Abstract
Burkholderia cepacia selective agar (BCSA) has previously been devised for isolation of B. cepacia from respiratory secretions of patients with cystic fibrosis and tested under research laboratory conditions. Here we describe a study in which BCSA, oxidation-fermentation polymyxin bacitracin lactose agar (OFPBL), and Pseudomonas cepacia agar (PCA) were compared in routine culture procedures for the ability to grow B. cepacia and inhibit other organisms. Three hundred twenty-eight specimens from 209 patients at two pediatric centers and 328 specimens from 109 adults were tested. Plates were inoculated, incubated, and read for quality and quantity of growth at 24, 48, and 72 h. Five (1.5%) specimens from 4 (1.9%) children and 75 (22.9%) specimens from 16 (14.7%) adults grew B. cepacia complex. At 24, 48, and 72 h, BCSA achieved 43, 93, and 100% detection, respectively; OFPBL achieved 26, 84, and 96%, respectively; and PCA achieved 33, 74, and 84% detection, respectively. Quality was assessed as pinpoint or good growth. At 24 h, most cultures growing B. cepacia complex had pinpoint colonies. By 48 and 72 h, 48 and 69% of B. cepacia complex cultures, respectively, had good growth on BCSA, while on OFPBL 19 and 30%, respectively, had good growth and on PCA 11 and 18%, respectively, had good growth. BCSA was superior to OFPBL and PCA in suppressing organisms other than B. cepacia complex; 40 non-B. cepacia complex organisms were isolated from BCSA, 263 were isolated from OFPBL, and 116 were isolated from PCA. We conclude that BCSA is superior to OFPBL and PCA in its ability to support the growth of B. cepacia complex and to suppress other respiratory organisms.
Burkholderia cepacia is an important pathogen in pulmonary infections of patients with cystic fibrosis (CF). Because of the resistance of B. cepacia to many antimicrobial agents, B. cepacia infections are difficult to eradicate once a patient has become infected, and studies have demonstrated that it can spread from patient to patient as well as cause substantial morbidity and mortality (7, 10, 12, 13, 15–17). In the clinical laboratory, B. cepacia can be difficult to isolate, as it usually grows more slowly than other organisms frequently found in respiratory secretions from CF patients and, consequently, in culture can be overgrown with bacteria such as mucoid Pseudomonas aeruginosa. Once isolated, B. cepacia is often difficult to identify, especially when isolated from a patient with longstanding colonization, because the organism can undergo phenotypic changes (1). These auxotrophic changes can cause the strain to no longer react as expected in basic identification tests, resulting in the strain of B. cepacia being misidentified as other members of the Burkholderia or Alcaligenes genus or as Ralstonia pickettii (8). Alternatively, these organisms, as well as Stenotrophomonas maltophilia, can be misidentified as B. cepacia (2, 8, 9).
A taxonomic study on B. cepacia-like bacteria and creation of a database comprising all presently known Burkholderia species and genomovars, Ralstonia species, and P. aeruginosa, based on whole-cell protein electrophoresis, have recently been performed (18). This work has indicated that the species previously called B. cepacia is actually a complex of closely related organisms that are genetically distinct from each other. The taxonomic studies revealed that whole-cell protein electrophoresis is a very useful method for genomovar differentiation within the B. cepacia complex. However, this method is laborious and technically demanding and therefore not available in most laboratories. A variety of DNA-based identification procedures are currently being developed and should allow differentiation of most genomovars within the B. cepacia complex in due course (4, 12, 14, 21). As yet, only two of the five members can be phenotypically separated from the group. The complex includes Burkholderia vietnamiensis (formerly genomovar V); Burkholderia multivorans (formerly genomovar II); and B. cepacia genomovars I, III, and IV (6, 18). Accurate discrimination by genomovar typing among members of the B. cepacia complex will enable determination of the prevalence of each genomovar in a given epidemiological niche.
