Abstract
Multiple-drug-resistant Mycobacterium tuberculosis (MDR-MTB) has been well studied in hospitals or health care institutions and in human immunodeficiency virus-infected populations. However, the characteristics of MDR-MTB in the community have not been well investigated. An understanding of its prevalence and circulation within the community will help to estimate the problem and optimize the strategies for control and prevention of its development and transmission. In this study, MDR-MTB isolates from Scotland collected between 1990 and 1997 were characterized, along with non-drug-resistant isolates. The results showed that they were genetically diverse, suggesting they were unrelated to each other and had probably evolved independently. Several new alleles of rpoB, katG, and ahpC were identified: rpoB codon 525 (ACC→AAC; Thr525Asn); katG codon 128 (CGG→CAG; Arg128Gln) and codon 291 (GCT→CCT; Ala291Pro); and the ahpC synonymous substitution at codon 6 (ATT→ATC). One of the MDR-MTB isolates from an Asian patient had an IS6110 restriction fragment length polymorphism pattern very similar to that of the MDR-MTB W strain and had the same drug resistance-related alleles but did not have any epidemiological connection with the W strains. Additionally, a cluster of M. tuberculosis isolates was identified in our collection of 715 clinical isolates; the isolates in this cluster had genetic backgrounds very similar to those of the W strains, one of which had already developed multiple drug resistances. The diverse population of MDR-MTB in Scotland, along with a low incidence of drug-resistant M. tuberculosis, has implications for the control of the organism and prevention of its spread.
Multiple-drug-resistant Mycobacterium tuberculosis (MDR-MTB), which is defined as isolates resistant to both isoniazid (INH) and rifampin (RIF)—the frontline antitubercular agents and the backbone of current antituberculosis treatment regimens—is worsening the global tuberculosis emergency and has caused deep concern worldwide. Understanding the causation, genetic mechanisms, and transmission of MDR-MTB will be of great value in optimizing the strategies to control and prevent its development and transmission. Recent molecular-genetic analyses of drug-resistant M. tuberculosis have disclosed a number of resistance mechanisms. For example, more than 95% of RIF-resistant M. tuberculosis isolates have mutations in rpoB, the gene encoding the RNA polymerase β-subunit (25). Three genes have been implicated in INH resistance: katG, encoding catalase-peroxidase, which transforms INH into its active form (41); inhA, encoding a putative mycolic acid synthesis enzyme involved in cell wall synthesis (2); and ahpC, encoding alkyl hydroperoxidase, which functions as a component of antioxidant reductase (32). The W strain of M. tuberculosis, first isolated in New York City in 1992, has resistance to INH, ethambutol (EMB), RIF, and streptomycin (STR) and has a characteristic IS6110 restriction fragment length polymorphism (RFLP) banding pattern; subsequently, variants with a few IS6110 RFLP differences were isolated (4, 13, 24) while other variants were found which were susceptible to antituberculosis drugs.
Unlike many bacterial species which can acquire antibiotic resistance genes by transduction, conjugation, or transformation, there is no evidence so far of the mobilization of genes between M. tuberculosis isolates; indeed, all resistances so far characterized have been genomically based. As a result, analysis of the alleles of drug resistance genes and of their associations will provide useful information on the generation and transmission of resistant isolates and will also provide insight into the population dynamics of M. tuberculosis in response to the selective pressure of antimycobacterial agents. So far the great majority of investigations of MDR-MTB have been of hospital- or health care institute-based outbreaks (references 1, 3, 4, 15, and 16 and reference 14 and references therein), and these have provided valuable information on MDR-MTB transmission and its vulnerable subjects. The few non-outbreak- or community-based investigations have tended to use IS6110 RFLP alone as a genetic marker for the characterization of the drug-resistant populations (27, 28, 38).
In order to have a genetic insight into the MDR-MTB population in Scotland, we have studied some of the drug resistance genes of the MDR-MTB isolates collected in Scotland between 1990 and 1997, including rpoB, katG, inhA, and ahpC. These isolates were also characterized by IS6110 RFLP and other genetic markers, together with drug-sensitive isolates collected at the same time. The results indicated that the MDR-MTB isolates in Scotland are genetically diverse. This, in combination with a low rate of primary drug resistance in Scotland, lead to some thoughts on the global control of MDR-MTB.
MATERIALS AND METHODS
Isolates.
