Consortium for Blood Pressure Genome‐Wide Association et al., 2011). Our analysis was based on the summary results from the UKB and ICBP GWAS that were calculated based on up to 757,601 participants and ~7.1 M genotyped and imputed single‐nucleotide polymorphisms (SNPs) with MAF ≥ 1% for variants present in both the UKB data and ICBP meta‐analysis for SBP, DBP, and pulse pressure (PP; defined as SBP − DBP).
2.2. Summary statistics of MVP
The BP summary statistics of MVP consists of 318,891 predominantly male multiethnic participants from Hispanic, non‐Hispanic Whites, Blacks, Asians, and Native Americans (Giri et al., 2019). There were 18.2 M genotyped and imputed SNPs in the summary statistics. The MVP data were used for replication analysis as well as meta‐analysis with UKB‐ICBP.
2.3. UKB individual‐level data
Participants in the UKB were genotyped using a custom Affymetrix UK Biobank Axiom array (Bycroft et al., 2018). Genotypes were imputed by the UKB using the Haplotype Reference Consortium reference panel (McCarthy et al., 2016); we retained variants with imputation Rsq > 0.3. Related individuals with pairwise kinship coefficients greater than 0.0884 (suggested by UKB) were removed from the analysis, resulting in 451,174 individuals of European, African, and Asian ancestries. The principal components were calculated by UKB with genotype data within each ancestry to account for population structure.
We analyzed three BP traits in UKB: SBP, DBP, and PP. We calculated the mean SBP and DBP values from two baseline BP measurements and added 15 and 10 mmHg to SBP and DBP, respectively, for individuals who took antihypertensive medications. Hypertensive cases were defined as either SBP ≥ 140 or DBP ≥ 90 or taking antihypertensive medications. CVD cases in UKB were defined using self‐reported baseline information and the ICD9 and ICD10 diagnostic codes on hospital admissions. The CVD cases includes ICD9 ("4109", "4119", "4129", "4139", "4140", "4141", "4148", "4149") and ICD10 (I210, I211, I212, I213, I214, I219, I21X, I220, I221, I228, I229, I230, I231, I232, I233, 234, I235, I236, I238, I240, I241, I248, I249, I250, I251, I252, I253, I254, I255, I256, I258, I259) codes. These data identified 35,968 CVD cases in subjects with European, African, and Asian ancestries. The study was approved by the Case Western Reserve University Institutional Review Board (STUDY20180592).
2.4. MR analysis
We performed a bidirectional MR analysis of SBP and DBP by applying the software IMRP (Zhu et al., 2021) and MRmix (Qi & Chatterjee, 2019), as well as estimated the causal contributions of BP on coronary artery disease (CAD), myocardial infarction (MI) and stroke. Considering SBP and DBP the following association model as described in Figure 1 was used:
| (1) |
where is a genetic instrumental variable (IV), and are the direct contributions of to SBP and DBP, and are the mutual causal effects between SBP and DBP, U represents confounding factors and and are error terms, respectively. MR analysis estimates the causal effect and through the genetic IV respective to SBP and DBP. A valid genetic IV satisfies and or vice visa, representing the genetic contribution to outcome through the mediation of exposure, where SBP and DBP can either be considered as exposure or outcome. We termed these variants as mediation variants. We define a horizontal pleiotropic variant as one with and , interpreted as the genetic contributions to SBP and DBP through two independent paths (or a pleiotropic path) (Figure 1). From now on, we will simply call horizontal pleiotropic variants as pleiotropic variants. IMRP is an iterative approach combining the pleiotropy test and the MR analysis. The iteration starts by performing MR‐Egger analysis (Egger et al., 1997) to estimate the causal effect of an exposure to outcome, followed by inverse variance‐weighted (IVW) (Borenstein et al., 2009; Burgess et al., 2013) analysis until the causal effect estimate converges. The causal effect is estimated by IVW after excluding all identified pleiotropic variants. At each iteration step, IMRP performs a pleiotropy test to update which genetic instrument variants show pleiotropy (p < 0.05) using the test:
| (2) |
where and are the estimated effect sizes of a genetic IV on exposure and the outcome, respectively, and is the causal estimate which is updated at each iterative step. We have previously shown that tests the null hypothesis when DBP (SBP) is an exposure (Zhu et al., 2021). In MR analysis, a valid IV satisfies (or depending on which causal direction in Figure 1, therefore, rejecting null hypothesis suggesting a pleiotropic effect. IMRP takes advantage of MR‐Egger, which is less biased, and IVW, which is more efficient. IMRP can be applied to GWAS summary statistics of an exposure and an outcome obtained with overlapping or unique samples. To ensure the causal estimate is robust, we also applied a substantially different MR approach MRmix (Qi & Chatterjee, 2019), an estimating equation approach that assumes follows a normal mixture model. MRmix usually shows a good trade‐off between bias and variance even with more than 50% invalid IVs (Qi & Chatterjee, 2019). MRmix requires standardized summary statistics and IMRP does not. For a continuous trait, the effect size is rescaled by , where z and n correspond to the z‐score for an IV and the sample size, respectively, with its standard error . For a binary trait, the effect size is rescaled by , where beta and p correspond to the effect size and minor allele frequency of an IV, respectively. This standardizing procedure has been used in MRmix (Qi & Chatterjee, 2019).
Figure 1.
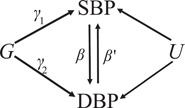
Blood pressure (BP) path diagram in Mendelian randomization and horizontal pleiotropic analysis, which corresponds to the model (1). To declare horizontal pleiotropy, we require both and 0. and are the mutual direct causal effects between and . DBP, diastolic BP; G, genetic instrumental variable; SBP, systolic BP; U, confounding factors
2.5. GWAS of pleiotropy analysis for SBP and DBP
After performing a bidirectional MR analysis of SBP and DBP and estimating the causal effects of SBP on DBP and DBP on SBP, we extended the pleiotropy test to all 7.1 Million SNPs by fixing the causal effects estimated from IMRP analysis in the two causal paths, using UKB‐ICBP summary statistics. This is equivalent to performing GWAS for two new traits: BPpleio1 = DBP SBP and BPpleio2 = SBP DBP, where and are the estimated causal effects of SBP on DBP  and DBP on SBP, respectively:
and DBP on SBP, respectively:
| (3) |
Equivalently, tests the null hypothesis . Unlike MR analysis where the IVs are selected to be associated with exposure, we required the pleiotropy test for both BPpleio1 and BPpleio2 to be significant to declare a horizontal pleiotropic variant. We performed the same analysis for replication using MVP summary statistics. Meta‐analysis of UKB‐ICBP and MVP was further performed to increase statistical power and to identify additional variants.
We applied the linkage disequilibrium (LD) score regression (LDSR) method (Bulik‐Sullivan et al., 2015) to test for genomic inflation in the GWAS pleiotropy analysis. It is expected that BPpleio1(2) will have a large genomic control inflation coefficient because of large sample sizes, genetic variants in high LD, and a large number of BP variants (Evangelou et al., 2018). We examined the degree of inflation from the intercept of LDSR.
2.6. Novel locus definition
Novel loci were defined as genome‐wide significant pleiotropy variants > 1 Mb away from known BP variants as well as LD r 2 < 0.1 with any known BP variants. Novel signals at a known locus were genome‐wide significant pleiotropy variants within 1 Mb of known BP variants as well as not being in LD with any known BP variants (r 2 < 0.1) at the locus. The 1000 Genome European ancestry data was used as the reference genetic data for LD calculation.
2.7. Functional annotations
We evaluated all sentinel SNPs at novel loci for evidence of mediation of expression quantitative trait loci (eQTL) and splicing quantitative trait loci (sQTL) in all 44 tissues using the Genotype‐Tissue Expression (GTEx) database. Following the method in Evangelou et al. (2018), a locus is annotated with a given eGene(sGene) only if the most significant eQTL(sQTL) SNP for the given eGene(sGene) is in high LD (r 2 ≥ 0.8) with the sentinel SNP. We performed overall enrichment tests using the mediation and pleiotropic variants separately. We used DEPICT (Data‐driven Expression Prioritized Integration for Complex Traits) (Pers et al., 2015) to identify tissues and cells that are highly expressed at genes within the BP mediation and pleiotropic loci. We also used DEPICT to test for enrichment in gene sets associated with GO ontologies, mouse knockout phenotypes, and the protein–protein interaction networks. We reported significant enrichments with a false discovery rate of 0.05. Analysis was done using the platform Complex‐Traits Genetics Virtual Lab (Cuellar‐Partida et al., 2019).
2.8. GRS and pleiotropic GRS (pGRS)
We constructed a traditional genetic risk score using independent genome‐wide significant BP variants from UKB‐ICBP. We first constructed SBP‐ and DBP‐weighted GRSs and then derived a single BP core GRS (cGRS) as the average of SBP and DBP GRSs. This approach has been previously used (Evangelou et al., 2018) to estimate the combined effect of BP variants on BP, HTN, and CVD. Analogously, we constructed a pGRS using the variants detected in pleiotropy analysis. We first constructed BPpleio1 and BPpleio2 weighted GRSs and next derived the pGRS as the difference of BPpleio1 and BPpleio2 GRSs. Because the genetic variants associated with BPpleio1 and BPpleio2 demonstrated horizontal pleiotropy evidence, we termed this the pleiotropy genetic risk score. Note that some SNPs contribute to both cGRS and pGRS because these variants are significantly associated with SBP or DBP, as well as BPpleio1 and BPpleio1. However, their weights represent their corresponding contributions to BP through the mediation and pleiotropy pathways. We performed linear regression analyses by jointly modeling cGRS and pGRS with and without adding the interaction of age–cGRS and age–pGRS on BP in the UKB data. We included the covariates of sex, age, BMI, geographical region, and 10 genetic principal components in the linear regression analysis. The heritability explained by the cGRS and pGRS was calculated by the adjusted R 2 in the linear regression adjusting out the covariates. Similarly, we performed logistic regression of cGRS and pGRS with and without age–cGRS and age–pGRS interactions on hypertension and cardiovascular events at baseline in the UKB data. The same covariates were included. We calculated Nagelkerke's R 2 to quantify the goodness‐of‐fit of the prediction by the cGRS and pGRS (Lee et al., 2012). We examined whether pGRS is able to predict additional variations of BP, HTN, and CVD after accounting for cGRS. We also examined the age‐varying effects of cGRS and pGRS by testing interaction effects. Our analysis included 386,752 unrelated individuals of European ancestry with phenotypes measured at baseline. For comparison, we further constructed the PP‐weighted GRS (ppGRS) and performed the above analysis.