In an effort to improve the speed and accuracy of the isolation of the B. cepacia complex, we had devised an enriched selective medium. B. cepacia selective agar (BCSA) contains 1% lactose and 1% sucrose in an enriched base of casein and yeast extract with 600 U of polymyxin per ml, 10 μg of gentamicin per ml, and 2.5 μg of vancomycin per ml (8). We compared BCSA to oxidation-fermentation polymyxin bacitracin lactose agar (OFPBL; oxidation-fermentation agar supplemented with lactose, 300 U of polymyxin per ml, and 0.2 U of bacitracin per ml) (20), and to Pseudomonas cepacia agar (PCA; DeCicco holding medium with 300 U of polymyxin per ml and 100 μg of ticarcillin per ml) (5), in growth and selection of a wide range of bacterial strains from our laboratory collection. In this paper, we describe a study comparing the same three media used in three hospitals in the routine setup procedures for respiratory specimens from CF patients. The participating laboratories received guidelines regarding comparison of the selective media but were allowed to incorporate the additional selective agars into their existing CF respiratory protocols as best suited the individual laboratory’s procedures. We felt that it was important to incorporate laboratory-to-laboratory and technologist-to-technologist variations into the study, as long as the main criterion was met; that the three selective media within each specimen were treated equally.
(The material reported in this paper was presented at the 12th Annual North American CF Conference, October 1998 [7a].)
MATERIALS AND METHODS
The laboratories participating in the study were at St. Paul’s Hospital (SPH), Vancouver, British Columbia, Canada; Children’s Hospital and Regional Medical Center (CHRMC), Seattle, Wash.; and The Children’s Hospital (TCH), Denver, Colo. Each hospital contributed the selective medium that they were using and was supplied with BCSA made by the University of British Columbia (UBC) laboratory. BCSA was made as preivously described (8), with the following clarification: the phenol red and crystal violet were prepared as 0.8 and 0.02% aqueous solutions, respectively, and 10 ml of each was added per liter. SPH required OFPBL, which was made and supplied by UBC. PCA was purchased from PML Microbiologicals (Tualatin, Oreg.) for SPH and CHRMC. CHRMC purchased prepared OFPBL plates from BBL, Cockeysville, Md. TCH purchased PCA and OFPBL from Remel LP (Lenexa, Kans.). The plates were included as part of the routine and selective media used by the individual laboratories for the culture of CF respiratory pathogens. At SPH, following the routine plates for sputum culture (blood, MacConkey, colistin-nalidixic acid, and inhibition mold agars) the laboratory’s usual B. cepacia isolation plate (PCA) was inoculated first, followed by the other two plates. TCH inoculated the plates in random order; their routine media included blood, MacConkey, mannitol salt, and Haemophilus influenzae isolation agars. CHRMC performed quantitative sputum analyses, and plates were inoculated in random order; their other media included MacConkey, Mycosel, DNase, mannitol salt, streptococcal selective, and H. influenzae selective agars. Quantitation of pathogens in sputum was performed by a modification of the technique of Wong et al. (3). Sputa were solubilized by mixing 0.5 g of sputum with 0.5 ml of Sputolysin (Calbiochem, La Jolla, Calif.) and vortexing thoroughly. Dilutions (10−1, 10−3, 10−5, and 10−7) were made in phosphate-buffered saline (pH 7.0) with 0.1% gelatin added. One-hundred-microliter aliquots of each of the first three dilutions were plated on media selective for B. cepacia complex. Throat swabs were processed by placing them in 0.99 ml of phosphate-buffered saline with gelatin and vortexing thoroughly. Processing of all throat swabs and any sputum samples of less than 0.1 g was only qualitative but was performed on selective media. The number of CFU per gram of sputum was quantified by colony counts on each plate and calculated by the formula (CFU × 2)/dilution. SPH and CHRMC incubated selective plates between 35 and 37°C (ambient air), and TCH incubated Burkholderia selective media at 30°C; observations were made after 1, 2, and 3 days of incubation.
