Abstract
Risk evaluation of lymph node metastasis (LNM) for endoscopically resected submucosal invasive (T1) colorectal cancers (CRC) is critical for determining therapeutic strategies, but interobserver variability for histologic evaluation remains a major problem. To address this issue, we developed a machine-learning model for predicting LNM of T1 CRC without histologic assessment. A total of 783 consecutive T1 CRC cases were randomly split into 548 training and 235 validation cases. First, we trained convolutional neural networks (CNN) to extract cancer tile images from whole-slide images, then re-labeled these cancer tiles with LNM status for re-training. Statistical parameters of the tile images based on the probability of primary endpoints were assembled to predict LNM in cases with a random forest algorithm, and defined its predictive value as random forest score. We evaluated the performance of case-based prediction models for both training and validation datasets with area under the receiver operating characteristic curves (AUC). The accuracy for classifying cancer tiles was 0.980. Among cancer tiles, the accuracy for classifying tiles that were LNM-positive or LNM-negative was 0.740. The AUCs of the prediction models in the training and validation sets were 0.971 and 0.760, respectively. CNN judged the LNM probability by considering histologic tumor grade.
Subject terms: Colorectal cancer, Machine learning
Introduction
Endoscopic resection for submucosal invasive (T1) colorectal cancer (CRC) has rapidly increased over the last decade1,2. Approximately 10% of T1 CRC can metastasize to lymph nodes, making histologic evaluation for T1 CRC critical for determining the indications for additional surgery3–5. Previous studies described five independent histologic risk factors for lymph node metastasis (LNM), as follows; depth of submucosal invasion (≥ 1000 µm), poorly differentiated or mucinous carcinoma, lymphatic invasion, venous invasion, and tumor budding3–5. The 2020 Japanese Society for Cancer of the Colon and Rectum guidelines recommend surgical intervention for T1 CRC patients after endoscopic resection when at least one histologic risk factor is present6. While this approach minimizes the risk for relapse after endoscopic resection, it may also result in overtreatment because many T1 CRC patients without LNM undergo additional surgery. Indeed, overall relapse rate after endoscopic resection was reported to be 2.3–7.3%7, while the incidence of LNM for additional surgery was reported to be 9.7–11.9%8–10. Many studies have reported the utility of various histologic risk factors for predicting LNM11–13, but the interobserver disagreement among pathologists cannot be completely resolved14. To address these issues, a more reproducible and precise prediction system must be developed.
The application of machine-learning techniques including neural networks has expanded in the field of medicine15–18. Several machine-learning models for histologic images have been proposed for histologic diagnosis or predicting prognosis19–25. Most of these models utilize convolutional neural networks (CNN) for classifying histologic images because of their strong and stable tissue classification ability. One concern, however, is the lack of clarity regarding the relationship between histologic features and the decision process of CNN. When predicting prognosis for cancer patients, it is important for both clinicians and patients to understand why the machine made such a decision. A previous study revealed the relationship between histologic images and prognostic risk stratification of renal cancer by CNN26, but no similar studies have been performed for CRC concerning the relationship between histologic patterns and the predictive values of artificial intelligence.
Here we introduce a machine-learning algorithm for predicting LNM for T1 CRC with H&E-stained histologic whole-slide images (WSI), and clarify the decision processes for the predictive values by combining CNN and random forests (RF).
Results
Neural network machine-learning models and characteristic tile images
The accuracy and loss of image classifier #1 for classifying cancer and others (2 classes) were 0.980 and 0.039, respectively (Supplementary Fig. 1). When classifying all histologic-type classes, the accuracy and loss were 0.920 and 0.247, respectively. The accuracy and loss of image classifier #2 were 0.740 and 0.524, respectively. (Supplementary Fig. 2).
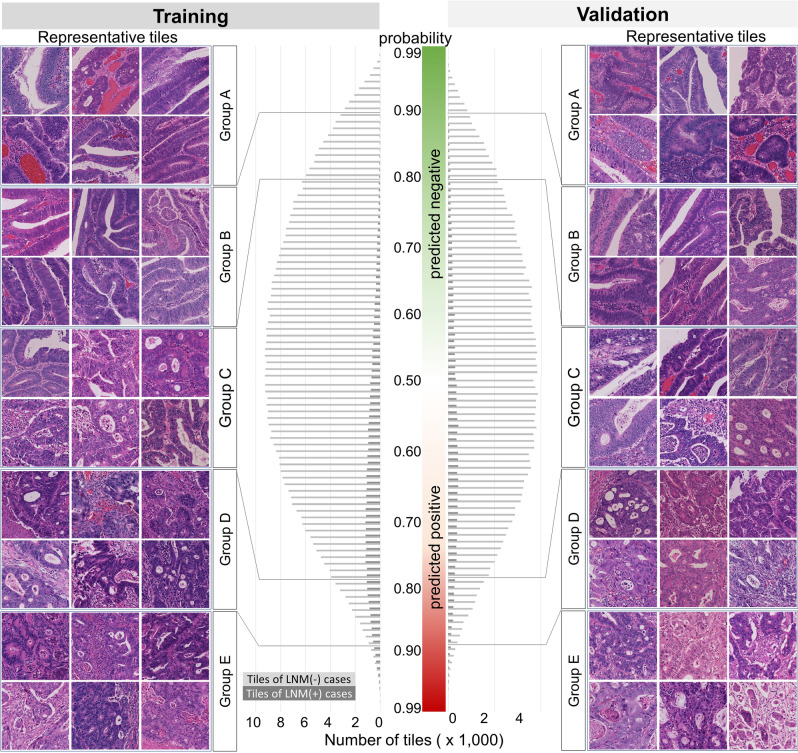
The number of tiles sorted by predictive probability for classifier #2 showed a normal distribution (Fig. 1), and the most frequent tile probabilities for the training and validation sets were 0.53 and 0.52 in LNM-negative cases, and 0.73 and 0.57 in LNM-positive cases, respectively (Fig. 1). Histologic characteristics of representative Group A tiles showed large cancer glands with little fibrotic stroma, and Group B tiles resembled those of Group A with more obvious structural atypia. In contrast, Group D tiles showed fused or cribriform cancer glands, and Group E tiles showed severe structural atypia with some isolated cancer cells embedded in desmoplastic stroma, resulting in an architectural complexity. In addition, the grade of nuclear atypia tended to be increased in Groups D and E compared with that in Groups A and B. Group C tiles, unconfidently judged by the classifier, showed a variety of histologic patterns. Importantly, both the training and validation datasets showed similar histologic trends, suggesting that classifier #2 returned the predictive value on the basis of an understandable histologic theory.
