Abstract
Klebsiella pneumoniae is an important pathogenic bacterium commonly associated with human healthcare and community-acquired infections. In recent years, K. pneumoniae has become a significant threat to global public and veterinary health, because of its high rates of antimicrobial resistance (AMR). Early diagnosis of K. pneumoniae infection and detection of any associated AMR would help to accelerate directed therapy and reduce the risk of the emergence of multidrug-resistant isolates. In this study, we identified three target genes (yhaI, epsL, and xcpW) common to K. pneumoniae isolates from both China and Europe and designed loop-mediated isothermal amplification (LAMP) assays for the detection of K. pneumoniae in clinical samples. We also designed LAMP assays for the detection of five AMR genes commonly associated with K. pneumoniae. The LAMP assays were validated on a total of 319 type reference strains and clinical isolates of diverse genetic backgrounds, in addition to 40 clinical human sputum samples, and were shown to be reliable, highly specific, and sensitive. For the K. pneumoniae–specific LAMP assay, the calculated sensitivity, specificity, and positive and negative predictive values (comparison with culture and matrix-assisted laser desorption/ionization–time of flight mass spectrometry) were all 100% on clinical isolates and, respectively, of 100%, 91%, and 90%, and 100% when tested on clinical sputum samples, while being significantly faster than the reference methods. For the bla KPC and other carbapenemases’ LAMP assays, the concordance between the LAMP results and the references methods (susceptibility tests) was 100%, on both pure cultures (n = 125) and clinical samples (n = 18). In conclusion, we developed highly sensitive and specific LAMP assays for the clinical identification of K. pneumoniae and detection of carbapenem resistance.
Keywords: rapid diagnostics, antimicrobial resistance (AMR), Klebsiella pneumoniae (K. pneumoniae), carbapenemases genes, LAMP (loop mediated isothermal amplification)
Introduction
Klebsiella pneumoniae is a Gram-negative bacterium belonging to the Enterobacteriaceae family within the Enterobacterales order (Adeolu et al., 2016). K. pneumoniae can be found in the environment, associated with plants, soil, and water, but can also colonize a wide range of animal and human hosts, resulting in clinical manifestations including liver abscess, respiratory tract, urinary tract, gut, skin, and systemic infections (Podschun and Ullmann, 1998; Li et al., 2014). Furthermore, K. pneumoniae is a common pathogen implicated in human healthcare-associated and community-acquired infections and is becoming a significant threat to global public and veterinary health, because of its high rates of antimicrobial resistance (AMR) (Wyres and Holt, 2016). The most common AMR K. pneumoniae lineages, associated with the occurrence of hospital outbreaks and deaths worldwide, are those producing the K. pneumoniae carbapenemase (KPC), conferring resistance to penicillins, most cephalosporins, and carbapenems (Nordmann et al., 2009; Palzkill, 2018). More than 100 distinct acquired AMR genes have been identified in K. pneumoniae, and historically, it is the organism where most AMR genes were first detected, before becoming widespread in other Gram-negative bacterial pathogens (Holt et al., 2015; Wyres and Holt, 2016). With the advent of high-throughput whole genome sequencing and core genome multilocus sequence typing in the last decade, several genomic studies investigated the population structure of K. pneumoniae and the evolution of AMR clones (Bialek-Davenet et al., 2014; Holt et al., 2015). These studies demonstrated the direct transfer of AMR plasmids between K. pneumoniae and other Enterobacterales in isolates recovered from hospital environments (Wyres and Holt, 2016), and the wide association of K. pneumoniae with carbapenemase-producing genes bla KPC, bla OXA-48-like, and bla NDM-1, and the extended-spectrum beta-lactamase (ESBL) gene bla CTX-M-15 (Munoz-Price et al., 2013; Pitout et al., 2015; Zowawi et al., 2015; Wyres and Holt, 2016). The dissemination of these carbapenemase-producing and ESBL genes and more recently plasmid-mediated colistin resistance gene mcr-1 (Liu et al., 2016) is of particular clinical concern. Currently, the choices for carbapenemase detection are phenotypic tests that require bacterial culture, such as the modified Hodge test (Bartolini et al., 2014) and disc diffusion tests using meropenem, phenyl boronic acid, or ethylenediaminetetraacetic acid (Bartolini et al., 2014; Sood, 2014); matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Ghebremedhin et al., 2016; Yu et al., 2018); carba NP test (Nordmann et al., 2012; Segawa et al., 2017) or its derivatives (Tamma and Simner, 2018); and the carbapenem inhibition test (van der Zwaluw et al., 2015; Kuchibiro et al., 2018). Molecular methods such as polymerase chain reaction (PCR) (Solanki et al., 2013; Moloney et al., 2019) and loop-mediated isothermal amplification (LAMP) (Solanki et al., 2013; Cheng et al., 2014; Nakano et al., 2015; Srisrattakarn et al., 2017; Funashima et al., 2018; Feng et al., 2021) have also been described, for more rapid diagnostics. Early intervention facilitated by rapid diagnostics would help to accelerate directed therapy, thus slowing down the dissemination of AMR and reducing the risk of the emergence of multidrug-resistant strains that have limited treatment options (Brogan and Mossialos, 2016; O’Neill, 2016; Piddock, 2016).
LAMP is a single-step nucleic acid amplification technique that is rapid, sensitive, and cost-effective. This technique can amplify a few copies of DNA into billions of copies within 30 min (Notomi et al., 2000; Diribe et al., 2014) and has been used to detect a wide range of viral, bacterial, and parasitic pathogens (Wong Y.-P. et al., 2018; Li et al., 2019; Tian et al., 2019; Vergara et al., 2020). LAMP is less sensitive to PCR inhibitors in poorly or nonprocessed samples, such as blood, sputum, and urine, making it suitable for use as a point-of-care diagnostic (Notomi et al., 2000; Nagamine et al., 2002; Kaneko et al., 2005; Parida et al., 2008; Francois et al., 2011; Dhama et al., 2014; Gadkar et al., 2018; Moehling et al., 2021). LAMP is performed at a constant isothermal temperature using specifically designed primers and a strand-displacement DNA polymerase (Notomi et al., 2000), and the amplified products can be detected by fluorometry, turbidity, and colorimetry (Wong Y.-P. et al., 2018; Moehling et al., 2021). LAMP amplification can also be detected by the measurement of by-products using hydroxynaphthol blue dye, calcein, or malachite green (Liu et al., 2012; Cheng et al., 2014; Kim et al., 2016; Lucchi et al., 2016; Srisrattakarn et al., 2017).
In this study, we identified target genes common to K. pneumoniae isolates from both China and Europe and designed LAMP assays for the detection of K. pneumoniae. We also designed LAMP assays for the detection of the most important classes of AMR genes in K. pneumoniae. The LAMP assays were validated on a total 319 strains (5 K. pneumoniae type strains, 167 K. pneumoniae clinical isolates, 32 non–K. pneumoniae type strains, and 115 non–K. pneumoniae clinical isolates) from the United Kingdom and China in addition to 40 clinical sputum samples. All the designed assays were shown to be reliable, highly specific, and sensitive.
Materials and Methods
Ethics Approval Statement
Ethical approval, including the waiver of informed consent of the clinical strains and samples collected in Ruijin Hospital, was approved by the Ethics Committee of Ruijin Hospital, School of Medicine, Shanghai Jiao Tong University, under approval no. RJ2019NO1-3. The research conformed to the principles of the Helsinki Declaration. The study involved no more than minimal risk to subjects, and no personal information was obtained.
Bacterial Isolates and Growth Conditions
In the United Kingdom, type strains of K. pneumoniae: NCTC 13809, NCTC 13438, and NCTC 13439 (Supplementary Table S1); Escherichia coli: NCTC 14321, NCTC 13441, and ATCC 25922; Enterobacter cloacae: NCTC 13380 and NCTC 14322; Salmonella enterica: ATCC 14028, NCTC 05776, NCTC 10705, NCTC 13346, and NCTC 10532; Acinetobacter baumannii: ATCC Ab19606; Acinetobacter lwoffii: NCTC 05866; Staphylococcus aureus: NCTC 12981; MRSA NCTC 12493, NCTC 8325; and MRSA BAA-1680/United States of America 300; Yersinia enterocolitica: NCTC 12982; and Streptococcus pyogenes: NCTC 12696 were purchased from Public Health England (PHE) culture collections (National Collection of Type Cultures [NCTC]) and American Type Culture Collection (ATCC) (Supplementary Table S2).
K. pneumoniae isolates (n = 47) were obtained from patient samples in University Hospitals of Leicester NHS Trust and Epsom and St. Helier University Hospitals, as well as from sink drain traps of a Lancashire NHS hospital (Supplementary Table S1). K. pneumoniae isolates were cultured aerobically at 37°C for 16 h on Klebsiella selective agar and then subcultured onto Luria–Bertani agar. S. enterica isolates (n = 5) from the Leicester hospital were cultured aerobically at 37°C on brilliant green agar for 16 h and then subcultured onto Luria–Bertani agar (Supplementary Table S2). S. aureus, Enterobacter spp., Pseudomonas spp. (P. putida KT2440 (Franklin et al., 1981), P. aeruginosa PAO1 (Jacobs et al., 2003)), Klebsiella oxytoca (n = 9, from sink drain traps of a Lancashire NHS hospital), and Acinetobacter spp. strains were cultured aerobically at 37°C for 16 h on Luria–Bertani agar before use in the LAMP development and validation (Supplementary Table S2).
In China, type strains of K. pneumoniae HS11286 and RJF999, E. coli DH5α, S. enterica ATCC14028, A. baumannii ATCC19606, P. aeruginosa PAO1, and S. aureus N315 were obtained from State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University. One hundred twenty K. pneumoniae clinical isolates and 110 non–K. pneumoniae clinical isolates (including 5 Klebsiella aerogenes, 5 K. oxytoca, 20 E. coli, 20 S. enterica, 20 A. baumannii, 20 P. aeruginosa, and 20 S. aureus) from China were collected and identified in Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine by MALDI-TOF and MS, using the Vitek 2 automated system (bioMérieux, Inc., Marcy l’Etoile, France) in accordance with the manufacturer’s instructions, and cultured aerobically at 37°C for 16 h on Luria–Bertani agar before use (Supplementary Table S2).
Identification of Gene Targets for the Detection of K. pneumoniae
A total of 8,638 K. pneumoniae genomes, from all over the world, were downloaded from online databases (FTP site, GenBank, NCBI) with their associated metadata, and 7,320 were selected based on assemblies metrics (Supplementary Table S3), using Quast version 4.5 (Gurevich et al., 2013). Contigs that comprised fewer than 200 nucleotides were excluded. Only genome assemblies with a total size between 4 and 6 Mbp, an N50 of >50 kb, and L50 <20 were included. To identify suitable discriminatory gene targets for K. pneumoniae, 100 (50 from Europe and 50 from Asia) of the K. pneumoniae genome assemblies were selected for the next stage of the study (Supplementary Table S4). The selected genomes assemblies were representative of the prevalent sequence types. In addition, 50 K. oxytoca genomes assemblies (Supplementary Table S5) were downloaded from online databases (FTP site, GenBank, NCBI). These 150 genomes assemblies were compared as detailed below. Genome assemblies were annotated using Prokka V.1.14.5 (Seemann, 2014), and annotated features were used for construction of a pangenome using Roary version 3.12.0 (Page et al., 2015) with a 95% identity cutoff.
