Abstract
The quantity of Borrelia burgdorferi organisms in tissue samples is an important determinant for infection studies in the mouse model of Lyme disease. This report presents the development of a rapid and sensitive external-standard-based PCR assay for the absolute quantification of B. burgdorferi in mouse tissue samples. The assay uses a double-stranded DNA dye to continuously monitor product formation and in less than an hour was able to quantify samples ranging up to 6 log units in concentration. The PCR efficiencies of the sample and the standard were matched by using a standard composed of purified B. burgdorferi chromosome mixed with tissue-matched mouse genome lacking bacterial DNA. Normalization of B. burgdorferi quantities to the mouse nidogen gene allowed comparison of B. burgdorferi numbers in samples isolated from different tissues and strains. PCR analysis of the chromosomal gene recA in cultured B. burgdorferi was consistent with a single recA per bacterium. The parameters defined in this assay should be applicable to quantification of other organisms, even infectious agents for which no ready source of DNA standard is available. In summary, this report presents a rapid external-standard-based PCR method for the quantification of B. burgdorferi in mouse DNA samples.
The mouse model for Lyme disease offers a useful tool for the characterization of host-pathogen interactions. The presence of bacteria in mouse tissues is an important component of disease development (22). Various assays for the detection of Borrelia burgdorferi in tissues have been developed, including culturing, histochemical and silver staining of thin sections, in situ hybridization, and PCR (1, 5, 7, 14, 22). Of the available techniques for the quantification of bacteria, PCR-based methods with limiting dilution or internal or external standards are the most sensitive and accurate. These techniques measure the number of bacteria indirectly by assuming that the number of bacterial DNA sequences in the sample is proportional to the number of bacteria. Studies demonstrating rapid loss of bacterial DNA following antibiotic treatment of mice indicate a direct correlation between B. burgdorferi DNA in tissues and live bacteria (13).
The choice of PCR quantification method depends on factors such as the number and quality of samples and the accuracy of quantification required. Our laboratory measures B. burgdorferi in thousands of mouse samples each year from a variety of mouse strains and tissue sources. We have utilized PCR-based methodology that is dependent upon a DNA isolation protocol that yields uniform and pure samples. This report focuses on the development of a rapid and sensitive external-standard PCR-based assay. This method makes use of continuous monitoring to enhance accuracy and increase the sampling dynamic range (9, 19). It employs the double-stranded DNA (dsDNA) dye SYBR green, which allows easy quantification of the product without isotope or gel electrophoresis. Comparisons of the numbers of bacteria in samples isolated from different mouse strains and tissues and at different postinfection times were made.
MATERIALS AND METHODS
Chemicals were purchased from Sigma, St. Louis, Mo., unless otherwise specified. Molecular biology and culture reagents were purchased from Gibco-BRL, Grand Island, N.Y.
Bacteria.
The N40 isolate of B. burgdorferi was provided by Stephen Barthold (University of California at Davis) at passage 3 from an infected mouse (4). Passage 4 cultures were maintained as 0.5-ml frozen stocks at −70°C. Fresh aliquots of frozen stocks were seeded in 15 ml of BSK-H medium containing 6% rabbit serum (Sigma) and cultured at 32°C for 3 to 5 days prior to injection.
Mice.
Male C3H/HeJNCr and C57BL/6NCr mice were obtained from the National Cancer Institute at 5 weeks of age. The mice were housed in the Animal Resource Center at the University of Utah Medical Center according to the guidelines of the National Institutes of Health for the care and use of laboratory animals.
Infection of mice with B. burgdorferi.
Spirochete concentrations of 3- to 5-day cultures were determined by dark-field microscopy with a Petroff-Hauser chamber. Dilutions were made with sterile culture medium to allow injection of 20 μl per animal. Mice 5 to 6 weeks of age were infected by intradermal injection with 2 × 103 B. burgdorferi organisms in the shaven back, a mode of infection reported to require the fewest spirochetes and to most closely mimic tick transmission (3, 15). Control mice were injected with an equal volume of sterile medium.
Preparation of DNA from infected tissues.