We assessed the association of cGRS, pGRS, and their interactions with age on BP in unrelated Africans (n = 7904) and South Asians (n = 8509) from the UKB to see whether BP‐associated SNPs identified from GWAS predominantly in Europeans are also associated with BP in populations of non‐European ancestry. Analysis was also performed for ppGRS. All analyses were performed using residuals after adjusting for sex, age, BMI, geographical region, and 10 genetic principal components.
Cross‐trait lookups of novel loci: We supplied the index SNPs at the novel loci observed in UKB‐ICBP pleiotropic analyses to FUMA (Watanabe et al., 2017) and GWAS catalog (MacArthur et al., 2017) to investigate pleiotropy with non‐BP traits, extracting all associations with p < 5 × 10−8 for all SNPs in high LD (r 2 ≥ 0.8).
3. RESULTS
We present a bidirectional MR analysis of SBP and DBP using 1125 and 1183 independent genome‐wide significant variants for SBP and DBP (p < 5 × 10−8) as genetic IVs obtained from the UKB‐ICBP GWAS (Evangelou et al., 2018). We standardized SBP and DBP and obtained an identical causal effect of SBP on DBP and DBP on SBP (0.864 ± 0.005 and 0.862 ± 0.005 by IMRP, respectively, Table S1), which is significantly larger than the observed trait correlation 0.738 between SBP and DBP in UKB European subjects, with an estimated 74.8% of the variation is the shared causal contribution between SBP and DBP. The causal estimates by MRmix were concordant (0.89 ± 0.012 and 0.90 ± 0.01, respectively, Table S1). Among the genetic IVs, 43% of the variants had pleiotropic effects on SBP and DBP.
We next extended the pleiotropic effect analysis to search for variants by performing two GWAS of BPpleio1 and BPpleio2; the Manhattan and QQ plots are presented in Figure 2. The GC lambda value was 1.533 and the LDSR intercept was 1.057 (0.013), with an inflation ratio of 4.23%, suggesting little inflation in the BPpleio1; a similar result was observed for BPpleio2. LDSR analysis (Bulik‐Sullivan et al., 2015) estimated 8.7% of the heritability arising from pleiotropic variants for BPpleio1. We calculated genetic correlations of BPpleio1 and BPpleio2 with SBP, DBP, and PP using summary statistics (Table S2). BPpleio1 and BPpleio2 are genetically highly correlated (r g = −0.902 ± 0.006), also highly correlated with PP (r g = −0.618 ± 0.016 and 0.897 ± 0.004) but less so with SBP or DBP. BPpleio1 is negatively correlated with SBP as is BPpleio2 with DBP; in contrast, PP is positively correlated with both SBP and DBP.
Figure 2.
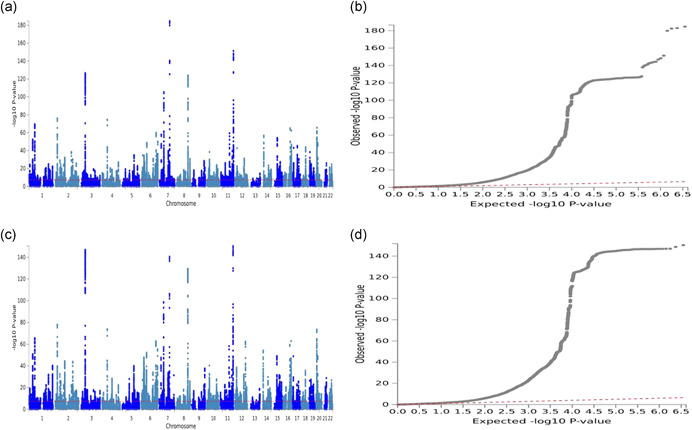
Manhattan and QQ plots for genome‐wide pleiotropy tests between SBP and DBP using UK Biobank‐ICBP summary statistics. The GWAS of pleiotropy tests is equivalent to performing GWAS for two new traits: BPpleio1 = DBP SBP and BPpleio2 = SBP DBP, where and are the estimated causal effects of SBP on DBP and DBP on SBP, respectively. (a, b) Manhattan and QQ plots for BPpleio2. (c, d) Manhattan and QQ plots for BPpleio1. DBP, diastolic BP; GWAS; genome‐wide association studies; ICBP, International Consortium for Blood Pressure; SBP, systolic BP; The horizontal line in Manhattans represents p = 5 × 10−8. The top and bottom Manhattan plots are highly similar, indicating the consistency of the two‐directional Mendelian Randomization analysis
and DBP on SBP, respectively. (a, b) Manhattan and QQ plots for BPpleio2. (c, d) Manhattan and QQ plots for BPpleio1. DBP, diastolic BP; GWAS; genome‐wide association studies; ICBP, International Consortium for Blood Pressure; SBP, systolic BP; The horizontal line in Manhattans represents p = 5 × 10−8. The top and bottom Manhattan plots are highly similar, indicating the consistency of the two‐directional Mendelian Randomization analysis
We observed 906 independent variants (r 2 < 0.1) reaching genome‐wide significance in either BPpleio1 or BPpleio2 (p < 5 × 10−8). To declare a variant as pleiotropic, we required one pleiotropy test p ≤ 5 × 10−8 and the other test p ≤ 0.05/906 by adjusting for multiple comparisons. We observed 815 independent pleiotropy variants with 91 variants genome‐wide significant for either SBP or DBP. Among them, 234 (or 29%) were not detected by the univariate GWAS analysis of SBP, DBP, or PP in the original UKB‐ICBP consortium (Evangelou et al., 2018) (Table S3). Among the 234 variants, 201 (or 86%) variants were not reported in any previous BP GWAS. In the set of associations, 163 variants in 124 loci were within a 1‐Mb region of previously reported known BP loci but were not in LD with known BP variants (r 2 < 0.1); the remaining 38 variants were at least 1 Mb away from the previous reported known BP loci and resided at 35 loci; the corresponding locus zoom plots are presented in Figure S1. We evaluated the associations of our sentinel SNPs at the 35 novel loci with other traits and diseases using the GWAS Catalog (MacArthur et al., 2017) and FUMA (Watanabe et al., 2017). The GWAS Catalog and FUMA search of published GWAS showed that 29 of the 35 novel loci are also significantly associated with other traits, including lipid levels, cardiovascular‐related outcomes, anthropometric traits, sleep traits, educational attainment, smoking, blood protein level, and schizophrenia (Table S4).
We defined the variants with p value of SBP < 5 × 10−8 and p value of BPpleio1 > 0.05/906 or p value of DBP < 5 × 10−8 and p value of BPpleio2 > 0.05/906 into mediation variants, which resulted in 1415 independent variants. We compared the SBP and DBP effect sizes for these mediation variants and observed that the mediation variants have the same effect directions for SBP and DBP (Figure 3). In comparison, 71% of pleiotropic variants show opposite effect directions for SBP and DBP, indicating new discoveries.
Figure 3.
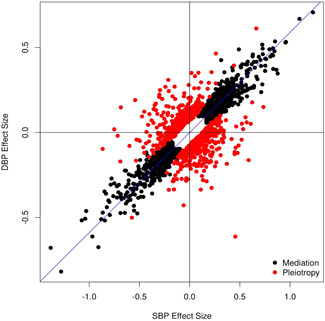
Comparison between the systolic blood pressure (SBP) and Diastolic blood pressure (DBP) effect sizes for mediation (black dots) and pleiotropic (red dots) variants in UK Biobank‐International Consortium for Blood Pressure
3.1. Replication of novel signals in MVP
Because the BP summary statistics in MVP were obtained from multiethnic populations of non‐Hispanic Whites, non‐Hispanic Blacks, Hispanics, non‐Hispanic Asians, and non‐Hispanic Native Americans, we performed MR analysis to estimate causal effect sizes between SBP and DBP in MVP, instead of using the causal effects estimated from UKB‐ICBP. With a fewer number of IVs in MVP, we obtained relatively smaller but similar causal effect sizes (0.692 and 0.724) between SBP and DBP as compared to UKB‐ICBP results, possibly due to the multiethnic samples in MVP (Table S1). We then performed the pleiotropy test among the 201 novel variants (seven were not available in MVP). We examined how many novel variants were significantly associated with BPpleio1(2) (p < 5 × 10−8) in UKB‐ICBP also showed replication with corresponding BPpleio1(2) at significance level p < 0.05/201. We were able to replicate 23 variants (Table S3), including two novel loci (rs12470661 and rs73937040, Table 1). When we released the replication significance level criterion at p = 0.05, we were able to replicate 118 variants including 19 variants at 18 novel loci (Table 1), with 84 variants having both BPpleio1 and BPpleio2 (p < 0.05) (Table S3) and all the 118 variants having the same effect direction between UKB‐ICBP and MVP; thus, our identified novel pleiotropic signals were replicable.
Table 1.