The K. pneumoniae and K. oxytoca pan-genomes were used to determine genes specifically associated with K. pneumoniae using Scoary (Brynildsrud et al., 2016). Genes were determined to be associated with K. pneumoniae if they occurred at a frequency of 100% in specific K. pneumoniae and of 0% in K. oxytoca. K. pneumoniae–associated genes were incorporated into a custom ABRicate (https://github.com/tseemann/abricate) database (Supplementary List S6 with ABRicate database) and used to screen a large collection of 7,320 K. pneumoniae genomes derived from all over the world (Supplementary Table S3), using a BLAST cutoff of 80% identity and 80% coverage. An 80% identity cutoff was chosen for BLAST to allow identification of discriminatory target genes while also detecting gene homologs or orthologs with high sequence identity, as potential homologs could confound diagnostic gene target selection. Following the ABRicate BLAST, an NCBI BLAST, with default parameters and excluding K. pneumoniae, was performed to find orthologs in other bacteria with sequence similarity, for each identified target gene.
Preparation of Target DNA
DNA from K. pneumoniae isolates was extracted using the Wizard® Genomic Purification Kit (Promega, United Kingdom). DNA concentration was measured using a Nanoquant plate on a Tecan 10M Spark microplate reader (Tecan, United Kingdom). For routine testing of K. pneumoniae by LAMP, two to three colonies of a pure culture of K. pneumoniae isolates were boiled in 200 µl of nuclease-free water to produce a lysate. Samples were stored at −20°C until use. For determining the limit of DNA concentration of detection of the optimized LAMP assay, DNA was 10-fold serially diluted from 1 ng/μl to 10 fg/μl in nuclease-free water. The limit of bacterial copies per LAMP reaction was also determined, using a protocol previously described in other studies (Goto et al., 2010; Manajit et al., 2018). For these studies, a 16 h broth culture of K. pneumoniae HS11286 (OD600 nm at 0.5) was serially diluted 10-fold dilutions in 0.85% of normal saline. 100 µl of each dilution was plated on Luria–Bertani agar plates, in triplicates, to enumerate the bacterial count, and another 1,000 µl was used for DNA preparation.
For each primer set, the colony-forming units (CFUs) per LAMP reaction were calculated, using the following formula:
LAMP Assays and Measures of Test Accuracy
LAMP primers were designed using Optigene LAMP Designer software. For each K. pneumoniae and carbapenemase target gene, a minimum of two sets of four to six primers; outer primers (F3 and B3), inner primers (FIP and BIP), and for some sets loop primers (LoopF and/or LoopB), recognizing six to eight distinct regions of target DNA, were designed (Table 1).
TABLE 1.
LAMP primer sets designed for this study, targeting the K. pneumoniae Inner membrane protein (yhaI) and carbapenemase genes (bla KPC, bla NDM, bla OXA-48-like, bla IMP, and bla VIM).
| Genes a | Primer | Sequence (5′ to 3′) |
|---|---|---|
| yhaI | yhaI-F3 | ATGTATTGATCGCCACCG |
| yhaI-B3 | GAGCCAGCGAAATAATTGC | |
| yhaI-FIP | TCAACAGCAGCCAGAGCGCTCAATACGGCATCCTCAG | |
| yhaI-BIP | CGGCGGATGCATGATATCGGAAACCACGGAATGATAACCC | |
| yhaI-LF | GGCAAAGGGAATGACAACAAA | |
| yhaI-LB | CCGGGATCTGGTTTGTGT | |
| bla KPC | KPC-F3 | CGTGACGGAAAGCTTACAA |
| KPC-B3 | GCTGTGCTTGTCATCCTT | |
| KPC-FIP | GCCGGTCGTGTTTCCCTTAAACTGACACTGGGCTCT | |
| KPC-BIP | ACTGGGCAGTCGGAGACAAGACGACGGCATAGTCAT | |
| KPC-LF | AGCCAATCAACAAACTGCTG | |
| KPC-LB | GAGTGTATGGCACGGCAA | |
| bla NDM | NDM-F3 | CATTAGCCGCTGCATTGATG |
| NDM-B3 | CCGCCATCCCTGACGATC | |
| NDM-FIP | ATCGCCAAACCGTTGGTCGCCCGGTGAAATCCGCCCG | |
| NDM-BIP | TGGTTTTCCGCCAGCTCGCAGCGACTGCCCCGAAAC | |
| NDM-LF | TCCATTTGCTGGCCAATCG | |
| NDM-LB | ACCGAATGTCTGGCAGCACA | |
| bla OXA-48-like | OXA48-like-F3 | ATCACAGGGCGTAGTTGT |
| OXA48-like-B3 | CGTCTGTCCATCCCACTT | |
| OXA48-like-FIP | TGCTTGGTTCGCCCGTTTAACTCTGGAATGAGAATAAGCAG | |
| OXA48-like-BIP | TACCCGCATCTACCTTTAAAATTCCAAGACTTGGTGTTCATCCT | |
| OXA48-like-LB | GCTTGATCGCCCTCGATTTG | |
| bla IMP | IMP-F3 | CACTTGGTTTGTGGAACG |
| IMP-B3 | AGCCAATAGTTAACCCCG | |
| IMP-FIP | TAAGCCACTCTATTCCGCCCAAAATAAAAGGCAGTATTTCCTCTC | |
| IMP-BIP | CATCCCCACGTATGCGTCTGCCAAATGAATTTTTAGCTTGAACC | |
| IMP-LB | CTAATGAGCTGCTTAAAAAAGACGG | |
| bla VIM | VIM-F3 | AATTCCGGTCGGAGAGGT |
| VIM-B3 | AAAGTGCGTGGAGACTGC | |
| VIM-FIP | CATTGGACGGGTAGACCGCGCCAGATTGCCGATGGTGT | |
| VIM-BIP | TGATTGATACAGCGTGGGGTGCACGGGAAGTCCAATTTGCT | |
| VIM-LF | CGTTGCGATATGCGACCAA |
The complete coding sequences of yhaI (Kp00840_02010 in Supplementary List S6); bla KPC (NC_023904.1), bla NDM-1 (FN396876), bla OXA-48 (AY236073), bla IMP-4 (AF244145), and bla VIM-1 (Y18050) were the reference sequences.
LAMP was initially performed using the fluorometric detection method. Individual LAMP reactions consisted of 15 µl of ISO-004 Isothermal Mastermix (OptiGene, Ltd., United Kingdom), 2.5 µl of 10× primer mix containing 8 µM of FIP/BIP, 2 µM of F3/B3 and 4 µM of LoopF/LoopB, 2.5 µl of nuclease-free water and 5 µl of DNA template, or nuclease-free water for negative controls. LAMP reactions were run at 65°C for 30 min, and the fluorescence of a DNA binding dye was evaluated every 15 s to generate amplification curves, using Genie II device (OptiGene, Ltd.). Amplicons’ annealing profiling was also performed by a temperature gradient from 98°C to 80°C at a rate of 0.05°C × s−1, using Genie II. Optimal primers for K. pneumoniae detection and differentiation were chosen based on accuracy and speed of amplification.
Once the best primers were chosen, LAMP was also performed using a colorimetric method of detection. Individual LAMP reactions consisted of 12.5 µl of WarmStart® Colorimetric LAMP 2× Mastermix (New England Biolabs Ltd., United Kingdom), 2.5 µl of 10× primers mix containing 16 μM FIP/BIP, 2 μM F3/B3 and 4 μM LoopF/LoopB, 5 µl of nuclease-free water and 5 µl of DNA template, or nuclease-free water for negative controls. Successful LAMP amplification induced a pH change, resulting from proton accumulation due to dNTP incorporation. This pH acidification was detected by phenol red, a pH-sensitive dye, which color changed from pink (negative) to yellow (positive). Photographs of LAMP reactions were taken after 30 min of amplification at 65°C with a MyBlock™ mini dry bath (Benchmark Scientific, United States).
For each primer set, contingency tables were constructed, and sensitivity, specificity, and positive and negative predictive values were calculated.
Testing of Clinical Sputum Samples
Clinical sputum samples were collected into a sterile sealed container according to routine procedure at Ruijin Hospital. A total of 40 clinical sputum samples (1–40) were selected for this study. According to culture and MALDI-TOF MS, 18 samples were positive for K. pneumoniae, and 22 samples were negative (Supplementary Table S7). Among the 22 negative samples, 17 contained other bacterial pathogens (Supplementary Table S7). All the sputum samples were decontaminated by adding an equal volume of 10% NaOH solution, vortexed thoroughly, and incubated at room temperature for 5 min. Following incubation, 400 μl of each sample was centrifuged for 5 min at 12,000 rpm at ambient temperature. Following centrifugation, the supernatants were discarded, and the pellets were used for DNA extraction.
Susceptibility to imipenem and meropenem was determined in the K. pneumoniae isolates recovered from the sputum samples, using the Vitek® 2 (bioMérieux, Inc.) automated system. Minimum inhibitory concentrations were classified according to breakpoints established by the Clinical and Laboratory Standards Institute (Wayne, 2020).
To assess the feasibility of blood samples testing with our diagnostics, the limit of bacterial copies per LAMP reaction was also determined in spiked defibrinated sheep blood samples, using a protocol previously described in other studies (Goto et al., 2010; Manajit et al., 2018). For these studies, a 16-h broth culture of K. pneumoniae HS11286 (OD600 nm at 0.5) was serially diluted 10-fold dilutions in 0.85% of normal saline; 100 µl of each dilution was plated on Luria–Bertani agar plates, in triplicates, to enumerate the bacterial count. Then, 20 µl of 10 bacterial cell suspensions, with concentrations ranging from of 0.38 CFUs/ml to 3.8 × 107 CFUs/ml, were used to spike 180 µl sterile defibrinated sheep blood samples each. All 10 spiked blood samples (200 µl each) were used for DNA extraction, and DNA was eluted in 100 µl. For each primer set, the CFUs per LAMP reaction were calculated (using the same formula than described in 2.4).
PCR Verification of the LAMP Assays
DNA extracted from the isolates and clinical samples was tested for the presence of the carbapenemase genes bla KPC, bla NDM, bla OXA-48-like, bla IMP, and bla VIM and of the yhaI gene by PCR. The PCR primers for yhaI (249bp size product) and bla KPC (240bp size product) genes were designed for this study using the Primer-Blast software (Ye et al., 2012) (Table 2). The PCR testing for the carbapenemase genes bla NDM , bla OXA-48-like , bla IMP , and bla VIM was performed with specific primers and conditions as previously described (Dallenne et al., 2010; Han et al., 2020) (Table 2).
