Abstract
L1 syndrome, a complex X-linked neurological disorder, is caused by mutations in the L1 cell adhesion molecule (L1CAM) gene. L1CAM molecule is a member of immunoglobulin (Ig) superfamily of neural cell adhesion molecules (CAMs), which plays a pivotal role in the developing nervous system. In this study, a L1CAM gene exonic missense variant (c.1108G > A, p.G370R) was identified in two induced fetuses (abnormal fetuses), who presented corpus callosum agenesis accompanied with hydrocephalus. Clinical data, published literature, online database, and bioinformatic analysis suggest that the single-nucleotide variant of L1CAM gene is a likely pathogenic mutation. In vitro assays were performed to evaluate the effects of this variant. Based on NSC-34/COS-7 cells transfected with wild-type (L1-WT) and mutated (L1-G370R) plasmids, the L1CAM gene exonic missense variant (c.1108G > A, p.G370R) reduced cell surface expression, induced partial endoplasmic reticulum retention, affected posttranslational modification, and reduced protein’s homophilic adhesive ability, but did not induce endoplasmic reticulum stress, which might probably associate with L1 syndrome. Finally, 35 isolated fetuses were screened for L1CAM gene variants by Sanger sequencing. These cases all prenatally suspected of corpus callosum agenesis accompanied with hydrocephalus, which may relate to L1 syndrome. Consequently, one L1CAM gene single missense variant (c.550C > T, p.R184W) was detected in one fetus. Our results provided evidence that the L1CAM gene missense variant (c.1108G > A, p.G370R) may relate to L1 syndrome. The findings of this study suggest a potential possibility of L1CAM gene screening for prenatal diagnoses for fetuses presented corpus callosum agenesis accompanied with hydrocephalus.
Supplementary Information
The online version contains supplementary material available at 10.1007/s43032-021-00828-4.
Keywords: L1 syndrome, L1 cell adhesion molecule, Prenatal diagnosis, Functional analysis
Introduction
L1 syndrome, a complex X-linked neurological disorder with 1:30,000 incidence in newborn males, is caused by mutations in the L1 cell adhesion molecule (L1CAM) gene [1, 2]. It comprises X-linked hydrocephalus due to stenosis of the aqueduct of Sylvius (HSAS; MIM #307,000), X-linked complicated corpus callosum agenesis (MIM #304,100), MASA syndrome (mental retardation, aphasia, shuffling gait, adducted thumbs; MIM #303,350), and X-linked complicated hereditary spastic paraplegia type 1 (MIM #303,350) [3].
The L1CAM gene consists of 29 exons, located in the Xq28 region, and encodes a neuronal cell adhesion molecule. L1CAM molecule is a member of the immunoglobulin (Ig) superfamily of neural cell adhesion molecules (CAMs), which plays pivotal roles in neurite outgrowth, pathfinding, and fasciculation, as well as neuronal cell migration and survival [4–11]. All L1-related molecules share a primary structural organization of six Ig-like motifs, followed by five fibronectin type III (FNIII)-like repeats at the extracellular surface, a hydrophobic transmembrane region, and a short cytoplasmic segment in the C-terminus [12–14]. In addition, the extracellular part of L1CAM protein is responsible for mediating homophilic interactions with the L1CAM protein itself and heterophilic interactions with several other cell adhesion molecules, extracellular matrix, and chondroitin sulfate proteoglycan [15–18].
At present, more than 270 different L1CAM gene mutations have been reported, and almost one-third of them are single missense mutations. Most of these missense variants are located in extracellular domains of L1CAM molecule, which generally result in a more severe phenotype than those affecting the cytoplasmic protein domain [19]. In addition, previous studies have suggested that such missense mutations may reduce cell surface expression and affect protein misfolding, homo- and heterophilic ligand binding, or intracellular processing. Finally, they might interfere with neurite growth and neurite branching.
Here, we reported one variant of L1CAM gene in two induced fetuses (abnormal fetuses) suspected of L1 syndrome, which is likely disease-causing. The study provided case presentation, protein functional analysis, and screening of L1CAM gene variants in isolated fetuses. Together, a prenatal diagnosis may be provided for fetuses presented corpus callosum agenesis accompanied with hydrocephalus.
Case Presentation
Clinical Summary
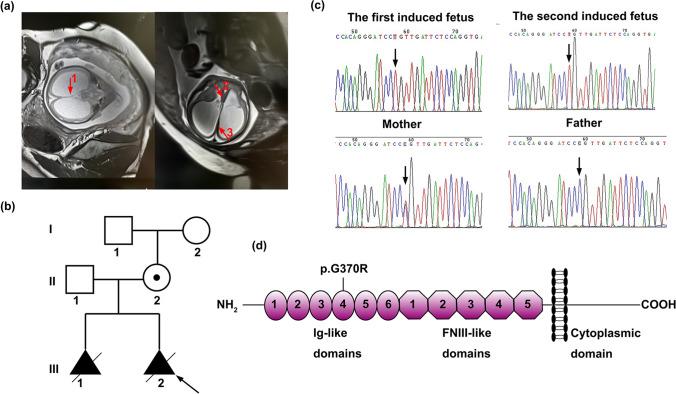
A 25-year-old, G2P0+2 woman (II.2) came to the genetic counseling clinic of West China Second University Hospital, Sichuan University (Chengdu, China). The woman informed the doctor that her first fetus presented corpus callosum agenesis through an ultrasound examination during the second trimester. Then, she required a termination of pregnancy. Her second fetus presented corpus callosum agenesis accompanied with hydrocephalus, narrowness of brain parenchyma, and the absence of the cavum septi pellucidi (CSP), from the reports of fetal ultrasound scan and magnetic resonance imaging (Fig. 1a). The woman was seen for genetic examination of the second induced fetus (abnormal fetus) and genetic counseling for the next pregnancy. The parents and maternal grandparents of the proband had no related clinical manifestations. The pedigree of this family is shown in Fig. 1b.
Fig. 1.
