Abstract
We examined potable water in Los Angeles, California, as a possible source of infection in AIDS and non-AIDS patients. Nontuberculous mycobacteria were recovered from 12 (92%) of 13 reservoirs, 45 (82%) of 55 homes, 31 (100%) of 31 commercial buildings, and 15 (100%) of 15 hospitals. Large-restriction-fragment (LRF) pattern analyses were done with AseI. The LRF patterns of Mycobacterium avium isolates recovered from potable water in three homes, two commercial buildings, one reservoir, and eight hospitals had varying degrees of relatedness to 19 clinical isolates recovered from 17 patients. The high number of M. avium isolates recovered from hospital water and their close relationship with clinical isolates suggests the potential threat of nosocomial spread. This study supports the possibility that potable water is a source for the acquisition of M. avium infections.
Members of the Mycobacterium avium complex (MAC), a group of opportunistic human pathogens, cause the most common disseminated bacterial infections in AIDS patients (1, 11, 30). As many as 40% of patients with advanced AIDS may develop MAC infection (12). Most patients with disseminated MAC (M. avium, M. intracellulare, and Mycobacterium sp. x [Mx]) disease are infected with M. avium. The term “Mx” refers to a number of mycobacteria which are DNA or RNA positive for MAC but negative for M. avium and M. intracellulare. MAC may also be invasive in patients with other immunocompromised conditions, such as patients with neoplastic disorders, patients with combined immunodeficiency disease, heart transplant recipients (19), and patients with no obvious predisposing factors (7, 10, 14, 22, 29).
Although there may be a decline in the numbers of M. avium infections in developed countries with the use of current AIDS therapy, the long-term effectiveness of these protocols remains to be seen. In addition, non-AIDS patients continue to exhibit infections caused by nontuberculous mycobacteria (NTM).
The environment is considered the most likely source of MAC infection since members of MAC are ubiquitous organisms that are recovered from water (2, 4, 5, 6), soil and dust (13), and animals (9). Currently, there is no evidence that documents person-to-person transmission. In collaborative studies with the Centers for Disease Control and Prevention and the U.S. Environmental Protection Agency, we reported the existence of MAC in water samples obtained by conventional culturing methods from reservoirs, residences, hospitals, and commercial structures in Los Angeles, California (2, 8).
The purpose of this study was to estimate the relatedness of M. avium strains in Los Angeles potable water to strains isolated from patients in the Los Angeles area. We previously compared a few M. avium isolates obtained from water and from AIDS patients for their relatedness by multilocus enzyme electrophoresis (2, 8). Small preliminary studies by pulsed-field gel electrophoresis (PFGE) demonstrated that the strains recovered from potable water and patients were identical (3, 28). In the present, expanded study, a portion of the large number of M. avium isolates obtained from water sources in Los Angeles and clinical isolates from both AIDS and non-AIDS patients were compared for their relatedness by looking at large restriction fragments (LRFs) by PFGE.
MATERIALS AND METHODS
Water collection.
Water samples were collected from 1991 to 1996 from the following sources throughout Los Angeles. (i) water was collected from the outlet lines of 13 Los Angeles reservoirs on four separate occasions during the year. These reservoirs supply 90% of the potable water to the city of Los Angeles. Reservoir water samples were collected, concurrently, by the Los Angeles Department of Water and Power for determination of coliform counts, heterotrophic plate counts, and total and free chlorine levels; these determinations were done at the Los Angeles Department of Water and Power Laboratory. (ii) Water was collected from the inlet lines to 15 hospitals, sink and shower water taps (both hot and cold water taps) in patients’ rooms, and the water sources for bedside carafes. (iii) Water was collected from 55 randomly selected homes. (iv) Water was collected from 31 commercial buildings (hose bibs and hot and cold water taps from both kitchen sinks and showers). Most homes, buildings, and hospitals were sampled only once. A subset of water samples was collected from the homes of non-AIDS patients exhibiting NTM infections. Extensive immunological studies were not done for most of these patients, but the patients had no prior history of immunodeficiency. The water lines were flushed for 1 min, and samples were collected in 1-liter sterile polypropylene bottles containing 1 ml of 10% sodium thiosulfate to neutralize the residual chlorine. The samples were transported to the laboratory immediately and, if they were not processed promptly, were stored at 2 to 8°C for not more than 24 h.