TABLE 2.
PCR primers for the K. pneumoniae inner membrane protein (yhaI) and carbapenemase genes (bla KPC, bla NDM, bla OXA-48-like, bla IMP, and bla VIM).
| Genes | Primer a | Sequence b (5′ to 3′) | Product size (bp) |
|---|---|---|---|
| yhaI | yhaI-F | ATTTGAGCGGCTGGAAAGAG | 249 |
| yhaI-R | AGCGGCCGATATCATGCAT | ||
| bla KPC | KPC-F | TTGTTGCTGAAGGAGTTGGG | 240 |
| KPC-R | GGTCGTGTTTCCCTTTAGCC | ||
| bla NDM | NDM-F | GGTTTGGCGATCTGGTTTTC | 621 |
| NDM-R | CGGAATGGCTCATCACGATC | ||
| bla OXA-48-like | OXA-48like-F | GCGTGGTTAAGGATGAACAC | 438 |
| OXA-48like-R | CATCAAGTTCAACCCAACCG | ||
| bla IMP | IMP-F | GGAATAGAGTGGCTTAAYTCTC | 232 |
| IMP-R | GGTTTAAYAAAACAACCACC | ||
| bla VIM | VIM-F | GATGGTGTTTGGTCGCATA | 390 |
| VIM-R | CGAATGCGCAGCACCAG |
The PCR primers of carbapenemase genes (bla KPC, bla NDM, bla OXA-48-like, bla IMP, and bla VIM) used were those as previously described by Dallenne et al. (2010), Han et al. (2020), respectively;
b Y = C or T.
Results
Identification of Gene Targets for the Detection of K. pneumoniae
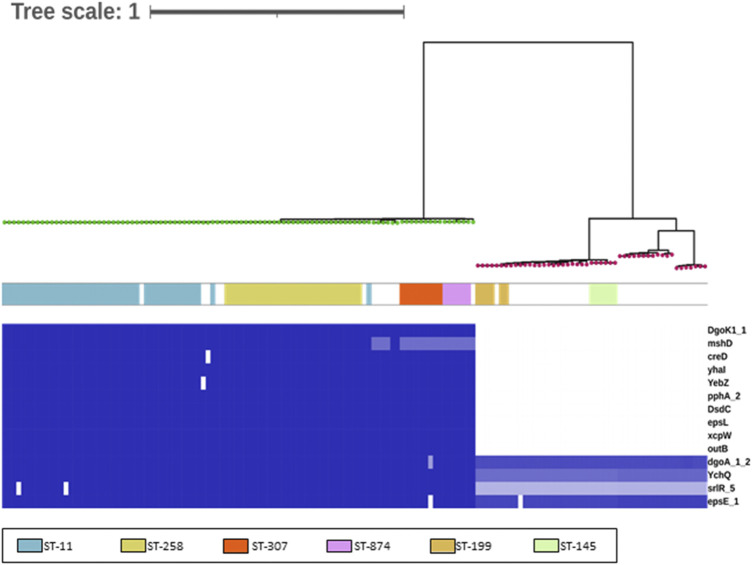
A total of 50 K. pneumoniae genomes from Europe, 50 K. pneumoniae genomes from China (Supplementary Table S4), and 50 K. oxytoca genomes (Supplementary Table S5) were selected to identify unique targets for K. pneumoniae. After annotation of genomes assemblies using Prokka and construction of the pan-genomes using Roary, the identification of genes specifically associated with K. pneumoniae was performed using Scoary. A total of nine genes were associated with K. pneumoniae at a frequency of 100% and 0% with K. oxytoca in the studied Klebsiella genomes (Table 3; Figure 1). These nine genes were incorporated into a custom ABRicate (Seemann, 2019) database (Supplementary List S6) and used to screen a large collection of 7,320 K. pneumoniae genomes derived from all over the world (Supplementary Table S3), using cutoffs of 80% identity and 80% coverage. Out of the nine genes, seven were found to be universally present in the 7,320 K. pneumoniae genomes (Table 3, Supplementary Table S8). NCBI BLAST searches, excluding K. pneumoniae, were then performed and revealed that three genes were exclusive to K. pneumoniae (Table 3; Figure 1). These three genes were yhaI (Kp00840_02010 in Supplementary List S6), encoding an inner membrane protein; xcpW (Kp00844_46720 in Supplementary List S6), encoding the type II secretion system protein J; and epsL (Kp00844_46740 in Supplementary List S6), encoding the type II secretion system protein L. These genes were selected as putative targets for design of a LAMP assay to rapidly detect K. pneumoniae in clinical samples.
TABLE 3.
List of gene targets identified for the detection of K. pneumoniae.
| Gene name | Protein | Sequence |
|---|---|---|
| yhaI | Inner membrane protein | Kp00840_02010 |
| epsL | Type II secretion system protein L | Kp00844_46720 |
| xcpW | Type II secretion system protein J | Kp00844_46740 |
| outB | General secretion pathway protein B | Kp00844_46830 |
| creD | Inner membrane protein | Kp00840_01750 |
| YebZ | Inner membrane protein | Kp00844_07280 |
| pphA_2 | Serine/threonine-protein phosphatase | Kp00844_07320 |
| DgoK1_1 | Putative 2-dehydro-3-deoxygalactonokinase | Kp00840_01120 |
| DsdC | d-Serine deaminase activator | Kp00844_23880 |
The Scoary analyses output a list of nine genes associated with K. pneumoniae at a frequency of 100 and 0% with K. oxytoca. Of the nine genes, seven (genes in bold) were found in the 7,320 K. pneumoniae genomes, using a custom ABRicate. NCBI BLAST searches identified three genes (in bold and underlined) as gene targets.
FIGURE 1.
Identification of discriminatory gene targets for K. pneumoniae. This matrix was generated using BLAST (Abricate) using an 80% identity and coverage threshold to identify potential variation. Intensity of the blue heat map indicates identity relative to query sequences (Supplementary File S2) (blue = 100%, white = 0%/absence). Genes encoded by both Klebsiella species are also listed, for reference (bottom 4). The genomes are ordered according to the branching produced using core genome SNP as identified using ParSNP and FastTree. The green nodes are K. pneumoniae genomes, and the red nodes, K. oxytoca genomes. The colors indicate the six sequence types (STs) identified in the Klebsiella included in the study.
Specificity, Sensitivity, and Limit of Detection of the K. pneumoniae LAMP Assay on Clinical Isolates
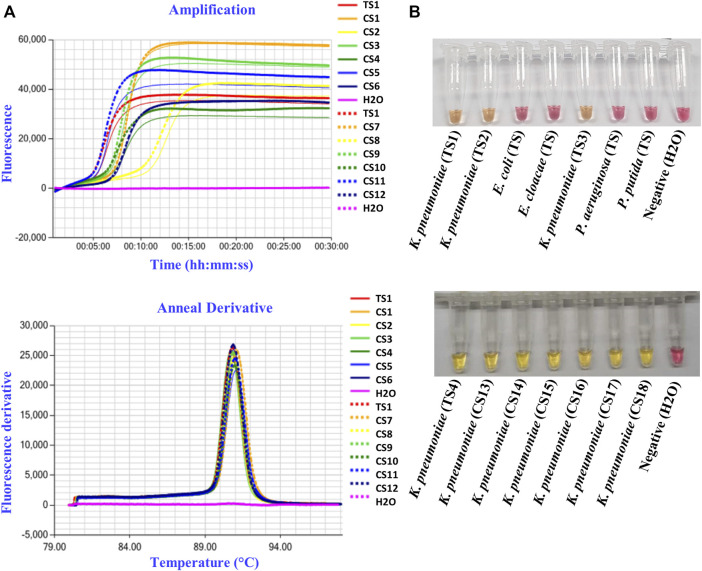
To detect K. pneumoniae, LAMP primers were designed for amplification of the three target genes identified using comparative genomics: yhaI, epsL, and xcpW. Each primer set was used to test a panel of 15 K. pneumoniae strains, with a fluorometric LAMP detection method (Genie II; Optigene Ltd., Horsham, England). Following screening on a panel of strains, the yhaI gene primer set that showed the best accuracy and speed of amplification was selected and subsequently used to amplify the genomic DNA of 45 K. pneumoniae strains from the United Kingdom, which it did in less than 15 min. The amplification of a single product in the LAMP reactions was confirmed, by analyzing the melting curves (Figure 2 and Table 4). A single sharp dissociation peak was observed for the yhaI gene primer set only in the LAMP reactions containing K. pneumoniae templates. The annealing temperature was 90.5°C ± 1°C. LAMP detection of K. pneumoniae was also evaluated with colorimetric detection, for use as a point-of-care diagnostic requiring minimal equipment. Using this detection method and the yhaI gene primers, a 15-min LAMP reaction was determined to be sufficient for the accurate detection of K. pneumoniae (Figure 2 and Table 4). Using colorimetric LAMP, 3 K. pneumoniae type strains, 47 K. pneumoniae clinical isolates from United Kingdom, and 2 K. pneumoniae type strains and 120 K. pneumoniae clinical isolates from China were detected within 15 min. No amplification and no annealing or color change were observed in the no-template control reactions, irrespective of the method of detection.
FIGURE 2.
K. pneumoniae LAMP detection in pure cultures. (A) Representative fluorometric LAMP results, targeting the yhaI gene, performed on type strain (TS1) and clinical isolates (CS1–12) of K. pneumoniae from the United Kingdom, with H2O as a negative control. (B) Representative colorimetric LAMP results, targeting the yhaI gene, performed on type strains (TS) and clinical isolates (CS) of K. pneumoniae from the United Kingdom (TS1–3) and from China (TS4 and CS13–18), as well as on type strains of E. coli, E. cloacae, Pseudomonas aeruginosa, and P. putida, with H2O as a negative control.
TABLE 4.
Summary of K. pneumoniae–specific LAMP results.
| Tested isolates | yhaI detection with colorimetric method (on 319 isolates) | yhaI detection with fluorometric method (on 88 isolates) |
|---|---|---|
| K pneumoniae type strains (PHE and ATCC) | 5/5 positives in 15 min | 5/5 positives in 10 min |
| United Kingdom K. pneumoniae clinical isolates (partner hospitals) | 47/47 positives in 15 min | 47/47 positives in 13 min |
| Ruijin Hospital K. pneumoniae clinical isolates | 120/120 in 15 min | |
| K. aerogenes (China) | 0/5 clinical isolates | |
| K. oxytoca (United Kingdom and China) | 0/9 type strains and 0/5 clinical isolates | 0/9 type strains |
| E. coli (United Kingdom and China) | 0/4 type strains and 0/20 clinical isolates | 0/4 type strains |
| S. enterica (United Kingdom and China) | 0/5 type strains and 0/25 clinical isolates | 0/5 type strains and 0/5 clinical isolates |
| Acinetobacter spp. (United Kingdom and China) | 0/2 type strains and 0/20 clinical isolates | 0/2 type strains |
| Pseudomonas spp. (United Kingdom and China) | 0/3 type strains and 0/20 clinical isolates | 0/2 type strains |
| S. aureus (United Kingdom and China) | 0/5 type strains and 0/20 clinical isolates | 0/5 type strains |
| E. cloacae (United Kingdom) | 0/2 positive | 0/2 type strains |
| Y. enterocolitica (United Kingdom) | 0/1 positive | 0/1 type strains |
| S. pyogenes (United Kingdom) | 0/1 positive | 0/1 type strains |
All results obtained by LAMP targeting the yhaI gene, performed on 319 strains: 5 K. pneumoniae type strains, 167 K. pneumoniae clinical isolates, 32 non–K. pneumoniae type strains, and 115 non–K. pneumoniae clinical isolates.