The control and infected mice were sacrificed at 2 or 4 weeks following infection, and rear ankle joint, bladder, ear, brain, and heart tissues were prepared as previously described (18). The tissues were placed in individual 15-ml polypropylene tubes containing 2.5 ml of a 0.1% collagenase A (Boehringer Mannheim, Indianapolis, Ind.) solution in phosphate-buffered saline (pH 7.4). Samples were digested with collagenase for 4 h at 37°C and then mixed with an equal volume of 0.2-mg/ml proteinase K (Boehringer Mannheim) in 200 mM NaCl, 20 mM Tris-HCl (pH 8.0), 50 mM EDTA, and 1% sodium dodecyl sulfate for 16 h at 55°C. DNA was recovered by extraction with an equal volume of phenol-chloroform and precipitation with ethanol. Following digestion with 1 mg of DNase-free RNase/ml, the DNA samples were subjected to a second extraction and precipitation and finally resuspended at 50 μg/ml in TE (0.5 mM EDTA and 5 mM Tris-HCl, pH 7.5). This protocol, which requires 3 days to complete, generates higher yields and more uniform DNA samples than column-based purification methods (data not shown).
Measurement of B. burgdorferi sequences by continuous monitoring of PCR.
PCR was performed in a fluorescence temperature cycler (LightCycler LC24; Idaho Technology, Idaho Falls, Idaho). Amplification was performed in a 10-μl final volume containing 3 mM MgCl2, 50 mM Tris (pH 8.3), 500 ng of bovine serum albumin/μl, 200 μM (each) deoxynucleoside triphosphate, 1:30,000 dilution of SYBR Green I (Molecular Probes, Eugene, Oreg.), 5 μM (each) primer, 0.05 U of Taq polymerase/μl, 11 ng of TaqStart antibody (ClonTech, Palo Alto, Calif.)/μl. All solutions were centrifuged prior to use in order to pellet particles that might interfere with the LightCycler optics. The oligonucleotide primers used to detect B. burgdorferi recA were nTM17.F (5′-GTG GAT CTA TTG TAT TAG ATG AGG CTC TCG-3′) and nTM17.R (5′-GCC AAA GTT CTG CAA CAT TAA CAC CTA AAG-3′). The oligonucleotide primers used to detect mouse nidogen were nido.F (5′-CCA GCC ACA GAA TAC CAT CC-3′) and nido.R (5′-GGA CAT ACT CTG CTG CCA TC-3′) (22). The standard amplification program included 40 cycles of three steps each, comprised of heating at 20°C/s to 95°C with a 1-s hold, cooling at 20°C/s to 60°C with a 1-s hold, and heating at 1°C/s to 84°C for recA and to 86°C for nidogen. Fluorescent product was detected at the last step of each cycle. After amplification, a melting curve was acquired by heating the product at 20°C/s to 95°C, cooling it at 20°C/s to 60°C, and slowly heating it at 0.2°C/s to 94°C with fluorescence collection at 0.2°C intervals. Melting curves were used to determine the specificity of the PCR (16).
Purification of PCR product standards.
Standard amplification mixtures were as described above, with the exception that the total reaction volume was 100 μl and contained 1 μg of C3H mouse DNA or 33 ng of purified B. burgdorferi DNA as a template source for the nidogen or recA reactions, respectively. Equal aliquots were divided among five glass capillary tubes (no. 1607; Idaho Technology) and temperature cycled in a rapid air thermal cycler (Rapid Cycler; Idaho Technology) for 25 cycles of 95°C for 1 s, 60°C for 1 s, and 72°C for 5 s. PCR products were purified with a QIAquick PCR purification kit (Qiagen, Chatsworth, Calif.). Part of each sample was examined for purity by gel electrophoresis. The copy number (copies/μl) for the PCR product standards was based on absorbance at 260 nm (Spectra Max 250; Molecular Devices, Sunnyvale, Calif.) and confirmed by limiting-dilution assay (see below). The recA PCR product was cloned into the pGEM-T Easy vector (Promega, Madison, Wis.) to generate pTM201.
Purification of B. burgdorferi recA DNA standard.
B. burgdorferi organisms were grown in culture to saturation (108 cells/ml) under the conditions described above. Genomic DNA was isolated with cetyltrimethylammonium bromide (2). Recovery of B. burgdorferi DNA during purification was determined by spiking whole cells with 32P-labeled B. burgdorferi DNA. The fraction of DNA recovered was estimated from the recovery of label. The recA concentration in B. burgdorferi DNA (copies/μl) was estimated by optical absorbance, assuming a 1.5-Mbp B. burgdorferi genome (8). Alternatively, the recA concentration was determined by limiting-dilution assay (described below) and confirmed by continuously monitored PCR with an external PCR product standard. Both quantification methods were used under the reaction conditions described below.
External standards.