Growth on each of the selective media was graded for quantity and quality of growth. Quantity of growth at SPH and TCH was determined as follows: scant, less than 10 colonies in the main inoculum area; 1+, more than 10 colonies in the main inoculum area; 2+, growth into the second quadrant; 3+, growth into the third or fourth quadrant. The quantitative counts supplied by CHRMC were converted in the following manner: 1 to 100 CFU/ml = scant; 100 to <104 CFU/ml = 1+; 104 to 106 CFU/ml = 2+; >106 CFU/ml = 3+. Quality of growth was determined as either pinpoint or good growth at each time point. At SPH and TCH, the technologist assigned to the CF bench for that day read plates. The technologists reading the cultures were familiar with their own selective medium but had not used the other two media before, and so they graded quality and quantity of growth in as unbiased a manner as possible. One technologist at CHRMC performed all quantitative counts and qualitative observations.
SPH sent most plates with any growth to UBC for further examination and identification. Initially, CHRMC sent pure cultures of all organisms isolated to UBC, but due to the large number of yeast and fungi isolated, the laboratory switched to sending only gram-negative organisms for confirmation of identification. TCH sent only gram-negative organisms for confirmation to UBC. Organisms were identified with the API 20 NE or API 20E (Biomerieux Vitek, Inc., Hazelwood, Mo.) and oxidative-fermentative sugars as previously described (8, 19). Organisms identified as Burkholderia species or unusual alkaline nonfermenting gram-negative bacilli were subjected to whole-cell protein electrophoresis as described by Vandamme et al. (18) for confirmation of identification and genomovar determination. At least one Burkholderia species per patient was tested by random amplified polymorphic DNA (RAPD) PCR (13) and compared to other isolates previously analyzed for that patient when available, as well as to other isolates from that center. RAPD fingerprinting was performed in order to determine whether the organism was the same clonal strain as isolated previously from that patient and to ascertain the number of distinct strains present in each center.
RESULTS
A total of 656 specimens were cultured; 328 cultures were from 109 patients attending the adult center (SPH) and 328 cultures were from 209 patients attending the two pediatric centers (Table 1). All specimens from adults were sputum cultures. From the pediatric clinics, the specimens were split between throat swabs and sputa (three bronchial alveolar lavage specimens were included with the sputum results). Cultures were examined for growth, and specimens were categorized as to whether there was growth on any of the selective agars, growth of organisms other than B. cepacia complex on any of the selective agars, or growth of B. cepacia complex. One hundred thirty-seven of 209 (65.6%) pediatric patients had no growth of any organisms on any agar selective for B. cepacia: only five (3.1%) sputum cultures from four (1.9%) children grew B. cepacia complex. B. cepacia complex was not isolated from any throat specimens. Of the 109 adults in the study, 61 (60.0%) had no growth of any organisms on the selective media and 16 (14.7%) had growth of B. cepacia complex from 75 (22.9%) sputum cultures.
TABLE 1.
Specimens of respiratory secretions submitted for analysis
| Category | No. | No growtha [no. (%)] | Growthb [no. (%)]
|
|
|---|---|---|---|---|
| Not B. cepacia | B. cepacia complex | |||
| Pediatric throat | 165 | 139 (84.2) | 26 (15.8) | 0 |
| Pediatric sputum | 163 | 77 (47.2) | 81 (49.7) | 5 (3.1) |
| Adult sputum | 328 | 144 (43.9) | 109 (33.2) | 75 (22.9) |
| Total specimens | 656 | 360 (54.9) | 216 (32.9) | 81 (12.3) |
No growth of organisms on any selective medium.
Growth of organisms on any selective medium.
Table 2 lists the organisms isolated on the B. cepacia selective media. The table is divided into three sections; organisms confirmed as B. cepacia complex, organisms that were not B. cepacia complex but were difficult to separate biochemically from B. cepacia complex, and all other organisms.
TABLE 2.