Concentration and decontamination.
Cetylpyridinium chloride, oxalic acid, and/or sodium hydroxide were used as decontaminating agents with 500-ml aliquots of water. These specimens were concentrated by vacuum filtration through 0.45-μm-pore-size HABG Millipore filters (2, 8). The filters were transferred to Middlebrook 7H10 agar plates containing 500 μg of cycloheximide per ml (7H10C), sealed in polyethylene bags, and incubated at 37°C.
Species identification of water isolates.
Filters were examined with a dissecting microscope (magnification, ×7 to ×10) at 3 weeks, and plates with no growth were examined again at 8 weeks. The various NTM colony types found on positive cultures were enumerated, and a representative of each colony type was transferred to 0.5 ml of Middlebrook 7H9 broth. After incubation at 37°C, cultures containing acid-fast coccobacilli were subcultured on 7H10C for purity. Nitrate- and Tween-negative isolates were analyzed with DNA probes (SNAP [Syngene, San Diego, Calif.] or AccuProbe [GenProbe, San Diego, Calif.]) for MAC isolates, and positive isolates were analyzed with probes specific for M. avium, M. intracellulare, and Mx. Probe-negative isolates were frozen at −70°C and later identified by biochemical procedures, with DNA probes, by high-performance liquid chromatography, and/or by PCR methods.
Clinical isolates.
MAC isolates from the blood, bone marrow, or sputum of AIDS patients were obtained from local hospitals and laboratories from 1991 to 1996. Clinical isolates were recovered from patients living in the city of Los Angeles. All isolates were subcultured on Wallenstein medium (Clinical Standard Laboratories, Dominguez Hills, Calif.) or 7H10C medium and were identified to the species level with DNA probes (SNAP or AccuProbe). MAC and non-MAC NTM recovered from human immunodeficiency virus-negative patients were acquired from hospitals and laboratories and included organisms recovered from sputa, respiratory tract lavage samples, or lymph node aspirates. The human immunodeficiency virus-negative patients were apparently immunocompetent. However, comprehensive immunological data were unavailable for these patients.
Scale-up of mycobacterial isolates for LRF analysis.
One M. avium colony was sequentially cultivated to a final volume of 180 ml in Middlebrook 7H9 containing 0.5 M sucrose. Following treatment with ampicillin, d-cycloserine, and d-threonine, when the turbidity reached an absorbance at 580 nm of 0.3 (17, 25), the colonies were centrifuged, and the centrifuged pellets were suspended in TS buffer (50 mM Tris, 0.5 M sucrose [pH 7.6]) and were stored in 1-ml aliquots at −70°C.
Preparation of DNA in agarose plugs.
The cryogenic vials containing frozen mycobacterial preparations were thawed on ice, diluted to an absorbance at 580 nm of 0.4 in TS buffer, and treated (25) for casting into InCert agarose (FMC Bioproducts, Rockland, Maine). Disposable plug molds (Bio-Rad, Richmond, Calif.) were used to cast the agarose plugs. Lysozyme-, RNase-, and proteinase K-treated plugs were stored at 5°C (17) and were processed for restriction enzyme digestion. Agarose plugs containing Staphylococcus aureus ATCC 8325 were prepared (17), digested with SmaI (New England Biolabs), and used as molecular weight standards for LRF analyses of M. avium isolates digested with AseI (New England Biolabs).
Restriction endonuclease enzyme digestion.