Copy number standards and samples were simultaneously amplified. The recA copy number standard contained purified B. burgdorferi DNA mixed with 200 ng of DNA isolated from uninfected mouse tissue (i.e., tissues lacking B. burgdorferi DNA). The nidogen standard used purified PCR product and lacked mouse DNA. Copy numbers for the mouse samples were calculated with the LightCycler software. The software first normalizes each sample by background subtraction. Then, a fluorescence threshold (≈5% of full scale) is used to determine fractional cycle numbers that correlate inversely to the log of the initial template concentration. The least-squares best fit of the standards is used to calculate the template copies initially present in the unknowns. Variations in sample load were corrected by normalization of the recA copies to 104 nidogen copies (≈104 nidogen copies/200 ng mouse DNA).
PCR-based limiting-dilution assay.
Dilutions of purified B. burgdorferi DNA were amplified under the standard conditions for continuously monitored PCR. The initial dilutions consisted of 10-fold serial dilutions that allowed a rough estimation of recA copies followed by six replicate samples of 2-fold serial dilutions that flanked an average of a single copy per reaction. The presence or absence of specific product was determined by observing the presence of an 85.5°C melting peak. The concentration of starting PCR product was estimated with the Macintosh program Quality (17).
Measurement of B. burgdorferi sequences by radiolabeled endpoint PCR.
Measurement of B. burgdorferi in tissue was performed as described previously (12). Briefly, chromosomal primers for mouse nidogen or the B. burgdorferi flagellum gene were used to generate radiolabeled PCR products from 150 ng of mouse DNA. Primer sequences and reaction conditions can be found in reference 22. The reactions were separated on a 6% polyacrylamide sequencing gel and subjected to phosphorimager analysis for quantification. DNA from each group of animals was amplified with nidogen-specific primers and analyzed on a single gel. Equal loading of tissue DNA was achieved by adjusting the samples to fall within a twofold range for nidogen product, which was confirmed by analysis of a second set of reactions. Amplification of B. burgdorferi sequences was performed with the flagellum gene primer at cycle numbers within the linear range for product. A standard curve for detection of B. burgdorferi DNA in mouse tissue was generated by spiking a constant number of freshly isolated mouse splenocytes with increasing numbers of B. burgdorferi organisms.
RESULTS
Previous methods for PCR-based quantification of B. burgdorferi in mouse tissues employed measurement of product formation by subjecting the radiolabeled PCR end product to gel electrophoresis and then visualizing the products by audioradiography of incorporated radiolabel (22). Though sensitive, this method was laborious, and the reliability of the data was very sensitive to reagent quality and operative handling. A newly available PCR quantification technology based on fluorescent monitoring of rapidly cycling PCR was examined to determine if it could improve upon the deficiencies of the radiolabeled-DNA protocol (21). With this technology, sample amplification and quantification takes place in a sealed tube in less than an hour, making this approach over 10-fold faster than the previous method.
Chromosome copy number.
PCR-based measurement of B. burgdorferi in tissues requires that the starting copy of the bacterial DNA target sequence accurately indicate the number of bacteria within a sample. For this study, the recA gene was selected (6). The recA gene is located on the chromosome and therefore should be present in every bacterium (8). Further, since recA has a 37% GC content, whereas the B. burgdorferi genome averages 28%, it should be easier to generate a highly specific primer set for this target sequence. Lastly, evolutionary conservation of recA might allow this primer set to detect other Borrelia species.
The accuracy of a quantitative primer set depends on its sensitivity, which arises from its PCR efficiency. PCR-based limiting-dilution assay requires single-copy sensitivity for accurate quantification. To determine if the recA primers had single-copy sensitivity, it was necessary to ascertain whether the concentration calculated by limiting dilution was equivalent to the concentration derived by physical measurement. The concentration of a recA PCR product cloned into the vector pGEM-T Easy had 3.0 × 1010 copies/μl by optical absorbance at 260 nm and 3.5 × 1010 ± 1.12 × 1010 copies/μl by limiting dilution. Purified B. burgdorferi DNA was also tested: it had 2.5 × 107 copies/μl by optical absorbance at 260 nm, assuming a 1.5-Mbp genome size, and 1.9 × 107 ± 0.44 × 107 copies/μl by limiting dilution. Both methods generated concentrations within 25% of each other, indicating that limiting dilution accurately predicted the concentration of two different DNA samples. Further, the limiting-dilution assay proved insensitive to the presence of 200 ng of mouse DNA, indicating that this primer set maintains its single-copy sensitivity under conditions that mimic mouse samples. These results indicate that the recA primer set should have single-copy sensitivity when measuring B. burgdorferi chromosomes in mouse DNA samples.