Clinical characteristics of family and schematic model of L1CAM molecule. a Magnetic resonance imaging of the second induced fetus (abnormal fetus). The absence of the cavum septi pellucidi is indicated by arrow 1 in the coronal plane. The corpus callosum agenesis is indicated by arrows 2 and 3 in the axial plane. b Pedigrees of the analyzed family in this study. Males are represented by squares, females are represented by circles, and triangle refers to abortion in early pregnancy. The proband is indicated by an arrow. c Sanger sequences of the L1CAM gene mutation (c.1108G > A). Hemizygous mutation detected in two induced fetuses (abnormal fetuses). The heterozygous mutation detected in these fetuses’ mother but not in these fetuses’ father. Mutation was indicated by arrow. d Schematic model of L1CAM molecule bearing domain structure: six immunoglobulin-like motifs (Ig-like), five fibronectin type III (FNIII)-like domains, a transmembrane region, and an intracellular domain (ICD). The location of the amino acid substitution (p.G370R) is indicated
Genetic Findings
DNA extracted from the muscle of the second induced fetus (abnormal fetus) and peripheral blood of parents was tested by trio whole-exome sequencing (Trio-WES). Candidate variant was validated using Sanger sequencing. Polymerase chain reaction (PCR) amplification was performed using primers (Supplementary Table 1) designed to cover variant identified by WES. In summary, exome capture sequencing was performed using Nano WES Human Exome V1 (Berry genomics) and Illunima NovaSeq6000 platform with 150-bp paired-end reads. Burrows-Wheeler Aligner software tool was used for aligning the sequencing reads with hg38/GRCh38. Local alignment and recalibration of the base quality of the Burrows-Wheeler aligned reads was performed by the GATK Indel Realigner and the GATK Base Recalibrator, respectively (broadinstitute.org/). Then, single-nucleotide variants (SNVs) and small insertions or deletions (InDels) were identified by GATK Unified Genotyper (broadinstitute.org). Variants were annotated and interpreted using ANNOVAR and the Enliven Variants Annotation Interpretation System (Berry genomics). Public databases used for filtering include gnomAD (http://gnomad.broadinstitute.org/), 1000 Genomes Project (1000G) (http://browser.1000genomes.org), etc. Pathogenicity of SNVs was evaluated based on the scientific medical literature and disease databases, such as PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), ClinVar (http://www.ncbi.nlm.nih.gov/clinvar), OMIM (http://www.omim.org), Human Gene Mutation Database (http://www.hgmd.org), and Human Genome Variation Society (http://www.hgvs.org/dblist/dblist.html).
A hemizygous variant (c.1108G > A, p.G370R) in L1CAM gene (NM_001278116.2) was detected, which was maternally inherited and validated by Sanger sequencing (Fig. 1c). The variant was also confirmed in the first abnormal fetus with corpus callosum agenesis. This mutation has been reported previously in one family with X-linked complicated spastic paraplegia, MASA syndrome, and HSAS [20]. However, there was no further functional analysis. The L1CAM gene variant Gly370 in our study lies in the Ig4 domain (Fig. 1d). The L1CAM gene variant (c.1108G > A, p.G370R) was predicted to be deleterious, according to PROVEAN, PolyPhen-2, CADD, etc. Thus, we performed a series of in vitro assays to evaluate the effects of this variant.
Materials and Methods
Subcloning of L1CAM cDNA and Mutagenesis
The construct which encoded the full-length human 3774 bp L1CAM cDNA (NM_001278116.2) with His-tag at the C-terminus and EGFP was subcloned into the pRP[Exp]-EGFP-EF1A > hL1CAM/6xHis vector (VectorBuilder, China). In addition, the full-length L1CAM cDNA with His-tag, but no EGFP, was subcloned into the pRP[Exp]-EF1A > hL1CAM/6xHis (VectorBuilder, China). The two constructs containing wild-type L1CAM cDNA were used as templates to generate the L1CAM cDNA variant. Replacing Gly-370 with Arg of L1CAM cDNA was performed by polymerase chain reaction (PCR) site-directed mutagenesis using gold medal Mix (#TSE101, Tsingke Biotechnology, China). L1-G370R variant was generated with the following primer sequence: Forward-5′-CTGGAGAATCAACAGGATCCCTGTGGAG-3′ and Reverse-5′-CTCCACAGGGATCCTGTTGATTCTCCAG-3′. The PCR product was digested using the DpnI restriction enzyme (#ER1702, Thermo Scientific, USA) and then added into DH5α (#CB101, Tiangen, China) to obtain a positive clone. The constructs of the wild-type(L1-WT) and p.G370R(L1-G370R) of L1CAM cDNA were verified by Sanger sequencing.
Cell Culture and Transfection
Mouse motor neuron (NSC-34) and African green monkey kidney fibroblast-like (COS-7) cells were purchased from Tongpai (Shanghai) Biotechnology Co., Ltd. The two cells were cultured in a high glucose Dulbecco’s Modified Eagle’s Medium (#D5796, Sigma-Aldrich, USA)/DMEM (#11,965,092, Gibco, USA) supplemented with 10% fetal bovine serum (#10,099,141, Gibco, USA) and 1% streptomycin/penicillin (#C0222, Beyotime Biotechnology, China) in an atmosphere of 5% CO2 at 37 °C. NSC-34/COS-7 cells were transfected with plasmids, using Lipofectamine™ 2000 transfection reagent (#11,668,019, Invitrogen, USA) in accordance with standard protocols. Briefly, NSC-34/COS-7 cells were plated at a density of 3 × 105 cells/9.6 cm2 dish, and NSC-34 cells were seeded at a density of 1.5 × 105 cells/35 mm onto poly-d-lysine-coated dishes. Twelve h later, 3 µg of plasmid DNA was added into 150 µL Opti-MEM (#31,985,070, Gibco, USA), and 9 µL of Lipofectamine™ 2000 was added to 150 µL Opti-MEM for each 6-well cell plate. For the 35-mm confocal plate, the plasmid DNA and Lipofectamine™ 2000 needed to be halved. After 5-min incubation at room temperature (RT), diluted DAN was added to diluted Lipofectamine™ 2000 reagent for another 5-min incubation. Then, cells were cultivated with DNA-lipid complex for 6 h, followed by the complete medium. Further incubation for 24 h, cells were processed for immunofluorescence and Western blotting.
Immunofluorescence Staining
The cells were fixed with 4% paraformaldehyde for 10 min, permeabilized for 15 min in 0.3% Triton X-100, and then blocked with 5% bovine serum albumin and 0.3% Triton X-100 in phosphate-buffered saline (PBS) for 1 h at RT. The samples were incubated with mouse monoclonal anti-6XHis-tag antibody (1:200, #ab18184, Abcam, UK) and rabbit monoclonal anti-KDEL antibody (1:200, #GXP282080, GenXspan, USA) at 4 °C overnight. After being washed with PBS, cells were subjected to fluorescent secondary antibodies for 1 h at RT, including Cy3-labeled Goat Anti-Mouse IgG (H + L) antibody (1:500, #A0521, Beyotime Biotechnology, China) and Alexa Fluor 488-labeled Goat Anti-Rabbit IgG(H + L) antibody (1:500, #A0423, Beyotime Biotechnology, China). Next, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI, 1:5000, #C1002, Beyotime Biotechnology, China). Finally, after being washed, the samples were imaged immediately on an FV-1000 confocal microscope (Olympus, Japan).
Western Blotting
The cells were lysed using a RIPA lysis buffer (#P0013B, Beyotime Biotechnology, China) and a cocktail of protease inhibitors (#P1005, Beyotime Biotechnology, China) on ice for 30 min. The protein concentration was measured by a BCA Protein Assay Kit (#P0012S, Beyotime Biotechnology, China). Then, the protein extracts were separated on 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and removed to nitrocellulose membranes. Next, the membranes were blocked in 5% nonfat dried milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) at RT for 1 h, followed by incubation with the mouse monoclonal anti-L1CAM antibody (1:1000, #ab24345, Abcam, UK), rabbit monoclonal anti-phosphorylated eukaryotic initiation factor 2 antibody (pEIF2α, 1:1000, #3398, CST, USA), and rabbit monoclonal anti-HSP90 antibody (1:1500, #AF1378, Beyotime Biotechnology, China), overnight at 4 °C. After being washed with TBST, the membranes were incubated with HRP-conjugated Affinipure Goat Anti-Mouse IgG (H + L) (1:5000, #SA00001-1, Proteintech, China) or HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H + L) (1:5000, #SA00001-2, Proteintech, China) for 1 h at RT. After being rewashed in TBST, the protein bands were analyzed using an ECL detection system (#P0018S, Beyotime Biotechnology, China), detected using the ChemiDoc™ MP Imaging System (Bio-Rad Laboratories Inc.) and quantified using ImageJ software (V1.8.0_112).