The 17 novel blood pressure (BP) loci identified by pleiotropic analysis
| SNP | CHR | BP | UKB‐ICBP | UKB‐ICBP | UKB‐ICBP | UKB‐ICBP | MVP | UKB‐ICBP–MVP | Genes |
|---|---|---|---|---|---|---|---|---|---|
| P_SBP | P_DBP | P_PP | p pleio a | p pleio a | p pleio a | ||||
| rs11162906 | 1 | 80500074 | 3.19 × 10−2 | 1.75 × 10−2 | 1.78 × 10−6 | 2.41 × 10−9 | 1.53 × 10−3 | 3.54 × 10−11 | AC098657.2 |
| rs17713879 | 2 | 254215 | 5.19 × 10−2 | 2.66 × 10−2 | 2.05 × 10−5 | 3.75 × 10−8 | 8.92 × 10−4 | 1.35 × 10−10 | SH3YL1 |
| rs3136302 | 2 | 48021379 | 4.08 × 10−1 | 5.84 × 10−5 | 9.62 × 10−6 | 2.96E−11 | 4.48 × 10−2 | 8.30 × 10−12 | MSH6 |
| rs6735304 | 2 | 101617631 | 4.18 × 10−2 | 2.38 × 10−2 | 1.43 × 10−6 | 1.47 × 10−8 | 6.21 × 10−4 | 6.36 × 10−11 | RPL31/TBC1D8 |
| rs12470661 | 2 | 232060050 | 5.09 × 10−2 | 3.07 × 10−3 | 6.68 × 10−7 | 5.33 × 10−11 | 1.97 × 10−4 | 4.55 × 10−14 | HTR2B/ARMC9 |
| rs10947978 | 6 | 41471608 | 7.01 × 10−1 | 9.89 × 10−6 | 1.44 × 10−5 | 2.65 × 10−11 | 1.84 × 10−2 | 3.26 × 10−12 | LINC01276 |
| rs56098119 | 6 | 90296727 | 7.63 × 10−2 | 1.38 × 10−2 | 6.81 × 10−6 | 1.69 × 10−8 | 2.89 × 10−2 | 1.77 × 10−9 | ANKRD6 |
| rs150953973 | 6 | 120780033 | 2.98 × 10−1 | 9.7 × 10−4 | 4.01 × 10−6 | 3.54 × 10−9 | 1.64 × 10−2 | 1.85 × 10−10 | RNU6‐214P |
| rs180271 | 7 | 93539479 | 6.11 × 10−1 | 1.72 × 10−4 | 2.63 × 10−5 | 3.69 × 10−9 | 2.05 × 10−3 | 2.92 × 10−11 | GNGT1 |
| rs11989271 | 8 | 122632611 | 1.51 × 10−1 | 8.47 × 10−4 | 7.80 × 10−7 | 1.14 × 10−10 | 9.37 × 10−3 | 1.38 × 10−11 | HAS2 |
| rs10868842 | 9 | 73119085 | 1.57 × 10−1 | 3.58 × 10−4 | 9.71 × 10−6 | 1.37 × 10−11 | 4.07 × 10−3 | 2.54 × 10−13 | LINC00583 |
| rs12768143 | 10 | 22808844 | 3.92 × 10−2 | 5.03 × 10−3 | 8.61 × 10−8 | 9.18 × 10−11 | 1.58 × 10−2 | 2.39 × 10−11 | PIP4K2A |
| rs1343676 | 12 | 33537387 | 7.19 × 10−1 | 2.03 × 10−7 | 8.65 × 10−7 | 9.56 × 10−15 | 4.04;× 10−4 | 3.04 × 10−16 | SYT10 |
| rs7322054 | 13 | 38246708 | 6.71 × 10−1 | 1.99 × 10−7 | 7.76 × 10−4 | 1.04 × 10−11 | 1.01 × 10−2 | 5.86 × 10−13 | TRPC4 |
| rs61972411 | 13 | 100602630 | 2.8 2× 10−4 | 6.59 × 10−1 | 1.87 × 10−7 | 1.45 × 10−8 | 1.89 × 10−2 | 2.23 × 10−9 | LOC101927437 |
| rs62621400 | 15 | 101718239 | 2.25 × 10−1 | 1.70 × 10−3 | 1.94 × 10−5 | 3.76 × 10−9 | 1.64 × 10−3 | 2.34 × 10−11 | CHSY1 |
| rs116643984 | 15 | 101791212 | 1.64 × 10−1 | 7.73 × 10−5 | 5.14 × 10−8 | 4.04 × 10−13 | 6.86 × 10−3 | 1.60 × 10−14 | CHSY1 |
| rs73937040 | 18 | 3258733 | 7.08 × 10−2 | 2.07 × 10−2 | 7.77 × 10−6 | 4.67 × 10−8 | 2.06 × 10−6 | 1.17 × 10−12 | MYL12A/MYL12B |
| rs146827176 | 20 | 35169916 | 6.48 × 10−1 | 3.10 × 10−6 | 1.01 × 10−3 | 2.13 × 10−9 | 5.08 × 10−3 | 3.82 × 10−11 | LOC101926987|MYL9 |
Abbreviations: CHR, chromosome; DBP, diastolic blood pressure; ICBP, International Consortium for Blood Pressure; MVP, Million Veteran Program; PP, pulse pressure; SBP, systolic blood pressure; SNP, single‐nucleotide polymorphism; UKB, UK Biobank.
The p values of pleiotropy test for BPpleio1 and BPpleio2 were consistent in general and we reported the lesser one here. The detailed summary statistics and corresponding p values were listed in Table S3.
We further examined the BPpleio1(2) effect size consistency of the 815 independent pleiotropic variants between UKB‐ICBP and MVP. For comparison, we also examined the SBP and DBP effect size consistency of the independent 1415 mediation variants between UKB‐ICBP and MVP. We observed that the BPpleio1(2) effect sizes have higher correlations for the pleiotropic variants than the mediation variants (Figure 4, correlation 0.90 vs. 0.74), even after the exclusion of two significant outliers (rs113081691 and rs17057329). We observed that 96% of pleiotropic variants have consistent effect directions between UKB‐ICBP and MVP. We then performed an inverse‐variance weighted meta‐analysis to combine the summary statistics of BPpleio1(2) for UKB‐ICBP and MVP. The combined horizontal pleiotropy evidence was further strengthened for the 118 novel variants (Tables 1 and S3).
Figure 4.

Comparison between the effect sizes between UKB‐ICBP and MVP. (a) SBP effect sizes of mediation variants. (b) DBP effect sizes of mediation variants. (c) BPpleio1 effect sizes for pleiotropic variants. (d) BPpleio2 effect sizes for pleiotropic variants. The two variants rs113081691 and rs17057329 in the red circles in (a) and (b) represent substantial different effect sizes of SBP and DBP between UKB‐ICBP and MVP, likely driven by multi‐ethnic samples from MVP. DBP, diastolic blood pressure; ICBP, International Consortium for Blood Pressure; MVP, Million Veteran Program; SBP, systolic blood pressure; UKB, UK Biobank.
3.2. Functional annotations
We performed expression quantitative trait locus (eQTL) analysis using GTEx data. Among the 35 novel loci listed in Table S3, we identified 26 with expression quantitative trait locus (eQTL) (Table S5) and 11 with Splicing Quantitative Trait Loci (sQTLs) (Table S6). The eQTLs were most often enriched in arterial tissues, followed by adipose, heart, and nerve tibial tissues. SNP rs17713879 is an eQTL affecting expression of the SH3YL1 and ACP1 genes in 34 tissues and is also a sQTL affecting splicing of these two genes in 50 tissues. SNP rs112500920 is an eQTL affecting expression of several genes, including EFL1 and AB3B2, in multiple tissues, notably adipose and arterial tissues. SNP rs12478520 is an eQTL affecting expression of multiple genes, including C2orf72, HTR2B, ARMC9, and PSMD1.
In the UKB‐ICBP data, we identified 815 independent pleiotropic variants. We also observed 1451 independent mediation variants. We assessed tissue enrichment of BP loci using DEPICT (Pers et al., 2015) at a false discovery rate (FDR) < 5% but separated 1415 mediation from 815 pleiotropic variants in the analysis. DEPICT identified enrichment across 43 and 51 tissues and cells using mediation and pleiotropic variants, respectively (Table S7). The enriched tissues are highly similar (correlation = 0.78) but there are also notable differences (Figure S2a). Enrichment was greatest for arteries in the cardiovascular system for both mediation and pleiotropic variants (p = 1.28 × 10−3 and 2.19 × 10−11, respectively). In general, enrichment observed for mediation variants was also observed for pleiotropic variants, but not vice versa. For example, heart‐related tissues, aortic valves, and atrial appendages were enriched for pleiotropic variants (p < 1.38 × 10−4) but not for mediation variants (Table S7). Pathway enrichments for mediation variants and pleiotropic variants were less well correlated (correlation = 0.51, Figure S2b). Pleiotropic variants were enriched in many molecular pathways that were missed by mediation variants, including response to hypoxia, oxygen levels, basement membrane, and renal system development (p < 8.66 × 10−7, Table S8). In contrast, negative regulation of transcription from RNA polymerase II promoter, histone deacetylase binding, hormone receptor binding, and Ras protein signal transduction, among others, were only enriched by mediation variants (p < 6.09 × 10−7, Table S8).
Evaluation of enriched mouse knockout phenotype terms by both mediation and pleiotropic variants implicated abnormal cardiovascular physiology, disorganized myocardium, abnormal vascular branching morphogenesis, and organogenesis, among others. However, pleiotropic variant‐enriched mouse phenotypes include abnormal kidney morphology, impaired wound healing, dilated heart right ventricle, abnormal aorta morphology, increased systemic arterial SBP, and increased body weight (p < 1.65 × 10−7). Mediation variants‐enriched mouse phenotypes include pericardial edema, wavy neural tube, and decreased systemic arterial BP (p < 1.83 × 10−4, Table S8). Common protein–protein interaction subnetwork enrichments for both mediation and pleiotropic variants include LAMA1, ITGB1, WNT1, and many SMAD subnetworks. Pleiotropic variant enriched top significant subnetworks include the HSPG2, TGM2, MMP9, FBN1, and DDR1 subnetworks (p < 1.76 × 10−7), as compared to mediation variant enriched subnetworks, SRC, YWHAQ, and RAPGEF1 (Table S9).