Recovery of organisms from BCSA, OFPBL, and PCA selective media from 296 respiratory specimens from patients with CF
| Organism | No. of isolates recovered on:
|
||
|---|---|---|---|
| BCSA | OFPBL | PCA | |
| B. cepacia complex (total) | 81 | 78 | 68 |
| Genomovar I | 1 | 0 | 1 |
| Genomovar III | 73 | 71 | 63 |
| B. multivorans (genomovar II) | 6 | 6 | 3 |
| B. vietnamiensis (genomovar V) | 1 | 1 | 1 |
| Organisms similar to B. cepacia complex (total) | 9 | 13 | 6 |
| B. gladioli | 1 | 5 | 1 |
| Probable Burkholderia sp. | 2 | 2 | 1 |
| Unidentified, not Burkholderia sp. | 1 | 1 | 1 |
| R. pickettii | 1 | 1 | 1 |
| Ralstonia species | 3 | 3 | 2 |
| Alcaligenes species | 1 | 1 | 0 |
| Other non-B. cepacia complex (total) | 31 | 250 | 110 |
| Achromobacter xylosoxidans | 2 | 3 | 2 |
| Alcaligenes denitrificans | 1 | 1 | 0 |
| P. aeruginosa | 3 | 22 | 26 |
| S. maltophilia | 0 | 7 | 10 |
| Flavobacterium indologenes | 5 | 3 | 2 |
| Other nonfermentersa | 0 | 2 | 4 |
| Coliformsb | 0 | 5 | 5 |
| Staphylococcus sp. | 0 | 23 | 8 |
| Other gram-positivec | 0 | 4 | 0 |
| Yeast | 3 | 138 | 18 |
| Fungus | 17 | 42 | 35 |
Other nonfermenters: Pseudomonas putida, one on OFPBL and two on PCA; Pseudomonas fluorescens, one on PCA; Sphingomonas paucimobilis, two on OFPBL and one on PCA.
Coliforms: Proteus species, three on OFPBL; Serratia marcescens, one on OFPBL; Serratia liquefaciens, one on PCA; Klebsiella pneumoniae, three on PCA; Serratia species, one on OFPBL and one on PCA.
Other gram-positive: diphtheroids and Streptococcus species.
Sixteen adult patients were infected with B. cepacia complex. Isolates from 14 adult patients were identified as B. cepacia genomovar III and were subtyped by RAPD PCR to groups 1, 2, 4, 6, and 16. One adult was transiently colonized with an organism that was most likely B. cepacia genomovar I. Two adults were infected with B. multivorans (genomovar II, different RAPD types), and one became cocolonized with B. cepacia RAPD group 6 (genomovar III). This patient was cocolonized with the two organisms for several months, after which only the B. cepacia organism was isolated. Fifteen specimens from one adult were received over an 18-week period; B. cepacia (RAPD group 02, genomovar III) failed to grow on two OFPBL and six PCA plates after 3 days of incubation. This organism was scored as good growth on all BCSA plates and as pinpoint growth on the remaining OFPBL and PCA plates.
Samples from only five pediatric patients yielded B. cepacia complex. Three children had isolates identified as B. multivorans with unique RAPD patterns. Of the two other children, one had B. vietnamiensis isolated from one culture and an unidentified nonfermenting gram-negative bacillus (UNFB) from another sputum 3 months later; the other child had a UNFB, most likely a Burkholderia species, but identification could not be confirmed. Sputa from four children at one center grew Burkholderia gladioli; each had a unique RAPD pattern. Quantitative analyses for B. gladioli ranged from 6.0 × 102 to 4.1 × 105 CFU/ml. The children’s specimens also produced most of the Ralstonia-like organisms and the organisms that were difficult to identify.