The plugs were washed twice with TE (10 mM Tris, 0.1 mM EDTA [pH 7.6]) on a rotator at a speed of 4 rpm for 2-h intervals and treated with 1 mM phenylmethylsulfonyl fluoride in TE at 5°C for 1 h, followed by two 0.5-h incubations in TE at 37°C with rotation. The plugs were transferred to microcentrifuge tubes containing restriction enzyme buffer, and after 30 min on ice, the plugs were transferred to fresh tubes containing restriction enzyme buffer and 20 U of AseI was added (17). Following several hours on ice, the tubes were transferred to a 37°C water bath overnight, and the plugs were rinsed with TE and incubated for 1 h at 37°C in 1 ml of TE containing 30 μg of proteinase K/ml. Following proteinase K digestion, the plugs were rinsed and stored in 0.5× TBE (0.045 M Tris-borate, 0.001 M EDTA) at 5°C until they were electrophoresed (23).
LRF analysis.
The restriction enzyme-digested agarose plugs (unknowns and molecular weight standards) were electrophoresed in 1.0% FastLane agarose (FMC Bioproducts) in 0.5× TBE buffer (23). The 1.0% FastLane agarose gels containing 0.5× TBE in a volume of 150 ml were cast in a Bio-Rad wide-long casting stand (21 by 24 cm) with a 21-cm-wide, 1.5-mm-thick, 15-well comb. The electrophoretic parameters used with the Bio-Rad CHEF DRIII system were an initial switch time of 30 s, a final switch time of 60 s, a 22-h run time, 6.0 V/cm, a 120° included angle, a 12.0°C chiller set temperature at the external probe, and a volume of 2.2 liters of 0.5× TBE in the gel box.
The electrophoresed gels were stained with 0.5 μg of ethidium bromide/ml in 0.5× TBE for 20 min and were visualized and documented on an UltraViolet Products Image Store 7500 (UltraViolet Products, Upland, Calif.) or an UltraLum gel documentation system (UltraLum, Paramount, Calif.). Documentation was stored as a thermal print and TIFF files for computer analysis. The LRFs in the gels and documented in TIFF files were sized and compared with DNA ProScan Pro-RFLP analysis software (DNA Proscan, Inc., Nashville, Tenn.). The LRF patterns were also analyzed by visual comparison.
Genetic relatedness of banding patterns.
Criteria for determination of the relatedness of a group of bacterial isolates when the DNA restriction patterns obtained by PFGE were interpreted were established as proposed by Tenover et al. (27). A minimum of 10 fragments must be resolved for each isolate for the criteria to be valid. The relatedness between isolates is based on genetic differences and is divided into four categories: indistinguishable, closely related, possibly related, and unrelated, with respective numbers of fragment differences of zero, two to three, four to six, and seven or more.
RESULTS
NTM were present in most reservoirs, homes, commercial buildings, and hospitals (90%) sampled; and 43 (42%) of the NTM (Table 1) were DNA probe positive for MAC. Hospitals had the highest incidence (93%) of M. avium (Table 1). The concentration of MAC in water samples ranged from 1 CFU/500 ml to too numerous to count (≈103/500 ml). MAC was not found predominantly in hot over cold water, as Du Moulin et al. (6) reported. The coliform count, heterotrophic plate count, and chlorine level determinations were performed at the Los Angeles Department of Water and Power Laboratory. No discernible relationship between the numbers of NTM recovered from reservoir water to coliform counts, heterotrophic plate counts, and total and free chlorine levels and in covered versus uncovered reservoirs existed.
TABLE 1.
Sources of MAC-positive water isolates
| Source | No. (%) of isolates
|
||||
|---|---|---|---|---|---|
| NTM | MAC | M. avium | M. intracellulare | Mx | |
| Hospitals (n = 15) | 15 (100) | 14 (93)a | 11 (79) | 1 (7) | 4 (29) |
| Homes (n = 55) | 45 (82) | 12 (22) | 3 (5) | 7 (13) | 2 (4) |
| Reservoirs (n = 13) | 12 (92) | 5 (38) | 2 (15) | 3 (23) | 0 |
| Commercial buildings (n = 31) | 31 (100) | 12 (39)b | 7 (23) | 5 (3) | 1 (3) |
Two hospitals had both M. avium and Mx.