BLAST queries of the primer binding regions from yhaI primer set against the NCBI nonredundant database, excluding K. pneumoniae, demonstrated extremely low sequence identity. Subsequent testing of the primer set on 10 other bacterial species (K. aerogenes, K. oxytoca, E. coli, S. enterica, Acinetobacter spp., Pseudomonas spp., S. aureus, E. cloacae, Y. enterocolitica, and S. pyogenes) showed no cross reactions (Table 4).
For yhaI gene detection, the calculated sensitivity, specificity, and positive and negative predictive values (Table 5, established using LAMP results obtained on the 319 strains described in Table 4) were all 100% on pure cultures.
TABLE 5.
Contingency table for K. pneumoniae–specific LAMP.
| LAMP test results | Reference method (MALDI-TOF and culture) | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 172 | 0 | 172 |
| Negative | 0 | 147 | 147 |
| Total | 172 | 147 | 319 |
Results obtained by LAMP targeting the yhaI gene, performed on 319 strains: 5 K. pneumoniae type strains, 167 K. pneumoniae clinical isolates, 32 non–K. pneumoniae type strains, and 115 non–K. pneumoniae clinical isolates.
To determine the limit of detection of the designed LAMP assay, the genomic DNAs of the type strains K. pneumoniae NCTC13809 and K. pneumoniae HS11286 were extracted and then diluted from 10 ng/μL to 1 fg/μl. Using fluorometric detection, the lowest concentration of K. pneumoniae NCTC13809 DNA amplified by the yhaI primers was 100 fg/μl (equivalent to 16.1 gene copies/µl). Using colorimetric detection, the limit of detection for yhaI in K. pneumoniae HS11286 was 10 pg/reaction, which is 100 times lower than the routine PCR method used at the Ruijin Hospital. When considering the viable CFU counts, used for DNA template preparation, the limit of detection for yhaI was found to be 3.8 CFUs/reaction.
LAMP Assays to Detect Carbapenem Resistance in Clinical Isolates
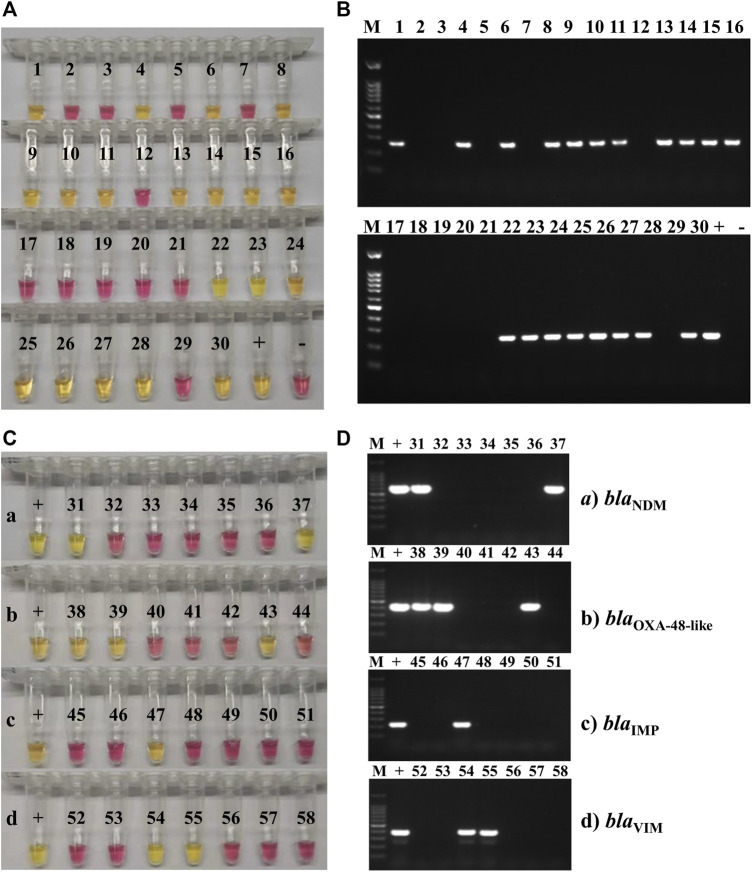
For the detection of carbapenemase (bla KPC )-producing K. pneumoniae, the set of LAMP primers that showed the best accuracy and speed of amplification was validated on a panel of 17 K. pneumoniae and K. oxytoca carbapenem-resistant strains from the United Kingdom. All 17 strains were bla KPC -positive using both fluorometric LAMP and bla KPC PCR detection. For LAMP, a single sharp dissociation peak was observed only in the LAMP reactions containing bla KPC -positive templates, at an annealing temperature of 92.5°C ± 0.5°C. For PCR, a single band was observed (approximately 240 bp). bla KPC primers were subsequently tested on K. pneumoniae type strain HS11286 and clinical isolates collected at Ruijin Hospital (Shanghai, China). K. pneumoniae type strain HS11286 and 66 of the 120 K. pneumoniae clinical isolates were bla KPC-positive within 15 min (Figure 3; Table 6), using colorimetric LAMP. The strains that were bla KPC-positive by LAMP were also positive by PCR and were carbapenem-resistant according to AST using Vitek® 2 automated system. No amplification and no annealing or color change were observed in the no-template control reactions, irrespective of the method of detection (Figure 3; Table 6). In addition, all the strains bla KPC negatives by PCR were also negative by bla KPC LAMP. For bla KPC gene detection, the calculated sensitivity, specificity, and positive and negative values (Table 6) were all 100%.
FIGURE 3.
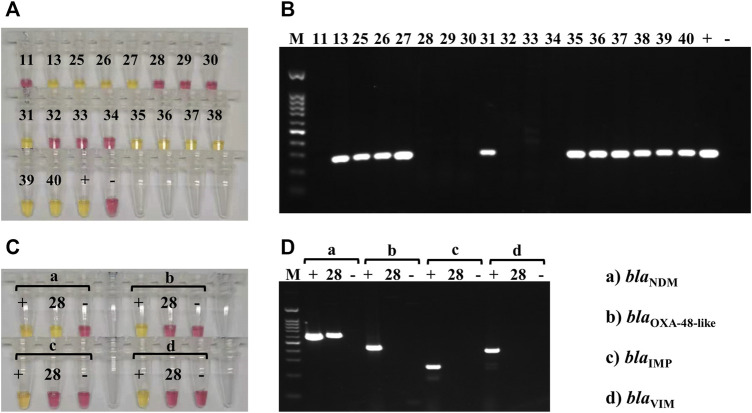
Carbapenemase-producing genes LAMP and PCR testing on clinical isolates. (A) Representative colorimetric LAMP results targeting the bla KPC gene performed on 30 of the 120 clinical isolates of K. pneumoniae collected at Ruijin Hospital (Shanghai, China); 19 isolates were bla KPC-positive (yellow; 1, 4, 6, 8–11, 13–16, 22–28, and 30), and 11 were negative (pink). (B) Representative results of endpoint PCR targeting the bla KPC gene performed on the same 30 clinical isolates of K. pneumoniae than the ones tested by LAMP. Nineteen isolates were bla KPC-positive (amplification band at 240 bp; 1, 4, 6, 8–11, 13–16, 22–28, and 30), and 11 were negative (no amplification band). The marker (M) used for these experiments was a 100-bp DNA ladder. (C) Representative colorimetric LAMP results targeting the bla NDM (a), bla OXA-48-like (b), bla IMP (c), and bla VIM, (d) carbapenemase-producing genes, obtained on 28 of the 120 clinical isolates of K. pneumoniae collected at Ruijin Hospital (Shanghai, China). For each AMR gene, the positive control (+) was a clinical isolate, collected from the Ruijin Hospital (Shanghai, China) and confirmed to be positive by both PCR and sequencing. Two K. pneumoniae clinical isolates were positive (yellow; 31 and 37) for bla NDM (a), three were positive (yellow; 38, 39, and 43) for bla OXA-48-like (b), one was positive (yellow; 47) for bla IMP (c), and two were positive (yellow; 54 and 55) for bla VIM (d) genes. (D) Representative PCR amplification results targeting the bla NDM (a), bla OXA-48-like (b), bla IMP (c), and bla VIM (d) genes, obtained on the same 28 clinical isolates of K. pneumoniae than the ones tested by LAMP. For each AMR gene, the positive control (+) was a clinical isolate, collected from the Ruijin Hospital (Shanghai, China) and confirmed to be positive by both PCR and sequencing. Two K. pneumoniae clinical isolates were positive (amplification band at 621 bp; 31 and 37) for bla NDM (a), three were positive (amplification band at 438 bp; 38, 39 and 43) for bla OXA-48-like (b), one was positive (amplification band at 232 bp; 47) for bla IMP (c), and two were positive (amplification band at 390 bp; 54 and 55) for bla VIM (d) genes. The marker (M) used for these experiments was a 100-bp DNA ladder.
TABLE 6.
Summary of carbapenem resistance–specific LAMP and PCR results.
| LAMP test results | Reference method (PCR) | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 68 | 0 | 68 |
| Negative | 0 | 57 | 57 |
| Total | 68 | 57 | 125 |
Contingency table displaying LAMP and PCR results targeting the bla KPC gene performed on 125 K. pneumoniae strains; 68 K. pneumoniae strains were bla KPC -positive by both LAMP and PCR, and 57 strains were negative by both LAMP and PCR.
To determine the limit of detection of the bla KPC LAMP, the genomic DNAs from the type strains K. pneumoniae NCTC13809 and K. pneumoniae HS11286 (both bla KPC -positive) were extracted and then diluted from 10 ng/μl to 1 fg/μl. The lowest concentration of K. pneumoniae NCTC13809 and HS11286 DNA amplified by the bla KPC primers was 10 pg/μl (equivalent to 1,609.2 gene copies/µl), using both fluorometric and colorimetric detection. When considering the viable CFU counts, used for DNA template preparation, the limit of detection was found to be 3.8 CFUs/reaction.