Most organisms other than B. cepacia complex that grew on the three selective media were graded as scant or 1+ (grew in the first quadrant of the main inoculum) and consisted of fungi, yeast, and gram-positive organisms. SPH identified the yeast and fungi to species level and identified most as Aspergillus fumigatus and Candida albicans. One adult had seven cultures that grew a mixture of Wangiella dermatitidis, C. albicans, Scedosporium apiospermum, A. fumigatus, Acremonium species, and Staphylococcus aureus on the selective plates. The cultures from this patient accounted for two of the fungi (S. apiospermum) isolated on BCSA, six yeasts and five fungi on OFPBL, and five fungi on PCA. OFPBL suppressed yeast and fungi poorly. OFPBL and PCA performed similarly in their inability to suppress growth of some gram-negative bacilli, notably P. aeruginosa and S. maltophilia. The gram-negative organisms and the Staphylococcus species frequently grew in clumps of mucus. Ten cultures positive for B. cepacia complex also grew other organisms such as P. aeruginosa, Serratia marcescens, Staphylococcus species, yeast, and fungi on OFPBL and PCA; a fungus grew on one BCSA plate. The presence of these contaminating organisms did not appear to hinder the detection of B. cepacia complex.
CHRMC used a quantitative sputum culture method on all CF sputa. Specimens from two patients yielded low counts of 20 to 700 CFU (per ml) of a UNFB resembling Burkholderia species and B. multivorans on BCSA and OFPBL and no growth on PCA. One of these patients also had 2.0 × 103 CFU (per ml) each of C. albicans on OFPBL and Staphylococcus sp. on PCA. A third patient had two cultures 10 weeks apart, growing Ralstonia species (between 1.6 × 104 and 1.0 × 106 CFU/ml). Two other patients had three specimens growing B. multivorans on all three plates (between 8.0 × 103 and 2.1 × 108 CFU/ml).
Analysis of quantity of growth of B. cepacia genomovar III for the adults (most of whom had been colonized for a number of years) was usually 2+ to 3+. BCSA demonstrated heavier growth more quickly than OFPBL or PCA. By 72 h, 66.7% of BCSA plates demonstrated 3+ growth compared to 61.7 and 48.1% for OFPBL and PCA, respectively.
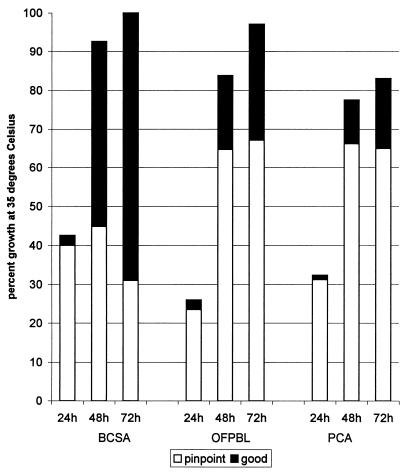
Quality of growth of B. cepacia complex (pinpoint or good growth) is displayed in Fig. 1. B. cepacia complex was visible on 42.5% of BCSA plates within the first 24 h. The detection rate improved to 92.6 and 100% after 2 and 3 days of incubation, respectively. B. cepacia complex initially appeared as pinpoint colonies, but the quality of growth improved to good growth for 69.0% of the positive cultures on BCSA by the third day. OFPBL and PCA had fewer plates demonstrating any growth after the first day’s incubation (26.0 and 32.6%, respectively). This rate improved with additional incubation, but by the third day, 67.1% of OFPBL and 64.9% of PCA plates still demonstrated only pinpoint colonies. Total growth at 72 h for the 81 cultures with B. cepacia complex was as follows: BCSA, 100%; OFPBL, 96.2%; and PCA, 83.9%.
FIG. 1.
Quality of growth for 81 cultures with B. cepacia complex, expressed as pinpoint or good growth over 3 days of incubation at 35°C. Not all selective plates were scored for quality of growth on each of the three days of incubation, and so the percentages reflect data only for the 67 to 81 plates for each of the three media scored each day.