One building had both M. avium and M. intracellulare.
LRF analysis of clinical and water M. avium isolates.
Two hundred forty-four clinical and 60 water isolates of M. avium were scaled up for analysis. Of these, 111 clinical isolates and 45 water isolates were successfully grown in quantities needed for this investigation. Ninety-seven of 111 clinical M. avium isolates in agarose plugs and 35 of 45 water M. avium isolates in agarose plugs liberated DNA in sufficient quantities and qualities for restriction analysis.
M. avium LRF analysis.
DNA fragments from clinical and water M. avium isolates were electrophoresed, and computer searches for potential matches of clinical and water isolates were conducted. Gels containing the water M. avium isolates which potentially matched clinical M. avium isolates were electrophoresed, and the plugs yielding the most well defined patterns and the greatest number of bands were selected. There was a reduction in the number of plugs to be compared on the bases of the locations and the distinct differences in the restriction patterns.
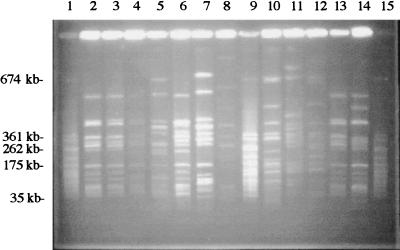
Sixteen water M. avium isolates with unique patterns were selected by Pro-RFLP software analysis for comparison with 30 clinical M. avium isolates in plugs (the 30 isolates were from 28 patients). Fifteen of the 16 water M. avium isolates demonstrated some relatedness to 19 of the 30 clinical isolates by their AseI restriction patterns (a representative gel is shown in Fig. 1).
FIG. 1.
PFGE of AseI restriction digests of clinical and water isolates of M. avium. The LRF pattern of the M. avium isolate from patient 33 (lane 13) was identical to those of water M. avium isolates from hospitals 4 (lane 2) and 5 (lane 3), closely related to those of isolates from hospitals 6 (lane 4) and 10 (lane 6), and possibly related to those of isolates from hospitals 7 (lane 5) and 14 (lane 7). The (lane 14) isolate from patient 53 was closely related to isolates from hospitals 4 (lane 2) and 5 (lane 3) and was possibly related to isolates from hospital 6 (lane 4). Isolates from patients 25 (lane 10), 29 (lane 11), and 32 (lane 12) were unrelated to all of the water M. avium isolates. Lanes 1, 9, and 15, SmaI restriction digests of S. aureus ATCC 8325 as molecular size markers. The reservoir 13 isolate (lane 8) was unrelated to any clinical isolate on the gel.
Numerous investigators (17, 18, 25, 27) have selected AseI as the most suitable restriction enzyme for LRF analysis of mycobacteria since it yields the most consistently clear LRF patterns. The relatedness of water and clinical M. avium isolates as determined by LRF analysis of AseI digests is provided in Table 2. Some strains appear to be common to the water systems of various buildings. Strains identical to clinical isolates CW15 and CW16 were found in four hospitals, one dwelling, and one reservoir. Strains identical to patient isolate 33 were found in four hospitals; and strains closely related to clinical isolate 33 were found in two hospitals, four dwellings, one commercial building, and one reservoir.
TABLE 2.