Figure 1.
The profiles of tile images in the training and validation datasets. Bars indicate the number of tiles for each probability rank. Representative tile images of the training set are shown on the left side, and those of the validation set are shown on the right side. Note the differences of histologic grade among the groups, which the tile images in Group A show well differentiated tubular adenocarcinoma, while those in Group E contain poorly differentiated cancer cell clusters with desmoplastic stroma. LNM, lymph node metastasis.
Predictive power of the random forest machine-learning model
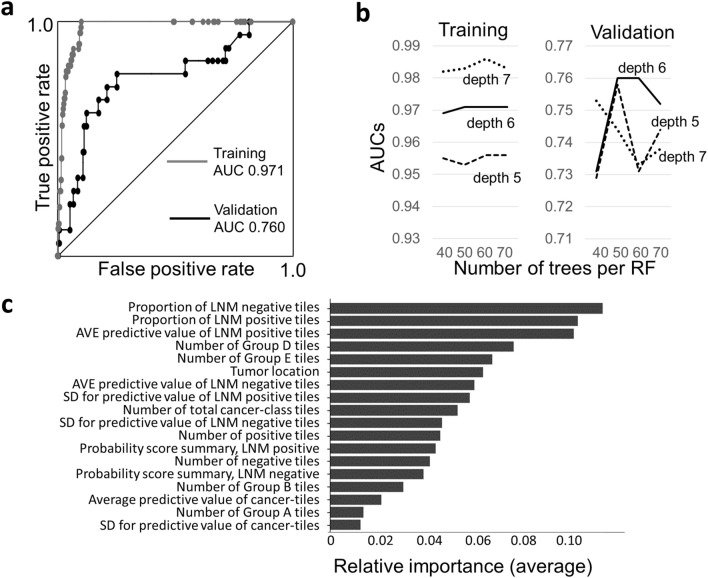
The area under the receiver operating characteristics curve (AUC) for predicting LNM in the training and validation sets were 0.971 and 0.760, respectively (Fig. 2a). The specificity and sensitivity were balanced when the RF score was 0.70, and the accuracy of the model for the training and validation sets was 0.91 and 0.75, respectively. The AUCs were highest when the depth and number of RF trees were 6 and 60, respectively (Fig. 2b). The important parameters for predicting LNM were the proportion of tiles predicted as LNM-negative or LNM-positive, average predictive probability of tiles, and number of Group D and E tiles (Fig. 2c). Tumor location was also important for predicting LNM. The differences in these parameters between LNM-negative and LNM-positive cases were statistically significant in the training set, but some differences were not statistically significant in the validation set (Table 1). The RF scores in the training set showed good risk stratification; 418 cases (76.3%) were classified as very-low risk with no metastatic cases (Table 2). In addition, 6 LNM-positive cases (5 training and 1 validation cases) with minimal conventional histologic risk factors (only deep submucosal invasion, > 1 mm) showed moderate to high risk, with RF scores ranging from 0.814 to 0.900. On the other hand, in the validation set, 4 LNM-positive cases were classified as very-low risk, resulting in a 2.4% LNM-positive rate (Table 2). For endoscopically resected cases, all of the LNM-positive cases in the training set (n = 5) were classified as moderate- or high-risk, while those in validation set (n = 1) were classified as very low-risk (Supplementary Table 1). These results indicated that the model successfully predicted LNM, but some validation cases were difficult to predict.
Figure 2.
Predictive accuracy of the random forest model. (a) Receiver operating characteristics (ROC) curves of the training and validation sets. (b) Area under the ROC curves (AUCs) for several conditions. The depth of decision trees for earning a maximum AUC was 7, while that of test set was 6, indicating overfitting of the training set with a depth of 7. RF, random forest. (c) Relative importance of 18 parameters. The importance was averaged for 20 random forests. LNM, lymph node metastasis; AVE, average; SD, standard deviation.
Table 1.
Comparison of important parameters for random forests.
| Training set | LNM(−) (n = 505) | LNM( +) (n = 43) | p-value |
|---|---|---|---|
| Proportion of LNM (−) tiles (%) | 50.18 ± 25.95 | 23.49 ± 15.32 | < 0.00001* |
| Proportion of LNM (+) tiles (%) | 49.82 ± 25.95 | 76.51 ± 15.32 | < 0.00001* |
| AVE predictive value of LNM (−) tiles | 0.644 ± 0.067 | 0.600 ± 0.025 | < 0.00001* |
| AVE predictive value of LNM (+) tiles | 0.640 ± 0.044 | 0.697 ± 0.052 | < 0.00001* |
| Number of Group D tiles | 32.67 ± 60.8 | 180.2 ± 253.6 | < 0.001* |
| Number of Group E tiles | 3.313 ± 9.738 | 32.68 ± 61.11 | 0.002* |
| Tumor location | |||
| C,A,T | 193 | 10 | 0.008** |
| D,S | 138 | 9 | |
| R | 174 | 24 | |
| Validation set | LNM(−) (n = 217) | LNM(+) (n = 18) | p-value |
|---|---|---|---|
| Proportion of LNM (−) tiles (%) | 46.06 ± 25.08 | 35.52 ± 25.38 | 0.057* |
| Proportion of LNM (+) tiles (%) | 53.94 ± 25.08 | 64.48 ± 25.38 | 0.057* |
| AVE predictive value of LNM (−) tiles | 0.638 ± 0.064 | 0.616 ± 0.050 | 0.058* |
| AVE predictive value of LNM (+) tiles | 0.644 ± 0.042 | 0.672 ± 0.056 | 0.031* |
| Number of Group D tiles | 32.36 ± 52.28 | 116.2 ± 172.4 | 0.031* |
| Number of Group E tiles | 3.152 ± 8.108 | 17.39 ± 29.87 | 0.033* |
| Tumor location | |||
| C,A,T | 69 | 4 | 1.00** |
| D,S | 77 | 8 | |
| R | 71 | 6 | |
Average ± standard deviation. LNM(−), lymph node metastasis negative; LNM(+), lymph node metastasis positive; AVE, average, C, cecum; A, ascending colon; T, transverse colon; D, descending colon; S, sigmoid colon; R, rectum. *Student t-test, ** non-rectum versus rectum, Fisher exact test.
