Abstract
Molecular analysis of Mycobacterium ulcerans has revealed two new insertion sequences (ISs), IS2404 and IS2606. IS2404 was identified by complete sequencing of a previously described repetitive DNA segment from M. ulcerans. This element is 1,274 bp long, contains 12-bp inverted repeats and a single open reading frame (ORF) potentially encoding a protein of 327 amino acids (aa), and apparently generates 7-bp direct repeats upon transposition. Amino acid similarity was found between the putative transposase and those encoded by ISs in other bacterial sequences from Aeromonas salmonicida (AsIs1), Escherichia coli (H repeat element), Vibrio cholerae (VcIS1), and Porphyromonas gingivalis (PGIS2). The second IS, IS2606, was discovered by sequence analysis of a HaeIII fragment of M. ulcerans genomic DNA containing a repetitive sequence. This element is 1,404 bp long, with 12-bp inverted repeats and a single ORF potentially encoding a protein of 445 aa. Database searches revealed a high degree of amino acid identity (70%) with the putative transposase of IS1554 from M. tuberculosis. Significant amino acid identity (40%) was also observed with transposases from several other microorganisms, including Rhizobium meliloti (ISRm3), Burkholderia cepacia (IS1356), Corynebacterium diphtheriae, and Yersinia pestis. PCR screening of DNA from 45 other species of mycobacteria with primers for IS2404 confirm that this element is found only in M. ulcerans. However, by PCR, IS2606 was also found in Mycobacterium lentiflavum, another slow-growing member of the genus Mycobacterium that is apparently genetically distinct from M. ulcerans. Testing the sensitivity of PCR based on IS2404 and IS2606 primers demonstrated the ability to detect 0.1 and 1 M. ulcerans genome equivalents, respectively. The ability to detect small numbers of cells by using two gene targets will be particularly useful for analyzing environmental samples, where there may be low concentrations of M. ulcerans among large numbers of other environmental mycobacteria.
Mycobacterium ulcerans is a slow-growing environmental mycobacterium which causes significant morbidity in many regions of the world, particularly rural West Africa (17). Infection with this organism can lead to severe, necrotizing skin lesions that require significant surgical excision as treatment (1). The status of M. ulcerans as an emerging pathogen was recently recognized by the World Health Organization in the establishment of the Global Buruli Ulcer Initiative (Yammousoukro Conference, Cote d’Ivoire, July 1998).
Previously, we identified a repetitive AluI DNA restriction fragment from M. ulcerans and used it as a template in a PCR assay for the diagnosis of human disease (25) and detection of the organism in the environment (24). The aims of this study were to characterize the AluI fragment more fully and also to characterize another repeated sequence that we isolated from a M. ulcerans clone library. We provide evidence that both sequences represent genetically distinct insertion sequence (IS) elements, which we have designated IS2404 and IS2606.
ISs are mobile genetic elements which perform no essential function for the cell but have the ability to modify gene expression, sequester genes, and promote genetic rearrangements (11). Until recently a total of 13 IS elements had been described from various mycobacteria (21). The completion of the Mycobacterium tuberculosis genome-sequencing project has revealed at least 30 different IS elements in that one species (8). These elements have been used for strain typing by allowing the detection of restriction fragment length polymorphisms (RFLPs) (12, 14, 30) or as templates for specific and sensitive diagnostic assays (28, 31). These studies take advantage of the high copy numbers and very restricted host ranges of ISs to develop sensitive targets for PCR amplification.
Multiple DNA targets for detection of M. ulcerans may be particularly useful, since PCR is the only method that has so far been able to identify this organism in the environment (22, 24). All attempts to culture M. ulcerans from the environment have failed (19) despite the strong epidemiological evidence that links the source of M. ulcerans to swamps and slow-flowing water (4, 32). Several PCR methods have recently been described for detection of this organism. These have been based on the 16S rRNA gene (20), the hsp65 gene (22), and IS2404 (25). The first two methods target genes with low copy numbers and high sequence conservation among all mycobacteria. They utilize a genus-specific first-round PCR followed by either a second-round PCR or high-stringency probe hybridization conditions to ensure sensitivity and specificity. In these situations the potential for false positives caused by chimera formation or primer cross-reactivity is likely to be high (15, 34). The use of two or more DNA targets is one approach to increase the confidence in positive data generated by PCR and to overcome the issues surrounding primer specificity.