Deglycosylation Assay, Endoplasmic Reticulum (ER) Stress Induction, and MG132 Assay
For deglycosylation of proteins, cell lysates of the NSC-34 and COS-7 cells were treated with endoglycosidase H (Endo H) according to the manufacturer’s instructions (#P0702S, New England Biolabs, USA). After transfection for 24 h, 60 µg of total protein was denatured in glycoprotein denaturing buffer at 100 °C for 10 min. Furthermore, GlycoBuffer 3 and 0.2 µL Endo H were added to the above reaction products and incubated for 1 h at 37 °C.
For ER stress induction, cells were exposed to 0.5 µM, 1 µM, 2 µM, and 3 µM thapsigargin for 4 h (#B6614, APExBIO, US).
A medium containing 0.2 µM or 1 µM MG132 (#T510313-0001, Sangon Biotech, China) was added to the NSC-34 cells to inhibit proteasomal degradation. Furthermore, the cells were incubated for 20 h at 37 °C. In parallel, the control samples were subjected to DMSO, which is the solvent of MG132.
Cell Aggregation Assay
NSC-34 cells were rinsed twice in PBS, detached with trypsin/EDTA, and mechanically triturated to a single cell suspension. Dissociated 1.5 × 105 cells were inoculated into 24-well plates, incubated at 37 °C on a rotary shaker at 60 rpm. After a 90-min rotation, cell suspension was fixed with an equal volume of 4% paraformaldehyde, centrifuged at 1000 × g for 5 min, and then gently resuspended in PBS and transferred onto a microscope slide. Control cultures were fixed before rotation. Fifteen images for each condition were visualized using an inverted bright field microscope (Olympus BX60, Japan) at 200 × magnification.
Screening of L1CAM Gene Variants in Isolated Cases with Prenatally Suspected of Corpus Callosum Agenesis Accompanied with Hydrocephalus
The 35 fetal genomic DNA samples, extracted from the amniotic fluid or fetal muscle between March 2016 and January 2020, were obtained from the Prenatal Diagnosis Center of West China Second University Hospital, Sichuan University (Chengdu, China). All cases shared corpus callosum agenesis accompanied with hydrocephalus and presented other phenotypic findings, including narrowness of brain parenchyma, brain hemorrhage, vermis of cerebellum agenesis, or the absence of cavum septi pellucidi. All DNA samples were previously detected by chromosome microarray analysis with CytoScan 750 K Array (Thermo Fisher Scientific, USA) and were found negative for clinically significant chromosomal abnormalities. All 29 exons and intron/exon boundaries of the L1CAM gene were sequenced by Sanger sequencing on an ABI 3500 Analyzer (Thermo Fisher Scientific, USA) to determine the L1CAM gene variants in one fetus. The sequences of the 12 pairs of primers were designed using Prime Primer 5 and were listed in Table 1. All sequencing results were compared with the L1CAM gene sequences (https://www.ncbi.nlm.nih.gov/nuccore/NC_000023.11?report=genbank&from=153861514&to=153886173&strand=true, https://www.ncbi.nlm.nih.gov/nuccore/NM_001278116.2) using the online NCBI Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). When one L1CAM gene variant was detected in the fetus, the L1CAM gene site of the parents was further validated using primers (Supplementary Table 1) designed to cover variant identified by Sanger sequencing. The pathogenicity of the L1CAM gene variant was evaluated according to the guidelines of the American Society of Medical Genetics and Genomics (ACMG, 2015).
Table 1.
Primers used for the amplification and sequencing of L1CAM cDNA
| Exon | Primer (5′-3′) | Product Size (bp) |
|---|---|---|
| 1 | Forward: ACAGCCGCTGCTGCCGCAG | 352 |
| Reverse: CCCCGCAGGTTACCCCTCACC | ||
| 2 | Forward: GGGCTTACCCAGATGTTAGTCACTA | 353 |
| Reverse: GGAGAAGGTGAGGAGGTAGAAGATA | ||
| 3–5 | Forward: CTTACTATGTCCCCTGCCATCTG | 1346 |
| Reverse: CACAAGAAACAAGATGGGCCA | ||
| 6–11 | Forward: CCCTAAGTGCTAGTCTCTGCTATGA | 1903 |
| Reverse: GACAGACTGGGAGTTAGGAGGTAAG | ||
| Reverse: CAAGCAGGACGAGCGGGTGACG | ||
| 12–19 | Forward: GGGGAGAGGTGACTGTCAGTTAG | 2552 |
| Forward: GGCTGCCAATGACCAAAACA | ||
| Reverse: CTCCAGAGTAGCCGATAGTGACCT | ||
| Reverse: GCTCCCCCTGGAAATTTGGA | ||
| 20–25 | Forward: AATTCGTCTTCTCTGTGTGTAGGGG | 1794 |
| Forward: GTGAGGCGCCGGGGGCCCGCCATCA | ||
| Reverse: AGGAGTGACAGGGACAGGGAAAA | ||
| Reverse: CCCGCCTGCCTTCCATTTGTTTAG | ||
| 26–29 | Forward: GTGAGGCGCCGGGGGCCCGCCATCA | 2716 |
| Forward: GGCCCCAGACAGCTTCCCAGACAGG | ||
| Forward: GGGCAGACATGGTGGGGTCTCCTCA | ||
| Reverse: CCCGCCTGCCTTCCATTTGTTTAG | ||
| Reverse: GGTGCTGCCAGAGTGCGATGC | ||
| Reverse: AGACCAAGCACAGGCATACAGGGA |
Statistical Analysis
Statistical analysis was performed using SPSS Statistics 24.0 (SPSS Inc., USA). Data were presented as mean ± standard deviations (SD). Differences between groups were compared using two-tailed Student’s t-test. P-values < 0.05 were considered as statistically significant.