Relatedness of MA water and clinical isolates using AseIa
| Patient | Source | AIDS status | Relatedness of isolates from the following sourcesb:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water from homes, commercial buildings, and reservoirs
|
Hospital water
|
||||||||||||||||
| D10 | D20-1 | D20-2 | B29 | D41 | R5 | B55 | Ho2 | Ho4 | Ho5 | Ho6 | Ho7 | Ho8 | Ho10 | Ho14 | |||
| 25 | Blood | AIDS | 2 | ||||||||||||||
| 28 | Blood | AIDS | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | ||||||
| 33 | Sputum | Non-AIDS | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 2 |
| 52 | Blood | AIDS | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | |||||||
| 52 | Sputum | AIDS | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | ||||
| 71 | Sputum | Non-AIDS | 1 | 1 | 1 | 2 | 1 | 2 | 2 | ||||||||
| 76 | Blood | AIDS | 1 | 2 | 2 | 2 | |||||||||||
| 80 | Blood | AIDS | 2 | ||||||||||||||
| 94 | Sputum | Non-AIDS | 1 | 1 | 2 | 1 | 1 | 1 | |||||||||
| 124 | Blood | AIDS | 2 | ||||||||||||||
| 216 | Blood | AIDS | 2 | 2 | 2 | ||||||||||||
| 274 | Sputum | Non-AIDS | 2 | 2 | |||||||||||||
| 295 | Blood | AIDS | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | |||
| 316 | Blood | AIDS | 1 | 1 | 2 | 1 | 1 | 1 | |||||||||
| C348, 25°C | Bone marrow | Non-AIDS | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||||
| C348, 37°C | Bone marrow | Non-AIDS | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |||||||
| C15 | Sputum | Non-AIDS | 0 | 0 | 0 | 0 | 2 | ||||||||||
| C16 | Sputum | Non-AIDS | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | |||||
| C32 | BAL | Non-AIDS | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | ||||||
Abbreviations: BAL, bronchoalveolar lavage fluid; B, building; D, home; Ho, hospital; R, reservoir.
Relatedness codes: 0, identical; 1, closely related; 2, possibly related; blank, unrelated.
The M. avium isolates from patient C16 and home 41 water yielded identical restriction fragment patterns for three different restriction digests (AseI, HindIII, and XbaI), and these patterns represent the closest relationship between clinical and water M. avium isolates (unpublished data). M. avium C348 (25°C) and C348 (37°C) were recovered from the bone marrow of a 2-year-old child. Extensive immunological testing showed that the child had no deficiencies, but the child succumbed to disseminated M. avium infection. These isolates grown at different temperatures were recovered from the same primary inoculum of the clinical specimen. Their AseI patterns were closely related but were not identical, indicating that this patient probably had a polyclonal infection. More genetic diversity was found among clinical isolates than among water isolates. Water isolates demonstrated 77% relatedness, whereas clinical isolates demonstrated 43% relatedness.
DISCUSSION
M. avium isolates recovered from potable water in three homes, two commercial buildings, one reservoir, and eight hospitals had various degrees of genetic relatedness to 19 clinical isolates recovered from 17 patients. Although this is significant, the actual number of M. avium isolates in Los Angeles potable water that are related to clinical isolates is probably much larger. There are several reasons for this and for the fact that there was less genetic diversity among the water isolates than among the clinical isolates. First, harsh decontamination procedures followed by growth on a selective medium are required for the isolation of M. avium from environmental samples (2, 3, 8). This leads to the loss of approximately half of the M. avium isolates from seeded samples and possibly to an even greater loss from environmental samples. Our methods may have selected only those strains that were the most resistant to the decontamination agents. Additional problems that could decrease the number of matches in this study were due to either the poor growth of some isolates or the insufficient release of DNA for LRF analysis.
The various numbers of M. avium isolates in water samples collected from the same location, the lack of repetitive sampling from most locations, and the limited number of sampling sites also suggest that the isolates used in this study were an incomplete representation of the M. avium isolates in Los Angeles potable water. Mycobacteria grow in the biofilms in water supply systems (24), and like other organisms in plumbing systems, they are probably shed in high numbers only periodically. This phenomenon of periodic shedding has been observed in studies of Legionella pneumophila, with the numbers of organisms isolated from individual sites varying from none to over 1,500 CFU per ml (26). More repetitive sampling could increase the yield of matching organisms from water.