In this paper we describe primer sets for the detection of IS2404 and IS2606. The primers were validated by screening three different strains of M. ulcerans, 45 other mycobacterial species, and 21 other organisms by PCR.
MATERIALS AND METHODS
Mycobacterial strains, culture conditions, and DNA extraction.
The origins of the mycobacterial strains and other organisms used in this study are listed in Table 1. Mycobacterial culture was performed as previously described (25), with the exception that M. ulcerans was cultured either on egg yolk agar or in 10 ml of Middlebrook 7H9 medium at 30°C for 6 to 10 weeks. The broth contained, per liter, 4.7 g of Middlebrook 7H9 base, 0.02 g of catalase, 5 g of bovine serum albumin, 1 g of casein, 200 μl of Tween 80, and 2.5 ml of glycerol. M. ulcerans cultured in this formulation of Middlebrook broth generally grew as a homogeneous, clump-free suspension.
TABLE 1.
Host ranges of IS2404 and IS2606 as determined by PCR
| Organism (strain and/or source)a | Presence of 492-bp IS2404 PCR productb | Presence of 332-bp IS2606 PCR productb |
|---|---|---|
| M. ulcerans (Chant strain; clinical isolate, Victoria, Australia) | + | + |
| M. ulcerans (13822/70; clinical isolate, Queensland, Australia) | + | + |
| M. ulcerans (5145; clinical isolate, Zaire, Africa) | + | + |
| M. ulcerans (9673; clinical isolate, Benin, Africa) | + | + |
| M. marinum (NCTC 2275) | − | − |
| M. kansasii (NCTC 10268) | − | − |
| M. tuberculosis (H37Rv) | − | − |
| M. avium (ATCC 25291) | − | − |
| M. simiae (ATCC 25295) | − | − |
| M. malmoense (NCTC 11298) | − | − |
| M. haemophilum (CAP strain E8) | − | − |
| M. szulgai (NCTC 10831) | − | − |
| M. gordonae (NCTC 10267) | − | − |
| M. nonchromogenicum (NCTC 10424) | − | − |
| M. terrae (NCTC 10856) | − | − |
| M. chelonae (ATCC 10269) | − | − |
| M. phlei (ATCC 11758) | − | − |
| M. agric | − | − |
| M. anthracenicumc | − | − |
| M. asiaticumc | − | − |
| M. aurumc | − | − |
| M. austroafricanumc | − | − |
| M. celatumc | − | − |
| M. chitaec | − | − |
| M. diernhoferic | − | − |
| M. duvallic | − | − |
| M. flavescensc | − | − |
| M. fortuitumc | − | − |
| M. gadiumc | − | − |
| M. gastric | − | − |
| M. gilvumc | − | − |
| M. haemophilumc | − | − |
| M. heidelbergensec | − | − |
| M. interjectumc | − | − |
| M. intermediumc | − | − |
| M. intracelluarec | − | − |
| M. lentiflavumc | − | +d |
| M. neoaurumc | − | − |
| M. obuensec | − | − |
| M. parafinicumc | − | − |
| M. parafortuitumc | − | − |
| M. pulverisc | − | − |
| M. rhodesiaec | − | − |
| M. scrofulaceumc | − | − |
| M. shimoideic | − | − |
| M. thermoresistablec | − | − |
| M. tokaiensec | − | − |
| M. triplexc | − | − |
| M. xenopic | − | − |
| Aeromonas sobria (environmental isolate) | − | − |
| Campylobacter jejuni (NCTC 11351) | − | − |
| Citrobacter freundii (ATCC 8090) | − | − |
| Clostridum perfringens (environmental isolate) | − | − |
| Enterobacter aerogenes (ATCC 13048) | − | − |
| Enterobacter agglomerans (environmental isolate) | − | − |
| Enterobacter cloacae (environmental isolate) | − | − |
| Enterococcus faecalis (ATCC 19433) | − | − |
| E. coli (ATCC 11775) | − | − |
| Klebsiella pneumoniae (environmental isolate) | − | − |
| Legionella pneumophila (ATCC43111) | − | − |
| Proteus mirabilis (environmental isolate) | − | − |
| Pseudomonas aeruginosa (ATCC 10145) | − | − |
| Salmonella typhimurium (environmental isolate) | − | − |
| Shigella sonnei (environmental isolate) | − | − |
| Streptococcus bovis (environmental isolate) | − | − |
| V. cholerae (environmental isolate) | − | − |
| Yersinia enterocolitica (clinical isolate) | − | − |
| Cryptosporidium parvum (clinical isolate) | − | − |
| Giardia intestinalis (clinical isolate) | − | − |
| Homo sapiens | − | − |
“M.” represents “Mycobacterium” throughout.