Results
L1-G370R Reduced Cell Surface Expression and Induced ER Retention
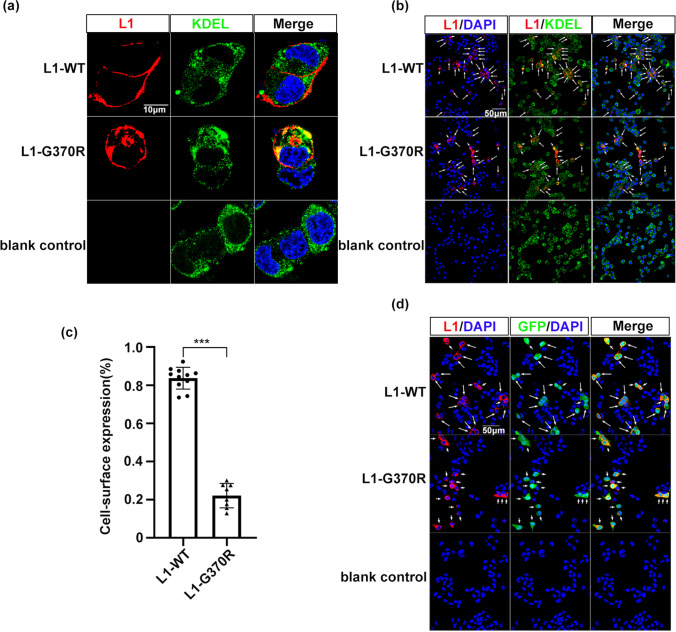
We performed L1CAM c.1108G > A mutation using PCR site-directed mutagenesis, to understand the pathogenic nature of L1CAM variant p.G370R on NSC-34 and COS-7 cells, and the sequences were checked by Sanger sequencing. The NSC-34 cells transfected with L1-WT or L1-G370R were double-stained with anti-6XHis-tag antibody that specifically recognizes exogenous L1CAM protein and the ER-retrieval motif KDEL antibody to investigate the changes in subcellular localization. A high percentage of L1-WT was localized in the cell membrane surface (83.66 ± 5.74%). However, we found only 22.11 ± 6.40% of L1-G370R expressed on the cell surface, presenting a redistribution to ER, as tested by co-localization with the ER-marker KDEL, resulting in 73.57% of reduction of cell surface L1CAM protein expression (P < 0.001, Fig. 2a–c).
Fig. 2.
L1CAM p.G370R induces ER retention. a, b Cells expression of L1CAM proteins in NSC-34 cells transfected with L1-WT and L1-G370R constructs using antibodies to 6XHis-tag (red), which exclusively recognizes exogenous L1CAM protein and the ER-marker KDEL (green). Nuclei were visualized by DAPI. Arrows depict L1CAM located on the cell surface. Arrowheads depict L1CAM co-localization with ER marker. c Diagram showing the percentage of cell surface expression of wild-type and mutant human L1CAM in NSC-34 cells. L1-WT, 83.66 ± 5.74%; L1-G370R, 22.11 ± 6.40%. Data represent means ± SD; ***P < 0.001. d Confocal images of NSC-34 cells co-expression of human L1CAM and EGFP constructs. GFP-positive cells presenting L1CAM cell surface expression were tagged by arrows, while barely presented L1CAM cell surface expressions were tagged by arrowheads. Scale: 10 μm (a), 50 μm (b, d)
NSC-34 cells were co-transfected L1CAM with GFP-expressing plasmids to examine transfection efficiencies and protein expression levels of L1-G370R. There were almost no differences in transfection efficiencies between the cells transfected with L1-WT or L1-G370R. Furthermore, all GFP-positive cells expressed L1CAM protein on the cell surface or redistributed in intracellular sites (Fig. 2d). This observation indicated that the differences in cell surface expression have not resulted from transfection efficiencies or L1CAM expression levels.
Posttranslational Modification Was Impaired for L1-G370R
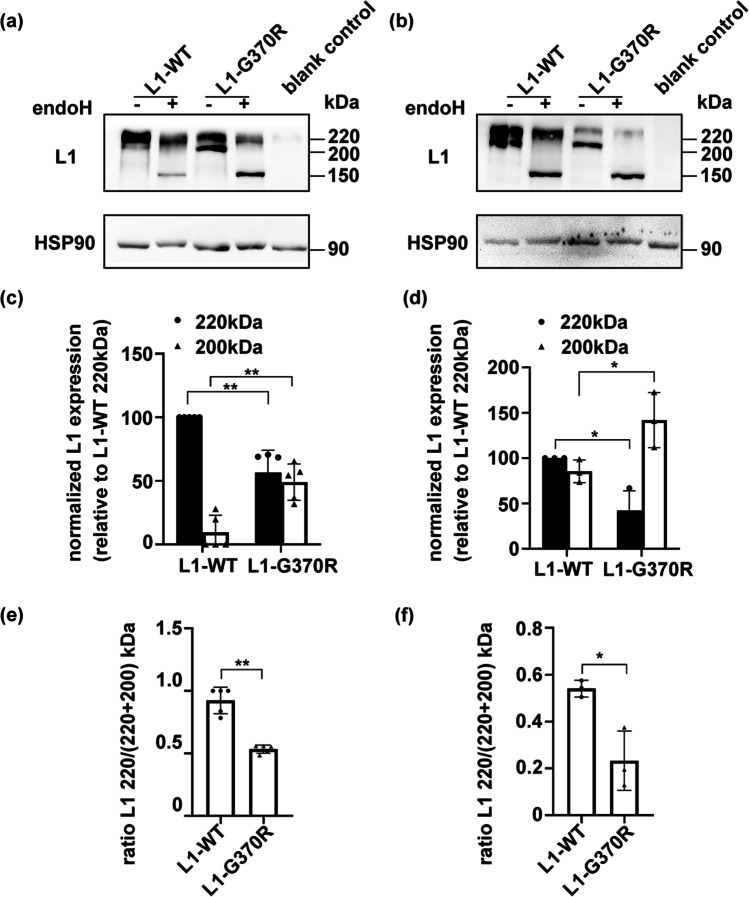
Cell lysates of transfected NSC-34 and COS-7 cells were subjected to SDS–PAGE and immunoblotting with anti-L1CAM antibody, revealing two bands identified at 220 and 200 kDa (Fig. 3a, b). The 220-kDa band corresponds to a full-length complex-mannose type of L1CAM located at the cell surface, generated through posttranslational modification in the Golgi apparatus. On the other hand, the 200-kDa band might represent an incompletely processed high-mannose type of L1CAM, which is generated only through co-translational modification in the ER [21, 22]. We found that in the NSC-34 and COS-7 cells, 220-kDa form, as well as the 220/(220 + 200) kDa ratio of L1-G370R, reduced, while the 200-kDa form increased, compared to L1-WT (P < 0.05, Fig. 3c–f). The cell lysates were digested with Endo H, which exclusively decomposes high-mannose oligosaccharides, but it is useless to decompose the complex-mannose oligosaccharides [23]. After incubation with Endo H, the 200-kDa form of L1-WT or L1-G370R was shifted to a 150-kDa fragment of L1CAM. In contrast, Endo H showed no significant effects on the 220-kDa band, representing a mature form of L1CAM (Fig. 3a, b). These findings demonstrated that the L1CAM 200-kDa form was likely to accumulate in the ER and was not transported through the Golgi organelle to the cell surface.
Fig. 3.