3.3. Improved prediction of BP, HTN, and CVD by including pleiotropic variants
Polygenic scores derived from multiple related traits can improve the prediction of outcomes (Inouye et al., 2018; Krapohl et al., 2018; Maier et al., 2018; Richardson et al., 2019). Chasman et al. (2020) decomposed a GRS into nearly independent components relative to biological mechanisms inferred from pleiotropic relationships, whereas Udler et al. (2018) factorized the genetic association matrix according to different classes of genetic variants relative to traits. However, these approaches do not use the discovered pleiotropic variants directly. Here, we construct a traditional BP GRS using all independent 1615 BP variants from the UKB‐ICBP, which we term cGRS (see Section 2). We further constructed a pGRS using the 906 variants associated with BPpleio1 or BPpleio2, which is a genetic risk score from pleiotropic variants. We jointly modeled cGRS and pGRS adjusting for age, gender, BMI, and 10 principal components, and observed that pGRS significantly predicted BP traits, as well as the risk of HTN and CVD, conditional on cGRS in all models (Table 2a and Figure 5) in the UKB European ancestry subjects. The cGRS captured 5.91%, 6.09%, and 2.23% SBP, DBP, and PP heritability excluding pGRS and 7.13%, 6.75%, and 7.27% including pGRS, or a 1.11‐ to 3.26‐fold increase. Similarly, Nagelkerke's R 2 for HTN and CVD was 4.71% and 0.47% excluding pGRS and 5.14% and 0.53% including pGRS, representing a 1.09‐ and 1.14‐fold increase. We observed odds ratios (ORs) of 1.64 and 1.15 for individual cGRS and pGRS on the risk of HTN (p < 1 × 10−300), respectively. The observed ORs of individual cGRS and pGRS for CVD were 1.21 and 1.06 (p = 1.71 × 10−231 and p = 2.74 × 10−25), respectively, and increased to 1.66 and 1.19 (p = 9.83 × 10−97 and p = 1.28 × 10−13) when comparing the upper versus lower quantiles of the cGRS and pGRS, respectively. However, the ORs were further increased to 6.78 and 2.44 (p < 1 × 10−300 and p = 1.15 × 10−39) for HTN and CAD, respectively, when comparing the top decile and quintile with bottom decile and quintile of cGRS and pGRS (Figure 5). We observed a clear advantage of including pGRS over cGRS only (Figure 5). It has been suggested that there are gene and age interactions that contribute to blood pressure and hypertension (Jiang et al., 2021; Shi et al., 2009; Simino et al., 2014). However, the detected interactions are limited because of low statistical power. We, thus, examined the interaction effects between age and cGRS and pGRS. After including the interactions of age and cGRS and pGRS the main effects for cGRS and pGRS on BP, HTN, and CVD were unchanged. However, we observed a significant interaction effect of age and cGRS for all BP traits and HTN but not CVD. The interaction of age and pGRS significantly contributed to all BP traits, HTN, and CVD (p value between 3.0 × 10−2 and 7.74 × 10−29, Table 2a).
Table 2.
Associations of the cGRS, pGRS, and their interactions with age on blood pressure traits, hypertension and cardiovascular events in unrelated populations in UK Biobank European, African, and Asian descents
| cGRS | pGRS | Heritabilitya | Age–cGRS | Age–pGRS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Effectb | 95% CI | P | Effectb | 95% CI | P | GRS only (%) | GRS + pGRS (%) | Fold change | Effectb | 95% CI | P | Effectb | 95% CI | P | N |
| (a) Europeans | ||||||||||||||||
| SBP | 4.56 | 4.51; 4.62 | <10 −300 | 2.01 | 1.96; 2.07 | <10 −300 | 5.91 | 7.13 | 1.21 | 0.011 | 0.004; 0.01 | 2.02 × 10 −3 | 0.068 | 0.014; 0.123 | 1.32 × 10 −2 | 385,549 |
| DBP | 2.44 | 2.41; 2.47 | <10 −300 | −0.83 | −0.86; −0.80 | <10 −300 | 6.09 | 6.75 | 1.11 | −0.026 | −0.030; −0.022 | 1.41 × 10 −39 | −0.101 | −0.132; −0.071 | 8.40 × 10 −11 | 385,560 |
| PP | 2.12 | 2.09; 2.16 | <10 −300 | 2.84 | 2.80; 2.88 | <10 −300 | 2.23 | 7.27 | 3.26 | 0.037 | 0.032; 0.04 | 1.40 × 10 −51 | 0.170 | 0.132; 0.20 | 1.05 × 10 −18 | 385,549 |
| HTN | 1.64 | 1.63; 1.66 | <10 −300 | 1.15 | 1.14; 1.16 | <10 −300 | 4.71 | 5.14 | 1.09 | 0.997 | 0.996; 0.99 | 1.50 × 10 −11 | 1.005 | 1.004; 1.00 | 7.74 × 10 −29 | 385,554 |
| CVD | 1.21 | 1.20; 1.23 | 1.71 × 10 −231 | 1.06 | 1.05; 1.08 | 2.74 × 10 −25 | 0.47 | 0.53 | 1.14 | 0.999 | 0.997; 1.00 | 4.25 × 10−1 | 1.002 | 1.00; 1.004 | 3.0 × 10 −2 | 385,554 |
| CVDc | 1.66 | 1.58; 1.74 | 9.83 × 10 −97 | 1.19 | 1.14; 1.25 | 1.28 × 10 −13 | 0.74 | 0.89 | 1.20 | 1.002 | 0.995; 1.00 | 6.14 × 10−1 | 1.004 | 0.996; 1.01 | 3.37 × 10−1 | 96,963 |
| (b) Africans | ||||||||||||||||
| SBP | 2.71 | 2.28; 3.13 | 1.45 × 10 −39 | 1.21 | 0.77; 1.65 | 8.34 × 10 −8 | 1.74 | 1.91 | 1.10 | 0.010 | −0.041; 0.061 | 7.0 × 10−1 | 0.045 | −0.007; 0.096 | 9.02 × 10−1 | 7802 |
| DBP | 1.53 | 1.27; 1.79 | 4.19 × 10 −31 | −0.35 | −0.62; −0.08 | 1.09 × 10 −2 | 1.59 | 1.73 | 1.09 | −0.015 | −0.046; 0.016 | 3.36 × 10−1 | 0.002 | −0.029; 0.033 | 9.0 × 10−1 | 7802 |
| PP | 1.18 | 0.91; 1.45 | 1.61 × 10 −17 | 1.56 | 1.28; 1.84 | 2.05 × 10 −27 | 0.75 | 1.83 | 2.44 | 0.025 | −0.007; 0.057 | 1.28 × 10−1 | 0.043 | 0.010; 0.07 | 1.1 × 10 −2 | 7802 |
| HTN | 1.32 | 1.25; 1.39 | 7.17 × 10 −27 | 1.04 | 0.99; 1.09 | 1.54 × 10−1 | 0.84 | 1.08 | 1.29 | 1.001 | 0.994; 1.00 | 8.26 × 10−1 | 1.005 | 0.999; 1.01 | 1.2 × 10−1 | 7807 |
| CVD | 1.14 | 1.03; 1.26 | 9.88 × 10 −3 | 1.07 | 0.97; 1.19 | 1.85 × 10−1 | 0.18 | 0.22 | 1.22 | 1.003 | 0.992; 1.01 | 6.04 × 10−1 | 1.000 | 0.989; 1.01 | 1.0 | 7807 |
| (c) Asians | ||||||||||||||||
| SBP | 2.92 | 2.54; 3.30 | 2.71 × 10 −50 | 1.25 | 0.86; 1.64 | 3.21 × 10 −10 | 2.53 | 3.00 | 1.18 | −0.011 | −0.056; 0.035 | 6.49 × 10−1 | 0.015 | −0.030; 0.060 | 5.12 × 10−1 | 8327 |
| DBP | 1.69 | 1.47; 1.91 | 5.95 × 10 −51 | −0.64 | −0.87; −0.42 | 1.69 × 10 −8 | 2.58 | 2.89 | 1.12 | −0.017 | −0.043; 0.009 | 1.96 × 10−1 | −0.047 | −0.073; 0.021 | 3.64 × 10 −4 | 8327 |
| PP | 1.23 | 0.97; 1.48 | 3.85 × 10 −21 | 1.89 | 1.63; 2.15 | 1.50 × 10 −46 | 1.00 | 3.33 | 3.33 | 0.007 | −0.023; 0.037 | 6.65 × 10−1 | 0.062 | 0.032; 0.092 | 4.71 × 10 −5 | 8327 |
| HTN | 1.37 | 1.30; 1.44 | 3.90 × 10 −36 | 1.10 | 1.05; 1.16 | 9.27 × 10 −5 | 1.84 | 2.08 | 1.13 | 0.998 | 0.992; 1.004 | 5.23 × 10−1 | 1.006 | 1.00; 1.012 | 3.86 × 10 −2 | 8332 |
| CVD | 1.18 | 1.10; 1.26 | 2.87 × 10 −6 | 1.08 | 1.01; 1.16 | 2.81 × 10 −2 | 0.19 | 0.39 | 2.01 | 1.003 | 0.995; 1.012 | 4.50 × 10−1 | 0.991 | 0.983; 1.0 | 4.27 × 10 −2 | 8332 |
Note: Bold values represent P < 0.05. Sex, age, body mass index, 10 genetic principal components, and geographic regions were adjusted. cGRS and pGRS were jointly modeled in the regression analyses.
Abbreviations: CI, confidence interval; cGRS, core genetic risk score; CVD, cardiovascular disease; DBP, diastolic blood pressure; pGRS, pleiotropic GRS; HTN, hypertension; PP, pulse pressure; SBP, systolic blood pressure.
The heritability for HTN and CVD represents Nagelkerke's R 2.
The effect represents the regression coefficient for SBP, DBP, and PP and represents the odds ratio for HTN and CVD.
The effect for CVD represents the odds ratio when comparing top quantile with bottom quantile of GRS and pGRS.
Figure 5.