Table 2.
Proportion of random forest (RF) scores.
| RF Scores | LNM-negative | LNM-positive | LNM (%) |
|---|---|---|---|
| Training set (n = 548) | |||
| 0–0.7 | 418 | 0 | 0 |
| 0.7–0.8 | 43 | 5 | 10.4 |
| 0.8–0.9 | 39 | 29 | 42.6 |
| 0.9– | 5 | 9 | 64.3 |
| Validation set (n = 235) | |||
| 0–0.7 | 162 | 4 | 2.4 |
| 0.7–0.8 | 26 | 3 | 10.3 |
| 0.8–0.9 | 27 | 9 | 25.0 |
| 0.9– | 2 | 2 | 50.0 |
LNM, lymph node metastasis.
Whole-slide mapping of representative cases
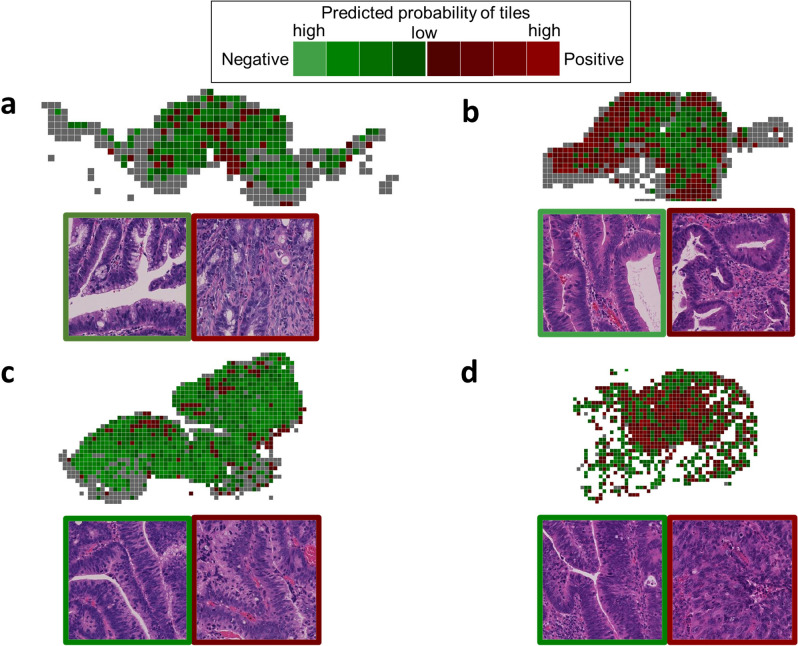
Cases with low RF scores had a small number of positive predicted tiles, while those with high RF scores had predominantly positive predicted tiles (Fig. 3). Cases that were LNM-negative but classified as high risk (i.e., false-positive) showed predominantly positive predicted tiles (Fig. 3b). In contrast, cases that were LNM-positive but classified as very-low risk (i.e., false-negative) tended to have many Group A and B tiles and few Group D and E tiles, resulting in low RF scores (Fig. 3c). Positive or negative predicted tiles tended to form clusters, but some single tiles existed within either cluster.
Figure 3.
Four representative validation cases. Color mapping of tiles and representative tile images are shown. (a) LNM negative case with RF score of 0.0000, (b) LNM negative case with RF score of 0.9117, (c) LNM positive case with RF score of 0.0001, (d) LNM positive case with RF score of 0.9143. The red tiles correspond to positive predicted, and green tiles correspond to negative predicted. The color brightness represents the probability of each tile: the brighter the tile, the higher the probability. Gray tiles are classified as non-tumor by classifier #1. Scales: 1 tile equals to 273-µm square.
Relationships between RF scores and conventional histologic risk factors
Cases were stratified by combining conventional histologic risk factors and their average RF scores are shown in Table 3. The lower the number of histologic risk factors, the lower the RF score. The important point is that the RF scores of LNM-positive cases with few conventional risk factors were consistently higher than those of LNM-negative cases, and some were statistically significant (Table 3). All of the LNM-positive cases harbor at least one risk factor, especially the deep submucosal invasion. There were 6 LNM-positive cases after endoscopic resection and all but one case showed moderate to high-risk (Supplementary Table 2A). On the other hand, LNM-positive cases with RF scores less than 0.8 harbor at least two histologic risk factors (Supplementary Table 2B). The average RF score was significantly higher in moderately and poorly differentiated tumors than in well differentiated tumors (Supplementary Table 3). When we compared cases with an RF score cutoff value of 0.7 and at least one histologic risk factor for conventional risk evaluation, the odds ratios for LNM were higher in both the training and validation sets than when using the conventional method (Supplementary Table 4A and 4B1). Though there were some false-negative cases in the machine learning model, the number of false-positive cases was low, resulting in better predictive odds. Similarly, the odds ratios of RF scores were higher than those when evaluating histologic-grade or poorly differentiated clusters. (Supplementary Table 4A, 4B2 and B3) Regarding conventional histologic-grade evaluation, interobserver disagreement was observed in some cancer images, while the machine learning model outputted correct predictive values for these cases (Supplementary Fig. 3).
Table 3.