Cell lysates from NSC-34 to COS-7 cells expressing wild-type (L1-WT) or mutant L1 (L1-G370R) were treated with endoglycosidase H (Endo H). a, b L1CAM immunoblot analysis showing Endo H specifically cleaved high-mannose type of L1CAM (200 kDa), whereas it had no work to complex-mannose type (220 kDa). c, d Histogram showing the 220-kDa form of L1-G370R was reduced, while the 200-kDa form was increased, compared to L1-WT. e, f Histogram showing the 220/(220 + 200) kDa ratio of L1-G370R was reduced relative to L1-WT. NSC-34 cells, a, c, e; COS-7 cells, b, d, f
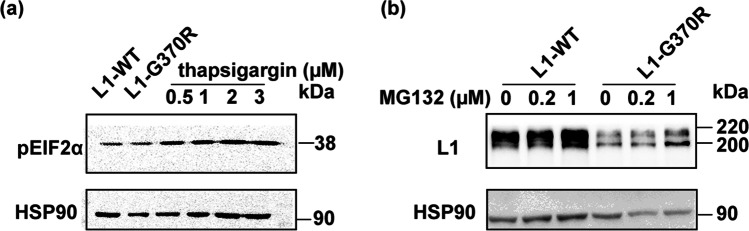
L1-G370R Did Not Induce ER Stress
Cells initiate the ER-associated degradation (ERAD) to avoid the excessive deposition of unfolded or misfolded proteins in the ER via the ubiquitin–proteasome pathway. If the load of the ER exceeds its capacity, cells may go into ER stress leading to cell death. To investigate whether ER dislocation of L1-G370R induces ER stress, we detected the pEIF2α level, the ERAD downstream molecule. There were almost no upgraded levels of pEIF2α in NSC-34 cells following overexpression of L1-G370R compared with the cells transfected with L1-WT, whereas treatment with the ER stress inducer thapsigargin increased the pEIF2α (Fig. 4a). When transfected NSC-34 cells were co-cultured with 0.2 µM or 1 µM proteasome inhibitor MG132 for 20 h, the expressing levels of L1-WT and L1-G370R improved, indicating L1-WT and L1-G370R were degraded by the ERAD (Fig. 4b). The collected data suggest impaired cell surface expression, rather than cytotoxicity caused by ER retained L1-G370R, is the key pathogenesis motif for the L1 syndrome.
Fig. 4.
L1-G370R does not induce ER stress. a pEIF2α expression levels from NSC-34 expressing L1-G370R were almost not raised compared with cells transfected with L1-WT. As a positive control, cultures were subjected to 0.5 µM, 1 µM, 2 µM, and 3 µM thapsigargin for 4 h. b The expression levels of L1-WT and L1-G370R were gradually upgraded after treatment with 0.2 µM and 1 µM proteasome inhibitor MG132 for 20 h
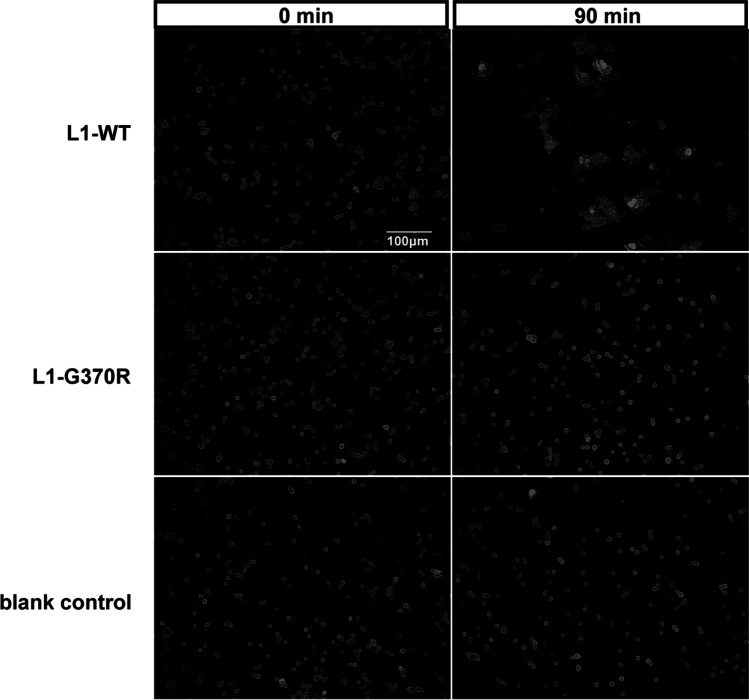
L1-G370R Impaired L1CAM-Dependent Cell–Cell Adhesion
L1CAM can promote cell–cell binding by homophilic adhesion in vitro [24]. Binding assays involving L1-WT and L1-G370R protein expressed on transfected NSC-34 cells were applied to analyze cell–cell binding ability. After the separation of transfected cells, they were given a chance to form aggregates for 90 min. Aggregation was completed within 90 min in NSC-34 cells transfected with the L1-WT construct. In contrast, cells expressing L1-G370R or blank control almost failed to form cell aggregates (Fig. 5). Thereby, L1CAM-mediated cell aggregation was disrupted by p.G370R. The collected data suggest L1CAM variant p.G370R was likely related to L1 syndrome (Table 2).
Fig. 5.
L1-G370R disrupted L1CAM-dependent cell–cell adhesion NSC-34 cells expressing L1-WT induced cell aggregation, but cells expressing L1-G370R and blank control almost failed to form cell aggregates. Scales: 100 μm
Table 2.
Summary of the results of functional experiments
| Nucleotide change | Exon | Amino acid change | Protein domain affected | Combinatorial prediction | |
|---|---|---|---|---|---|
| c.1108G > A | 9 | p.G370R | Ig4 | Affects function | |
| Protein maturation | Surface expression | Posttranslational modification | ER stress | Cell–cell adhesion | |
| Incomplete | Reduced | Impaired | Uneffected | Impaired |
Screening for L1CAM Gene Mutations
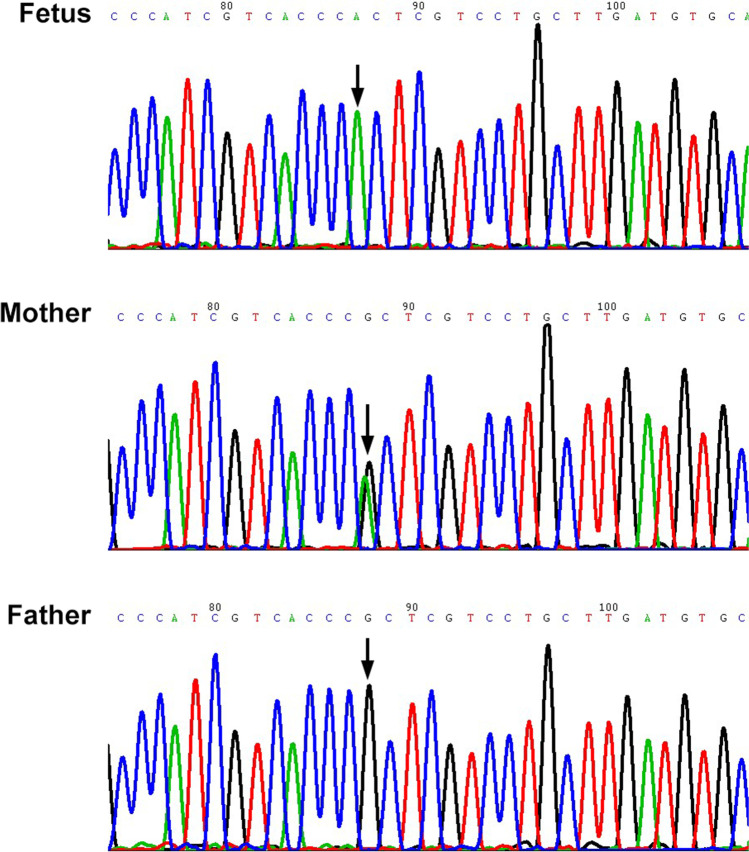
In families suspected of L1 syndrome, the determination of L1CAM gene mutations would give a chance of prenatal diagnosis, ultimately avoiding the congenital disabilities relevant to L1CAM gene mutation. In the prenatal, ultrasound attention should be paid to hydrocephalus and agenesis of the corpus callosum related to L1 syndrome. We used Sanger sequencing to detect L1CAM gene variants in 35 fetal genomic DNA samples. They all shared corpus callosum agenesis accompanied with hydrocephalus. After blasting sequencing results to L1CAM gene sequences (NC_000023.11), one single missense nucleotide variation of L1CAM gene (c.550C > T, p.R184W) was detected in one fetus and validated by Sanger sequencing (Fig. 6). Through clinical data, published literature, and bioinformatic analysis, the L1CAM gene single-nucleotide variant (p.R184W) is a likely pathogenic mutation [21, 22, 25, 26].