For the detection of other carbapenem-resistance genes bla NDM, bla OXA-48-like, bla IMP, and bla VIM, LAMP primers were designed and tested on K. pneumoniae or other species of clinical isolates from China. All clinical isolates were also tested by PCR for the bla NDM, bla OXA-48-like, bla IMP, and bla VIM genes (Figure 3). Seven of 18 isolates were bla NDM-positive, 7 of 18 isolates were bla OXA-48-like-positive, 3 of 18 isolates were bla IMP-positive, and 4 of 18 isolates were bla VIM-positive, using both colorimetric LAMP and PCR. The strains that were positive by both LAMP and PCR were also carbapenem-resistant according to AST using Vitek® 2 automated system.
Testing of Clinical Sputum Samples and Spiked Blood
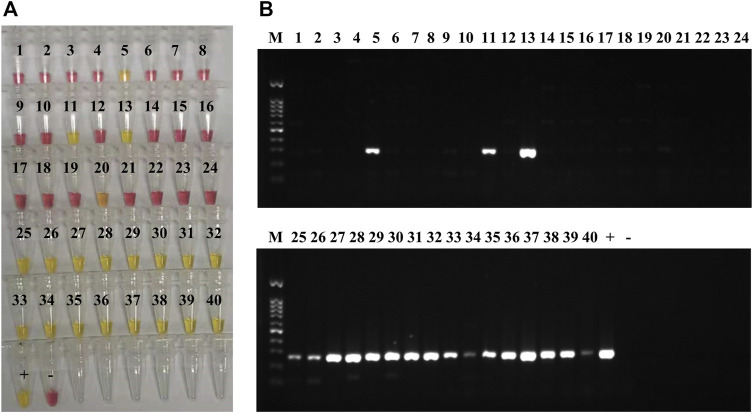
According to MALDI-TOF MS and culture, 18 of the clinical sputum samples were positive for K. pneumoniae (11, 13, 25–40; Supplementary Table S7), and 22 samples were negative for Kp (1–10, 12, and 14–24; Supplementary Table S7). Among the 22 Kp-negative samples, 17 contained other bacterial pathogens (Supplementary Table S7). All the 18 samples positive for Kp by culture and MALDI-TOF were also positive by LAMP and PCR assays targeting the yhaI gene (11, 13, 25–40; Figure 4 and Supplementary Table S7). Of the 22 samples negative for Kp by culture and MALDI-TOF, 20 were also negative by LAMP (1–4, 6–10, 12, 14–19, and 21–24; Figure 4 and Supplementary Table S7), and 21 were negative by PCR (1–4, 6–10, 12, 14–24; Figure 4 and Supplementary Table S7). According to MALDI-TOF, sample 5 contained only P. aeruginosa, whereas it was positive by both LAMP and PCR for the yhaI gene (Figure 4 and Supplementary Table S7). Sample 20, containing Serratia marcescens and P. aeruginosa according to MALDI-TOF, was slightly positive by LAMP and negative by PCR (Figure 4 and Supplementary Table S7). The calculated sensitivity, specificity, and positive and negative predictive values of the Kp LAMP assay (yhaI gene) were 100%, 91%, and 90% and 100%, respectively, when tested on clinical sputum samples, in comparison with culture and MALDI-TOF methods.
FIGURE 4.
K. pneumoniae LAMP and PCR detection in clinical sputum samples. (A) Colorimetric LAMP results, targeting the yhaI gene, obtained on DNA extracted from 40 clinical sputum samples; 20 of the samples were positive (yellow; 5, 11, 13, 20, and 25–40), and 20 were negative (pink; 1–4, 6–10, 12, 14–19, and 21–24). DNA extracted from K. pneumoniae HS11286 was used as positive control (+), and the negative control (−) was molecular H2O. (B) PCR results, for yhaI gene amplification, obtained on DNA extracted 40 clinical sputum samples (same than A); 19 of the samples were positive (amplification bands; 5, 11, 13, and 25–40), and 21 were negative (no band; 1–4, 6–10, 12, and 14–24). DNA extracted from K. pneumoniae HS11286 was used as positive control (+) and the negative control (−) was molecular H2O. The marker (M) was a 100-bp DNA ladder.
Susceptibility tests to imipenem and meropenem (using the Vitek® 2 automated system; bioMérieux, Inc.) were performed on the 18 K. pneumoniae isolates from sputum samples and showed that 12 of the isolates were resistant to these carbapenems (13, 25–28, 31, and 35–40; Supplementary Table S7). Both LAMP and PCR assays, performed on the DNA directly extracted from the sputum samples, showed that 11 of 12 carbapenem-resistant K. pneumoniae isolates carried the bla KPC gene (13, 25–27, 31 and 35–40; Figure 5 and Supplementary Table S7) and that one isolate harbored the bla NDM gene (28; Figure 5 and Supplementary Table S7).
FIGURE 5.
Carbapenemase-producing genes LAMP and PCR testing on clinical sputum samples. (A) Colorimetric LAMP results, targeting the bla KPC gene, obtained on DNA extracted from the 18 clinical sputum samples that contained K. pneumoniae (according to MALDI-TOF); 11 of the samples were bla KPC -positive (yellow; 13, 25–27, 31, and 35–40), and 7 were negative (pink). DNA extracted from K. pneumoniae HS11286 was used as a positive control (+), and the negative control (−) was molecular-grade H2O. (B) PCR results, for bla KPC amplification, obtained on DNA extracted from the 18 clinical sputum samples that contained K. pneumoniae (same as A); 11 of the samples were bla KPC -positive (amplification bands; 13, 25–27, 31, and 35–40), and 7 were negative (no band). DNA extracted from K. pneumoniae HS11286 was used as positive control (+), and the negative control (−) was molecular-grade H2O. The marker (M) was a 100-bp DNA ladder. (C) Colorimetric LAMP results, targeting the bla NDM (a), bla OXA-48like (b), bla IMP (c), and bla VIM (d) genes, obtained on the DNA extracted from sample 28. Positive controls (+) and negative controls (−) were used for each assay. Sample 28 was positive (yellow) for bla NDM (a) and negative (pink) for the other carbapenemase-producing genes (b–d). (D) PCR results, targeting the bla NDM (a), bla OXA-48like (b), bla IMP (c), and bla VIM (d) genes, obtained on the DNA extracted from sample 28. Positive controls (+) and negative controls (−) were used for each assay. Sample 28 was positive (amplification band at 621 bp) for bla NDM (a) and negative (no band) for the other carbapenemase-producing genes. The marker (M) was a 100-bp DNA ladder.
To assess the feasibility of blood samples testing with our diagnostics, the limit of bacterial copies per LAMP reaction was also determined in spiked defibrinated sheep blood samples. A total of 10 bacterial cell suspensions, with concentrations ranging from of 0.38 CFUs/ml to 3.8 × 107 CFUs/ml, were used to spike sterile defibrinated sheep blood samples that were then extracted using a commercial DNA extraction kit. The minimal detectable concentration for LAMP was 7.6 CFUs/reaction for both yhaI and bla KPC detection in colorimetric LAMP.
Discussion
Public health systems around the world would greatly benefit from an accurate, rapid, and user-friendly method to diagnose pathogenic microorganisms and their potential AMR. In this study, we identified specific markers for the rapid and accurate detection of K. pneumoniae, using comparative genomics tools. These markers were found in more than 7,000 K. pneumoniae genomes from all over the world and absent from other species of Klebsiella and other bacterial genera; they were then chosen as target genes for K. pneumoniae–specific LAMP diagnostics. We also developed LAMP assays to rapidly and accurately detect the most common carbapenemase-producing genes: bla KPC, bla NDM, bla OXA-48-like, bla IMP, and bla VIM.
The LAMP assays developed herein can achieve sensitive and accurate detection of K. pneumoniae and carbapenemase genes, with performance comparable to gold-standard clinical methodologies that are time-consuming or based on expensive specialized instruments. Indeed, when compared with culture-based methods or PCR, LAMP results achieved sensitivity, specificity, and positive and negative values of 100% in both pure cultures and spiked blood samples. In clinical sputum samples, the calculated sensitivity, specificity, and positive and negative values of the yhaI LAMP (K. pneumoniae detection) were 100%, 91%, and 90% and 100%, respectively, when compared with MALDI-TOF MS and culture. Only 2 clinical samples out of 40 were yhaI-positive by LAMP, although not identified as containing K. pneumoniae by MALDI-TOF MS; one sample contained P. aeruginosa, and the second one contained S. marcescens and P. aeruginosa according to MALDI-TOF. As our primers have been extensively tested on non–K. pneumoniae isolates (including 23 Pseudomonas spp. strains), this is likely to be due to the high sensitivity of our LAMP assay, with LAMP having previously been described as suitable for the early diagnosis of infectious diseases (Garg et al., 2021). The bla KPC LAMP assay showed a 100% concordance with PCR and antibiotics susceptibility, even in clinical samples. Furthermore, the assays described here demonstrated a limit of detection 100 times lower than the routine PCR method used in Ruijin Hospital, and LAMP results were obtained in a much shorter time frame: 1 h from sample processing to results versus 2 h 30 min for PCR detection and 24–48 h for culture-based methods. Several LAMP assays have been developed for the detection of K. pneumoniae (Kang et al., 2012; Dong et al., 2015; Barnes et al., 2018; Li et al., 2019; Tian et al., 2019; Vergara et al., 2020) using different target genes, including KPN_04473 fimD (Kang et al., 2012; Barnes et al., 2018; Vergara et al., 2020), ure1 (Li et al., 2019), celB (Tian et al., 2019), and rscA (Dong et al., 2015). These different target genes were chosen following bibliographic reviews or sequences alignment and online BLAST searches. The target gene (yhaI) described in this work was chosen following a comprehensive comparative genomics study that demonstrated the specificity of yhaI to K. pneumoniae and its presence in >7,000 Kp genomes distributed globally. The designed yhaI LAMP assay was validated with high sensitivity and specificity on 5 Kp type strains, 167 Kp clinical isolates, 32 non-Kp type strains, and 115 non-Kp clinical isolates from both the United Kingdom and China. Previous studies have also shown the efficiency (high sensitivity, specificity, and concordance with PCR) of LAMP assays for the detection of the carbapenemase families: bla KPC, bla NDM, bla OXA-48-like, bla VIM, and bla IMP (Qi et al., 2012; Solanki et al., 2013; Cheng et al., 2014; Nakano et al., 2015; Kim et al., 2016; Srisrattakarn et al., 2017; Funashima et al., 2018; Feng et al., 2021). However, most of these studies used different detection methods, such as fluorimetry, turbidity, and gel electrophoresis, which require specific equipment or repeated lid opening operations (potential cause of cross-contamination) (Qi et al., 2012; Solanki et al., 2013; Cheng et al., 2014; Nakano et al., 2015; Kim et al., 2016; Funashima et al., 2018). LAMP assays, developed using hydroxynaphthol blue dye for colorimetric detection, showed high sensitivity and specificity but used an in-house LAMP Mastermix for which operational factors during reagents preparation may increase the unreliability of the results (Srisrattakarn et al., 2017). A recent study used the same the commercial colorimetric LAMP Mastermix than in our study (Feng et al., 2021) to detect carbapenemase-producing genes; however, their assays were not validated on clinical samples.