Correlation between conventional histologic risk factors and RF scores.
| Histologic factors | Training set (n = 548) | p* | Validation set (n = 235) | p* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LNM | Average RF Score | LNM | Average RF Score | |||||||
| (-) | ( +) | LNM(-) | LNM( +) | (-) | ( +) | LNM(-) | LNM( +) | |||
| (None) | 111 | 0 | 0.0921 | NA | NA | 44 | 0 | 0.25269 | NA | NA |
| SM | 172 | 7 | 0.2559 | 0.8597 | < 0.001 | 65 | 1 | 0.23585 | 0.71252 | NA |
| Ly | 5 | 0 | 0.0960 | NA | NA | 2 | 0 | 0.00000 | NA | NA |
| V | 10 | 0 | 0.0855 | NA | NA | 4 | 0 | 0.17455 | NA | NA |
| Por | 0 | 0 | NA | NA | NA | 2 | 0 | 0.50594 | NA | NA |
| BD | 2 | 0 | 0.0000 | NA | NA | 0 | 0 | NA | NA | NA |
| SM, Ly | 17 | 1 | 0.2263 | 0.9371 | NA | 15 | 1 | 0.24825 | 0.53768 | NA |
| SM, V | 76 | 5 | 0.4162 | 0.8327 | < 0.001 | 29 | 3 | 0.47607 | 0.59520 | 0.359 |
| SM, Por | 7 | 0 | 0.3492 | NA | NA | 1 | 0 | 0.89556 | NA | NA |
| SM, BD | 18 | 0 | 0.3193 | NA | NA | 18 | 0 | 0.32070 | NA | NA |
| Ly, V | 0 | 0 | NA | NA | NA | 2 | 0 | 0.48984 | NA | NA |
| Ly, Por | 1 | 0 | 0.0000 | NA | NA | 0 | 0 | NA | NA | NA |
| Ly, BD | 1 | 0 | 0.0000 | NA | NA | 0 | 0 | NA | NA | NA |
| V, BD | 0 | 1 | NA | 0.9317 | NA | 0 | 0 | NA | NA | NA |
| Por, BD | 1 | 0 | 0.0667 | NA | NA | 0 | 0 | NA | NA | NA |
| SM, Ly, V | 13 | 6 | 0.5098 | 0.8708 | NA | 2 | 1 | 0.57474 | 0.91431 | NA |
| SM, Ly, Por | 3 | 2 | 0.4094 | 0.8615 | 0.066 | 1 | 0 | 0.72192 | NA | NA |
| SM, Ly, BD | 14 | 2 | 0.3477 | 0.8778 | < 0.001 | 5 | 2 | 0.19339 | 0.42831 | 0.335 |
| SM, V, Por | 2 | 0 | 0.0000 | NA | NA | 0 | 0 | NA | NA | NA |
| SM, V, BD | 17 | 2 | 0.4578 | 0.9114 | < 0.001 | 9 | 1 | 0.51281 | 0.00010 | NA |
| SM, Por, BD | 2 | 1 | 0.3222 | 0.8200 | NA | 3 | 0 | 0.28134 | NA | NA |
| Ly, Por, BD | 0 | 1 | NA | 0.8839 | NA | 2 | 0 | 0.34964 | NA | NA |
| V, Por, BD | 1 | 0 | 0.7717 | NA | NA | 0 | 0 | NA | NA | NA |
| SM, Ly, V, Por | 3 | 0 | 0.6002 | NA | NA | 1 | 0 | 0.82216 | NA | NA |
| SM, Ly, V, BD | 10 | 6 | 0.4314 | 0.8566 | 0.004 | 4 | 2 | 0.70850 | 0.79865 | 0.121 |
| SM, Ly, Por, BD | 9 | 1 | 0.5763 | 0.8752 | NA | 5 | 1 | 0.19467 | 0.75696 | NA |
| SM, V, Por, BD | 5 | 1 | 0.2986 | 0.8440 | NA | 1 | 0 | 0.28337 | NA | NA |
| SM, Ly, V, Por, BD | 5 | 7 | 0.5555 | 0.8759 | 0.052 | 3 | 5 | 0.50640 | 0.69660 | 0.274 |
LNM, lymph node metastasis; RF, random forest; SM, deep submucosal invasion; Ly, lymphatic invasion; V, venous invasion; Por, poorly differentiated clusters; BD, high-grade tumor budding; NA, not applicable. *Student t-test.
Discussion
We present an LNM prediction model for T1 CRC with explanatory histologic images for visualization of the basis of the predictive values. We combined CNN and RF to visualize the learning and deciding algorithms for predicting LNM, providing persuasive logic for pathologists and clinicians.
Prognostic prediction of CRC by artificial intelligence has been attempted using several approaches. Some researchers developed a machine-learning algorithm based on the conventional clinicopathologic information, including histologic factors of CRC27–29. These studies were performed using several machine-learning algorithms. A possible advantage of utilizing conventional clinicopathologic information is the readability of the analysis results, which clinicians can use to explain to patients why the machine returned certain results. This point is critical for the application of machine-learning techniques in the field of clinical medicine because understandable evidence is the most important factor for both patients and clinicians in making a clinical decision. On the other hand, diagnostic reproducibility is also critical for a stable prediction model. In particular, interobserver disagreement of histopathologic risk factors for predicting LNM of T1 CRC has remained problematic and may not be completely resolved. Because artificial intelligence can manage a huge amount of information, direct analysis of histologic images may contribute to developing a stable prediction model. Interestingly, our LNM prediction model mimicked histologic-grade evaluation for tile images. Since the interobserver disagreement for histologic grades is difficult to solve as described in previous reports30–32, and we also demonstrated in Supplementary Fig. 3, our model can provide more stable and reliable prediction results by skipping this evaluation step.
CNN is an ideal method for classifying histologic images into various categories that has been used for classifying histologic findings in many previous studies to predict gene alterations and patient prognosis18,21,23,33. Supervised machine-learning with appropriately labeled data enables complex histologic analyses by extracting features of colored images. Skrede et al.25 revealed that a combination of 2 neural networks for tile images can predict the prognosis of CRC through analyses of a large multicenter cohort. Most histologic analyses with CNN have been performed for small tile images with hundreds of pixels squares. Though it is difficult to understand the thorough process of CNN for returning its values, a visualizing method such as Grad-CAM may help to understand the histologic machine-learning theory33,34. This method may clarify the importance of the pixels within a tile image. Because most carcinomas, however, including CRC, show intra-tumoral histologic heterogeneity35, it is not enough to understand whole tumor characteristics by analyzing each tile. Learning how to extract features from a set of tile images is important to give clinicians and patients valuable information, such as a cancer prognosis.