+, present; −, absent.
Reference strains from the culture collection of the Queensland Diagnostic and Reference Laboratory for Mycobacterial Diseases.
PCR product identity confirmed as IS2606 by nucleotide sequencing. Organism identity was confirmed by partial 16S rDNA nucleotide sequencing.
DNA was extracted from 50 mg (wet weight) of cell pellets by the method of Böddinghaus et al. (5). The DNA concentration was estimated by spectrophotometry at 260 nm. For experiments to test PCR sensitivity, the DNA was heated to 100°C for 10 min to produce sheared molecules and then diluted as required to represent 100, 10, 1, and 0.1 mycobacterial genomes, assuming 1 mycobacterial genome is equivalent to 4 to 5 fg of DNA (16).
Oligonucleotides.
All oligonucleotides used in this study are listed in Table 2. Specificity was checked with the BLAST algorithm (2). The internal stabilities and compatibilities of the PCR primers were checked with Amplify PCR software (Bill Engels, Department of Genetics, University of Wisconsin, Madison).
TABLE 2.
PCR primers used in this study
| Oligo-nucleotide | Sequence (5′→3′) | Position in gene | Function |
|---|---|---|---|
| MU3 | CGCGTGGGTCCCTCGGGTCT | 145–126 | MU3 and MU4 are outward PCR primers for amplifying between tandem copies of IS2404 |
| MU4 | ATCGCCGAAGCCTGCCGGAT | 1116–1135 | |
| MU5 | AGCGACCCCAGTGGATTGGT | 383–401 | MU5 and MU6, 492-bp PCR product from IS2404 |
| MU6 | CGGTGATCAAGCGTTCACGA | 852–871 | |
| MU7 | GGCCTGGCGGATTGCTCAAGG | 213–233 | MU7 and MU8, 332-bp PCR product from IS2606 |
| MU8 | CGTAGATGTGGGCGAAATGG | 542–523 | |
| MYCGENF | AGAGTTTGATCCTGGCTCAG | 16–35a | MYCGENF and MYCGENR, 1,030-bp PCR product from all 16S rRNA gene in (3) |
| MYCGENR | TGCACACAGGCCACAAGGGA | 1046–1027a |
Numbering is based on the E. coli 16S rRNA gene.
Molecular cloning.
Two different strategies were employed to generate clone libraries of M. ulcerans genomic DNA, and both techniques have been described in detail elsewhere (10, 25). Briefly, the first approach used genomic DNA digested with the restriction endonuclease HaeIII, cloned into bacteriophage M13mp18, and then transformed into Escherichia coli NM522. The second strategy used PvuII-digested genomic DNA cloned into pUC18 and then transformed into E. coli DH12S. The libraries were screened by plaque and colony hybridization, respectively, to identify clones containing the desired DNA fragments. General methods used for DNA manipulation and nucleotide sequencing of clones and PCR products have been described previously (10).