Increasing the number of sampling sites could also lead to the isolation of a greater variety of M. avium strains from Los Angeles potable water. The Los Angeles water supply comprises water mixed from a variety of sources including the Colorado River and a number of reservoirs in different areas of the state of California, and it should therefore contain a wide variety of M. avium isolates from these diverse environments. It is likely that many of these strains are capable of amplifying in the distribution system or within building plumbing systems.
The lower level of diversity in water M. avium isolates found in this study does not necessarily indicate that water is an insignificant source of infection. It is likely a reflection of the limited number of sampling sites, the limited number of colonies that we were able to analyze, and the probable loss of a number of strains due to harsh decontamination procedures. With all of these complications it is surprising that isolates identical to three different clinical strains were found at multiple locations. Evidence from studies of L. pneumophila showed that specific sampling sites are unique microenvironments and that the conditions at each site select for a particular dominant strain. Distinct predominant Legionella strains were isolated from each of six hospitals supplied with water from the same distribution system (15), from two adjoining hospitals supplied by the same municipal main (21), and even from different taps within the same hospital (16). It is reasonable to expect similar diversity with respect to M. avium isolations from different locations within the distribution system and even within individual buildings.
Strains classified as closely related cannot be ruled out as the infecting strains. Growth conditions in the body are quite different from those in the distribution or plumbing systems, and it is possible that a subpopulation of organisms slightly different from the original infecting strain will predominate after passage through a host. Pestel-Caron and Arbeit (20) observed significant variability in the locations and copy numbers of IS1245 elements among independent patient isolates representing the same strain. These changes could easily alter the position of one or more of the restriction fragments of AseI digests.
The larger number of isolates found in hospital water suggests the possibility of the nosocomial spread of M. avium to immunocompromised patients and to AIDS patients in particular (3, 28). Nosocomial spread could be verified by performing a prospective epidemiological study, but such a study should not be performed until better methods for the isolation of M. avium from water and for genetic typing are developed. Too many strains either are not recoverable by the isolation techniques that are currently available or are not typeable by LRF analysis due to insufficient growth or the insufficient release of DNA upon lysis.
This study proposes that potable water is probably a source of M. avium infection in humans. A comparative analysis of other clinical and water NTM isolates by PCR and LRF methods will be discussed in future papers. Food may also be a potential source of infection with NTM. We are currently isolating NTM from foods (fresh fruits, vegetables, unpasteurized juices, etc.) and are comparing these isolates to clinical isolates.
ACKNOWLEDGMENTS
We thank the following for generous assistance: Kenneth C. Jones and Chip Harlow, Genetic Information Services, Chatsworth, Calif.; Nancy H. Bishop, Lance Risen, Sean Yoder, and Claudia Argueta, California State University, Northridge; Theresa Sase, Chief Librarian, Northridge Hospital Medical Center, Northridge, Calif.; Ron Ritzman, Neotherapeutics, Irvine, Calif.; J. O. Falkinham III and Laura Via, Virginia Polytechnic Institute and State University, Blacksburg; Robert D. Arbeit, Boston University School of Medicine; David Mintzer, Clinical Standards Laboratories, Dominguez Hills, Calif.; Jeremy Cole; Linda Croad; and Adolph Surtshin.
This work was supported by the U.S. Environmental Protection Agency (cooperative agreement CR-818918) and the Los Angeles Department of Water and Power (grant VC-35261005).