RF has been used for predicting the prognosis of various cancers on the basis of radiologic and histopathologic images15,17,20. The classification ability of the RF score depends on the given parameters, such as number of tile images and predictive probabilities, and differs from that of classification by CNN, which requires only input images. RF can output the importance of parameters of any type, which plays critical role for converting characteristics of various tile images into a simple score through the understandable logic of decision trees. Histologic WSIs contain a lot of information, and most solid tumors exhibit histologic heterogeneity; thus, visualization of the decision process for predicting prognosis may provide a novel approach for histologic classification. In this study, CNN calculated and extracted characteristic images affecting the risk of metastasis (Group D and E), which contained poorly differentiated cancer clusters and desmoplastic stroma. Furthermore, RF indicates the proportion of LNM-positive or LNM-negative tiles, and their average probabilities were important for predicting LNM for cases. RF scores of our model tended to be higher in the LNM positive cases, indicating that this method may produce different results from the conventional method. Moreover, RF can calculate variables other than image features, such as tumor location, which provides researchers some additional options to improve the accuracy of the model. Tumor location is a risk stratification factor for T1 CRC13,36,37. Rectal cancers are more likely to metastasize than colon cancers36, and our results revealed similar tendency (Table 1). According to this fact, in our prediction model, RF indicated the importance of the tumor location, but no local predilection was demonstrated in the validation set (Table 1). This phenomenon may be attributed to the small number of LNM-positive cases in the validation set, having a negative effect on the LNM prediction. It is important to note that the large amount of outcome data for predicting LNM cases with RF was much smaller than the amount of data from millions of tile images, which require oversampling the data of LNM-positive cases to fill the tenfold gap against those of LNM-negative cases and avoid inappropriate predilection to the majority group. In contrast, the cancer tile images for the LNM-positive group numbered nearly 50 thousand, which seemed to be enough to avoid overfitting in CNN without over- or under-sampling, as indicated by the learning curves (Supplementary Fig. 1 and 2).
Because most T1 CRC patients are curable by surgical treatment even if overtreated, it could be worse if the tumor relapsed locally or by distant metastases7. In this context, false-negative cases should be minimized to avoid life-threatening relapse. In this study, we successfully developed a prediction model extracting 76.3% of truly negative case with low RF scores in the training set. On the other hand, 4 LNM-positive cases in the validation set were predicted to be very-low risk. All 4 of these cases contained many negative-prediction tiles; therefore, the most probable cause of the false-negative would be a lack of training tile images of LNM-positive cases with the features of these cases. To solve this matter, additional images of LNM-positive T1 CRC should be provided. The variability of input data among different institutions is also important. The quality of histological images depends on several factors such as the resection procedure, preparation of paraffin sections, staining procedure, and digitizing steps. In this study, we analyzed assembled tile images originating from H&E stained whole-slide images. The image quality or color balance may differ from that in other institutions, even if the staining procedures and digitizing steps are fully automated. These factors may decrease the performance of the prediction model and should be carefully evaluated before applying this method in clinical practice. Because the factors are quite complex and some of their variabilities are difficult to resolve, a multicenter validation study should be performed to clarify the differences of the factors, and to understand how to maintain the precision of the model in any situation. These are limitations of this single institutional study, and further investigation should be performed in a large-scale multicentral cohort study. Another limitation of this study is that the followed up patients with no LNM by periodic colonoscopy and/or abdominal computed tomography scanning for 5 years after initial resection may be misdiagnosed relapsing LNM, will result in false-negative or false-positive that the ground truth data can be changed in such cases.
In conclusion, we introduced a machine-learning model for predicting LNM of T1 CRC, which enabled risk-stratification of cases without any conventional histologic risk assessment. Our method will be a useful tool for clinicians and patients to understand the decision process of artificial intelligence through visualizing characteristic histologic images.
Methods
Case recruitment
A total of 855 consecutive T1 CRC resected either endoscopically or surgically at The Cancer Institute Hospital between 2005 and 2015, were initially recruited in this study. Of these 855 cases, 68 were excluded because of inadequate sample conditions (e.g., non-enbloc resections), and 787 cases underwent further analyses. Among the patients, 61 cases (7.8%) showed metastasis in dissected lymph nodes (LNM-positive). Among the patients treated with endoscopic resection, 119 patients (43.1%) underwent additional intestinal resection with lymph node dissection, and 157 (56.9%) patients were followed up for 5 years by periodic colonoscopy and/or abdominal computed tomography scanning, based on the guidelines of Japanese Society for Cancer of the Colon and Rectum6. The cases were randomly sorted by computer to divide the cases into a training set (n = 551) and test set (n = 236). The LNM rate was equivalent for both datasets. Patient characteristics is described in Supplementary Table 5. To develop the histologic-type classifier, we randomly recruited 22 T1 CRC cases and an additional 100 T2/3 CRC cases that were surgically resected in 2008 and 2009.
Histologic evaluation and digitizing
Serial 2-mm- to 5-mm-thick tissue sections of the whole lesion were cut from resected specimens fixed with 20% buffered formalin and embedded in paraffin; 3-μm-thick sections were then prepared for staining. Each section was stained with H&E. For each case, all cancer-containing sections showing submucosal invasion were included. Conventional histologic evaluation of the depth of submucosal invasion (≥ 1000 µm), poorly differentiated or mucinous carcinoma, lymphatic invasion, venous invasion, and tumor budding was performed by 2 pathologists (M.T, H.K), to disclose the clinicopathologic characteristics of the cases (Supplementary Table 5), basically according to the Japanese Society for Cancer of the Colon and Rectum criteria6. In addition, we also evaluated histologic grades considering predominant tumor component, as follows: well, moderately and poorly differentiated. We also performed histologic grading by 3 pathologists (M.T, N.Y, H.K) for representative cases to demonstrate interobserver disagreement. Deep submucosal invasion was defined as tumor invasion of the submucosal layer with a depth of at least 1000 µm. The presence of poorly differentiated clusters was evaluated according to the criteria of Ueno et al.38.