Fig. 6.
Sanger sequences of the L1CAM gene mutation (c.550C > T). Hemizygous mutation detected in the fetus presented corpus callosum agenesis accompanied with hydrocephalus. The heterozygous mutation detected in the fetus’ mother but not in the fetus’ father. Mutation was indicated by arrow
By only detecting 29 exons and intron/exon boundaries using Sanger sequencing, our data indicated that the detection rate of L1CAM gene mutant was 3% (1/35) in isolated fetuses prenatally suspected of corpus callosum agenesis accompanied with hydrocephalus.
Discussion
Here, a L1CAM gene exonic missense variant (c.1108G > A, p.G370R) was identified in two induced fetuses (abnormal fetuses), who presented corpus callosum agenesis accompanied with hydrocephalus. Clinical data, published literature, online database, and online tools suggest the L1CAM gene single-nucleotide variant is a likely pathogenic mutation. In vitro assays indicated that this variant might relate to L1 syndrome. The L1CAM gene exonic missense variant (c.1108G > A, p.G370R) reduced cell surface expression, induced partial ER accumulation, affected posttranslational processing, and, finally, impaired the L1CAM homophilic adhesive ability and, thus, loss of function. Thirty-five isolated cases prenatally suspected of corpus callosum agenesis accompanied with hydrocephalus were screened to acquire detection rate of L1CAM gene variant. Consequently, we found one L1CAM gene single missense nucleotide variation (c.550C > T, p.R184W) in one fetus. Our data indicated that the detection rate of L1CAM gene mutant was 3% (1/35) in isolated fetuses prenatally suspected of corpus callosum agenesis accompanied with hydrocephalus, using Sanger sequencing to detect 29 exons and intron/exon boundaries. Taken together, our results provided evidence that the L1CAM gene missense variant (c.1108G > A, p.G370R) may relate to L1 syndrome. The findings of this study suggest a potential possibility of L1CAM gene screening for prenatal diagnoses for fetuses presented corpus callosum agenesis accompanied with hydrocephalus.
Previous studies have shown that Ig1-Ig4 domains of L1CAM compose a horseshoe conformation that mediates homophilic adhesion function [27, 28]. The L1CAM gene variant Gly370 in our study lies in the Ig4 domain, a key amino acid domain. Fransen et al. suggested that extracellular missense mutations affecting key domains of amino acids might cause a more devastating phenotype than those affecting surface sites [29]. Also, Carlos Ruiz et al. reported the G370R mutation in a family with X-linked complicated spastic paraplegia, MASA syndrome, and HSAS [20]. Although it is tempting to consider this L1CAM gene missense variant (c.1108G > A, p.G370R) as pathogenic, it is particularly important to further identify the functional effects before drawing any firm conclusions. Therefore, we performed this study.
When NSC-34 cells were transiently transfected with L1-G370R plasmids, significantly reducing the amounts of cell surface protein and inducing redistribution to ER that co-immunostained to KDEL were observed in most transfected cells, compared with L1-WT. Confocal images of NSC-34 cells transfected with L1CAM wild type or mutant with GFP-expressing plasmids revealed similar transfection efficiencies and protein expression levels for the two constructs. These results revealed that the differences of cell surface expression of L1-G370R were not caused by transfection efficiencies or protein expression levels. Bateman et al. have predicted that L1CAM gene variant p.G370R might alter the conformation of this protein [30]. Accumulation in ER of misfolded transmembrane proteins has been shown to be pathological for several genetic disorders, for example, cystic fibrosis and Kallmann syndrome [31, 32]. Therefore, the ER accumulation of misfolded L1-G370R might be pathogenic.
The glycoprotein L1CAM passes through ER co-translational processing, Golgi complex posttranslational processing, and is finally transferred to the cell surface [33]. We thus studied whether ER retainability of L1-G370R may be immature protein. By immunoblotting, we found that the 220-kDa form and the 220/(220 + 200) kDa ratio of L1-G370R were reduced, but the 200-kDa form increased compared to L1-WT. Also, when treated with Endo H, the molecular mass of 200-kDa form of L1-WT or L1-G370R was shifted to 150-kDa. These findings indicated that the 200-kDa form of L1-G370R accumulated in the ER did not complete Golgi apparatus posttranslational modification, thus impairing the cell surface 220-kDa L1CAM corresponding to the mature form. Moulding et al. have demonstrated that the R184Q and D598N L1CAM proteins impair cell surface expression and incomplete posttranslational processing in astrocytes, Vero, COS-7, and CHO cells [21]. Furthermore, the L1CAM gene missense mutation (p.W635C) accumulated in the ER also interferes with posttranslational modification [34]. These results align with our results that p.G370R causes defects in the trafficking of L1CAM to the cell surface.
In the process of normal protein synthesis, unfolded or misfolded proteins are often produced. Cells initiate ERAD to remove unfolded or misfolded proteins that are retained in the ER by cytosolic proteosomes. If this process fails, it may induce ER stress that may render cells to death. Several neurological disorders are caused by ER stress resulting from ER accumulation of mutated protein [35, 36]. However, our study indicated that NSC-34 cells overexpressing L1-G370R did not induce ER stress. This may be because mutant L1CAM protein is efficiently degraded by ERAD [37]. Consistent with our hypothesis, our experiment revealed that L1-WT and L1-G370R proteins were upgraded when proteasomal degradation was inhibited. In line with our data, the L1CAM gene variants R184Q and W1036L have been identified as two pathogenic mutations but do not induce ER stress in NSC-34 cells [22]. These findings demonstrated that impaired cell surface expression rather than cytotoxicity caused by ER retained L1-G370R is the critical pathogenesis for the L1 syndrome.