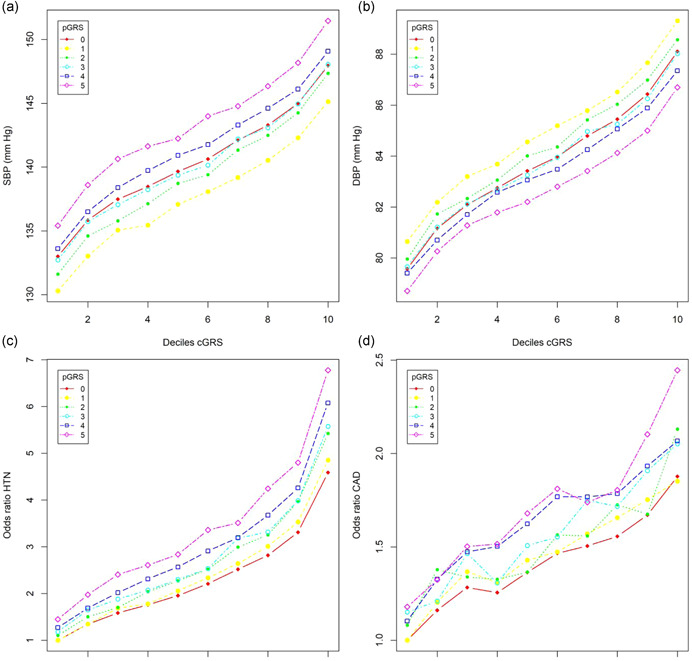
Relationship the core genetic risk score (cGRS) and pleiotropic genetic risk score (pGRS) with blood pressure (BP), risk of hypertension (HTN), and cardiovascular disease (CVD) in UK Biobank. (a) Sex‐adjusted mean systolic BP (SBP); (b) sex‐adjusted mean diastolic BP (DBP); (c) odds ratios of HTN; (d) odds ratios of CVD. cGRS was calculated in every decile and pGRS was calculated in every quintile. Odds ratios were calculated by comparing each of the cGRS deciles and pGRS quintiles with the lowest decile and 20th. The curves with pGRS = 0 represent the models without including pGRS
3.4. Extension to other ancestries
We examined associations with BP and CVD of the above‐defined European cGRS and pGRS in unrelated African (N = 7904) and South Asian (N = 8509) subjects in the UKB (Table 2b,c). Although sample sizes were much smaller than among UKB European subjects, the cGRS is significantly associated with SBP, DBP, PP, HTN, and CVD in both the UKB African and Asian ancestry subjects. In the UKB African ancestry individuals, including the pGRS results in a 1.10‐ to 2.44‐fold increase of SBP, DBP, and PP heritability. Nagelkerke's R 2 for HTN and CVD has 1.23‐ and 1.22‐fold increases, respectively. Similar increments in the UKB Asian cohort are also observed (Table 2c). Significant interactions of age and pGRS were again observed for BP traits and CVD in UKB Asians (Table 2c).
Comparison between BP pleio1 (BP pleio2 ) and PP: We noted that PP is genetically positively correlated with both SBP and DBP but BPpleio1 or BPpleio2 is negatively correlated with SBP and DBP (Table S2). Among the 815 pleiotropic variants, 274 were genome‐wide significant with either SBP or DBP and 467 with PP. We then calculated the GRS of PP (GRSPP) in UKB using the independent variants associated with PP by the effect sizes estimated in the UBK‐ICBP data. We observed that the GRSPP is positively correlated with both the GRSs of SBP and DBP. In comparison, the pGRS is again oppositely correlated with the GRSs of SBP and DBP (Table S2). We next compared BP variances explained by the GRSPP and pGRS in UKB. In general, GRSPP and pGRS can account for similar amounts of BP, HTN, and CVD variation (Table S9). However, when we included cGRS, pGRS and GRSPP in a regression model, we observed that all three genetic risk scores significantly predicted variation in BP, HTN, and CVD in UKB European subjects (Table S9), suggesting pGRS and GRSPP identify different aspects of trait variations.
3.5. MR of BP on CAD, MI, and stroke
We downloaded published GWAS summary statistics for CAD (Nikpay et al., 2015), MI (Webb et al., 2017), and stroke (Malik et al., 2018) and performed MR analysis of SBP, DBP, PP, BPpleio1, and BPpleio2 on CAD, MI, and stroke (Table 3). For SBP and DBP, IVs were the genetic variants genome‐wide significantly associated with SBP and DBP but with no pleiotropic evidence. For BPpleio1 and BPpleio2, IVs were the genetic variants associated with BPpleio1 and BPpleio2. For PP, we selected all variants independently associated with PP as the IVs. The effect size and standard error of an IV for BPpleio1 or BPpleio2 were the corresponding numerator and denominator of the test statistic in Equation (2). As expected, SBP, DBP, and PP causally contributed to CAD, MI, and stroke. The derived trait—BPpleio—also causally contributed to CAD, MI, and stroke, suggesting a primary causal pleiotropy pathway that is not associated with the BP mediation pathway directly contributing to the outcomes (Table 3). The estimated ORs ranged from 1.66 to 1.85 per SD unit increase in BP on the three outcomes using mediation variants and ranged from 1.13 to 1.48 using pleiotropic variants. Our analysis identified 1%–22% IVs demonstrating pleiotropic effects for BP and the three clinical outcomes (Table 3).
Table 3.
Using mediation variants and pleiotropy variants separately in Mendelian Randomization analysis with outcomes: CAD, MI, and stroke
| Outcome | Exposure | # of IVsa | IMRP | # of PVsc | MRmix | ||||
|---|---|---|---|---|---|---|---|---|---|
| Causal effectb | 95% CI | p | Causal effectb | 95% CI | p | ||||
| CAD | SBP | 758 | 1.85 | 1.73; 1.97 | 3.73 × 10−78 | 103 | 1.80 | 1.56; 2.09 | 2.68 × 10−15 |
| MI | SBP | 756 | 1.77 | 1.65; 1.90 | 1.60 × 10−57 | 89 | 1.72 | 1.42; 2.08 | 3.57 × 10−8 |
| stroke | SBP | 758 | 1.66 | 1.55; 1.76 | 1.37 × 10−57 | 77 | 1.70 | 1.29; 2.24 | 1.80 × 10−4 |
| CAD | BPpleio1 | 730 | 1.48 | 1.34; 1.63 | 2.64× 10−14 | 150 | 1.49 | 1.06; 2.10 | 0.022 |
| MI | BPpleio1 | 729 | 1.32 | 1.18; 1.47 | 7.11 × 10−7 | 125 | 1.43 | 1.09; 1.88 | 9.41 × 10−3 |
| stroke | BPpleio1 | 730 | 1.42 | 1.30; 1.55 | 1.98 × 10−14 | 92 | 1.16 | 0.81; 1.67 | 0.417 |
| CAD | DBP | 774 | 1.85 | 1.74; 1.97 | 1.16 × 10−80 | 105 | 1.86 | 1.60; 2.16 | 5.38 × 10−16 |
| MI | DBP | 771 | 1.85 | 1.73; 1.98 | 3.22 × 10−68 | 91 | 1.86 | 1.51; 2.29 | 5.65 × 10−9 |
| Stroke | DBP | 773 | 1.67 | 1.57; 1.77 | 2.77 × 10−60 | 79 | 1.88 | 1.51; 2.33 | 8.84 × 10−9 |
| CAD | BPpleio2 | 730 | 1.37 | 1.24; 1.50 | 4.63 × 10−11 | 157 | 1.34 | 1.02; 1.75 | 0.037 |
| MI | BPpleio2 | 729 | 1.13 | 1.02; 1.25 | 0.017 | 134 | 1.31 | 1.05; 1.63 | 0.015 |
| Stroke | BPpleio2 | 730 | 1.28 | 1.18; 1.39 | 5.07 × 10−9 | 90 | 1.13 | 0.80; 1.59 | 0.491 |
| CAD | PP | 899 | 1.53 | 1.45; 1.62 | 3.76 × 10−50 | 22 | 1.61 | 1.43; 1.82 | 6.11 × 10−15 |
| MI | PP | 898 | 1.41 | 1.33; 1.50 | 3.31 × 10−29 | 17 | 1.46 | 1.31; 1.64 | 4.03 × 10−11 |
| Stroke | PP | 899 | 1.46 | 1.38; 1.54 | 2.98 × 10−44 | 8 | 1.20 | 0.97; 1.51 | 0.095 |
Abbreviations: BP, blood pressure; CAD, coronary artery disease; CI, confidence interval; DBP, diastolic BP; IV, instrumental variable; MI, myocardial infarction; PV, pleiotropic variants; SBP, systolic BP.
Number of genetic instrumental variables that are genome‐wide significantly associated with exposure. For SBP and DBP, IVs were the genetic variants associated with SBP and DBP but with no pleiotropic evidence of SBP and DBP, respectively. For BPpleio1 and BPpleio2, IVs were the genetic variants associated with BPpleio1 and BPpleio2, respectively.
Odds ratio.
Number of pleiotropic variants detected by IMRP among the IVs.
3.6. Meta‐analysis of UKB‐ICBP and MVP
We performed the IVW meta‐analysis of pleiotropy test to combine UKB‐ICBP and MVP. We observed an additional 219 novel signals with either BPpleio1 or BPpleio2 at a significance level 5 × 10−8, including an additional 40 novel loci (Table S10).