In order to estimate the number of B. burgdorferi organisms per sample, the average number of recA genes per bacterium was determined. B. burgdorferi were grown in culture and counted by dark-field microscopy with a Petroff-Hauser chamber. The B. burgdorferi genomic DNA was purified from cultured organisms, and the concentration of recA sequences was measured by PCR-based limiting-dilution assay. The recovery of radiolabeled B. burgdorferi DNA during purification was 60%. This allowed calculation of a 1.3 ± 0.41 ratio between recA starting copies and bacteria (Table 1). Thus, one recA copy per bacterium is most consistent with a single chromosome per bacterium.
TABLE 1.
B. burgdorferi chromosome copy number
| Sample | Quantification method | na | Ratiob |
|---|---|---|---|
| Purified B. burgdorferi | Limiting-dilution assay | 24 | 1.3 ± 0.41 |
| recA PCR product as external standard | 3 | 0.22 ± 0.030 | |
| Limiting dilution assay in presence of 200 ng of joint DNA | 24 | 0.92 ± 0.26 | |
| Culture-spiked spleen | B. burgdorferi DNA as external standard in presence of 200 ng of spleen DNA | 2 | 1.0 ± 0.12 |
n, number of samples.
Ratio, number of recA molecules per estimated number of bacteria in sample. The error values are standard deviations of replicate samples.
Development of external standard for PCR quantification.
To allow higher sample throughput, quantification of B. burgdorferi in mouse tissue samples was performed with an external-standard-based PCR. The accuracy of external standards depends on matching the PCR efficiency of the target sequence in both standard and sample. To enhance this match, the standard consisted of a known amount of purified B. burgdorferi DNA mixed with mouse DNA. The mouse DNA was isolated from the same tissue type as the sample but lacked bacterial DNA.
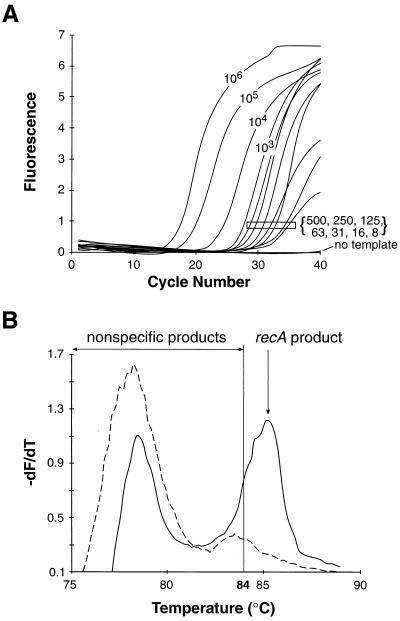
The dynamic range of this assay was then tested by measuring the amplification of recA from samples containing B. burgdorferi DNA serially diluted with mouse joint DNA. Cycle-by-cycle collection of fluorescence generated a series of sigmoidal amplification profiles (Fig. 1A). For each sample, the fluorescent signal demonstrated three phases: at early cycles there was not enough dsDNA product to raise the fluorescence above background; at log-linear phase, exponential replication was detected; and at plateau phase, the PCR was saturated. With fewer starting copies, more cycles were required for detection of the log-linear phase. The data depict the ability of a single PCR experiment to detect differences in starting copy numbers ranging over 5 log units.
FIG. 1.
Fluorescent-amplification profiles and melting curves for B. burgdorferi recA PCR product in the presence of SYBR Green I. Serially diluted samples of purified B. burgdorferi DNA containing an estimated 106, 105, 104, 103, 500, 250, 125, 63, 31, 16, 8, or 0 starting templates of recA in 200 ng of C3H/HeNCr mouse joint DNA were prepared and amplified for 40 cycles as described in Materials and Methods. (A) Background-subtracted fluorescent emissions of samples plotted as a function of cycle number. The number associated with each curve is the starting recA copy number, with the bracketed numbers corresponding to the boxed curves. (B) PCR end products for samples containing either two (solid line) or zero (dashed line) starting recA templates. Following amplification, dsDNA fluorescence was measured as the temperature was increased at 0.2°C/s from 65 to 94°C. The rate of fluorescence change with changing temperature (−dF/dT) was plotted as a function of temperature. The recA product Tm was 85.5°C and, nonspecific products melted between 75 and 87°C.