Our assays are also comparable to the commercialized CE-IVD LAMP Eazyplex SuperBug kits (complete A, complete B, and CRE; Amplex-Diagnostics GmbH, Germany), designed to detect carbapenemase-producing genes and selected ESBL genes in up to 30 min using the portable Genie II device (Vergara et al., 2014; Findlay et al., 2015; Hinic et al., 2015; García-Fernández et al., 2016; Fiori et al., 2019; Moreno-Morales et al., 2020; Sekowska et al., 2020; Vergara et al., 2020; Holma et al., 2021). Sensitivity and specificity of the Eazyplex SuperBug kits, assessed on clinical isolates of Enterobacteriaceae (Vergara et al., 2014; Findlay et al., 2015), were greater than 95%. These kits were also tested on bronchoalveolar lavage fluid samples spiked with Enterobacteriaceae and carbapenem-resistant Acinetobacter spp. (Moreno-Morales et al., 2020; Vergara et al., 2020), clinical urine samples (Hinic et al., 2015), and rectal swab samples (Holma et al., 2021), with 80%–100% sensitivity, a specificity greater than 95%, and a limit of detection between 102 and 103 CFUs/ml (Hinic et al., 2015; Moreno-Morales et al., 2020; Vergara et al., 2020; Holma et al., 2021). Other CE-IVD marked portable molecular diagnostics for carbapenemases, which are cartridge-based PCR assays and require minimal sample preparation, such as Xpert Carba-R (Tenover et al., 2013; Anandan et al., 2015; Findlay et al., 2015; Vergara et al., 2020) and Novodiag CarbaR + combining real-time PCR and microarray technologies (Girlich et al., 2019; Holma et al., 2021), have been recently commercialized. Both these diagnostic methods detect and differentiate the most prevalent carbapenemases gene families in less than 2 h, from samples to results, and showed high sensitivity and specificity in clinical samples with a limit of detection of 102 CFUs/ml (Tenover et al., 2013; Anandan et al., 2015; Findlay et al., 2015; Girlich et al., 2019; Vergara et al., 2020; Holma et al., 2021). These rapid molecular diagnostics are all highly sensitive and specific but require the use of specialized costly devices and reagents. Our designed assays are as sensitive and specific, while being a lot less expensive as it can be performed using only a dry bath heating block. It could be used at point of care as it only requires a heating block and could easily be adapted into different diagnostics platforms, with a potential for mobile connectivity (Barnes et al., 2018; Rohaim et al., 2020). The reagents also have the potential to be lyophilized for use in resource limited areas where refrigeration is impractical.
The emergence of carbapenem-resistant K. pneumoniae (Wong M. H. Y. et al., 2018; Zhou et al., 2020) has created a new challenge in rapid detecting and combating this already dangerous pathogen. Early diagnosis and intervention could be beneficial in a number of clinical contexts. In urinary tract infection cases (among the most common types of infection) (Flores-Mireles et al., 2015) and pneumonia cases (Beutz and Abraham, 2005); early diagnosis and targeted treatment could help to avoid progression into more severe and potentially fatal conditions such as sepsis (Beutz and Abraham, 2005; Simerville et al., 2005; Seymour et al., 2017). Sepsis cases are difficult to diagnose and constitute a medical emergency, as they progress rapidly, and delay in effective antimicrobial treatment has been associated with an increase in in-hospital mortality (Gaieski et al., 2010; Ferrer et al., 2014; Garnacho-Montero et al., 2014; Martin-Loeches et al., 2015). To improve the survival rates of patients with sepsis, treatment has to be administered within the first few hours (0–6 h) of diagnosis (Gaieski et al., 2010; Ferrer et al., 2014; Garnacho-Montero et al., 2014; Martin-Loeches et al., 2015). Antibiotic therapy usually starts with the administration of broad-spectrum antibiotics, until the source of infection has been identified, at which point a more targeted agent can be used. Broad-spectrum agents are not always very effective, driving emergence and spread of AMR, which is currently one of the biggest global public health threats facing humanity. It is estimated that by 2050, 10 million lives a year and a cumulative US $100 trillion of economic output are at risk from AMR (Brogan and Mossialos, 2016; O’Neill, 2016; Piddock, 2016). Antibiotic effectiveness needs to be preserved as a further increase in AMR would lead to significant risks linked to key medical procedures (such as invasive surgeries, chemotherapy, intensive care, etc.) and infectious diseases management. Rapid identification of the causative pathogen and its AMR, using the assays detailed in this study, would aid in the selection of appropriate treatment, thus reducing the unnecessary use of antibiotics and thereby slowing down dissemination of AMR (Brogan and Mossialos, 2016; O’Neill, 2016; Piddock, 2016), as well as improving the prognosis of septic patients. Our current LAMP assay has been developed for the detection of K. pneumoniae. Additional investigations are being pursued to design similar LAMP tests for E. coli, a particular concern for AMR (Alvarez-Uria et al., 2018) and a microorganism commonly responsible for sepsis (Restrepo-Alvarez et al., 2019).
Acknowledgments
We would like to express our gratitude to respective hospitals for providing the clinical isolates for the assay validation. Thanks also to Dr Jennifer Ritchie who supplied hospital environment isolates, which were obtained as part of an MRC iCASE studentship to Nadia Mohammed (MR/P01643X/1).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Ruijin Hospital, School of Medicine, Shanghai Jiao Tong University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AP, DK, and BS conceived, planned and performed the majority of experiments with support from JM, JQ, and RL. KB helped carrying out the experiments and analyze the data. JM, AP, and AV performed the comparative genomics analysis and interpretation of the data. JL, CT, XW, WM, DJ, and JC collected clinical isolates and patients’ samples. JM, JQ, and RL conceived the idea of the presented work and gave final approval of the version to be published. AP wrote the manuscript with input from all authors. All authors reviewed and approved the final manuscript.
Funding
This study was funded under the National Key R&D Program of China (2018YFE0102400), Innovate United Kingdom (104993/105002-610286), Cultivation Project of Shanghai Major Infectious Disease Research Base (20dz2210500), Key Laboratory of Emergency Prevention, Diagnosis and Treatment of Respiratory Infectious Diseases, Shanghai (20dz2261100), the Science and Technology Commission of Shanghai Municipality (19JC1413000, 19JC1413004), and National Natural Science Foundation of China (82000011).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.794961/full#supplementary-material
References
- Adeolu M., Alnajar S., Naushad S., S. Gupta R. (2016). Genome-based Phylogeny and Taxonomy of the 'Enterobacteriales': Proposal for Enterobacterales ord. Nov. Divided into the Families Enterobacteriaceae, Erwiniaceae Fam. nov., Pectobacteriaceae Fam. nov., Yersiniaceae Fam. nov., Hafniaceae Fam. nov., Morganellaceae Fam. nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 66 (12), 5575–5599. 10.1099/ijsem.0.001485 [DOI] [PubMed] [Google Scholar]
- Alvarez-Uria G., Gandra S., Mandal S., Laxminarayan R. (2018). Global Forecast of Antimicrobial Resistance in Invasive Isolates of Escherichia coli and Klebsiella pneumoniae . Int. J. Infect. Dis. 68, 50–53. 10.1016/j.ijid.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandan S., Damodaran S., Gopi R., Bakthavatchalam Y. D., Veeraraghavan B. (2015). Rapid Screening for Carbapenem Resistant Organisms: Current Results and Future Approaches. Jcdr 9 (9), DM01–03. 10.7860/JCDR/2015/14246.6530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes L., Heithoff D. M., Mahan S. P., Fox G. N., Zambrano A., Choe J., et al. (2018). Smartphone-based Pathogen Diagnosis in Urinary Sepsis Patients. EBioMedicine 36, 73–82. 10.1016/j.ebiom.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini A., Frasson I., Cavallaro A., Richter S. N., Palù G. (2014). Comparison of Phenotypic Methods for the Detection of Carbapenem Non-susceptible Enterobacteriaceae. Gut Pathog. 6, 13. 10.1186/1757-4749-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutz M. A., Abraham E. (2005). Community-acquired Pneumonia and Sepsis. Clin. Chest Med. 26 (1), 19–28. 10.1016/j.ccm.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Bialek-Davenet S., Criscuolo A., Ailloud F., Passet V., Jones L., Delannoy-Vieillard A.-S., et al. (2014). Genomic Definition of Hypervirulent and Multidrug-ResistantKlebsiella pneumoniaeClonal Groups. Emerg. Infect. Dis. 20 (11), 1812–1820. 10.3201/eid2011.140206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogan D. M., Mossialos E. (2016). A Critical Analysis of the Review on Antimicrobial Resistance Report and the Infectious Disease Financing Facility. Globalization and Health 12, 8. 10.1186/s12992-016-0147-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsrud O., Bohlin J., Scheffer L., Eldholm V. (2016). Erratum to: Rapid Scoring of Genes in Microbial Pan-genome-wide Association Studies with Scoary. Genome Biol. 17 (1), 262. 10.1186/s13059-016-1132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Zheng F., Rui Y. (2014). Rapid Detection of blaNDM, blaKPC, blaIMP, and blaVIM Carbapenemase Genes in Bacteria by Loop-Mediated Isothermal Amplification. Microb. Drug Resist. 20 (6), 533–538. 10.1089/mdr.2014.0040 [DOI] [PubMed] [Google Scholar]
- Dallenne C., Da Costa A., Decré D., Favier C., Arlet G. (2010). Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 (3), 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Dhama K., Karthik K., Chakrabort S., Tiwari R., Kapoor S., Kumar A., et al. (2014). Loop-mediated Isothermal Amplification of DNA (LAMP): a New Diagnostic Tool Lights the World of Diagnosis of Animal and Human Pathogens: a Review. Pakistan J. Biol. Sci. 17 (2), 151–166. 10.3923/pjbs.2014.151.166 [DOI] [PubMed] [Google Scholar]
- Diribe O., North S., Sawyer J., Roberts L., Fitzpatrick N., La Ragione R. (2014). Design and Application of a Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Staphylococcus Pseudintermedius. J. Vet. Diagn. Invest. 26 (1), 42–48. 10.1177/1040638713516758 [DOI] [PubMed] [Google Scholar]
- Dong D., Liu W., Li H., Wang Y., Li X., Zou D., et al. (2015). Survey and Rapid Detection of Klebsiella pneumoniae in Clinical Samples Targeting the rcsA Gene in Beijing, China. Front. Microbiol. 6, 519. 10.3389/fmicb.2015.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Niu S., Chang Y., Jia X., Huang S., Yang P. (2021). Design of Rapid Detection System for Five Major Carbapenemase Families (blaKPC, blaNDM, blaVIM, blaIMP and blaOXA-48-like) by Colorimetric Loop-Mediated Isothermal Amplification. Idr 14, 1865–1874. 10.2147/IDR.S301757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer R., Martin-Loeches I., Phillips G., Osborn T. M., Townsend S., Dellinger R. P., et al. (2014). Empiric Antibiotic Treatment Reduces Mortality in Severe Sepsis and Septic Shock from the First Hour: Results from a Guideline-Based Performance Improvement Program. Crit. Care Med. 42 (8), 1749–1755. 10.1097/CCM.0000000000000330 [DOI] [PubMed] [Google Scholar]