A total of 715 isolates of M. tuberculosis, including 10 MDR-MTB isolates, were studied; all of them were received by the Scottish Mycobacteria Reference Laboratory in Edinburgh, Scotland, from 1990 to 1997.
Antimicrobial susceptibility test.
Drug sensitivity testing was performed with a Bactec radiometric system (Becton Dickinson, Paramus, N.J.) with the following antimicrobial agents: INH, 0.1 μg/ml; EMB, 2.0 μg/ml; RIF, 0.5 μg/ml; pyrazinamide (PZA), 100 μg/ml; ciprofloxacin, 2.0 μg/ml; rifabutin, 0.2 μg/ml; amikacin, 2.0 μg/ml; STR, 4.0 μg/ml; capreomycin, 8.0 μg/ml; clofazimine, 0.8 μg/ml; prothionamide, 2.0 μg/ml; clarithromycin, 4.0 μg/ml; and sparfloxacin, 2.0 μg/ml (33). All 715 isolates were tested for susceptibility to INH, RIF, EMB, and PZA. In the case of isolates resistant to any of these, further drug susceptibility tests were performed.
PCR-based single-strand conformational polymorphism (PCR-SSCP) analysis.
Conventional PCR was used to amplify rpoB, katG, inhA, and ahpC genes, and the products were checked on agarose gels. One microliter of the PCR product was mixed with 2 μl of denaturing buffer (95% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol, 20 mM EDTA), and the mixture was heated in a heated-lid thermocycler (WellTemp, Cambridge, United Kingdom) for 5 min at 95°C and then immediately cooled in ice water for 5 min. The denatured PCR products were separated via electrophoresis on a PhastSystem (Pharmacia Biotech AB, Uppsala, Sweden) on PhastGel 8 to 25% gradient gel (Pharmacia Biotech) with PhastGel DNA buffer strips (Pharmacia Biotech). The separation program has two steps: a sample application step (100 V; 4 mA; 1.0 W; 15°C; 10 Vh) and a sample separation step (400 V; 10 mA; 2.0 W; 15°C; 500 Vh). After separation, the gels were stained with PhastGel DNA silver-staining kit (Pharmacia Biotech) and air dried for about 3 h, according to the manufacturer’s instructions.
DNA sequencing and DNA sequence analysis.
DNA was sequenced with an Applied Biosystems (Warrington, United Kingdom) 377A automated DNA sequencer with a Prism-Ready mix kit based on Ampli-Taq CS and polymerase. The programs in the Genetics Computer Group package (version 8.1) used in this study for DNA sequence analysis were GAP (26) and BESTFIT (34).
IS6110 RFLP analysis.
IS6110 RFLP analysis was performed according to the recommended method with some modifications (11, 37). Briefly, the M. tuberculosis genomic DNA was digested with PvuII, subjected to agarose gel electrophoresis, and subsequently blotted onto a nylon membrane. After hybridization with a digoxigenin (DIG)-labelled IS6110 DNA probe, the hybridization bands were detected by the DIG detection procedure. The IS6110 probe was labelled in a PCR with DIG-dUTP (Boehringer Mannheim GmbH, Mannheim, Germany). PvuII-digested supercoiled DNA ladder (Gibco-BRL, Life Technologies Ltd. Paisley, United Kingdom) and φX174-HaeIII DNA (Advanced Biotechnologies, London, United Kingdom) were DIG labelled with a random-primed DNA-labelling method. Pairwise similarities of IS6110 fingerprint patterns were calculated by the Dice coefficient of similarity with GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium).
Polymorphism analysis of codon 463 in katG.
The polymorphism analysis was conducted by PCR-based RFLP with primers K5 and K6 and endonuclease restriction enzyme NciI (Boehringer Mannheim GmbH).
RESULTS
Susceptibility patterns.
Ten of the 12 MDR-MTB isolates and their susceptibility patterns are shown in Table 1.
TABLE 1.