Another feature of this assay was its ability to specifically track the formation of recA PCR product. For the recA primer set, the melting point (Tm) for the recA product was 85.5°C while the majority of nonspecific products melt below 84°C (Fig. 1B). Acquisition of fluorescent data at 84°C causes the nonspecific products to melt, resulting in a loss of binding to the dsDNA-specific dye. Thus, the nonspecific products do not contribute to the total fluorescent signal. The selective acquisition of specific product was best observed in the no-template reaction (Fig. 1A). Despite the presence of nonspecific products (Fig. 1B) the absence of any specific product was accurately reflected in the lack of fluorescent signal when data was collected at 84°C.
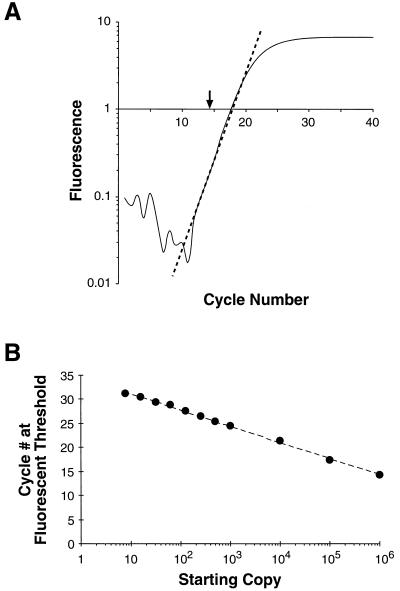
The most reliable point for the quantification of the sample starting copy number is found by determining the cycle number at which the product fluorescence becomes greater than a defined threshold. This point, termed the fluorescent threshold, represents the cycle number at which there are enough PCR products bound to dye to generate a signal approximately 3 standard deviations above background (0.2 fluorescent units). The fluorescent threshold is calculated from the crossover point between the log-linear portion of an amplification curve with 0.2 fluorescent units (Fig. 2A). The more starting copies in a sample, the fewer PCR cycles are required to reach this threshold (20). When fluorescent thresholds were plotted as a function of the log of the starting copy number, a standard curve was generated (Fig. 2B). The linearity of this plot extends over a 105-fold range. The variation between PCR experiments was less than one cycle for samples containing greater than 10 copies (data not shown). Below 10 copies, stochastic effects and reduced PCR efficiency interfere with precise quantification.
FIG. 2.
B. burgdorferi recA standard curve generated by PCR and SYBR Green I fluorescence. (A) Log-linear plot of the amplification profile for the 106-recA-starting-copy sample presented in Fig. 1. The dashed line represents a linear-regression fit to the log-linear portion of the amplification profile. The fluorescence threshold (arrow) is the fractional cycle number indicated by the crossover point of the dashed line with 0.2 fluorescence units. Panel B depicts the fluorescence threshold for the samples presented in Fig. 1A plotted as a function of the starting copy number. The dashed line indicates a linear regression fit to this data (r2 = 0.998).
As a possible substitute for purified B. burgdorferi DNA, the accuracy of a PCR product-based external standard was also tested. A recA PCR product was purified, and its concentration was calculated by absorbance at 260 nm and confirmed by limiting-dilution assay (data not shown). The purified recA PCR product was then used as an external standard to measure the recA sequences in a purified B. burgdorferi DNA sample (Table 1). The PCR product standard indicated that the sample had one-fifth the predicted number of recA sequences. This underestimate likely arose from greater efficiency of amplification for the smaller PCR product than for the same target sequence within the 1-Mbp B. burgdorferi chromosome. For purposes of accuracy, only the purified B. burgdorferi DNA was used as an external standard.
Quantification of mouse samples.
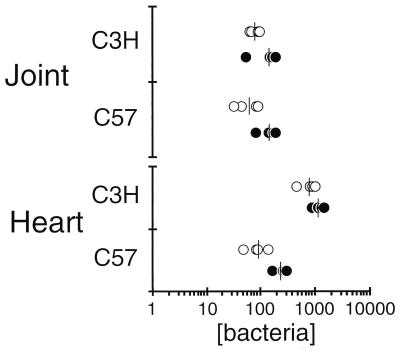
As a way of validating this new technique, samples from mice sacrificed 2 weeks postinfection were measured for their B. burgdorferi chromosome content by two methods: the new continuously monitored PCR technique and the standard radiolabeled endpoint PCR of this laboratory (Fig. 3). Both methods generated the expected trends: the two strains of mice have approximately the same number of B. burgdorferi organisms in their joints, and the C3H/HeN heart had an average of eightfold more Borrelia organisms than C57BL/6N hearts (12). There were two important issues raised by this comparison. First, to generate this data, the radiolabeled endpoint method required about four labor-intensive days, whereas the continuously monitored PCR method required only 2 h. Second, because of differences in experimental design, this comparison required sample normalization by optical absorbance (i.e., starting copies per 200 ng of total DNA). However, DNA purity and PCR efficiency varies between DNAs isolated from different tissues; therefore, comparisons of numbers of bacteria between tissues would be likewise affected.