Hybridization analysis.
Hybond N+ nylon membranes (Amersham Corp.) were used for all blots. For Southern analysis, M. ulcerans genomic DNA was digested with the appropriate restriction enzyme and separated by electrophoresis through 1.0% agarose gels (3). DNA was transferred to the nylon membrane overnight in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) following the protocol of the membrane manufacturer (Amersham Corp.). Plaque lifts were performed as described previously (25), while alkaline lysis and microwave immobilization were used for colony blots (6). Probes were prepared by PCR amplification of the 492- and 332-bp products from IS2404 and IS2606, respectively, with a deoxynucleoside triphosphate (dNTP) labelling mix containing digoxigenin-11-dUTP. All hybridizations were performed at 65°C with high-stringency posthybridization washes. Detection of DNA was achieved with CDP-Star, following the protocols of the manufacturer (Boehringer Mannheim).
Statistical analysis.
Maximum parsimony was used to analyze the phylogenetic relationships between IS2404, IS2606, and related sequences. The amino acid sequences of the transposases from each IS were aligned by the Pileup algorithm (9) and then subjected to maximum-parsimony analysis (Joe Felsenstein, Department of Genetics, University of Washington, Seattle) with software contained in the Phyllip package (2a).
PCR.
To identify IS2404 tandem repeats, approximately 100 ng of genomic M. ulcerans DNA was amplified in a total reaction volume of 50 μl containing a buffer supplied by the manufacturer of Taq polymerase (Promega, Madison, Wis.), 1 U of Taq polymerase, 1 mM primers, 1.5 mM MgCl2, and 200 mM dNTPs. The reactions were performed in an automated thermal cycler (MJ Research, Watertown, Mass.). After an initial denaturation at 94°C for 2 min, the DNA was amplified by 35 cycles of 1-min steps at 94, 55, and 72°C. The DNA bands were excised from 1% agarose gels and purified with a Sephaglas BandPrep kit (Pharmacia, Uppsala, Sweden). The purified PCR product was then sequenced with primers MU-3 and MU-4 in the direct-incorporation protocol of the Fmol DNA-sequencing system (Promega).
The reaction conditions used for amplification of IS2404, IS2606, and the 16S rRNA gene were as follows. Each PCR mixture (20 μl) contained 1× PCR buffer II (10× PCR buffer II contained 500 mM KCl, 100 mM Tris-HCl [pH 8.3]), 2.5 mM MgCl2, 1 mM dNTPs (1 mM [each] dATP, dTTP, dCTP, and dGTP), 0.5 μM (each) primer, 1 U of Ampli-Taq Gold DNA polymerase (Perkin-Elmer), and 5 μl of pyrocarbonic acid diethyl ester-treated water containing DNA. Hot-start PCR was performed in an FTS-960 thermal sequencer (Corbett Research, Sydney, Australia) with the following protocol: activation of the polymerase at 94°C for 10 min and then five cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min; 30 cycles of 95°C for 20 s, 58°C for 30 s, and 72°C for 45 s; and a final extension step at 72°C for 5 min. The PCR products were held at 4°C until they were analyzed and detected by either 1.5% agarose gel electrophoresis with ethidium bromide staining or 7.5% polyacrylamide gel electrophoresis with silver staining (7).
Nucleotide sequence accession numbers.
The nucleotide sequences of IS2404 and IS2606 have been allocated Genbank accession no. AF003002 and AF082836, respectively.
RESULTS
Characterization of IS2404.