4. DISCUSSION
Our analysis of GWAS summary statistics from over 1 million subjects has, here, revealed important aspects of the genetic architecture of the two principal and highly correlated BP traits, SBP and DBP. The bidirectional MR analysis of SBP and DBP demonstrated that 1 SD unit increase of SBP leads to 0.86 SD unit increase of DBP, and vice versa, indicating that SBP and DBP share 74.8% of its variation. We assume these arise from common genetic factors and common biological mechanisms between SBP and DBP. This shared causal contribution is substantially higher than the 55.6% estimated by phenotype correlation analysis of SBP and DBP in UKB European ancestry subjects. We then identified the genetic variants that impact SBP and DBP through two different paths: (1) mediation path (either from SBP to DBP or vice versa) and departure from the mediation path (pleiotropic path, Figure 1). We defined the variants contributing to BP traits through the pleiotropic path as pleiotropic variants, which have different biological processes from the BP variants through the mediation path. We observed that most of the BP variants identified through SBP or DBP univariate associations were mediation variants and would be expected to be discovered in either the SBP or DBP GWAS. By examining the variants departing from the mediation paths, we identified 815 independent variants demonstrating horizontal pleiotropy evidence in the original UK Biobank + ICBP consortium data (Evangelou et al., 2018), of which 201 were undetected by univariate GWAS of SBP, DBP, or PP in literature. Replication analysis in the MVP confirmed 118 of the 201 novel variants, including the 18 novel loci (Tables 1 and S3). Pleiotropic variants often demonstrated an effect size opposite in direction for SBP and DBP and yet contributed 8.71% heritability of the newly defined BP trait (Figure 3). The effect sizes of pleiotropic variants also demonstrated a higher correlation than that of the mediation variants between UKB‐ICBP and MVP, suggesting that the pleiotropic variants may be more transferable across ethnic populations. BPpleio1 and BPpleio2 were highly correlated with PP (ρ ≥ 0.62) in UKB Whites, taken as an indicator of arterial stiffness, and considered as an independent risk factor for CVD (Franklin, 2004). However, BPpleio1 and BPpleio2 were less correlated than PP with either SBP or DBP, which is consistent with pGRS being less correlated than GRSPP with either SBP or DBP‐defined GRS (Table S2). Thus, BPpleio1 or BPpleio2 represent a different risk factor of CVD from PP. This is consistent with the finding that GRSs of SBP, DBP, PP, and BPpleio all contribute to the risk of CAD, stroke, and MI in the MR analysis (Table 3). Thus, our results clearly suggest that a substantial fraction of BP variants affect both SBP and DBP through pleiotropic effects. By combining UKB‐ICBP and MVP, we identified an additional novel 219 variants with horizontal pleiotropic evidence, including 40 novel loci, although independent replication of these latter results are warranted (Table S10). Thus, pleiotropic variant searches in existing datasets can identify many new BP genes.
In addition to the traditional BP GRS (Evangelou et al., 2018; International Consortium for Blood Pressure Genome‐Wide Association et al., 2011), which we termed the core genetic risk score cGRS, the pleiotropic genetic risk score pGRS independently predicted BP, HTN, and CVD outcomes (Table 2) in the UK Biobank European ancestry subjects. Additionally, including the pGRS led to substantial increments in heritability explained for BP traits (Table 2 and Figure 5). Although we observed consistent opposite directional effects of pGRS for SBP and DBP in UK Biobank European, African, and Asian ancestry subjects, the prediction of HTN and CVD risk was significantly improved by including pGRS (Figure 5c,d). The cGRS and pGRS defined in European participants both consistently and significantly predicted BP, HTN, and CVD in UK Biobank Africans and Asians, suggesting that pGRS is able to improve prediction accuracy across populations. Recent studies have suggested that cGRS models alone have a modest improvement of predictive accuracy for CAD (Elliott et al., 2020; Khan et al., 2020; Mosley et al., 2020). The principal outcome of this set of analyses, therefore, demonstrates that adding pGRS significantly improves the prediction model over cGRS alone. This approach of constructing polygenic risk scores is conceptually different from existing approaches using multiple related traits (Inouye et al., 2018; Krapohl et al., 2018; Maier et al., 2018; Richardson et al., 2019) and can be generalized to other diseases by incorporating multiple disease‐related traits through pleiotropy analysis.
In our analysis, the UK Biobank Europeans were a part of data for identifying BP variants and pleiotropic variants. We then constructed the cGRS and pGRS in UK Biobank data by using the estimated effect sizes of the independent genome‐wide significant variants from UKB‐ICBP summary statistics as the weights. This procedure was used in Evangelou et al. (2018) and was not involved in a model selection. However, there is a potential winner's curse effect as suggested by Evangelou et al. When we applied the cGRS and pGRS to the UK Biobank Africans and Asians, we again observed the improved R 2 (Table S9), suggesting that adding cGRS improves the prediction power.
Our analysis avoided an examination of the interaction of individual variants and age because of insufficient power. We were able to observe interaction effects of both cGRS and pGRS with age in UKB Europeans for SBP, DBP, PP, and HTN although the interaction for CVD was only significant for pGRS (Table 2). Age–pGRS interactions were also replicated in Asians despite a substantially smaller sample size. We observed that the interaction contribution to phenotype variation was consistently small (0.1%–0.3% BP heritability in both UKB Europeans and Asians). The negative interaction contributions of both cGRS and pGRS to DBP may partially explain the decline of DBP after 60 years older (Franklin, 1999). In comparison, the interaction of age and cGRS was positive for SBP, suggesting genetic effects on SBP increases in older individuals. As noted, the cGRS interaction effects in UKB Europeans could not be observed in UKB Africans or Asians, likely as a result of the latter's smaller sample size. In comparison, the age‐modulated interaction of pGRS was observed in both UKB European and Asian subjects, indicating stronger pGRS interactions than for the GRS. In our functional annotation analysis, we observed a wider range of BP‐related tissues and biological pathways for the BP pleiotropic variants than mediation variants, which implies that pleiotropic variants are influenced by a wider range of environmental factors and, therefore, continue to make genetic contributions over the life span.
Our analysis approach bears some similarities with the recently developed GWAS‐by‐subtraction (Demange et al., 2021). However, there are significant differences. (1) The GWAS‐by‐subtraction is based on a genomic structure equation model (genomic‐SEM) and our method is based on MR. (2) GWAS‐by‐subtraction assumes that all genetic effects on one trait affect the other. In contrast, our method does not make this assumption. Instead, our method estimates the causal effects in both directions. (3) The GWAS‐by‐subtraction tests the null hypothesis = 0, where and are the estimated effect sizes of an SNP on two traits (trait 1 and 2), is the genetic covariance between the two traits and is the heritability of trait 2. In contrast, our method tests the null hypothesis = 0 and = 0, where are the two causal effects estimated from MR. Note that is not the same as causal effect when pleiotropic variants play a significant contribution for two traits such as in our study. The GWAS‐by‐subtraction includes the pleiotropic variants in estimating genetic covariance and our method intends to remove these variants. Therefore, our method is less affected by pleiotropic variants. However, further research is warranted in understanding these two methods better.
Traditionally pleiotropy has been studied for two different phenotypes. In comparison, our analysis was performed on two highly correlated and similar BP traits, in which we defined mediation and pleiotropy variants. We were able to identify a “core” mediation pathway shared by both SBP and DBP, and a pleiotropy pathway that has different effects on SBP and DBP. Our method is, therefore, useful in analyzing different symptoms of a disease for understanding the biological mechanism of the disease. However, our approach can also be applied to less genetically correlated or different phenotypes to identify pleiotropic variants. In this case, we will less likely to identify “core” phenotypes as we observed in BP traits.
Our study also supports an omnigenic model for complex traits (Boyle et al., 2017; Chakravarti & Turner, 2016; X. Liu et al., 2019). In fact, it could be inferred that pleiotropic variants act on multiple peripheral genes to impact the expression of core genes. As a result, pleiotropic variants have weak effects on a phenotype and are more difficult to detect in a traditional BP GWAS that focuses on single‐trait analysis, as observed here. In comparison, mediation variants may be more likely to occur in core genes. We acknowledge that the data presented here can only provide suggestive rather than conclusive evidence for that hypothesis.
In conclusion, our new findings here include identifying 815 independent BP pleiotropic variants—201 of which were not previously identified in BP GWAS in the UKB‐ICBP study; of these, 118 were confirmed including 18 novel BP loci. In addition, 219 novel BP signals and 40 novel loci were identified by combining UKB‐ICBP and MVP. Our way to construct a polygenic risk score represents a substantial advance in understanding the genetic architecture of the highly correlated SBP and DBP.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Xiaofeng Zhu conceived and designed the study. Xiaofeng Zhu, Luke Zhu, and Heming Wang performed the analyses. Xiaofeng Zhu drafted the initial manuscript and Aravinda Chakravarti edited the manuscript. All authors critically revised and approved the manuscript. We thank the reviewers’ constructive comments that substantially improved the manuscript.
WEB RESOURCES
CTG‐View: https://view.genoma.io/
DEPICT: https://data.broadinstitute.org/mpg/depict/
GTEx: www.gtexportal.org
FUMA: https://fuma.ctglab.nl/
IMRP: https://github.com/XiaofengZhuCase/IMRP
MRmix: https://github.com/gqi/MRMix
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
We thank Dr. Jacklyn Hellwege for providing the list of published BP loci. Data on coronary artery disease/myocardial infarction have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG'. Data on coronary artery disease/myocardial infarction have been contributed by the Myocardial Infarction Genetics and CARDIoGRAM Exome investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG. This study was supported by grant HG011052 (to Xiaofeng Zhu) from the National Human Genome Research Institute (NHGRI) and HL086694 (Aravinda Chakravarti) from the National Heart, Lung, and Blood Institute. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html.
Zhu, X. , Zhu, L. , Wang, H. , Cooper, R. S. , & Chakravarti, A. (2022). Genome‐wide pleiotropy analysis identifies novel blood pressure variants and improves its polygenic risk scores. Genetic Epidemiology, 46, 105–121. 10.1002/gepi.22440
DATA AVAILABILITY STATEMENT
Full summary statistics related to UKB‐ICBP were obtained through a request to the authors of UKB‐ICBP Paul Elliott or Mark Caulfield. Summary statistics relating to the Million Veteran Program (MVP) are publically available with the accession code phs001672.v1.p1: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v1.p1. The UK Biobank data are available upon application to the UK Biobank (https://www.ukbiobank.ac.uk). The coronary artery disease and myocardial infarction summary statistics can be downloaded at http://www.cardiogramplusc4d.org/data-downloads/; stroke summary statistics can be downloaded at: http://megastroke.org/privacy.html.