FIG. 3.
Comparison of quantitative PCR methods. Two weeks following infection of C3H/HeNCr or C57BL/6NCr mice with 2,000 B. burgdorferi organisms, DNA samples from joint and heart tissues were prepared as described in Materials and Methods. Each DNA sample (200 ng) was subjected to two quantitative methods: radiolabeled endpoint PCR with flagellum gene primers (open circles; see text for more details) or continuously monitored PCR with recA primers (solid circles) as described in Materials and Methods. The values assume one gene per bacterium. The bars indicate the averages of the data sets.
For accurate comparison of B. burgdorferi in different tissue samples, the amount of total mouse DNA in each sample requires normalization. As a first step in normalizing the DNA for PCR, DNA samples were diluted to a 260-nm optical absorbance of 1.0. Further normalization refinement relied on the PCR quantification of a single-copy mouse gene, nidogen, within each sample. The previous PCR-based method required at least two rounds of radiolabeled endpoint PCR for this normalization (22). After the first round of PCR, load volumes were adjusted and the samples were retested until all samples had equivalent amounts of radiolabel incorporated into nidogen product. Once the DNA was normalized, the number of B. burgdorferi organisms in a sample was assayed. For the new method reported here, nidogen and recA copies per sample were measured with an external standard. Because each mouse sample of 200 ng of DNA contains an average of 104 nidogen copies, normalization of recA for each sample was performed arithmetically by multiplying the measured recA copies by 104 and dividing by the number of nidogen copies. The old method did not normalize with a fixed amount of nidogen, therefore only the samples analyzed on the same gel could be accurately compared. Normalizing to a fixed nidogen concentration simplifies the assay and allows comprehensive comparisons of mouse strain, time postinfection, and tissue type to bacterial numbers.
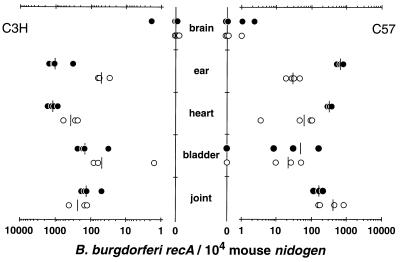
The results in Fig. 4 depict the quantification of B. burgdorferi numbers in a broad range of sample sources. These results are in agreement with trends observed from earlier quantification studies that used radiolabel endpoint PCR analysis (22). The data for this figure required about 8 h to collect, whereas the old quantification method would have required over a week of labor to collect the same data. Further, the data in Fig. 4 should represent a more accurate comparison of B. burgdorferi numbers in different tissues than those obtained from the old method.
FIG. 4.
Number of B. burgdorferi organisms present in different strains of mice harvested at different times postinfection. DNA samples from the indicated tissues of C3H/HeNCr (left) or C57BL/6NCr (right) mice infected for 2 weeks (solid circles) or 4 weeks (open circles) with 2,000 B. burgdorferi organisms were prepared as described in Materials and Methods. Each DNA sample (200 ng) was measured for mouse nidogen and B. burgdorferi recA copy numbers by continuously monitored PCR (see Materials and Methods for details). The sample load was corrected by presenting the recA copy number normalized to 104 nidogen copies. Samples negative for recA are plotted at zero on the y axis. The bars represent the averages of the samples.
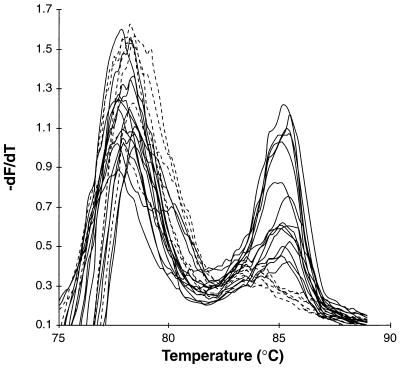
To test the detection limits of this assay, a single-copy detection assay was performed. The measurement of small numbers of bacteria requires an approach that is more qualitative than quantitative because low-copy-number amplification is sensitive to stochastic and high-cycle PCR effects. Therefore, for this assay, either the presence or absence of a recA-specific product was scored. Figure 5 depicts the endpoint products for 21 samples diluted to an average of 1.9 copies per reaction in the presence of mouse joint DNA. Based on Poisson distribution, 85% of these samples were expected to contain one or more recA starting copies while 15% of the samples were expected to lack recA. The observed frequency of positive samples was 71%. In combination with other serial dilutions, the limiting-dilution assay gave approximately the same recA concentration as the same assay performed in the absence of mouse DNA (Table 1). Similar tests and results were observed with DNA prepared from uninfected brain, bladder, heart, and skin tissue, indicating that these mouse DNA preparations lack significant PCR inhibitors that could reduce the sensitivity of the assay (data not shown). Therefore, the negative results for the brain and bladder samples in Fig. 4 likely arose from the absence of recA sequences in the reaction.