Patterns of antimicrobial susceptibility of MDR-MTB isolates and corresponding genetic mutations detected
| Drug or mutation | Susceptibility or characteristics of isolatea:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 9002 | 9202 | 9208 | 9214 | 9219 | 9310 | 9511 | 9604 | 2022 | 1238 | |
| INH | R | R | R | R | R | R | R | R | R | R |
| EMB | R | s | s | R | s | s | R | s | R | s |
| RIF | R | R | R | R | R | R | R | R | R | R |
| PZA | R | s | s | s | s | s | R | R | R | s |
| Rifabutin | R | R | R | R | s | R | R | R | R | s |
| Ciprofloxacin | R | s | s | s | s | s | s | s | s | |
| Amikacin | R | s | s | s | s | |||||
| STR | R | s | s | R | R | |||||
| Capreomycin | s | s | s | s | ||||||
| Clofazimine | s | R | s | s | ||||||
| Prothionamide | R | R | R | s | s | |||||
| Clarithromycin | R | R | R | R | s | R | R | |||
| Sparfloxacin | s | |||||||||
| katGb | X | n | n | n | n | X | X | X | n | X |
| Codon 463 in katG | Arg | Arg | Arg | Arg | Arg | Arg | Leu | Leu | Arg | Arg |
| inhA | n | n | X | n | X | n | n | n | n | n |
| aphC | n | X | n | n | n | n | n | n | X | n |
| rpoBc | X | X | X | X | X | X | X | X | X | X |
IS6110 RFLP patterns.
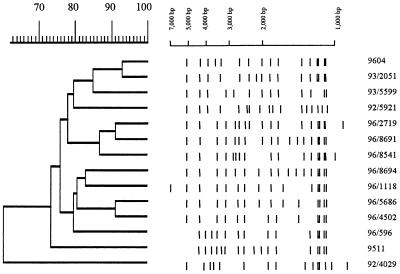
A total of 715 clinical isolates of M. tuberculosis collected from 1990 to 1997, including the 10 MDR-MTB isolates, were subjected to IS6110 RFLP analysis. The patterns of the MDR-MTB isolates were diverse (Fig. 1): two pairs of isolates (isolates 9208 and 9219 and isolates 9511 and 9604) had more than 80% similarity by the Dice coefficient (8) but still had three and five differences in IS6110-hybridizing bands within each pair, respectively. IS6110 RFLP patterns can be used to efficiently group together genetically related isolates of M. tuberculosis (11). A dendrogram based on the RFLP pattern similarities of the 715 isolates also indicated that the 10 MDR-MTB isolates did not cluster together but were generally scattered throughout the dendrogram, except isolates 9511 and 9604, suggesting that they were indeed not genetically closely related strains but had evolved independently. Using the published IS6110 pattern of the W strain (4, 24) as a reference, isolate 9511 was found to be very similar, and this isolate was also resistant to INH, EMB, RIF, and STR (Table 1). In the 715-isolate dendrogram, isolates 9511 and 9604 clustered together with 11 other isolates at greater than 75% similarity (Fig. 2).
FIG. 1.
IS6110 RFLP patterns of the 10 MDR-MTB isolates.
FIG. 2.
Schematic presentation of the IS6110 RFLP patterns and dendrogram of the subcluster containing isolates 9511 and 9604 from the database containing 715 M. tuberculosis isolates. Isolate 92/4029 from an adjacent subcluster was used as an out group.
rpoB polymorphism.
The causative mutation of many RIF-resistant mutants has been mapped to the rpoB gene, the vast majority of these being in the region from codon 505 to codon 353 (25). Accordingly, the DNA sequence of this region was determined for all 10 of the isolates from a PCR product amplified across the region (primers TR7 and RpoB). Subsequent sequencing revealed that all 10 of the MDR-MTB isolates had missense mutations in the region (Tables 2 and 3).
TABLE 2.
Mutations detected in rpoB of the MDR-MTB isolates
| Codona | Nucleotide mutation | Amino acid changes | Isolate |
|---|---|---|---|
| 505 | TCC→TTG | Phe→Leu | 9208 |
| 511 | CTG→CCG | Leu→Pro | 9002 |
| 516 | GAC→GCC | Asp→Ala | 9002 |
| 525 | ACC→AAC | Thr→Asn | 9202 |
| 526 | CAC→TAC | His→Tyr | 9310 |
| 526 | CAC→CTC | His→Leu | 1238 |
| 531 | TCG→TGG | Ser→Trp | 9202, 9214, 9511, 9604, 2022 |
| 533 | CTG→CCG | Leu→Pro | 9219 |
TABLE 3.