REFERENCES
- Borenstein, M. , Hedges, L. , Higgins, J. , & Rothstein, H. (2009). Generality of the basic inverse‐variance method, Introduction to Meta‐analysis. Wiley. [Google Scholar]
- Boyle, E. A. , Li, Y. I. , & Pritchard, J. K. (2017). An expanded view of complex traits: From polygenic to omnigenic. Cell, 169, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik‐Sullivan, B. K. , Loh, P. R. , Finucane, H. K. , Ripke, S. , Yang, J. , consortium of Schizophrenia Working Group of the Psychiatric Genomics , Patterson, N. , Daly, M. J. , Price, A. L. , & Neale, B. M. (2015). LD score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nature Genetics, 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S. , Butterworth, A. , & Thompson, S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genetic Epidemiology, 37, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft, C. , Freeman, C. , Petkova, D. , Band, G. , Elliott, L. T. , Sharp, K. , Motyer, A. , Vukcevic, D. , Delaneau, O. , O'Connell, J. , Cortes, A. , Welsh, S. , Young, A. , Effingham, M. , McVean, G. , Leslie, S. , Allen, N. , Donnelly, P. , & Marchini, J. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature, 562, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti, A. , & Turner, T. N. (2016). Revealing rate‐limiting steps in complex disease biology: The crucial importance of studying rare, extreme‐phenotype families. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 38, 578–586. [DOI] [PubMed] [Google Scholar]
- Chasman, D. I. , Giulianini, F. , Demler, O. V. , & Udler, M. S. (2020). Pleiotropy‐Based Decomposition Of Genetic Risk Scores: Association and Interaction Analysis For Type 2 Diabetes and CAD. American Journal of Human Genetics, 106, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar‐Partida, G. , Lundberg, M. , Kho, P. F. , D'Urso, S. , Gutierrez‐Mondragon, L. , & Hwang, L. (2019). Complex‐traits genetics virtual lab: A community‐driven web platform for post‐GWAS analyses. bioRxiv. https://www.biorxiv.org/content/10.1101/518027v2.full.pdf [Google Scholar]
- Demange, P. A. , Malanchini, M. , Mallard, T. T. , Biroli, P. , Cox, S. R. , Grotzinger, A. D. , Tucker‐Drob, E. M. , Abdellaoui, A. , Arseneault, L. , van Bergen, E. , Boomsma, D. I. , Caspi, A. , Corcoran, D. L. , Domingue, B. W. , Harris, K. M. , Ip, H. F. , Mitchell, C. , Moffitt, T. E. , Poulton, R. , … Nivard, M. G. (2021). Investigating the genetic architecture of noncognitive skills using GWAS‐by‐subtraction. Nature Genetics, 53, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu, L. , Brown‐Gentry, K. , Goodloe, R. , Glenn, K. , Yang, W. , Kornegay, N. , Pui, C. H. , Relling, M. V. , & Crawford, D. C. (2011). Evidence for age as a modifier of genetic associations for lipid levels. Annals of Human Genetics, 75, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger, M. , Davey Smith, G. , Schneider, M. , & Minder, C. (1997). Bias in meta‐analysis detected by a simple, graphical test. BMJ, 315, 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret, G. B. , Ferreira, T. , Chasman, D. I. , Jackson, A. U. , Schmidt, E. M. , Johnson, T. , Thorleifsson, G. , Luan, J. , Donnelly, L. A. , Kanoni, S. , Petersen, A. K. , Pihur, V. , Strawbridge, R. J. , Shungin, D. , Hughes, M. F. , Meirelles, O. , Kaakinen, M. , Bouatia‐Naji, N. , Kristiansson, K. , … Komulainen, P. (2016). The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nature Genetics, 48, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, J. , Bodinier, B. , Bond, T. A. , Chadeau‐Hyam, M. , Evangelou, E. , Moons, K. G. M. , Dehghan, A. , Muller, D. C. , Elliott, P. , & Tzoulaki, I. (2020). Predictive accuracy of a polygenic risk score‐enhanced prediction model vs a clinical risk score for coronary artery disease. Journal of the American Medical Association, 323, 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou, E. , Warren, H. R. , Mosen‐Ansorena, D. , Mifsud, B. , Pazoki, R. , Gao, H. , Ntritsos, G. , Dimou, N. , Cabrera, C. P. , Karaman, I. , Ng, F. L. , Evangelou, M. , Witkowska, K. , Tzanis, E. , Hellwege, J. N. , Giri, A. , Velez Edwards, D. R. , Sun, Y. V. , Cho, K. , … Lind, L. (2018). Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nature Genetics, 50, 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini, N. , Fox, E. , Zhang, Z. , Edwards, T. L. , Nalls, M. A. , Sung, Y. J. , Tayo, B. O. , Sun, Y. V. , Gottesman, O. , Adeyemo, A. , Johnson, A. D. , Young, J. H. , Rice, K. , Duan, Q. , Chen, F. , Li, Y. , Tang, H. , Fornage, M. , Keene, K. L. , … Zhu, X. (2013). Genome‐wide association analysis of blood‐pressure traits in African‐ancestry individuals reveals common associated genes in African and non‐African populations. American Journal of Human Genetics, 93, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, S. S. (1999). Ageing and hypertension: The assessment of blood pressure indices in predicting coronary heart disease. Journal of Hypertension Supplement: Official Journal of the International Society of Hypertension, 17, S29–S36. [PubMed] [Google Scholar]
- Franklin, S. S. (2004). Pulse pressure as a risk factor. Clinical and Experimental Hypertension, 26, 645–652. [DOI] [PubMed] [Google Scholar]
- Giri, A. , Hellwege, J. N. , Keaton, J. M. , Park, J. , Qiu, C. , Warren, H. R. , Torstenson, E. S. , Kovesdy, C. P. , Sun, Y. V. , Wilson, O. D. , Robinson‐Cohen, C. , Roumie, C. L. , Chung, C. P. , Birdwell, K. A. , Damrauer, S. M. , DuVall, S. L. , Klarin, D. , Cho, K. , Wang, Y. , … Edwards, T. L. (2019). Trans‐ethnic association study of blood pressure determinants in over 750,000 individuals. Nature Genetics, 51, 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, T. J. , Theusch, E. , Haldar, T. , Ranatunga, D. K. , Jorgenson, E. , Medina, M. W. , Kvale, M. N. , Kwok, P. Y. , Schaefer, C. , Krauss, R. M. , Iribarren, C. , & Risch, N. (2018). A large electronic‐health‐record‐based genome‐wide study of serum lipids. Nature Genetics, 50, 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye, M. , Abraham, G. , Nelson, C. P. , Wood, A. M. , Sweeting, M. J. , Dudbridge, F. , Lai, F. Y. , Kaptoge, S. , Brozynska, M. , Wang, T. , Ye, S. , Webb, T. R. , Rutter, M. K. , Tzoulaki, I. , Patel, R. S. , Loos, R. J. F. , Keavney, B. , Hemingway, H. , Thompson, J. , … Samani, N. J. (2018). Genomic risk prediction of coronary artery disease in 480,000 adults: Implications for primary prevention. Journal of the American College of Cardiology, 72, 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome‐Wide Association, S. , Ehret, G. B. , Munroe, P. B. , Rice, K. M. , Bochud, M. , Johnson, A. D. , Chasman, D. I. , Smith, A. V. , Tobin, M. D. , Verwoert, G. C. , Hwang, S. J. , Pihur, V. , Vollenweider, P. , O'Reilly, P. F. , Amin, N. , Bragg‐Gresham, J. L. , Teumer, A. , Glazer, N. L. , Launer, L. , … Stringham, H. M. (2011). Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature, 478, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Holmes, C. , & McVean, G. (2021). The impact of age on genetic risk for common diseases. PLOS Genetics, 17, e1009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. S. , Cooper, R. , & Greenland, P. (2020). Do polygenic risk scores improve patient selection for prevention of coronary artery disease? Journal of the American Medical Association, 323, 614–615. [DOI] [PubMed] [Google Scholar]
- Krapohl, E. , Patel, H. , Newhouse, S. , Curtis, C. J. , von Stumm, S. , Dale, P. S. , Zabaneh, D. , Breen, G. , O'Reilly, P. F. , & Plomin, R. (2018). Multi‐polygenic score approach to trait prediction. Molecular Psychiatry, 23, 1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky‐Su, J. , Lyon, H. N. , Emilsson, V. , Heid, I. M. , Molony, C. , Raby, B. A. , Lazarus, R. , Klanderman, B. , Soto‐Quiros, M. E. , Avila, L. , Silverman, E. K. , Thorleifsson, G. , Thorsteinsdottir, U. , Kronenberg, F. , Vollmert, C. , Illig, T. , Fox, C. S. , Levy, D. , Laird, N. , … Lange, C. (2008). On the replication of genetic associations: Timing can be everything! American Journal of Human Genetics, 82, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. H. , Goddard, M. E. , Wray, N. R. , & Visscher, P. M. (2012). A better coefficient of determination for genetic profile analysis. Genetic Epidemiology, 36, 214–224. [DOI] [PubMed] [Google Scholar]