REFERENCES
- 1.Armstrong D, Gold J W M, Dryjanski J, Whimbey E, Polsky B, Hawkins C, Brown A E, Bernard E, Kiehn T F. Treatment of infections in patients with the acquired immunodeficiency syndrome. Ann Intern Med. 1985;103:738–743. doi: 10.7326/0003-4819-103-5-738. [DOI] [PubMed] [Google Scholar]
- 2.Aronson T W, Holtzman A, Glover N, Froman S, Berlin O G W, Dominquez P, Kunkel K, Overturf G, Stelma G, Smith C, Yakrus M. Abstracts of the 93rd General Meeting of the American Society for Microbiology 1993. Washington, D.C: American Society for Microbiology; 1993. Potable water—a source of Mycobacterium avium complex (MAC) infection, abstr. U-80; p. 183. [Google Scholar]
- 3.Aronson T, Holtzman A, Glover N, Boian M, Tran T, Froman S, Berlin O G W, Hill H, Stelma G, Yakrus M. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Molecular epidemiologic matching of nontuberculous mycobacteria (NTM) isolates recovered from AIDS and non-AIDS patients and from potable water, abstr. U-152; p. 128. [Google Scholar]
- 4.Collins F M, Yates M D. Mycobacteria in water. J Appl Bacteriol. 1984;57:736–739. doi: 10.1111/j.1365-2672.1984.tb01384.x. [DOI] [PubMed] [Google Scholar]
- 5.Du Moulin G C, Stottmeier K D. Waterborne mycobacteria: an increasing threat to health. ASM News. 1986;52:525–529. [Google Scholar]
- 6.Du Moulin G C, Stottmeier K D, Pelletier P A, Tsang A Y, Hedley-Whyte J. Concentration of Mycobacterium avium by hospital hot water systems. JAMA. 1988;260:1599–1601. doi: 10.1001/jama.260.11.1599. [DOI] [PubMed] [Google Scholar]
- 7.Fergie J E, Milligan T W, Henderson B M, Stafford W W. Intrathoracic Mycobacterium avium complex infection in immunocompetent children: case report and review. Clin Infect Dis. 1997;24:250–253. doi: 10.1093/clinids/24.2.250. [DOI] [PubMed] [Google Scholar]
- 8.Glover N, Holtzman A, Aronson T, Froman S, Berlin O G W, Dominguez P, Kunkel K, Overturf G, Stelma G, Smith C, Yakrus M. The isolation and identification of Mycobacterium avium complex (MAC) recovered from Los Angeles potable water, a possible source of infection in AIDS patients. Int J Environ Health Res. 1994;4:63–72. [Google Scholar]
- 9.Grange J M, Yates M D, Boughton E. The avian tubercle bacillus and its relatives. J Appl Bacteriol. 1990;68:411–431. doi: 10.1111/j.1365-2672.1990.tb02892.x. [DOI] [PubMed] [Google Scholar]
- 10.Grange J M, Yates M D, Pzniak A. Bacteriologically confirmed non-tuberculous mycobacterial lymphadenitis in south east England: a recent increase in the number of cases. Arch Dis Child. 1995;72:516–517. doi: 10.1136/adc.72.6.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene M B, Gurdip S S, Sidhu S, Lewin S, Levine J F, Masur H, Simberkoff M S, Nicholas P, Good R C, Zolla-Pazner S B, Pollock A A, Tapper M L, Holzman R S. Mycobacterium avium-intracellulare: a cause of disseminated life-threatening infections in homosexuals and drug abusers. Ann Intern Med. 1982;97:539–546. doi: 10.7326/0003-4819-97-4-539. [DOI] [PubMed] [Google Scholar]
- 12.Horsburgh C R. Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N Engl J Med. 1991;324:1332–1338. doi: 10.1056/NEJM199105093241906. [DOI] [PubMed] [Google Scholar]
- 13.Ichiyama S, Shimokata K, Tsukamura M. The isolation of Mycobacterium avium complex from soil, water and dust. Microbiol Immunol. 1988;32:733–739. doi: 10.1111/j.1348-0421.1988.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 14.Iseman M D. Mycobacterium avium complex and the normal host. N Engl J Med. 1989;321:896–898. doi: 10.1056/NEJM198909283211310. [DOI] [PubMed] [Google Scholar]