DNA primers used in this study
| Primer | Sequence | Descriptiona |
|---|---|---|
| TR7 | 5′-GGGAGCGGATGACCACCCA-3′ | nt 2266–2284 of L27989 |
| RpoB2 | 5′-CGATCGGCGATTGGCCTGTG-3′ | nt 2689–22709 in the complementary strand of L27989 |
| Kat1 | 5′-ACTACGGGCCGCTGTTTATCC-3′ | nt 2268–2288 of X68081 |
| Kat2 | 5′-TACCGCTGTAACGCTCATCGC-3′ | nt 2595–2615 of the complementary strand of X68081 |
| Kat3 | 5′-GCGGTCACACTTTCGGTAAGA-3′ | nt 2781–2801 |
| Kat4 | 5′-ACCCGCAGCGAGAGGTCAGTG-3′ | nt 3115–3135 of the complementary strand of X68081 |
| Kat5 | 5′-GGGCATCGGGATTGACTGTCT-3′ | nt 3366–3386 |
| Kat6 | 5′-CCGCCTTTGCTGCTTTCTCTA-3′ | nt 3633–3653 of the complementary strand of X68081 |
| Kat7 | 5′-CAACATCACGGTGCCCTTCAC-3′ | nt 3661–3681 |
| Kat8 | 5′-GGCGAGGGCTCCCAGGTGATA-3′ | nt 3973–3993 of the complementary strand of X68081 |
| InhA1 | 5′-GCTGAGTCACACCGACAAACG-3′ | nt 34–54 of U41388 |
| InhA2 | 5′-CCAGGACTGAACGGGATACGA-3′ | nt 200–220 of the complementary strand of U41388 |
| InhA3 | 5′-GCAAAACGAGGAGCACCTGGC-3′ | nt 1119–1139 of U41388 |
| InhA4 | 5′-AATACGCCGAGATGTGGATGC-3′ | nt 1280–1300 of the complementary strand of U41388 |
| AhpC1 | 5′-CTTGCGGCACTGCTGAACCAC-3′ | nt 543–563 of U16243 |
| AhpC2 | 5′-ACAGGTCACCGCCGATGAGAG-3′ | nt 786–806 of the complementary strand of U16243 |
nt, nucleotide.
Polymorphism in katG, inhA and ahpC.
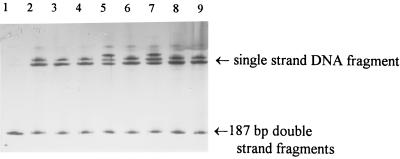
PCR-SSCP was performed on all of the PCR products from katG, inhA, and ahpC to identify mutant sequences, which were then subjected to DNA sequencing. An example of the PCR-SSCP polymorphic pattern from inhA is illustrated in Fig. 3.
FIG. 3.
PCR-SSCP patterns of the partial inhA locus of isolates with primers InhA1 and InhA2. Lane 1, no denaturing DNA of drug-susceptible isolate 8 as nondenatured DNA control; lane 2, denaturing DNA of isolate 8 as denatured DNA control; lanes 3 to 9, DNA from isolates 9002, 9202, 9208, 9214, 9219, 9310, and 9511. Clear shifts of single-stranded DNA can be seen in lanes 5 and 7, which were confirmed by DNA sequencing to be due to single-nucleotide mutations (see the text for details).
Four pairs of primers (Table 3) were designed to span the region of the katG coding region that has been reported to harbor most of the mutations. The mutations found in the isolates had several features (Table 4). Firstly, 5 of the 10 isolates were found to have mutations in katG; all of them were missense substitutions and three of them were at codon 315, a preponderance which has been noted by others (references 9 and 25 and references therein) and which has been demonstrated to confer high INH resistance (29). Secondly, two new mutations were identified, one of them at codon 128 (CGG→CAG; Ala128Gln) and the other at codon 291 (GCT→CCT; Ala291Pro).
TABLE 4.
Mutations detected in katG of MDR-MTB isolates
The Arg-Leu polymorphism at codon 463 of the katG gene has been demonstrated not to confer INH resistance but has been proposed as an evolutionarily stable genetic marker (36). Two isolates (isolates 9511 and 9604) had the codon for leucine; the others had the arginine codon when the 10 isolates were screened by the PCR-based RFLP.
inhA was examined with one primer pair (InhA1-InhA2) amplifying across the putative inhA regulatory region and another primer pair (InhA3-InhA4) amplifying across codon 280. Among the 10 isolates, none was found to have a mutation at codon 280. Two isolates (isolates 9208 and 9219) were identified that had a single-nucleotide substitution 1 nucleotide upstream of the putative ribosome binding site (nucleotide 148 [EMBL accession no. U41388]), and these two isolates did not have any mutations in their katG genes.