Whole-slide images (WSIs) of the sections were obtained by a digital slide scanner (NanoZoomer, Hamamatsu Photonics, Hamamatsu, Japan). The WSIs were cut into non-overlapping small tile images of 299-pixel squares (equal to a 273-µm square). To ensure that the image contained at least one nucleated cell, each tile image was temporarily converted to gray-scale (0–255), and the image containing the minimum pixel value of 110 was adopted for further image analysis (pixel thresholding, Fig. 4b).
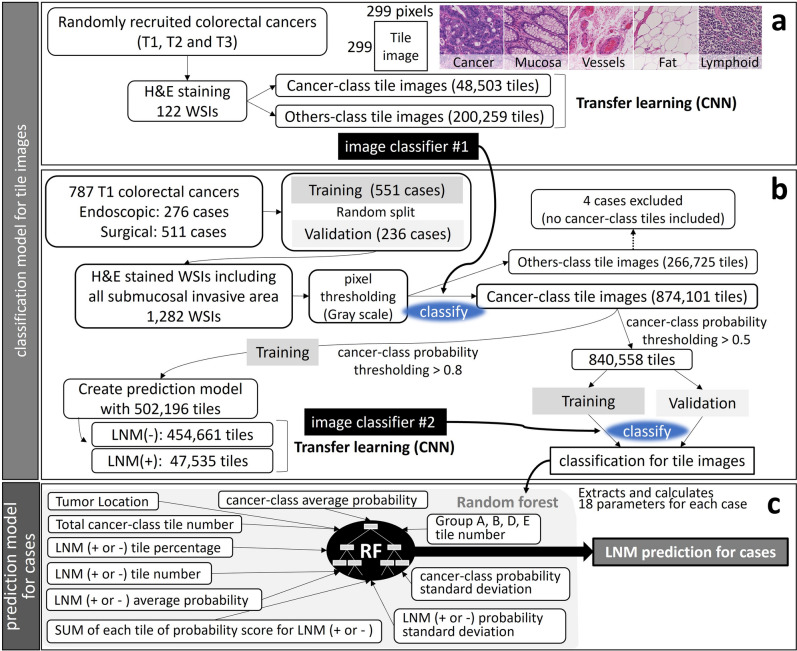
Figure 4.
Study workflow for developing lymph node metastasis prediction model for tile images and for the cases using 2 convolutional neural networks (CNN) and a random forest (RF) algorithm. (a) Creating image classifier #1 by a neural network with cancer or other tissue labeled tile images. (b) Creating image classifier #2 by re-training the neural network with lymph node metastasis positive or negative labeling based on patient outcome. LNM(−), negative for lymph node metastasis; LNM(+), positive for lymph node metastasis; SUM, summary. (c) Random forest classifier determines the predictive values for lymph node metastasis based on several parameters of the tile images.
Statistical analysis
The primary endpoint of this study was LNM, and 5-year disease-free survival was defined as no metastases for cases without surgical treatment. Receiver operating characteristics (ROC) curves were used to estimate the predictive power of the model. The balanced error rate of the ROC curve was considered an optimal cut-off value for determining the accuracy of the model. The area under the ROC curve (AUC) was defined as the predictive ability of the models. Qualitative factors were analyzed with the Fisher exact test and odds ratios were calculated. Quantitative factors were analyzed with the Student t-test. A P-value of less than 0.05 was considered statistically significant in both analyses. We conducted all analyses using R version 3.6.239.
Neural network machine-learning models
First, we created a histologic-type classifier by CNN to enable the efficient selection of cancer tiles. The tile images from 122 WSIs of randomly selected CRCs were labeled with “cancer” or 9 other labels, including non-tumoral mucosa, hyperplastic mucosa, adenoma, lymphoid tissue, smooth muscle, vessels, fat, nerve, and non-material background. The number of cancer-labeled images was 48,503, and that of the others was 200,259. A deep-learning pre-trained model, MobileNet V240, was re-trained to recognize 10 different tissue types to develop image classifier #1 (Fig. 4a). Labeling of tile images originating from WSIs was performed by 1 pathologist (M.T), using OpenSlide library running on Python 3.741.
Second, the tile images from 1282 WSIs of T1 CRC underwent pixel thresholding and were classified with image classifier #1. Each image was re-labeled by the classifier with the probable histologic type and probability for classifying. Four cases were excluded because no cancer-class tiles were detected, therefore 548 training cases and 235 validation cases were applied to further analyses. Among the cancer-class images, we adopted images with a probability > 0.8 to create the LNM prediction model. These tile images were re-labeled again with the primary endpoint and re-trained to develop image classifier #2, which classified whether or not a cancer tile image was likely to metastasize.
Then, cancer-class tile images of both the training set and validation set with a probability > 0.5 (840 558 tiles) were classified by image classifier #2 and re-labeled with the probable LNM status and probability for metastasis. We defined the tile images according to the probability as follows: those with non-metastatic probability > 0.9 as Group A, those with non-metastatic probability between 0.8 and 0.9 as Group B, those with metastatic probability between 0.8 and 0.9 as Group D, those with metastatic probability > 0.9 as Group E, and those with either probability < 0.8 as Group C. We also defined probability score summary of CNN, which can be obtained by multiplying the cancer-class probability with the metastatic probability for each tile, and summarized for case. We tuned the classifiers by learning iteration and learning rate to obtain maximum accuracy and minimum loss while re-training. The loss was evaluated by cross entropy. The workflow from WSIs to complete classifying tile images is shown in Fig. 4b.