In a previous study we identified a highly repeated 1,109-bp AluI fragment that appeared specific for M. ulcerans (25), but the full characterization of this fragment was not undertaken. Amplification of M. ulcerans genomic DNA with primers MU3 and MU4 produced three DNA bands of approximately 200, 350, and 500 bp (data not shown). Sequencing of the 350-bp fragment revealed terminal sequences from the beginning and the end of the 1,109-bp AluI repeat, with 115 bp of unknown flanking sequence in the middle. This suggested that the band represented a product derived from amplification between two tandem repeats of the element. Sequence obtained from the other two bands represented sequence from either end of the element. Alignment of all three sequences identified the positions of sequence divergence and thus indicated that the length of the complete repeated element was 1,274 bp, flanked by 7-bp direct repeats. There were 12-bp terminal inverted repeats and a single large open reading frame extending from nucleotide 163 to 1146 of the repeat, potentially encoding a protein of 328 amino acids. These data suggested that the repeated element may constitute an IS element. Inverted and terminal direct repeats are features shared by most IS elements, which typically harbor inverted repeats of 8 to 30 bp (11). The presence of a single open reading frame which codes for a potential transposase enzyme involved in the process of IS transposition events is another consistent feature. A gapped BLAST search with the deduced amino acid sequence revealed significant similarity with the putative transposases from AsIS1 of Aeromonas salmonicida (13), the H repeat of the Rhs elements of E. coli (36), the IS element PGIS2 of Porphyrimonas gingivalis (33), and the IS element VcIS1 of Vibrio cholerae (27). This repeat has therefore been named IS2404.
Isolation and characterization of IS2606.
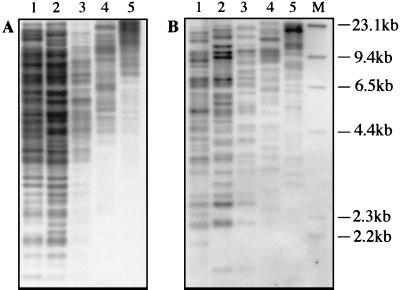
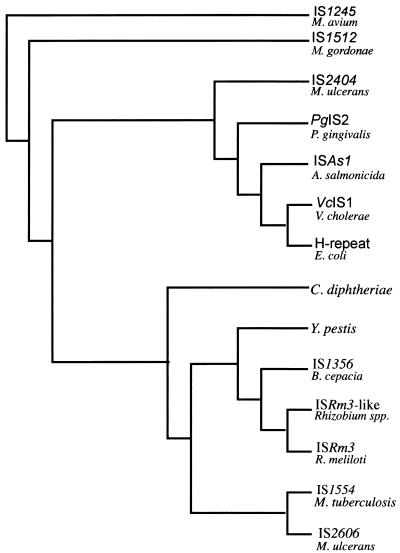
A genomic M13 phage library of HaeIII-digested M. ulcerans DNA produced a clone (L4-3) containing a 450-bp insert that hybridized strongly to a M. ulcerans genomic DNA probe but not with a genomic probe from the closely related Mycobacterium marinum. The strong intensity of the hybridization signal was suggestive of a repeated sequence. This was confirmed by Southern hybridization (Fig. 1B). Based on these observations, further characterization of L4-3 was undertaken. Nucleotide sequence analysis revealed the 5′ region of a putative transposase gene, so an additional genomic library was produced by digesting M. ulcerans DNA with PvuII and cloning fragments into pUC18. Two different clones were obtained, designated pJKD2216 and pJKD2222, which contained single, but different and intact, copies of the entire element. The clones were sequenced, and the boundaries of the element were identified based on the points of sequence divergence between the clones. The element was found to be 1,404 bp long, incorporating terminal 12-bp inverted repeats. A single large open reading frame extended from nucleotides 50 to 1384 and potentially encoded a protein of 445 amino acids. Amino acid similarity searches with the BLAST algorithm showed 81% identity with a putative transposase from IS1554 in M. tuberculosis (8). Significant identity (60%) was also observed with transposases from insertion sequences in Rhizobium meliloti (ISRm3) (35), Burkholderia cepacia (IS1356) (29), Corynebacterium diphtheriae (accession no. P35879), and Yersinia pestis (accession no. AF053947). The element was therefore identified as an insertion sequence and designated IS2606. Maximum-parsimony analysis was conducted with the amino acid sequences for the putative transposases from IS2404 and IS2606 and the other high-scoring sequences from the databases (Fig. 2). This demonstrated that IS2606 and IS2404 are unrelated and distinct from other known mycobacterial ISs, such as IS1245 (Mycobacterium avium) (12) and IS1512 (Mycobacterium gordonae) (18).