Earlier studies concluded that defects in L1CAM homophilic binding are likely to contribute to L1 syndrome [25]. Reducing cell surface expression would impair L1CAM mediate homophilic binding and signaling in cells; thus we performed a cell aggregation assay. Compared to NSC-34 cells overexpressing L1-WT, L1CAM-dependent cell–cell adhesion was impaired in NSC-34 cells overexpressing L1-G370R, which agrees with the research showing that L1-G370R dramatically reduces ligand binding through fluorescent microspheres assay [25]. Also, Mariola et al. have shown that L1CAM gene variants p.W635C and p.V768I affect L1CAM-dependent homophilic binding [34, 38].
Congenital corpus callosum agenesis accompanied with hydrocephalus is a rare and exceedingly heterogeneous condition that can result from multiple causes. It can be detected by antenatal ultrasound but poses a great challenge to prenatal diagnosis as the outcome is variable. An online search for “corpus callosum agenesis and hydrocephalus” on Online Mendelian Inheritance in Man (OMIM) resulted in 93 entries. Some condition of corpus callosum agenesis accompanied with hydrocephalus has recognizable syndromes, including L1 syndrome (MIM #304,100), Chudley-McCullough syndrome (MIM #604,213), agenesis of corpus callosum, cardiac, ocular, and genital syndrome (MIM #618,929), MASA syndrome (MIM #303,350), and so on. However, some individuals with corpus callosum agenesis accompanied with hydrocephalus do not have a clearly inherited cause [39, 40]. The identifiable genes for the above identifiable syndromes, respectively, are L1CAM, G-protein signaling modulator (GPSM2), and cadherin (CDH2). We screened 35 unrelated fetuses prenatally suspected of corpus callosum agenesis accompanied with hydrocephalus for L1CAM gene variants to acquire a detection rate of L1CAM gene variant in these cases. Consequently, we found one single missense nucleotide variation L1CAM gene (c.550C > T, p.R184W) in one fetus. The L1CAM gene variant (c.550C > T, p.R184W) was predicted to be deleterious, according to PROVEAN, PolyPhen-2, MUpro, CADD, etc. Furthermore, the amino acid change at this site (R184) was pathogenic which have been published by Schäfer et al. [21, 22, 25, 26]. Our data indicated that the detection rate of L1CAM gene mutant was 3% (1/35) in isolated fetuses prenatally suspected of corpus callosum agenesis accompanied with hydrocephalus, using Sanger sequencing to detect 29 exons and intron/exon boundaries. A previous report indicated that approximately 5% of children with congenital hydrocephalus was X-chromosomal hydrocephalus caused by L1CAM gene mutations [41]. Another report indicated that the detection rate of L1CAM gene mutant was 16% in X-chromosomal hydrocephalus without a family history [39, 40]. Due to low incidence of this condition, detection method application, and selection bias in the reported study, the actual prevalence is difficult to estimate. In the future, we will further increase the sample size and apply other genetic methods for search. L1CAM gene mutation detection can be offered to fetuses presented corpus callosum agenesis accompanied with hydrocephalus.
In conclusion, we provided two induced fetuses (abnormal fetuses) suspected of L1 syndrome with L1CAM gene variant (c.1108G > A, p.G370R). Clinical data, published literature, online database, and online tools suggest the L1CAM gene single-nucleotide variant is a likely pathogenic mutation. In addition, in vitro analyses indicated that the L1CAM gene mutation (c.1108G > A, p.G370R) reduced cell surface expression, induced partial ER retention, affected posttranslational modification, reduced protein’s homophilic adhesive ability, but did not induce ER stress, which was likely to be associated with the L1 syndrome. The molecular mechanisms involving in L1 syndrome should be further studied. Ultimately, we screened 35 unrelated fetuses prenatally suspected of corpus callosum agenesis accompanied with hydrocephalus for L1CAM gene variants by Sanger sequencing. Consequently, one L1CAM gene single missense variant (c.550C > T, p.R184W) was detected in one fetus. Our results provide evidence that the L1CAM gene missense variant (c.1108G > A, p.G370R) may relate to L1 syndrome. The findings of this study suggest a potential possibility of L1CAM gene screening for prenatal diagnoses for fetuses presented corpus callosum agenesis accompanied with hydrocephalus.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2021YFC1005300) and the Science and Technology Department of Sichuan Province, China (2021YFS0078).
Author Contribution
Conceptualization: SL, MY, HW, PW; methodology: PW, HL, QW, HX; formal analysis and investigation: PW, HL, QW, HX; writing — original draft preparation: PW; writing — review and editing: SL, MY, HW; funding acquisition: SL; supervision: SL, MY.
Funding
Key Technologies Research and Development Program, 2021YFC1005300,Shanling Liu,Department of Science and Technology of Sichuan Province,2021YFS0078,Shanling Liu
Code Availability
Not applicable.
Declarations
Ethics Approval
Our study was approved by the Ethical Committee of West China Second University Hospital, Sichuan University. In addition, the family signed written informed consent.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Mei Yang and Shanling Liu contributed equally to this work.
Contributor Information
Mei Yang, Email: 562545550@qq.com.
Shanling Liu, Email: sunny630@126.com.
References
- 1.Lyonnet S, Pelet A, Royer G, et al. The gene for X-linked hydrocephalus maps to Xq28, distal to DXS52. Genomics. 1992;14(2):508–510. doi: 10.1016/s0888-7543(05)80254-x. [DOI] [PubMed] [Google Scholar]
- 2.Schrander-Stumpel C, Legius E, Fryns JP, Cassiman JJ. MASA syndrome: new clinical features and linkage analysis using DNA probes. J Med Genet. 1990;27(11):688–692. doi: 10.1136/jmg.27.11.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vos YJ, Hofstra RM. An updated and upgraded L1CAM mutation database. Hum Mutat. 2010;31(1):E1102–1109. doi: 10.1002/humu.21172. [DOI] [PubMed] [Google Scholar]
- 4.Chang S, Rathjen FG, Raper JA. Extension of neurites on axons is impaired by antibodies against specific neural cell surface glycoproteins. J Cell Biol. 1987;104(2):355–362. doi: 10.1083/jcb.104.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bixby JL, Lilien J, Reichardt LF. Identification of the major proteins that promote neuronal process outgrowth on Schwann cells in vitro. J Cell Biol. 1988;107(1):353–361. doi: 10.1083/jcb.107.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seilheimer B, Schachner M. Studies of adhesion molecules mediating interactions between cells of peripheral nervous system indicate a major role for L1 in mediating sensory neuron growth on Schwann cells in culture. J Cell Biol. 1988;107(1):341–351. doi: 10.1083/jcb.107.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagenaur C, Lemmon V. An L1-like molecule, the 8D9 antigen, is a potent substrate for neurite extension. Proc Natl Acad Sci U S A. 1987;84(21):7753–7757. doi: 10.1073/pnas.84.21.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stallcup WB, Beasley L. Involvement of the nerve growth factor-inducible large external glycoprotein (NILE) in neurite fasciculation in primary cultures of rat brain. Proc Natl Acad Sci U S A. 1985;82(4):1276–1280. doi: 10.1073/pnas.82.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seilheimer B, Persohn E, Schachner M. Antibodies to the L1 adhesion molecule inhibit Schwann cell ensheathment of neurons in vitro. J Cell Biol. 1989;109(6 Pt 1):3095–3103. doi: 10.1083/jcb.109.6.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood PM, Schachner M, Bunge RP. Inhibition of Schwann cell myelination in vitro by antibody to the L1 adhesion molecule. J Neurosci. 1990;10(11):3635–3645. doi: 10.1523/JNEUROSCI.10-11-03635.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samatov TR, Wicklein D, Tonevitsky AG. L1CAM: Cell adhesion and more. Prog Histochem Cytochem. 2016;51(2):25–32. doi: 10.1016/j.proghi.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Hortsch M. The L1 family of neural cell adhesion molecules: old proteins performing new tricks. Neuron. 1996;17(4):587–593. doi: 10.1016/s0896-6273(00)80192-0. [DOI] [PubMed] [Google Scholar]