Mutations were found at the ahpC locus in 2 of the 10 isolates (isolates 9202 and 2022 [Table 1]). Both isolates had substitutions in the intergenic region between ahpC and oxyR, at position −9 and position −46, respectively (positions are as designated in reference 40). Interestingly, a previously unreported synonymous substitution at codon 6 (ATT→ATC) was also found to be present in isolate 9202.
DISCUSSION
Resistance to RIF.
Seven different mutations were identified among the 10 isolates (Table 2); several features of the RIF resistance phenotype and their genetic bases were disclosed by this study. Firstly, isolate 9002 had substitutions at both codon 511 (CTG→CCG; Leu511Pro) and codon 516 (GAC→GCC; Asp516Ala); although each of these has been reported previously, this is the first report of them in combination. Secondly, codon 531 is known to be a hot spot in rpoB for mutational change in M. tuberculosis (25), and half (five) of the isolates here also carried this mutation. Thirdly, like RIF, rifabutin is a derivative of rifamycin B (17), and it is normally used for prophylaxis against Mycobacterium avium complex infection in patients with AIDS (31). However, since a proportion of RIF-resistant M. tuberculosis isolates are susceptible to it (6), and as it can achieve a relatively high concentration in human lung tissue (12), it can be a useful alternative to treatment with RIF. The relationship between rpoB mutations and resistance to RIF and rifabutin was examined. While all 10 isolates were resistant to RIF by definition, 2 (isolates 1238 and 9219) of them were susceptible to rifabutin (Table 2). As noted by Bodmer et al. (6), who examined the susceptibility of 14 different rpoB mutants to several rifamycin B derivatives, it seems likely that the mutations in codons 511, 516, and 531 confer cross-resistance to both RIF and rifabutin, and in this list we would also include the codon 505 mutation. The codon 533 mutation only rendered isolate 9219 resistant to RIF and not to rifabutin, and this mutation is also in addition to those investigated by Bodmer et al. (6). More intriguingly, although various mutations have been noted which render resistance to both RIF and rifabutin (6) as in the His526Tyr substitution, this seems not to be the case for the His526Leu substitution, suggesting that different substitutions of a codon may have different effects.
Resistance to INH.
Although katG, inhA, and ahpC have been associated with INH resistance in M. tuberculosis, the mechanism(s) of INH action and of resistance to it are not completely understood. For instance, it is uncertain whether ahpC mutations confer resistance to INH directly or only secondarily as compensation for the decreased ability of the organism to survive oxidative-stress environments due to KatG alterations (7, 19, 22, 32, 36). In the 10 isolates of MDR-MTB studied here, 1 had no detectable mutations in the three genes examined, implying either that there was a resistance mutation outside the investigated regions of these three genes or that change was located elsewhere in the genome. Of the other nine isolates of MDR-MTB examined, all had a mutation in one, but not more than one, of these three genes, providing further supporting evidence that these genes are implicated in the majority of INH resistances. The absence of double katG-ahpC mutations here supports the hypothesis that mutation of ahpC independently confers INH resistance rather than as a secondary response to the mutation in katG.
Clones and drug resistance in M. tuberculosis.
Isolate 9511 is particularly interesting in that it was isolated from a patient of Pakistani nationality and has an IS6110 RFLP pattern very similar to that of the W strain. In addition, it had the same alleles of katG and inhA as the W strain, i.e., katG polymorphism at codon 463, a katG drug resistance mutation at Ser315Thr, and no mutations in inhA. However, it had a different mutation in rpoB; the W strain has mutations in rpoB of either His526Tyr or Ser531Leu (4), while isolate 9511 had a Ser531Trp mutation. This all suggests that isolate 9511 is closely related to the W strains isolated in the United States. No IS6110 patterns very similar to isolate 9511 could be found in our database, and as it was isolated from a Pakistani patient who had never been in the United States before he was diagnosed, it is likely that it is derived from Asia (20). Isolate 9604 was recovered from a Scottish patient; it is located in the same IS6110 RFLP subcluster as isolate 9511 (Fig. 2) and had the same drug resistance-related mutations and katG 463 polymorphism as isolate 9511. As several closely related isolates from Scottish patients have been identified in our database (Fig. 2), this suggests that this isolate is also related to the W strain but has probably evolved in Scotland.