Random-forest machine learning model
To establish the LNM prediction model for the cases, we loaded data from the tile images of the training set onto a machine-learning tool, the Scikit-learn on Python 3.6. We selected a random forest (RF) classifier machine-learning algorithm to minimize the effect of overfitting. Because the number of LNM-positive (n = 43) and LNM-negative (n = 505) cases in the training dataset had a tenfold gap, we randomly oversampled LNM-positive cases to equalize the number of cases applied to the RF model. The data contained 18 parameters as follows: tumor location; total number of cancer-class tiles; number of tiles classified as metastatic or non-metastatic; number of Group A, B, D, E tiles; percentages of tiles classified as metastatic or non-metastatic; average probabilities; standard deviations of cancer-class probabilities and metastatic or non-metastatic probabilities; and probability score summary for each tile (Fig. 4c). Tumor locations were grouped into 3 parts as follows; (1) cecum, ascending and transverse colon, (2) descending and sigmoid colon, and (3) rectum. The RF algorithm used was based on a previous report42,43. In brief, the RF randomly collected these parameters for creating dozens of decision trees. Teacher data of the primary endpoint were given to the RF, and the importance of parameters that minimize the impurity for separating LNM-positive or LNM-negative cases was calculated. The importance of each parameter was averaged in all the trees, and finally, the output predictive value of LNM for the case was determined. To reduce false-negative metastatic cases, we established 500 RFs to obtain a higher AUC for each RF and adopted the top 20 RFs for case analyses. The importance of the parameters of these RFs was averaged to show how the algorithm determined the score. The RFs returned values of metastatic probability between 0 and 1, and the maximum value of each case was adopted as the RF score. We tuned the RF to maximize the AUC for predicting LNM in all cases by adjusting the number and depth of the trees. We defined the RF scores as follows: 0–0.7, very-low risk; 0.7–0.8, low risk; 0.8–0.9, moderate risk; and 0.9–1.0, high risk.
Ethics
The experimental protocols in this study were approved by the Ethics Committee of the Cancer Institute, Japanese Foundation for Cancer Research (approved number: 2020-1045). The written informed consent was obtained from all participants. This study was performed in accordance with the Declaration of Helsinki.
Supplementary Information
Acknowledgements
The authors thank Tomoyo Kakita, Motoyoshi Iwakoshi, and Hiroshi Yoko-o for their excellent technical assistance.
Author contributions
M.T. conceived the study; and managed the data acquisition, digitizing whole-slides, and data labeling. N.Y. and M.T. contributed to design the machine learning program and methodology, classifier model, data analysis, and manuscript drafting. H.K. and K.N. supervised the conventional histologic assessments. S.S. and Y.F. provided clinical information including patients’ outcome. K.T., N.Y. and H.K. supervised all the analyses and contributed to manuscript editing. All authors approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07038-1.
References
- 1.Urabe Y, Tanaka S, Saito Y, et al. Impact of revisions of the JSCCR guidelines on the treatment of T1 colorectal carcinomas in Japan. Z. Gastroenterol. 2015;53:291–301. doi: 10.1055/s-0034-1385764. [DOI] [PubMed] [Google Scholar]
- 2.Marin-Gabriel JC, Fernandez-Esparrach G, Diaz-Tasende J, Herreros de Tejada A. Colorectal endoscopic submucosal dissection from a Western perspective: today's promises and future challenges. World J. Gastrointest. Endosc. 2016;8:40–55. doi: 10.4253/wjge.v8.i2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tateishi Y, Nakanishi Y, Taniguchi H, Shimoda T, Umemura S. Pathological prognostic factors predicting lymph node metastasis in submucosal invasive (T1) colorectal carcinoma. Mod. Pathol. 2010;23:1068–1072. doi: 10.1038/modpathol.2010.88. [DOI] [PubMed] [Google Scholar]
- 4.Kawachi H, Eishi Y, Ueno H, et al. A three-tier classification system based on the depth of submucosal invasion and budding/sprouting can improve the treatment strategy for T1 colorectal cancer: a retrospective multicenter study. Mod. Pathol. 2015;28:872–879. doi: 10.1038/modpathol.2015.36. [DOI] [PubMed] [Google Scholar]
- 5.Egashira Y, Yoshida T, Hirata I, et al. Analysis of pathological risk factors for lymph node metastasis of submucosal invasive colon cancer. Mod. Pathol. 2004;17:503–511. doi: 10.1038/modpathol.3800030. [DOI] [PubMed] [Google Scholar]
- 6.Hashiguchi Y, Muro K, Saito Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int. J. Clin. Oncol. 2020;25:1–42. doi: 10.1007/s10147-019-01485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitoh Y, Inaba Y, Sasaki T, et al. Management of colorectal T1 carcinoma treated by endoscopic resection. Dig. Endosc. 2016;28:324–329. doi: 10.1111/den.12503. [DOI] [PubMed] [Google Scholar]
- 8.Nishimura T, Oka S, Tanaka S, et al. Clinical significance of immunohistochemical lymphovascular evaluation to determine additional surgery after endoscopic submucosal dissection for colorectal T1 carcinoma. Int. J. Colorectal. Dis. 2021;36:949–958. doi: 10.1007/s00384-020-03795-5. [DOI] [PubMed] [Google Scholar]
- 9.Iguchi K, Mushiake H, Aoyama T, et al. Additional surgical resection after endoscopic resection for patients with high-risk T1 colorectal cancer. In Vivo. 2019;33:1243–1248. doi: 10.21873/invivo.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita K, Oka S, Tanaka S, et al. Preceding endoscopic submucosal dissection for T1 colorectal carcinoma does not affect the prognosis of patients who underwent additional surgery: a large multicenter propensity score-matched analysis. J. Gastroenterol. 2019;54:897–906. doi: 10.1007/s00535-019-01590-w. [DOI] [PubMed] [Google Scholar]
- 11.Harris EI, Lewin DN, Wang HL, et al. Lymphovascular invasion in colorectal cancer: an interobserver variability study. Am. J. Surg. Pathol. 2008;32:1816–1821. doi: 10.1097/PAS.0b013e3181816083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puppa G, Senore C, Sheahan K, et al. Diagnostic reproducibility of tumour budding in colorectal cancer: a multicentre, multinational study using virtual microscopy. Histopathology. 2012;61:562–575. doi: 10.1111/j.1365-2559.2012.04270.x. [DOI] [PubMed] [Google Scholar]