- Findlay J., Hopkins K. L., Meunier D., Woodford N. (2015). Evaluation of Three Commercial Assays for Rapid Detection of Genes Encoding Clinically Relevant Carbapenemases in Cultured Bacteria. J. Antimicrob. Chemother. 70 (5), 1338–1342. 10.1093/jac/dku571 [DOI] [PubMed] [Google Scholar]
- Fiori B., D'Inzeo T., Posteraro B., Menchinelli G., Liotti F. M., De Angelis G., et al. (2019). Direct Use of Eazyplex SuperBug CRE Assay from Positive Blood Cultures in Conjunction with Inpatient Infectious Disease Consulting for Timely Appropriate Antimicrobial Therapy in Escherichia coli and Klebsiella pneumoniae Bloodstream Infections. Idr 12, 1055–1062. 10.2147/IDR.S206323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Mireles A. L., Walker J. N., Caparon M., Hultgren S. J. (2015). Urinary Tract Infections: Epidemiology, Mechanisms of Infection and Treatment Options. Nat. Rev. Microbiol. 13 (5), 269–284. 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois P., Tangomo M., Hibbs J., Bonetti E.-J., Boehme C. C., Notomi T., et al. (2011). Robustness of a Loop-Mediated Isothermal Amplification Reaction for Diagnostic Applications. FEMS Immunol. Med. Microbiol. 62 (1), 41–48. 10.1111/j.1574-695X.2011.00785.x [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. (1981). Molecular and Functional Analysis of the TOL Plasmid pWWO from Pseudomonas Putida and Cloning of Genes for the Entire Regulated Aromatic Ring Meta Cleavage Pathway. Proc. Natl. Acad. Sci. 78 (12), 7458–7462. 10.1073/pnas.78.12.7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funashima Y., Sugahara K., Hirata Y., Kato K., Sato K., Sasaki Y., et al. (2018). A Trial of Simple and Rapid Carbapenemase Big Five Gene Analysis Using LAMP Method. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 28 (2), 77–83. [PubMed] [Google Scholar]
- Gadkar V. J., Goldfarb D. M., Gantt S., Tilley P. A. G. (2018). Real-time Detection and Monitoring of Loop Mediated Amplification (LAMP) Reaction Using Self-Quenching and De-quenching Fluorogenic Probes. Sci. Rep. 8 (1), 5548. 10.1038/s41598-018-23930-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaieski D. F., Mikkelsen M. E., Band R. A., Pines J. M., Massone R., Furia F. F., et al. (2010). Impact of Time to Antibiotics on Survival in Patients with Severe Sepsis or Septic Shock in Whom Early Goal-Directed Therapy Was Initiated in the Emergency Department. Crit. Care Med. 38 (4), 1045–1053. 10.1097/CCM.0b013e3181cc4824 [DOI] [PubMed] [Google Scholar]
- García-Fernández S., Morosini M.-I., Gijón D., Beatobe L., Ruiz-Garbajosa P., Domínguez L., et al. (2016). Detection of Carbapenemase Production in a Collection of Enterobacteriaceae with Characterized Resistance Mechanisms from Clinical and Environmental Origins by Use of Both Carba NP and Blue-Carba Tests. J. Clin. Microbiol. 54 (2), 464–466. 10.1128/JCM.02580-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N., Sahu U., Kar S., Ahmad F. J. (2021). Development of a Loop-Mediated Isothermal Amplification (LAMP) Technique for Specific and Early Detection of Mycobacterium leprae in Clinical Samples. Sci. Rep. 11 (1), 9859. 10.1038/s41598-021-89304-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnacho-Montero J., Gutiérrez-Pizarraya A., Escoresca-Ortega A., Corcia-Palomo Y., Fernández-Delgado E., Herrera-Melero I., et al. (2014). De-escalation of Empirical Therapy Is Associated with Lower Mortality in Patients with Severe Sepsis and Septic Shock. Intensive Care Med. 40 (1), 32–40. 10.1007/s00134-013-3077-7 [DOI] [PubMed] [Google Scholar]
- Ghebremedhin B., Halstenbach A., Smiljanic M., Kaase M., Ahmad-Nejad P. (2016). MALDI-TOF MS Based Carbapenemase Detection from Culture Isolates and from Positive Blood Culture Vials. Ann. Clin. Microbiol. Antimicrob. 15, 5. 10.1186/s12941-016-0120-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlich D., Bernabeu S., Grosperrin V., Langlois I., Begasse C., Arangia N., et al. (2019). Evaluation of the Amplidiag CarbaR+MCR Kit for Accurate Detection of Carbapenemase-Producing and Colistin-Resistant Bacteria. J. Clin. Microbiol. 57 (3), e01800–18. 10.1128/JCM.01800-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M., Shimada K., Sato A., Takahashi E., Fukasawa T., Takahashi T., et al. (2010). Rapid Detection of Pseudomonas aeruginosa in Mouse Feces by Colorimetric Loop-Mediated Isothermal Amplification. J. Microbiol. Methods 81 (3), 247–252. 10.1016/j.mimet.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 29 (8), 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R., Shi Q., Wu S., Yin D., Peng M., Dong D., et al. (2020). Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated from Adult and Children Patients in China. Front. Cell. Infect. Microbiol. 10, 314. 10.3389/fcimb.2020.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinic V., Ziegler J., Straub C., Goldenberger D., Frei R. (2015). Extended-spectrum β-lactamase (ESBL) Detection Directly from Urine Samples with the Rapid Isothermal Amplification-Based Eazyplex SuperBug CRE Assay: Proof of Concept. J. Microbiol. Methods 119, 203–205. 10.1016/j.mimet.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Holma T., Antikainen J., Haiko J. (2021). Evaluation of Three Molecular Carbapenemase Tests: Eazyplex SuperBug Complete B, Novodiag CarbaR+, and Amplidiag CarbaR+MCR. J. Microbiol. Methods 180, 106105. 10.1016/j.mimet.2020.106105 [DOI] [PubMed] [Google Scholar]
- Holt K. E., Wertheim H., Zadoks R. N., Baker S., Whitehouse C. A., Dance D., et al. (2015). Genomic Analysis of Diversity, Population Structure, Virulence, and Antimicrobial Resistance inKlebsiella Pneumoniae, an Urgent Threat to Public Health. Proc. Natl. Acad. Sci. USA 112 (27), E3574–E3581. 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., et al. (2003). Comprehensive Transposon Mutant Library of Pseudomonas aeruginosa . Proc. Natl. Acad. Sci. 100 (24), 14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Iida T., Aoki K., Ohno S., Suzutani T. (2005). Sensitive and Rapid Detection of Herpes Simplex Virus and Varicella-Zoster Virus DNA by Loop-Mediated Isothermal Amplification. J. Clin. Microbiol. 43 (7), 3290–3296. 10.1128/JCM.43.7.3290-3296.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Deng R., Wang C., Deng T., Peng P., Cheng X., et al. (2012). Etiologic Diagnosis of Lower Respiratory Tract Bacterial Infections Using Sputum Samples and Quantitative Loop-Mediated Isothermal Amplification. PLoS One 7 (6), e38743. 10.1371/journal.pone.0038743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Kim H. S., Lee J. M., Yoon S. S., Yong D. (2016). Rapid Detection of Pseudomonas aeruginosa and Acinetobacter Baumannii Harboring blaVIM-2, blaIMP-1 and blaOXA-23 Genes by Using Loop-Mediated Isothermal Amplification Methods. Ann. Lab. Med. 36 (1), 15–22. 10.3343/alm.2016.36.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchibiro T., Komatsu M., Yamasaki K., Nakamura T., Nishio H., Nishi I., et al. (2018). Evaluation of the Modified Carbapenem Inactivation Method for the Detection of Carbapenemase-Producing Enterobacteriaceae. J. Infect. Chemother. 24 (4), 262–266. 10.1016/j.jiac.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Li B., Zhao Y., Liu C., Chen Z., Zhou D. (2014). Molecular Pathogenesis of Klebsiella pneumoniae . Future Microbiol. 9 (9), 1071–1081. 10.2217/fmb.14.48 [DOI] [PubMed] [Google Scholar]
- Li C., Fu G., Shi Y., Zhang A.-M., Xia X., Fang Y., et al. (2019). Rapid, Specific, and Sensitive Detection of the ureR_1 Gene in Klebsiella pneumoniae by Loop-Mediated Isothermal Amplification Method. Braz. J. Med. Biol. Res. 52 (3), e8186. 10.1590/1414-431X20198186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zou D., Li Y., Wang X., He X., Wei X., et al. (2012). Sensitive and Rapid Detection of the new Delhi Metallo-Beta-Lactamase Gene by Loop-Mediated Isothermal Amplification. J. Clin. Microbiol. 50 (5), 1580–1585. 10.1128/JCM.06647-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-Y., Wang Y., Walsh T. R., Yi L.-X., Zhang R., Spencer J., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: a Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16 (2), 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Lucchi N. W., Ljolje D., Silva-Flannery L., Udhayakumar V. (2016). Use of Malachite Green-Loop Mediated Isothermal Amplification for Detection of Plasmodium Spp. Parasites. PLoS One 11 (3), e0151437. 10.1371/journal.pone.0151437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manajit O., Longyant S., Sithigorngul P., Chaivisuthangkura P. (2018). Development of Uracil-DNA-Glycosylase-Supplemented Loop-Mediated Isothermal Amplification Coupled with Nanogold Probe (UDG-LAMP-AuNP) for Specific Detection of Pseudomonas�aeruginosa. Mol. Med. Rep. 17 (4), 5734–5743. 10.3892/mmr.2018.8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Loeches I., Levy M., Artigas A. (2015). Management of Severe Sepsis: Advances, Challenges, and Current Status. Dddt 9, 2079–2088. 10.2147/DDDT.S78757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehling T. J., Choi G., Dugan L. C., Salit M., Meagher R. J. (2021). LAMP Diagnostics at the Point-of-Care: Emerging Trends and Perspectives for the Developer Community. Expert Rev. Mol. Diagn. 21 (1), 43–61. 10.1080/14737159.2021.1873769 [DOI] [PubMed] [Google Scholar]
- Moloney E., Lee K. W., Craig D., Allen A. J., Graziadio S., Power M., et al. (2019). A PCR-Based Diagnostic Testing Strategy to Identify Carbapenemase-Producing Enterobacteriaceae Carriers upon Admission to UK Hospitals: Early Economic Modelling to Assess Costs and Consequences. Diagn. Progn Res. 3, 8. 10.1186/s41512-019-0053-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Morales J., Vergara A., Kostyanev T., Rodriguez-Baño J., Goossens H., Vila J. (2020). Evaluation of a Loop-Mediated Isothermal Amplification Assay to Detect Carbapenemases Directly from Bronchoalveolar Lavage Fluid Spiked with Acinetobacter Spp. Front. Microbiol. 11, 597684. 10.3389/fmicb.2020.597684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price L. S., Poirel L., Bonomo R. A., Schwaber M. J., Daikos G. L., Cormican M., et al. (2013). Clinical Epidemiology of the Global Expansion of Klebsiella pneumoniae Carbapenemases. Lancet Infect. Dis. 13 (9), 785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. (2002). Accelerated Reaction by Loop-Mediated Isothermal Amplification Using Loop Primers. Mol. Cell Probes 16 (3), 223–229. 10.1006/mcpr.2002.0415 [DOI] [PubMed] [Google Scholar]
- Nakano R., Nakano A., Ishii Y., Ubagai T., Kikuchi-Ueda T., Kikuchi H., et al. (2015). Rapid Detection of the Klebsiella pneumoniae Carbapenemase (KPC) Gene by Loop-Mediated Isothermal Amplification (LAMP). J. Infect. Chemother. 21 (3), 202–206. 10.1016/j.jiac.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Cuzon G., Naas T. (2009). The Real Threat of Klebsiella pneumoniae Carbapenemase-Producing Bacteria. Lancet Infect. Dis. 9 (4), 228–236. 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Dortet L. (2012). Rapid Detection of Carbapenemase-producingEnterobacteriaceae. Emerg. Infect. Dis. 18 (9), 1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated Isothermal Amplification of DNA. Nucleic Acids Res. 28 (12), 63e–63. 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. (2016). The Review on Antimicrobial Resistance. Available at: https://amr-review.org/home.html .