Taken together these results suggest that there is a worldwide cluster of M. tuberculosis isolates which includes the W strain, isolate 9511, and isolate 9604. The isolates of this cluster have much genomic similarity (common IS6110 RFLPs, Leu codon 463 in katG). Not all isolates in these different lineages have antimicrobial resistances; some W strains are pansusceptible to the antimycobacterial agents (4), and all but one of the isolates closely related to 9511 and 9604 were pansusceptible, too (Fig. 2). The drug resistance mutations seen in this cluster are not unique to it and are often found in other unrelated isolates, such as codon 315 of katG in isolate 1238, codon 526 of rpoB in isolate 9310, codon 531 of rpoB in isolate 9202, and other mutations reported elsewhere (reference 25 and references therein). It would be interesting to perform a more extensive genetic survey of isolates in this cluster from around the world to chart the evolutionary links among them and their acquisition of resistances. Along these lines, we have identified a large cluster of isolates with very similar genetic makeups in our collection, many, but not all, of which have developed INH resistance (our unpublished data). As a result, we speculate that some clones of M. tuberculosis which occur worldwide have a tendency to become drug resistant.
Heterogeneity of the MDR-MTB isolates and its implications.
Since the late 1980s there have been outbreaks of MDR-MTB infection around the world, especially in association with patients with AIDS; epidemiological analysis of these MDR-MTB infection outbreaks has suggested that MDR-MTB is transmitted more rapidly and that this is of itself contributing to the increased incidence of tuberculosis in the world (1, 14–16, 27, 28). However, our study of MDR-MTB isolates from Scotland, at both genomic and resistance gene levels, suggests that rather than the epidemic spread of one strain there is a heterogeneous MDR-MTB population. This observation is consistent with two other facts.
Firstly, the incidence of primary resistance (the presence of a drug-resistant strain in a patient who has never received antituberculosis treatment), which is widely used as a parameter to evaluate the efficacy of a tuberculosis control program (10, 39), is low in Scotland. The Scottish incidence of isolates with resistance to one or more antimycobacterial drugs, which was 4.0% from 1990 to 1997, contrasts with the much higher rates of 8.4% in Germany in 1995, 11% in Korea in 1994, 14% in the United States in 1991, and 24% in Morocco in 1995. Indeed, the primary MDR-MTB isolation rate of 0.3% in Scotland during the same period contrasts with 3.5% in the United States in 1991 (5, 10, 19, 27). Secondly, the incidence of tuberculosis in Scotland has steadily declined up to the mid-1980s; since then, although it has leveled out (30), there have been few recorded significant outbreaks of the disease. Despite human immunodeficiency virus infection in Scotland since this period, there is a very low tuberculosis incidence in these patients (21) and there was no overrepresentation of MDR-MTB cases in this group. The causes of the low incidences of tuberculosis, both of sensitive and resistant strains, in Scotland are presently under study. Conceivably, more rigorous control measures or perhaps differences in the human population in Scotland have prevented such an outbreak to date.
In conclusion, it is apparent that the general incidence of MDR-MTB in Scotland is low, paralleling the low incidence of single-drug resistances in Scotland, and that the origins of these MDR-MTB isolates are diverse. Some have apparently been acquired from outside Scotland, while others have apparently evolved within Scotland, probably over a number of decades. It is important to remember that drug-resistant tuberculosis first became prevalent when chemotherapy was introduced in the 1940s and that by the late 1950s and early 1960s many industrialized countries had isolates resistant to both INH and STR. It was only with a combination of enhanced implementation of tuberculosis control programs and the introduction of RIF-containing short-course chemotherapy (35) that this situation was alleviated. While the incidence of drug-resistant tuberculosis is currently low in Scotland, it is apparent that such isolates can be acquired from both exogenous and endogenous sources. Tight controls must be maintained for the treatment of patients with tuberculosis in Scotland, most particularly to prevent the emergence of a multiple-drug-resistant strain derived from isolates which may be more host adapted to the Scottish population, as may be the case with the W strain in the United States, than one derived from outside this country. Past treatment failures may have left a legacy of future potential MDR-MTB.
ACKNOWLEDGMENTS
We thank P. Carter, and K. Reay for DNA sequencing and synthesis of the oligonucleotide primers. DNA sequence analysis benefited from SEQNET, the BBSRC facility (Daresbury, United Kingdom). We also thank three anonymous referees for their critical comments and suggestions to improve this paper.