- 13.Takamatsu M, Kawachi H, Yamamoto N, et al. Immunohistochemical evaluation of tumor budding for stratifying T1 colorectal cancer: optimal cut-off value and a novel computer-assisted semiautomatic method. Mod. Pathol. 2019;32:675–683. doi: 10.1038/s41379-018-0189-1. [DOI] [PubMed] [Google Scholar]
- 14.Kai K, Aishima S, Aoki S, et al. Cytokeratin immunohistochemistry improves interobserver variability between unskilled pathologists in the evaluation of tumor budding in T1 colorectal cancer. Pathol. Int. 2016;66:75–82. doi: 10.1111/pin.12374. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Zhu G, Jiang X, et al. Survival prediction in gallbladder cancer using CT based machine learning. Front. Oncol. 2020;10:604288. doi: 10.3389/fonc.2020.604288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klauschen F, Müller KR, Binder A, et al. Scoring of tumor-infiltrating lymphocytes: From visual estimation to machine learning. Semin. Cancer Biol. 2018;52:151–157. doi: 10.1016/j.semcancer.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Cheng L, Li L, Wang L, et al. A random forest classifier predicts recurrence risk in patients with ovarian cancer. Mol. Med. Rep. 2018;18:3289–3297. doi: 10.3892/mmr.2018.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bychkov D, Linder N, Turkki R, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018;8:3395. doi: 10.1038/s41598-018-21758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma B, Guo Y, Hu W, et al. Artificial intelligence-based multiclass classification of benign or malignant mucosal lesions of the stomach. Front. Pharmacol. 2020;11:572372. doi: 10.3389/fphar.2020.572372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montazeri M, Montazeri M, Montazeri M, Beigzadeh A. Machine learning models in breast cancer survival prediction. Technol. Health Care. 2016;24:31–42. doi: 10.3233/THC-151071. [DOI] [PubMed] [Google Scholar]
- 21.Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA. 2018;115:E2970–E2979. doi: 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen PC, Gadepalli K, MacDonald R, et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med. 2019;25:1453–1457. doi: 10.1038/s41591-019-0539-7. [DOI] [PubMed] [Google Scholar]
- 25.Skrede OJ, De Raedt S, Kleppe A, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 26.Wulczyn E, Steiner DF, Xu Z, et al. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE. 2020;15:e0233678. doi: 10.1371/journal.pone.0233678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudo SE, Ichimasa K, Villard B, et al. Artificial intelligence system to determine risk of T1 colorectal cancer metastasis to lymph node. Gastroenterology. 2020;160:1075. doi: 10.1053/j.gastro.2020.09.027. [DOI] [PubMed] [Google Scholar]
- 28.Jiang D, Liao J, Duan H, et al. A machine learning-based prognostic predictor for stage III colon cancer. Sci. Rep. 2020;10:10333. doi: 10.1038/s41598-020-67178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Ju L, Tong J, Zhou CM, Yang JJ. Machine learning algorithms for predicting the recurrence of stage IV colorectal cancer after tumor resection. Sci. Rep. 2020;10:2519. doi: 10.1038/s41598-020-59115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komuta K, Batts K, Jessurun J, et al. Interobserver variability in the pathological assessment of malignant colorectal polyps. Br. J. Surg. 2004;91:1479–1484. doi: 10.1002/bjs.4588. [DOI] [PubMed] [Google Scholar]
- 31.Ueno H, Kajiwara Y, Shimazaki H, et al. New criteria for histologic grading of colorectal cancer. Am. J. Surg. Pathol. 2012;36:193–201. doi: 10.1097/PAS.0b013e318235edee. [DOI] [PubMed] [Google Scholar]
- 32.Barresi V, Bonetti LR, Ieni A, et al. Histologic grading based on counting poorly differentiated clusters in preoperative biopsy predicts nodal involvement and pTNM stage in colorectal cancer patients. Hum. Pathol. 2014;45:268–275. doi: 10.1016/j.humpath.2013.07.046. [DOI] [PubMed] [Google Scholar]
- 33.Iizuka T, Fukasawa M, Kameyama M. Deep-learning-based imaging-classification identified cingulate island sign in dementia with Lewy bodies. Sci. Rep. 2019;9:8944. doi: 10.1038/s41598-019-45415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai G, He W, Xu L, et al. Exploring the effect of hypertension on retinal microvasculature using deep learning on East Asian population. PLoS ONE. 2020;15:e0230111. doi: 10.1371/journal.pone.0230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones HG, Jenkins G, Williams N, et al. Genetic and epigenetic intra-tumour heterogeneity in colorectal cancer. World J. Surg. 2017;41:1375–1383. doi: 10.1007/s00268-016-3860-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aytac E, Gorgun E, Costedio MM, et al. Impact of tumor location on lymph node metastasis in T1 colorectal cancer. Langenbecks Arch. Surg. 2016;401:627–632. doi: 10.1007/s00423-016-1452-x. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi H, Mochizuki H, Morita T, et al. Characteristics of recurrence after curative resection for T1 colorectal cancer: Japanese multicenter study. J. Gastroenterol. 2011;46:203–211. doi: 10.1007/s00535-010-0341-2. [DOI] [PubMed] [Google Scholar]
- 38.Ueno H, Hashiguchi Y, Kajiwara Y, et al. Proposed objective criteria for "grade 3" in early invasive colorectal cancer. Am. J. Clin. Pathol. 2010;134:312–322. doi: 10.1309/AJCPMQ7I5ZTTZSOM. [DOI] [PubMed] [Google Scholar]
- 39.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2013).
- 40.Mark Sandler, A.H., Menglong, Z., Andrey, Z., Liang-Chieh, C. MobileNetV2: inverted residuals and linear bottlenecks. In The IEEE Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018).
- 41.OpenSlide library for Python, version 1.1.2. https://openslide.org/download/. 2020.
- 42.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 43.Takamatsu M, Yamamoto N, Kawachi H, et al. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput. Methods Prog. Biomed. 2019;178:155–161. doi: 10.1016/j.cmpb.2019.06.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.