FIG. 5.
An overlay of melting curve profiles following PCR with a calculated average of 1.9 recA starting copies. recA template was diluted into 21 samples containing 200 ng of C3H mouse DNA and a calculated average of 1.9 recA copies per reaction. The samples were amplified, and melting curves were generated by the protocol described in the legend to Fig. 1. The melting profiles fit into two categories: nonspecific products with no recA product peak (dashed lines; n = 6) and recA product peaks with nonspecific product peaks (solid lines; n = 15).
Another possible reason for negative recA copies in a sample could arise from loss of B. burgdorferi DNA during sample purification. To test for such loss, cultured bacteria were added to mouse spleen cells. Mouse and bacterial DNA were isolated by tissue DNA preparation, and the amount of recA in each sample was measured. Based on back calculations, the number of recA molecules per sample corresponded to the expected bacterial equivalence in each sample (Table 1). These results indicate that there was no significant loss of bacterial DNA during preparation of tissue DNA.
DISCUSSION
This report describes the development of a new technique for external-standard PCR quantification of B. burgdorferi organisms in mouse tissue samples. To measure absolute numbers of bacteria per sample, it was necessary to measure the number of recA copies per bacterium. Analysis of B. burgdorferi isolated from culture is most consistent with a single chromosome per bacterium. This differs from a closely related species, Borrelia hermsii, which has an average of 4 chromosomes per cultured bacterium and 16 chromosomes per bacterium when isolated directly from a mouse (11). Currently, it is not possible to determine whether the B. burgdorferi chromosome copy number is altered when the bacteria are colonizing the mouse.
The B. burgdorferi plasmids LP54, LP16, and CP26 were demonstrated by Hinnebusch and Barbour to be in equal ratio with the chromosome (10). Taken together with our findings, these results indicate that these plasmids should also have a single copy per bacterium. Further, B. burgdorferi may have evolved a low-copy-number replication and segregation mechanism for maintenance of all DNA. Additional study will be required to support this hypothesis.
In a single experiment, this assay has been shown to accurately measure samples containing 10 to 106 bacteria. A recA-specific product peak was detected in melting curves for samples containing as few as a single bacterium. The use of the dsDNA dye SYBR Green I simplified quantification because sample amplification and analysis were performed simultaneously. Further, quantification of DNA samples was possible in 1 h.
The dynamics of B. burgdorferi dissemination can be followed by this technique. For example, the highest concentration of B. burgdorferi, in tissue at 2 weeks was in the skin. At 4 weeks, higher levels were found in the ankle joints. The prevalence in skin is expected, as this is the site at which ticks acquire the bacteria from infected mice. The prevalence in the joint is associated with the pathology resulting from persistent infection. The brain appears to be less favorable for colonization, since most of these samples were negative for recA. Importantly, in these DNA samples, no tissue-specific inhibitors of PCR were detected that could cause underestimation of recA copy numbers.
The assay presented in this report should be applicable to the quantification of other pathogens. The parameters defined here include the use of an external standard for quantification, detection threshold calculation, sample normalization by quantification of a mouse gene, and tissue-to-tissue comparison. The use of the dsDNA dye SYBR Green I for detection of product should allow this protocol to be readily adaptable to already-existing pathogen primer sets used for quantification. For infectious agents in which a source of pure DNA is unavailable, a highly infected sample source could be diluted with an uninfected sample to generate an external standard. Alternatively, fluorescent threshold or PCR product standard alone should provide reliable estimates for rapid semiquantitative determinations.
ACKNOWLEDGMENTS
We thank Kathy Seiler for preparation of mouse DNA samples, Wai Mun Huang and Marianne Schroeder for assistance in designing recA primers, Sherwood Casjens for helpful discussion, and Carl Wittwer and his laboratory for assistance with the LightCycler quantification.