This study was financially supported by The Scottish Office Department of Health; Chest, Heart and Stroke Scotland; and a Milner Scholarship from the University of Aberdeen.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R., Jr inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 3.Beck-Sague C, Dooley S W, Hutton M D, Otten J, Breeden A, Crawford J T, Pitchenik A E, Woodley C, Cauthen G, Jarvis W R. Hospital outbreak of multidrug-resistant Mycobacterium tuberculosis infections. Factors in transmission to staff and HIV-infected patients. JAMA. 1992;268:1280–1286. doi: 10.1001/jama.1992.03490100078031. [DOI] [PubMed] [Google Scholar]
- 4.Bifani P J, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lutfey M L, Moghazeh S L, Eisner W, Daniel T M, Kaplan M H, Crawford J T, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–457. [PubMed] [Google Scholar]
- 5.Bloch A B, Cauthen G M, Onorato I M, Dansbury K G, Kelly G D, Driver C R, Snider D E., Jr Nationwide survey of drug-resistant tuberculosis in the United States. JAMA. 1994;271:665–671. [PubMed] [Google Scholar]
- 6.Bodmer T, Zurcher G, Imboden P, Telenti A. Mutation position and type of substitution in the beta-subunit of the RNA polymerase influence in-vitro activity of rifamycins in rifampicin-resistant Mycobacterium tuberculosis. J Antimicrob Chemother. 1995;35:345–348. doi: 10.1093/jac/35.2.345. [DOI] [PubMed] [Google Scholar]
- 7.Deretic V, Philipp W, Dhandayuthapani S, Mudd M H, Curcic R, Garbe T, Heym B, Via L E, Cole S T. Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene: implications for sensitivity to isoniazid. Mol Microbiol. 1995;17:889–900. doi: 10.1111/j.1365-2958.1995.mmi_17050889.x. [DOI] [PubMed] [Google Scholar]
- 8.Dice L R. Measures of amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 9.Dobner P, Rusch-Gerdes S, Bretzel G, Feldmann K, Rifai M, Loscher T, Rinder H. Usefulness of Mycobacterium tuberculosis genomic mutations in the genes katG and inhA for the prediction of isoniazid resistance. Int J Tuberc Lung Dis. 1997;1:365–369. [PubMed] [Google Scholar]
- 10.el Baghdadi J, Lazraq R, Ibrahimy S, Bouayad Z, Guinet R, Benslimane A. Survey of primary drug resistance of Mycobacterium tuberculosis in Casablanca, Morocco. Int J Tuberc Lung Dis. 1997;1:309–313. [PubMed] [Google Scholar]
- 11.Fang Z, Morrison N, Watt B, Doig C, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farr B M. Rifamycins. In: Mandell G L, Douglas J, Bennett J E, editors. Principles and practices of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone, Inc.; 1994. pp. 317–328. [Google Scholar]
- 13.Frieden T R, Sherman L F, Maw K L, Fujiwara P I, Crawford J T, Nivin B, Sharp V, Hewlett D, Jr, Brudney K, Alland D, Kreisworth B N. A multi-institutional outbreak of highly drug-resistant tuberculosis: epidemiology and clinical outcomes. JAMA. 1996;276:1229–1235. [PubMed] [Google Scholar]
- 14.Fujiwara P I, Sherman L F. Multidrug-resistant tuberculosis: many paths, same truth. Int J Tuberc Lung Dis. 1997;1:297–298. [PubMed] [Google Scholar]
- 15.Gordin F M, Nelson E T, Matts J P, Cohn D L, Ernst J, Benator D, Besch C L, Crane L R, Sampson J H, Bragg P S, El-Sadr W. The impact of human immunodeficiency virus infection on drug-resistant tuberculosis. Am J Respir Crit Care Med. 1996;154:1478–1483. doi: 10.1164/ajrccm.154.5.8912768. [DOI] [PubMed] [Google Scholar]
- 16.Goyal M, Ormerod L P, Shaw R J. Epidemiology of an outbreak of drug-resistant tuberculosis in the U.K. using restriction fragment length polymorphism. Clin Sci. 1994;86:749–751. doi: 10.1042/cs0860749. [DOI] [PubMed] [Google Scholar]
- 17.Heifets L B, Iseman M D. Determination of in vitro susceptibility of mycobacteria to ansamycin. Am Rev Respir Dis. 1985;132:710–711. doi: 10.1164/arrd.1985.132.3.710. [DOI] [PubMed] [Google Scholar]
- 18.Heym B, Stavropoulos E, Honore N, Domenech P, Saint-Joanis B, Wilson T M, Collins D M, Colston M J, Cole S T. Effects of overexpression of the alkyl hydroperoxide reductase AhpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect Immun. 1997;65:1395–1401. doi: 10.1128/iai.65.4.1395-1401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S J, Bai G H, Hong Y P. Drug-resistant tuberculosis in Korea, 1994. Int J Tuberc Lung Dis. 1997;1:302–308. [PubMed] [Google Scholar]
- 20.Leen, C. Personal communication.