DISCUSSION
Three selective agars were compared in clinical laboratory settings for their ability to isolate B. cepacia complex from respiratory secretions of patients with CF. B. cepacia complex grew more quickly and to a larger colony size on BCSA than on OFPBL or PCA. Quantitative analysis indicated that as few as 20 CFU of B. multivorans per ml could be detected on BCSA and OFPBL, demonstrating excellent sensitivity of the media. Several strains of B. cepacia genomovar III and B. multivorans did not grow, or grew poorly, on PCA. Similarly, one strain each of B. cepacia genomovars III and I did not grow on OFPBL. If either OFPBL or PCA had been used as the sole selective medium, potentially up to four patients with B. cepacia complex could have missed being detected. BCSA was able to support the growth of different clones within genomovars of the B. cepacia complex, since all B. multivorans isolates had DNA fingerprints that were different from each other, and the B. cepacia genomovar III isolates from 14 adult patients had five distinct RAPD patterns. Barth and Pitt (1) described auxotrophy in isolates of B. cepacia from CF patients; it may be that some strains from patients who have been colonized for a long time can no longer grow on minimal media. Carbon assimilation studies performed in an effort to identify to species level within the B. cepacia complex have demonstrated auxotrophy (8a). BCSA is more enriched than OFPBL or PCA, as it contains yeast extract, not found in OFPBL or PCA, and five times the pancreatic digest of casein that is found in OFPBL (PCA does not contain casein). Yeast extract and casein provide a rich variety of ingredients to overcome the nutritional deficiencies that may prevent some strains of B. cepacia from growth on other selective media.
As described previously (2, 8), we had observed isolates of S. maltophilia that had been sent to us, identified as B. cepacia; in these clinical trials, no isolates of S. maltophilia grew on BCSA. As well, BCSA inhibited significantly more non-B. cepacia complex organisms than did OFPBL and PCA. Several organisms that we were unable to identify were isolated but were not B. cepacia complex, as determined by whole-cell protein electrophoresis. These were more commonly found in children, or in adults who had not had B. cepacia complex before. The appearance of these organisms seemed to be transient, as they were not present in subsequent cultures. It is important that new isolates of suspected B. cepacia complex be sent to a reference laboratory experienced in the phenotypic and genotypic identification of this group.
The in-hospital laboratory study demonstrated that, for the culture of CF respiratory specimens, BCSA was superior to OFPBL and PCA for rapidity and quality of recovery of B. cepacia complex and was more inhibitory toward organisms other than B. cepacia complex. These features would make BCSA an important addition to CF sputum culture protocols.
ACKNOWLEDGMENTS
This study was supported by grants from the Canadian Cystic Fibrosis Foundation. P.V. is indebted to the Fund for Scientific Research, Vlaanderen (Belgium), for a postdoctoral research fellowship.
We are grateful for the technical assistance provided by the participating laboratories. We wish to thank Gary Probe for his assistance in the preparation of RAPD PCR data.
REFERENCES
- 1.Barth A L, Pitt T L. Auxotrophy of Burkholderia (Pseudomonas) cepacia from cystic fibrosis patients. J Clin Microbiol. 1995;33:2192–2194. doi: 10.1128/jcm.33.8.2192-2194.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burdge D R, Noble M A, Campbell M E, Krell V L, Speert D P. Xanthomonas maltophilia misidentified as Pseudomonas cepacia in cultures of sputum from patients with cystic fibrosis: a diagnostic pitfall with major clinical implications. Clin Infect Dis. 1994;20:445–448. doi: 10.1093/clinids/20.2.445. [DOI] [PubMed] [Google Scholar]
- 3.Burns J L, Emerson J, Stapp J R, Yim D L, Krzewsinski J, Louden L, Ramsey B W, Clausen C R. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- 4.Coenye T, Govan J R W, Kersters K, Vandamme P. Proceedings of the 12th North American Cystic Fibrosis Conference. 1998. Automated AFLP analysis for the identification of Burkholderia species and genomovars occurring in CF patients, abstr. 428. [Google Scholar]
- 5.Gilligan P H, Gage P A, Bradshaw L M, Schidlow D V, DeCicco B T. Isolation medium for the recovery of Pseudomonas cepacia from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 1985;22:5–8. doi: 10.1128/jcm.22.1.5-8.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillis M, Van T V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 7.Govan J R, Brown P H, Maddison J, Doherty C J, Nelson J W, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 7a.Henry D, Campbell M, McGimpsey C, Clarke A, Louden L, Burns J L, Roe M H, Vandamme P, Speert D. Proceedings of the 12th North American Cystic Fibrosis Conference, abstr. 435. 1998. [Google Scholar]
- 8.Henry D A, Campbell M E, LiPuma J J, Speert D P. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Henry, D. A. Unpublished data.