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T. G., et al. (2015). Roary: Rapid Large-Scale Prokaryote pan Genome Analysis. Bioinformatics 31 (22), 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzkill T. (2018). Structural and Mechanistic Basis for Extended-Spectrum Drug-Resistance Mutations in Altering the Specificity of TEM, CTX-M, and KPC β-lactamases. Front. Mol. Biosci. 5, 16. 10.3389/fmolb.2018.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Sannarangaiah S., Dash P. K., Rao P. V. L., Morita K. (2008). Loop Mediated Isothermal Amplification (LAMP): a New Generation of Innovative Gene Amplification Technique; Perspectives in Clinical Diagnosis of Infectious Diseases. Rev. Med. Virol. 18 (6), 407–421. 10.1002/rmv.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. V. (2016). Reflecting on the Final Report of the O'Neill Review on Antimicrobial Resistance. Lancet Infect. Dis. 16 (7), 767–768. 10.1016/S1473-3099(16)30127-X [DOI] [PubMed] [Google Scholar]
- Pitout J. D. D., Nordmann P., Poirel L. (2015). Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob. Agents Chemother. 59 (10), 5873–5884. 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podschun R., Ullmann U. (1998). Klebsiella Spp. As Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 11 (4), 589–603. 10.1128/cmr.11.4.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Du Y., Zhu X., Bai H., Luo Y., Liu Y. (2012). A Loop-Mediated Isothermal Amplification Method for Rapid Detection of NDM-1 Gene. Microb. Drug Resist. 18 (4), 359–363. 10.1089/mdr.2011.0220 [DOI] [PubMed] [Google Scholar]
- Restrepo-Álvarez C. A., Bernal E., Ascuntar-Tello J., Jaimes F. (2019). Análisis clínico y microbiológico de la sepsis grave y el choque séptico por Escherichia coli en Medellín, Colombia. Rev. Chil. Infectol. 36 (4), 447–454. 10.4067/S0716-10182019000400447 [DOI] [PubMed] [Google Scholar]
- Rohaim M. A., Clayton E., Sahin I., Vilela J., Khalifa M. E., Al-Natour M. Q., et al. (2020). Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2. Viruses 12 (9), 972. 10.3390/v12090972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2019). Abricate. Github. Available at: https://github.com/tseemann/abricate .
- Seemann T. (2014). Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 30 (14), 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Segawa T., Matsui M., Suzuki M., Tsutsui A., Kuroda M., Shibayama K., et al. (2017). Utilizing the Carba NP Test as an Indicator of Expression Level of Carbapenemase Genes in Enterobacteriaceae. J. Microbiol. Methods 133, 35–39. 10.1016/j.mimet.2016.12.015 [DOI] [PubMed] [Google Scholar]
- Sękowska A., Bogiel T., Gospodarek-Komkowska E. (2020). Evaluation of Eazyplex SuperBug CRE Test for Beta-Lactamase Genes Detection in Klebsiella Spp. And P. aeruginosa Strains. Curr. Microbiol. 77 (1), 99–103. 10.1007/s00284-019-01806-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour C. W., Gesten F., Prescott H. C., Friedrich M. E., Iwashyna T. J., Phillips G. S., et al. (2017). Time to Treatment and Mortality during Mandated Emergency Care for Sepsis. N. Engl. J. Med. 376 (23), 2235–2244. 10.1056/NEJMoa1703058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerville J. A., Maxted W. C., Pahira J. J. (2005). Urinalysis: a Comprehensive Review. Am. Fam. Physician 71 (6), 1153–1162. [PubMed] [Google Scholar]
- Solanki R., Vanjari L., Ede N., Gungi A., Soory A., Vemu L. (2013). Evaluation of LAMP Assay Using Phenotypic Tests and Conventional PCR for Detection of Bla NDM-1 and Bla KPC Genes Among Carbapenem-Resistant Clinical Gram-Negative Isolates. J. Med. Microbiol. 62 (Pt 10), 1540–1544. 10.1099/jmm.0.059907-0 [DOI] [PubMed] [Google Scholar]
- Sood S. (2014). Identification and Differentiation of Carbapenemases in Klebsiella pneumoniae: a Phenotypic Test Evaluation Study from Jaipur, India. Jcdr 8 (7), DC01–03. 10.7860/JCDR/2014/7027.4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisrattakarn A., Lulitanond A., Wilailuckana C., Charoensri N., Wonglakorn L., Saenjamla P., et al. (2017). Rapid and Simple Identification of Carbapenemase Genes, Bla NDM, Bla OXA-48, Bla VIM, Bla IMP-14 and Bla KPC Groups, in Gram-Negative Bacilli by In-House Loop-Mediated Isothermal Amplification with Hydroxynaphthol Blue Dye. World J. Microbiol. Biotechnol. 33 (7), 130. 10.1007/s11274-017-2295-5 [DOI] [PubMed] [Google Scholar]
- Tamma P. D., Simner P. J. (2018). Phenotypic Detection of Carbapenemase-Producing Organisms from Clinical Isolates. J. Clin. Microbiol. 56 (11). 10.1128/JCM.01140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Canton R., Kop J., Chan R., Ryan J., Weir F., et al. (2013). Detection of Colonization by Carbapenemase-Producing Gram-Negative Bacilli in Patients by Use of the Xpert MDRO Assay. J. Clin. Microbiol. 51 (11), 3780–3787. 10.1128/JCM.01092-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Wang L., Zhang J., Han Q., Xia X.-s., Song Y., et al. (2019). CelB Is a Suitable Marker for Rapid and Specific Identification of Klebsiella pneumoniae by the Loop-Mediated Isothermal Amplification (LAMP) Assay. Braz. J. Microbiol. 50 (4), 961–967. 10.1007/s42770-019-00144-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaluw K., de Haan A., Pluister G. N., Bootsma H. J., de Neeling A. J., Schouls L. M. (2015). The Carbapenem Inactivation Method (CIM), a Simple and Low-Cost Alternative for the Carba NP Test to Assess Phenotypic Carbapenemase Activity in Gram-Negative Rods. PLoS One 10 (3), e0123690. 10.1371/journal.pone.0123690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara A., Boutal H., Ceccato A., López M., Cruells A., Bueno-Freire L., et al. (2020). Assessment of a Loop-Mediated Isothermal Amplification (LAMP) Assay for the Rapid Detection of Pathogenic Bacteria from Respiratory Samples in Patients with Hospital-Acquired Pneumonia. Microorganisms 8 (1), 103. 10.3390/microorganisms8010103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara A., Zboromyrska Y., Mosqueda N., Morosini M. I., García-Fernández S., Roca I., et al. (2014). Evaluation of a Loop-Mediated Isothermal Amplification-Based Methodology to Detect Carbapenemase Carriage in Acinetobacter Clinical Isolates. Antimicrob. Agents Chemother. 58 (12), 7538–7540. 10.1128/AAC.03870-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne P. A. (2020). “Performance Standards for Antimicrobial Susceptibility Testing,” in CLSI Supplement M100. Editor C.a.L.S. Institute. 30th ed. (Wayne: CLSI; ). [Google Scholar]
- Wong M. H. Y., Shum H.-P., Chen J. H. K., Man M.-Y., Wu A., Chan E. W.-C., et al. (2018a). Emergence of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae . Lancet Infect. Dis. 18 (1), 24. 10.1016/S1473-3099(17)30629-1 [DOI] [PubMed] [Google Scholar]
- Wong Y.-P., Othman S., Lau Y.-L., Radu S., Chee H.-Y. (2018b). Loop-mediated Isothermal Amplification (LAMP): a Versatile Technique for Detection of Micro-organisms. J. Appl. Microbiol. 124 (3), 626–643. 10.1111/jam.13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres K. L., Holt K. E. (2016). Klebsiella pneumoniae Population Genomics and Antimicrobial-Resistant Clones. Trends Microbiol. 24 (12), 944–956. 10.1016/j.tim.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. (2012). Primer-BLAST: a Tool to Design Target-specific Primers for Polymerase Chain Reaction. BMC Bioinformatics 13, 134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Liu J., Li Y., Yu J., Zhu W., Liu Y., et al. (2018). Rapid Detection of Carbapenemase Activity of Enterobacteriaceae Isolated from Positive Blood Cultures by MALDI-TOF MS. Ann. Clin. Microbiol. Antimicrob. 17 (1), 22. 10.1186/s12941-018-0274-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Xiao T., David S., Wang Q., Zhou Y., Guo L., et al. (2020). Novel Subclone of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 with Enhanced Virulence and Transmissibility, China. Emerg. Infect. Dis. 26 (2), 289–297. 10.3201/eid2602.190594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zowawi H. M., Forde B. M., Alfaresi M., Alzarouni A., Farahat Y., Chong T.-M., et al. (2015). Stepwise Evolution of Pandrug-Resistance in Klebsiella pneumoniae . Sci. Rep. 5, 15082. 10.1038/srep15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.