- 21.Leitch A G, Rubilar M, Watt B, Laing R, Willcocks L, Brettle R P, Leen C L. Why disease due to Mycobacterium tuberculosis is less common than expected in HIV-positive patients in Edinburgh. Respir Med. 1995;89:495–497. doi: 10.1016/0954-6111(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 22.Lynch A S, Lin E C C. Responses to molecular oxygen. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1526–1538. [Google Scholar]
- 23.Miller L P, Crawford J T, Shinnick T M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss A R, Alland D, Telzak E, Hewlett D, Jr, Sharp V, Chiliade P, LaBombardi V, Kabus D, Hanna B, Palumbo L, Brudney K, Weltman A, Stoeckle K, Chirgwin K, Simberkoff M, Moghazeh S, Eisner W, Lutfey M, Kreiswirth B. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int J Tuberc Lung Dis. 1997;1:115–121. [PubMed] [Google Scholar]
- 25.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. . (Review.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 27.Niemann S, Rusch-Gerdes S, Richter E. IS6110 fingerprinting of drug-resistant Mycobacterium tuberculosis strains isolated in Germany during 1995. J Clin Microbiol. 1997;35:3015–3020. doi: 10.1128/jcm.35.12.3015-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigouts L, Kubin M, Havelkova M, Portaels F. DNA fingerprint analysis of drug resistant Mycobacterium tuberculosis strains isolated in the Czech Republic. Cent Eur J Public Health. 1994;2:58–59. [PubMed] [Google Scholar]
- 29.Rouse D A, DeVito J A, Li Z, Byer H, Morris S L. Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol Microbiol. 1996;22:583–592. doi: 10.1046/j.1365-2958.1996.00133.x. [DOI] [PubMed] [Google Scholar]
- 30.Scottish Centre for Infection and Environmental Health. Tuberculosis. Scottish Centre for Infection and Environmental Health Weekly Report. 1995. 29(95/43):1. [PubMed] [Google Scholar]
- 31.Sesin G P, Manzi S F, Pacheco R. New trends in the drug therapy of localized and disseminated Mycobacterium avium complex infection. Am J Health Syst Pharm. 1996;53:2585–2590. doi: 10.1093/ajhp/53.21.2585. . (Review.) (Erratum, 54:442, 1997.) [DOI] [PubMed] [Google Scholar]
- 32.Sherman D R, Mdluli K, Hickey M J, Arain T M, Morris S L, Barry C E, Stover C K. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 33.Siddiqi S. Bactect TB systems: product and procedure manual. Paramus, N.J: Becton Dickinson and Co.; 1989. [Google Scholar]
- 34.Smith T F, Waterman M S. Comparison of biosequences. Adv Appl Math. 1981;2:482–489. [Google Scholar]
- 35.Snider J. Global burden of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: ASM Press; 1994. pp. 3–59. [Google Scholar]
- 36.Sreevatsan S, Pan X, Zhang Y, Deretic V, Musser J M. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997;41:600–606. doi: 10.1128/aac.41.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Victor T C, Warren R, Butt J L, Jordaan A M, Felix J V, Venter A, Sirgel F A, Schaaf H S, Donald P R, Richardson M, Cynamon M H, van Helden P D. Genome and MIC stability in Mycobacterium tuberculosis and indications for continuation of use of isoniazid in multidrug-resistant tuberculosis. J Med Microbiol. 1997;46:847–857. doi: 10.1099/00222615-46-10-847. [DOI] [PubMed] [Google Scholar]
- 39.Weyer K, Kleeberg H H. Primary and acquired drug resistance in adult black patients with tuberculosis in South Africa: results of a continuous national drug resistance surveillance programme involvement. Tuberc Lung Dis. 1992;73:106–112. doi: 10.1016/0962-8479(92)90064-Q. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Dhandayuthapani S, Deretic V. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc Natl Acad Sci USA. 1996;93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]