- 9.Kiska L D, Kerr A, Jones M C, Caracciolo J A, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan P H. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–891. doi: 10.1128/jcm.34.4.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipuma J J, Marks-Austin K A, Holsclaw D S, Winnine G B, Gilligan P H, Stull T L. Inapparent transmission of Pseudomonas (Burkholderia) cepacia among patients with cystic fibrosis. Pediatr Infect Dis J. 1994;13:716–719. doi: 10.1097/00006454-199408000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Lipuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Proceedings of the 12th North American Cystic Fibrosis Conference. 1998. Polymerase chain reaction identification of species within the Burkholderia cepacia complex, abstr. 367. [Google Scholar]
- 12.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia stains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by random amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahenthiralingam E, Bichof J, Byrne S K, Vandamme P. Proceedings of the 12th North American Cystic Fibrosis Conference. 1998. Molecular speciation of Burkholderia cepacia complex strains recovered from patients with cystic fibrosis, abstr. 368. [Google Scholar]
- 15.Pegues D A, Carson L A, Tablan O C, FitzSimmons S C, Roman S B, Miller J M, Jarvis W R the Summer Camp Study Group. Acquisition of Pseudomonas cepacia at summer camps for patients with cystic fibrosis. J Pediatr. 1994;124:694–702. doi: 10.1016/s0022-3476(05)81357-5. [DOI] [PubMed] [Google Scholar]
- 16.Revets H, Vandamme P, Van Zeebroeck A, De Boeck K, Struelens M J, Verhaegen J, Ursi J P, Verschraegen G, Franckx H, Malfroot A, Dab I, Lauwers S. Burkholderia (Pseudomonas) cepacia and cystic fibrosis: the epidemiology in Belgium. Acta Clin Belg. 1996;51:222–230. doi: 10.1080/22953337.1996.11718514. [DOI] [PubMed] [Google Scholar]
- 17.Smith D L, Gumery L B, Smith E G, Stableforth D E, Kaufmann M E, Pitt T L. Epidemic of Pseudomonas cepacia in an adult cystic fibrosis unit: evidence of person-to-person transmission. J Clin Microbiol. 1993;31:3017–3022. doi: 10.1128/jcm.31.11.3017-3022.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman T, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 19.Von Graevenitz A. Acinetobacter, Alcaligenes, Moraxella, and other nonfermentative gram-negative bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 520–532. [Google Scholar]
- 20.Welch D F, Muszynski M J, Pai C H, Marcon M J, Hribar M M, Gilligan P H, Matsen J M, Ahlin P G, Hilman B C, Chartrand S A. Selective and differential medium for recovery of Pseudomonas cepacia from the respiratory tracts of patients with cystic fibrosis. J Clin Microbiol. 1987;25:1730–1734. doi: 10.1128/jcm.25.9.1730-1734.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitby P W, Carter K B, Coenye T, Vandamme P, LiPuma J J, Stull T L. Proceedings of the 12th North American Cystic Fibrosis Conference. 1998. Identification of Burkholderia cepacia genomovars I, III and IV by nested polymerase chain reactions, abstr. 429. [Google Scholar]