- 13.Williams AF, Barclay AN. The immunoglobulin superfamily–domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 14.Hlavin ML, Lemmon V. Molecular structure and functional testing of human L1CAM: an interspecies comparison. Genomics. 1991;11(2):416–423. doi: 10.1016/0888-7543(91)90150-d. [DOI] [PubMed] [Google Scholar]
- 15.Morales G, Hubert M, Brümmendorf T, et al. Induction of axonal growth by heterophilic interactions between the cell surface recognition proteins F11 and Nr-CAM/Bravo. Neuron. 1993;11(6):1113–1122. doi: 10.1016/0896-6273(93)90224-f. [DOI] [PubMed] [Google Scholar]
- 16.Buchstaller A, Kunz S, Berger P, et al. Cell adhesion molecules NgCAM and axonin-1 form heterodimers in the neuronal membrane and cooperate in neurite outgrowth promotion. J Cell Biol. 1996;135(6 Pt 1):1593–1607. doi: 10.1083/jcb.135.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenfeld DP, Hynes MA, Skoler KM, Furley AJ, Jessell TM. TAG-1 can mediate homophilic binding, but neurite outgrowth on TAG-1 requires an L1-like molecule and beta 1 integrins. Neuron. 1994;12(3):675–690. doi: 10.1016/0896-6273(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 18.Grumet M, Flaccus A, Margolis RU. Functional characterization of chondroitin sulfate proteoglycans of brain: interactions with neurons and neural cell adhesion molecules. J Cell Biol. 1993;120(3):815–824. doi: 10.1083/jcb.120.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasaki M, Thompson P, Lemmon V. CRASH syndrome: mutations in L1CAM correlate with severity of the disease. Neuropediatrics. 1997;28(3):175–178. doi: 10.1055/s-2007-973696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz JC, Cuppens H, Legius E, et al. Mutations in L1-CAM in two families with X linked complicated spastic paraplegia, MASA syndrome, and HSAS. J Med Genet. 1995;32(7):549–552. doi: 10.1136/jmg.32.7.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moulding HD, Martuza RL, Rabkin SD. Clinical mutations in the L1 neural cell adhesion molecule affect cell-surface expression. J Neurosci. 2000;20(15):5696–5702. doi: 10.1523/JNEUROSCI.20-15-05696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schafer MK, Nam YC, Moumen A, et al. L1 syndrome mutations impair neuronal L1 function at different levels by divergent mechanisms. Neurobiol Dis. 2010;40(1):222–237. doi: 10.1016/j.nbd.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 23.Rothman JE, Bursztyn-Pettegrew H, Fine RE. Transport of the membrane glycoprotein of vesicular stomatitis virus to the cell surface in two stages by clathrin-coated vesicles. J Cell Biol. 1980;86(1):162–171. doi: 10.1083/jcb.86.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaraj K, Kristiansen LV, Skrzynski A, Castiella C, Garcia-Alonso L, Hortsch M. Pathogenic human L1-CAM mutations reduce the adhesion-dependent activation of EGFR. Hum Mol Genet. 2009;18(20):3822–3831. doi: 10.1093/hmg/ddp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Angelis E, Watkins A, Schafer M, Brummendorf T, Kenwrick S. Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum Mol Genet. 2002;11(1):1–12. doi: 10.1093/hmg/11.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Li YT, Chen JS, Jian W, et al. L1CAM mutations in three fetuses diagnosed by medical exome sequencing. Taiwan J Obstet Gynecol. 2020;59(3):451–455. doi: 10.1016/j.tjog.2020.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Haspel J, Grumet M. The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes. Front Biosci. 2003;8:s1210–1225. doi: 10.2741/1108. [DOI] [PubMed] [Google Scholar]
- 28.Wei CH, Ryu SE. Homophilic interaction of the L1 family of cell adhesion molecules. Exp Mol Med. 2012;44(7):413–423. doi: 10.3858/emm.2012.44.7.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Angelis E, MacFarlane J, Du JS, et al. Pathological missense mutations of neural cell adhesion molecule L1 affect homophilic and heterophilic binding activities. EMBO J. 1999;18(17):4744–4753. doi: 10.1093/emboj/18.17.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bateman A, Jouet M, MacFarlane J, Du JS, Kenwrick S, Chothia C. Outline structure of the human L1 cell adhesion molecule and the sites where mutations cause neurological disorders. EMBO J. 1996;15(22):6050–6059. [PMC free article] [PubMed] [Google Scholar]
- 31.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83(1):121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 32.Monnier C, Dodé C, Fabre L, et al. PROKR2 missense mutations associated with Kallmann syndrome impair receptor signalling activity. Hum Mol Genet. 2009;18(1):75–81. doi: 10.1093/hmg/ddn318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff JM, Rathjen FG, Frank R, Roth S. Biochemical characterization of polypeptide components involved in neurite fasciculation and elongation. Eur J Biochem. 1987;168(3):551–561. doi: 10.1111/j.1432-1033.1987.tb13453.x. [DOI] [PubMed] [Google Scholar]
- 34.Marx M, Diestel S, Bozon M, et al. Pathomechanistic characterization of two exonic L1CAM variants located in trans in an obligate carrier of X-linked hydrocephalus. Neurogenetics. 2012;13(1):49–59. doi: 10.1007/s10048-011-0307-4. [DOI] [PubMed] [Google Scholar]
- 35.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 36.Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13(3):385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- 37.Brodsky JL. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation) Biochem J. 2007;404(3):353–363. doi: 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Angelis E, Watkins A, Schäfer M, Brümmendorf T, Kenwrick S. Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum Mol Genet. 2002;11(1):1–12. doi: 10.1093/hmg/11.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Vos YJ, de Walle HE, Bos KK, et al. Genotype-phenotype correlations in L1 syndrome: a guide for genetic counselling and mutation analysis. J Med Genet. 2010;47(3):169–175. doi: 10.1136/jmg.2009.071688. [DOI] [PubMed] [Google Scholar]
- 40.Finckh U, Schröder J, Ressler B, Veske A, Gal A. Spectrum and detection rate of L1CAM mutations in isolated and familial cases with clinically suspected L1-disease. Am J Med Genet. 2000;92(1):40–46. doi: 10.1002/(sici)1096-8628(20000501)92:1<40::aid-ajmg7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 41.Schrander-Stumpel C, Fryns JP. Congenital hydrocephalus: nosology and guidelines for clinical approach and genetic counselling. Eur J Pediatr. 1998;157(5):355–362. doi: 10.1007/s004310050830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.