- Liang, J. , Le, T. H. , Edwards, D. R. V. , Tayo, B. O. , Gaulton, K. J. , Smith, J. A. , Lu, Y. , Jensen, R. A. , Chen, G. , Yanek, L. R. , Schwander, K. , Tajuddin, S. M. , Sofer, T. , Kim, W. , Kayima, J. , McKenzie, C. A. , Fox, E. , Nalls, M. A. , Young, J. H. , … Franceschini, N. (2017). Single‐trait and multi‐trait genome‐wide association analyses identify novel loci for blood pressure in African‐ancestry populations. PLOS Genetics, 13, e1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Kraja, A. T. , Smith, J. A. , Brody, J. A. , Franceschini, N. , Bis, J. C. , Rice, K. , Morrison, A. C. , Lu, Y. , Weiss, S. , Guo, X. , Palmas, W. , Martin, L. W. , Chen, Y. D. , Surendran, P. , Drenos, F. , Cook, J. P. , Auer, P. L. , Chu, A. Y. , … Chasman, D. I. (2016). Meta‐analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nature Genetics, 48, 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Li, Y. I. , & Pritchard, J. K. (2019). Trans effects on gene expression can drive omnigenic inheritance. Cell, 177, 1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur, J. , Bowler, E. , Cerezo, M. , Gil, L. , Hall, P. , Hastings, E. , Junkins, H. , McMahon, A. , Milano, A. , Morales, J. , Pendlington, Z. M. , Welter, D. , Burdett, T. , Hindorff, L. , Flicek, P. , Cunningham, F. , & Parkinson, H. (2017). The new NHGRI‐EBI Catalog of published genome‐wide association studies (GWAS Catalog). Nucleic Acids Research, 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, R. M. , Zhu, Z. , Lee, S. H. , Trzaskowski, M. , Ruderfer, D. M. , Stahl, E. A. , Ripke, S. , Wray, N. R. , Yang, J. , Visscher, P. M. , & Robinson, M. R. (2018). Improving genetic prediction by leveraging genetic correlations among human diseases and traits. Nature Communications, 9, 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, R. , Chauhan, G. , Traylor, M. , Sargurupremraj, M. , Okada, Y. , Mishra, A. , Rutten‐Jacobs, L. , Giese, A. K. , van der Laan, S. W. , Gretarsdottir, S. , Anderson, C. D. , Chong, M. , Adams, H. , Ago, T. , Almgren, P. , Amouyel, P. , Ay, H. , Bartz, T. M. , Benavente, O. R. , … Elliott, P. (2018). Multiancestry genome‐wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature Genetics, 50, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, S. , Das, S. , Kretzschmar, W. , Delaneau, O. , Wood, A. R. , Teumer, A. , Kang, H. M. , Fuchsberger, C. , Danecek, P. , Sharp, K. , Luo, Y. , Sidore, C. , Kwong, A. , Timpson, N. , Koskinen, S. , Vrieze, S. , Scott, L. J. , Zhang, H. , Mahajan, A. , … Haplotype Reference, C. (2016). A reference panel of 64,976 haplotypes for genotype imputation. Nature Genetics, 48, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley, J. D. , Gupta, D. K. , Tan, J. , Yao, J. , Wells, Q. S. , Shaffer, C. M. , Kundu, S. , Robinson‐Cohen, C. , Psaty, B. M. , Rich, S. S. , Post, W. S. , Guo, X. , Rotter, J. I. , Roden, D. M. , Gerszten, R. E. , & Wang, T. J. (2020). Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. Journal of the American Medical Association, 323, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpay, M. , Goel, A. , Won, H. H. , Hall, L. M. , Willenborg, C. , Kanoni, S. , Saleheen, D. , Kyriakou, T. , Nelson, C. P. , Hopewell, J. C. , Webb, T. R. , Zeng, L. , Dehghan, A. , Alver, M. , Armasu, S. M. , Auro, K. , Bjonnes, A. , Chasman, D. I. , Chen, S. , … Samani, N. J. (2015). A comprehensive 1,000 Genomes‐based genome‐wide association meta‐analysis of coronary artery disease. Nature Genetics, 47, 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pers, T. H. , Karjalainen, J. M. , Chan, Y. , Westra, H. J. , Wood, A. R. , Yang, J. , Lui, J. C. , Vedantam, S. , Gustafsson, S. , Esko, T. , Frayling, T. , Speliotes, E. K. , Genetic Investigation of ANthropometric Traits (GIANT), C. , Boehnke, M. , Raychaudhuri, S. , Fehrmann, R. S. , Hirschhorn, J. N. , & Franke, L. (2015). Biological interpretation of genome‐wide association studies using predicted gene functions. Nature Communications, 6, 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, G. , & Chatterjee, N. (2019). Mendelian randomization analysis using mixture models for robust and efficient estimation of causal effects. Nature Communications, 10, 1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapsomaniki, E. , Timmis, A. , George, J. , Pujades‐Rodriguez, M. , Shah, A. D. , Denaxas, S. , White, I. R. , Caulfield, M. J. , Deanfield, J. E. , Smeeth, L. , Williams, B. , Hingorani, A. , & Hemingway, H. (2014). Blood pressure and incidence of twelve cardiovascular diseases: Lifetime risks, healthy life‐years lost, and age‐specific associations in 1.25 million people. Lancet, 383, 1899–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, T. G. , Harrison, S. , Hemani, G. , & Davey Smith, G. (2019). An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. eLife, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillaci, G. , & Pucci, G. (2010). The dynamic relationship between systolic and diastolic blood pressure: Yet another marker of vascular aging? Hypertension Research: Official Journal of the Japanese Society of Hypertension. 33, 659–661. [DOI] [PubMed] [Google Scholar]
- Shi, G. , Gu, C. C. , Kraja, A. T. , Arnett, D. K. , Myers, R. H. , Pankow, J. S. , Hunt, S. C. , & Rao, D. C. (2009). Genetic effect on blood pressure is modulated by age: The Hypertension Genetic Epidemiology Network Study. Hypertension, 53, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simino, J. , Shi, G. , Bis, J. C. , Chasman, D. I. , Ehret, G. B. , Gu, X. , Guo, X. , Hwang, S. J. , Sijbrands, E. , Smith, A. V. , Verwoert, G. C. , Bragg‐Gresham, J. L. , Cadby, G. , Chen, P. , Cheng, C. Y. , Corre, T. , de Boer, R. A. , Goel, A. , Johnson, T. , … Rao, D. C. (2014). Gene‐age interactions in blood pressure regulation: A large‐scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. American Journal of Human Genetics, 95, 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. D. , & Ebrahim, S. (2003). 'Mendelian randomization': Can genetic epidemiology contribute to understanding environmental determinants of disease? International Journal of Epidemiology, 32, 1–22. [DOI] [PubMed] [Google Scholar]
- Sung, Y. J. , de Las Fuentes, L. , Winkler, T. W. , Chasman, D. I. , Bentley, A. R. , Kraja, A. T. , Ntalla, I. , Warren, H. R. , Guo, X. , Schwander, K. , Schwander, K. , Manning, A. K. , Brown, M. R. , Aschard, H. , Feitosa, M. F. , Franceschini, N. , Lu, Y. , Cheng, C. Y. , … Loh, M. (2019). A multi‐ancestry genome‐wide study incorporating gene‐smoking interactions identifies multiple new loci for pulse pressure and mean arterial pressure. Human Molecular Genetics, 28, 2615–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, Y. J. , Winkler, T. W. , de Las Fuentes, L. , Bentley, A. R. , Brown, M. R. , Kraja, A. T. , Schwander, K. , Ntalla, I. , Guo, X. , Franceschini, N. , Franceschini, N. , Lu, Y. , Cheng, C. Y. , Sim, X. , Vojinovic, D. , Marten, J. , Musani, S. K. , Li, C. , Feitosa, M. F. , … Lehne, B. (2018). A large‐scale multi‐ancestry genome‐wide study accounting for smoking behavior identifies multiple significant loci for blood pressure. American Journal of Human Genetics, 102, 375–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran, P. , Drenos, F. , Young, R. , Warren, H. , Cook, J. P. , Manning, A. K. , Grarup, N. , Sim, X. , Barnes, D. R. , Witkowska, K. , Staley, J. R. , Tragante, V. , Tukiainen, T. , Yaghootkar, H. , Masca, N. , Freitag, D. F. , Ferreira, T. , Giannakopoulou, O. , Tinker, A. , … Skaaby, T. (2016). Trans‐ancestry meta‐analyses identify rare and common variants associated with blood pressure and hypertension. Nature Genetics, 48, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler, M. S. , Kim, J. , von Grotthuss, M. , Bonàs‐Guarch, S. , Cole, J. B. , Chiou, J. , Anderson, C. A. on behalf of METASTROKE and the ISGC, Boehnke, M. , Laakso, M. , Atzmon, G. , Glaser, B. , Mercader, J. M. , Gaulton, K. , Flannick, J. , Getz, G. , & Florez, J. C. (2018). Type 2 diabetes genetic loci informed by multi‐trait associations point to disease mechanisms and subtypes: A soft clustering analysis. PLOS Medicine, 15, e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, H. R. , Evangelou, E. , Cabrera, C. P. , Gao, H. , Ren, M. , Mifsud, B. , Ntalla, I. , Surendran, P. , Liu, C. , Cook, J. P. , Kraja, A. T. , Drenos, F. , Loh, M. , Verweij, N. , Marten, J. , Karaman, I. , Lepe, M. P. S. , O'Reilly, P. F. , Knight, J. , … Elliott, P. (2017). Genome‐wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nature Genetics, 49, 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K. , Taskesen, E. , van Bochoven, A. , & Posthuma, D. (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, T. R. , Erdmann, J. , Stirrups, K. E. , Stitziel, N. O. , Masca, N. G. , Jansen, H. , Kanoni, S. , Nelson, C. P. , Ferrario, P. G. , König, I. R. , Eicher, J. D. , Johnson, A. D. , Hamby, S. E. , Betsholtz, C. , Ruusalepp, A. , Franzén, O. , Schadt, E. E. , Björkegren, J. L. , Weeke, P. E. , … Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia, I. (2017). Systematic evaluation of pleiotropy identifies 6 further loci associated with coronary artery disease. Journal of the American College of Cardiology, 69, 823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Feng, T. , Tayo, B. O. , Liang, J. , Young, J. H. , Franceschini, N. , Smith, J. A. , Yanek, L. R. , Sun, Y. V. , Edwards, T. L. , Chen, W. , Nalls, M. , Fox, E. , Sale, M. , Bottinger, E. , Rotimi, C. , COGENT BP, C. , Liu, Y. , McKnight, B. , … Redline, S. (2015). Meta‐analysis of correlated traits via summary statistics from GWASs with an application in hypertension. American Journal of Human Genetics, 96, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Li, X. , Xu, R. , & Wang, T. (2021). An iterative approach to detect pleiotropy and perform Mendelian Randomization analysis using GWAS summary statistics. Bioinformatics, 37, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
Full summary statistics related to UKB‐ICBP were obtained through a request to the authors of UKB‐ICBP Paul Elliott or Mark Caulfield. Summary statistics relating to the Million Veteran Program (MVP) are publically available with the accession code phs001672.v1.p1: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v1.p1. The UK Biobank data are available upon application to the UK Biobank (https://www.ukbiobank.ac.uk). The coronary artery disease and myocardial infarction summary statistics can be downloaded at http://www.cardiogramplusc4d.org/data-downloads/; stroke summary statistics can be downloaded at: http://megastroke.org/privacy.html.