- 15.Marrie T J, Haldane D, Bezanson G, Peppard R. Each water outlet is a unique niche for Legionella pneumophila. Epidemiol Infect. 1992;108:261–270. doi: 10.1017/s0950268800049736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrie T J, Green P, Burbridge S, Bezanson G, Neale S, Hoffman P S, Haldane D. Legionella in the potable water of Nova Scotia hospitals and Halifax residences. Epidemiol Infect. 1994;112:143–150. doi: 10.1017/s0950268800057502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maslow J N, Slutsky A M, Arbeit R D. The application of pulsed-field gel electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular biology. Washington, D.C: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 18.Mazurek G H, Hartman S, Zhang Y, Brown B A, Hector J S R, Murphy D, Wallace R J., Jr Large DNA restriction fragment polymorphism in the Mycobacterium avium-Mycobacterium intracellulare complex: a potential epidemiologic tool. J Clin Microbiol. 1993;31:390–394. doi: 10.1128/jcm.31.2.390-394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novick R J, Moreno-Cabral C E, Stinson E B, Oyer P E, Starnes V A, Hunt S A, Shumway N E. Nontuberculous mycobacterial infections in heart transplant recipients: a seventeen year experience. J Heart Transplant. 1990;9:357–363. [PubMed] [Google Scholar]
- 20.Pestel-Caron M, Arbeit R D. Characterization of IS1245 for strain typing of Mycobacterium avium. J Clin Microbiol. 1998;36:1859–1863. doi: 10.1128/jcm.36.7.1859-1863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plough J F, Para M F, Maher W E, Hackman B, Webster L. Subtypes of Legionella pneumophila serogroup 1 associated with different attack rates. Lancet. 1983;ii:649–650. doi: 10.1016/s0140-6736(83)92531-x. [DOI] [PubMed] [Google Scholar]
- 22.Prince D S, Peterson D D, Steiner R M, Gottlieb J E, Scott R, Israel H L, Figueroa W G, Fish J E. Infections with Mycobacterium avium complex in patients without predisposing conditions. N Engl J Med. 1989;321:863–868. doi: 10.1056/NEJM198909283211304. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed., book 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. p. 128. [Google Scholar]
- 24.Schulze-Robbecke B, Janning B, Fischeder R. Occurrence of mycobacteria in biofilm samples. Tubercle Lung Dis. 1992;73:141–144. doi: 10.1016/0962-8479(92)90147-C. [DOI] [PubMed] [Google Scholar]
- 25.Slutsky A, Arbeit R D, Barber T W, Rich J, Reyn C F, Pieciak W, Barlow M A, Maslow J N. Polyclonal infections due to Mycobacterium avium complex in patients with AIDS detected by pulsed-field gel electrophoresis of sequential clinical isolates. J Clin Microbiol. 1994;32:2233–2238. doi: 10.1128/jcm.32.7.1773-1778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stout J E, Yu V L, Yee Y C, Vaccarello S, Diven W, Lee T C. Legionella pneumophila in residential water supplies: environmental surveillance with clinical assessment for Legionnaires disease. Epidemiol Infect. 1992;109:49–57. [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;32:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Reyn F C, Maslow J N, Barber T W, Falkinham III J, Arbeit R D. Persistent colonization of potable water as a source of Mycobacterium avium infection in AIDS. Lancet. 1994;343:1137–1141. doi: 10.1016/s0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 29.Wright J E. Non-tuberculous mycobacterial lymphadenitis. Aust N Z J Surg. 1996;66:225–228. doi: 10.1111/j.1445-2197.1996.tb01170.x. [DOI] [PubMed] [Google Scholar]
- 30.Zackowski P, Fligiel S, Berlin O G W, Johnson L., Jr Disseminated Mycobacterium avium-intracellulare infection in homosexual men dying of acquired immunodeficiency. JAMA. 1982;248:2980–2982. doi: 10.1001/jama.1982.03330220024029. [DOI] [PubMed] [Google Scholar]