This work was supported by Public Health Service grants AI-32223 and AI-43521 to J.J.W., AI-24158 to J.H.W., and 5P30-CA-42014 to the University of Utah. The project described was also supported in part by an award from the American Lung Association (J.H.W.).
REFERENCES
- 1.Armstrong A L, Barthold S W, Persing D H, Beck D S. Carditis in Lyme disease susceptible and resistant strains of laboratory mice infected with Borrelia burgdorferi. Am J Trop Med Hyg. 1992;47:249–258. doi: 10.4269/ajtmh.1992.47.249. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. Current protocols in molecular biology. Wiley Press, New York, N.Y.
- 3.Barthold S W. Infectivity of Borrelia burgdorferi relative to route of inoculation and genotype in laboratory mice. J Infect Dis. 1991;163:419–420. doi: 10.1093/infdis/163.2.419. [DOI] [PubMed] [Google Scholar]
- 4.Barthold S W, Beck D S, Hansen G M, Terwilliger G A, Moody K D. Lyme borreliosis in selected strains and ages of laboratory mice. J Infect Dis. 1990;162:133–138. doi: 10.1093/infdis/162.1.133. [DOI] [PubMed] [Google Scholar]
- 5.Barthold S W, Persing D H, Armstrong A L, Peeples R A. Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am J Pathol. 1991;139:263–273. [PMC free article] [PubMed] [Google Scholar]
- 6.Dew-Jager K, Yu W Q, Huang W M. The recA gene of Borrelia burgdorferi. Gene. 1995;167:137–140. doi: 10.1016/0378-1119(95)00646-x. [DOI] [PubMed] [Google Scholar]
- 7.Duray P H, Kusnitz A, Ryan J. Demonstration of the Lyme disease spirochete by a modified Dieterle stain method. Lab Med. 1985;16:685–687. [Google Scholar]
- 8.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch J, Barbour A G. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J Bacteriol. 1992;174:5251–5257. doi: 10.1128/jb.174.16.5251-5257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitten T, Barbour A G. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics. 1992;132:311–324. doi: 10.1093/genetics/132.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y, Seiler K P, Eichwald E J, Weis J H, Teuscher C, Weis J J. Distinct characteristics of resistance to Borrelia burgdorferi-induced arthritis in C57BL/6N mice. Infect Immun. 1998;66:161–168. doi: 10.1128/iai.66.1.161-168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malawista S E, Barthold S W, Persing D H. Fate of Borrelia burgdorferi DNA in tissues of infected mice after antibiotic treatment. J Infect Dis. 1994;170:1312–1316. doi: 10.1093/infdis/170.5.1312. [DOI] [PubMed] [Google Scholar]
- 14.Nocton J J, Dressler F, Rutledge B J, Rys P N, Persing D H, Steere A C. Detection of Borrelia burgdorferi DNA by polymerase chain reaction in synovial fluid from patients with Lyme arthritis. N Engl J Med. 1994;330:229–234. doi: 10.1056/NEJM199401273300401. [DOI] [PubMed] [Google Scholar]
- 15.Pachner A R, Delaney E, Ricalton N S. Murine Lyme borreliosis: route of inoculation determines immune response and infectivity. Reg Immunol. 1992;4:345–351. [PubMed] [Google Scholar]
- 16.Ririe K M, Rasmussen R P, Wittwer C T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigo A G, Goracke P C, Rowhanian K, Mullins J I. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retroviruses. 1997;13:737–742. doi: 10.1089/aid.1997.13.737. [DOI] [PubMed] [Google Scholar]
- 18.Seiler K P, Vavrin Z, Eichwald E, Hibbs J, Jr, Weis J J. Nitric oxide production during murine Lyme disease: lack of involvement in host resistance or pathology. Infect Immun. 1995;63:3886–3895. doi: 10.1128/iai.63.10.3886-3895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–131. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 20.Wittwer C T, Ririe K, Rasmussen R. Fluorescence monitoring of rapid cycle PCR for quantification. In: Ferre F, editor. Gene quantification. New York, N.Y: Birkhauser; 1998. pp. 129–144. [Google Scholar]
- 21.Wittwer C T, Ririe K M, Andrew R V, David D A, Gundry R A, Balis U J. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 22.Yang L, Weis J H, Eichwald E, Kolbert C P, Persing D H, Weis J J. Heritable susceptibility to severe Borrelia burgdorferi-induced arthritis is dominant and is associated with persistence of large numbers of spirochetes in tissues. Infect Immun. 1994;62:492–500. doi: 10.1128/iai.62.2.492-500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]