FIG. 1.
Southern blot hybridization analysis of selected M. ulcerans strains probed with IS2404 (A) and IS2606 (B). Lane 1, strain 5142; lane 2, strain 13822/70; lanes 3 to 5, Chant strain. Genomic DNA was digested with PvuII (lanes 1 to 3), NcoI (lane 4), and EcoRI (lane 5); lane M, λ HindIII molecular size marker (Boehringer Mannheim).
FIG. 2.
Phylogenetic relationships among transposase amino acid sequences from IS2404, IS2606, and selected IS elements as determined by maximum-parsimony analysis.
Nucleotide sequence analysis showed that there were not PvuII, NcoI, or EcoRI sites within IS2404 or IS2606. Southern blot analysis of M. ulcerans genomic DNA, restricted with these enzymes and probed with each IS, was used to estimate copy numbers (Fig. 1). As demonstrated in previous work (25), IS2404 produced a complex banding pattern, indicating in excess of 50 copies per genome. In addition, RFLP was demonstrated among different strains (Fig. 1). IS2606 also produced a complex banding pattern, suggesting 30 to 40 copies of the element, with significant RFLP evident among the strains (Fig. 1).
Specificity testing of PCR primers for IS2404 and IS2606.
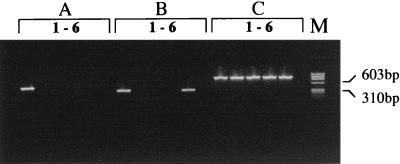
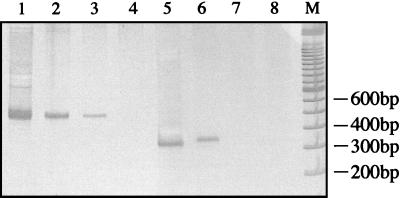
The PCR primers MU1 and MU2 have previously been described for amplification of a 568-bp product from the AluI fragment within IS2404 (25). However, because of occasional spurious banding with MU1 and MU2, we designed new primers, MU5 and MU6, to amplify a 492-bp region of IS2404. Primers MU7 and MU8 were designed to amplify a 332-bp product from IS2606. BLAST analysis of each primer indicated no significant homology with any other sequences in the databases. The correct-size PCR products were obtained for both IS2404 and IS2606 in M. ulcerans isolates from different regions of Australia and Africa, demonstrating nucleotide sequence conservation within each element despite the RFLPs observed among strains (Fig. 1). For IS2404, there were no PCR products of any size detected when primers MU5 and MU6 were used in PCRs with 10 ng of DNA from a panel of 45 other mycobacteria and 21 other microorganisms (Table 1). Thus IS2404 appears specific for M. ulcerans. However when the primer pair for IS2606, MU7 and MU8, was tested against the same panel of organisms, a PCR product of the correct size and nucleotide sequence was obtained from Mycobacterium lentiflavum, suggesting that the host range of IS2606, while restricted, is not specific for M. ulcerans (Table 1 and Fig. 3). Isolates were also subjected to a mycobacterial genus-specific PCR based on the 16S rRNA gene (Table 2) to demonstrate that each DNA preparation contained amplifiable template (Fig. 3).
FIG. 3.
PCR analysis of selected mycobacteria designed to detect IS2404 (A), IS2606 (B), and 16S rDNA (C), as described in Materials and Methods and in Table 2. Lanes 1, M. ulcerans; lanes 2, M. marinum; lanes 3, Mycobacterium haemophilum; lanes 4, M. tuberculosis; lanes 5, M. lentiflavum; lanes 6, no-template negative control; lane M, φx174 HaeIII molecular size marker (Promega).
IS2404 and IS2606 PCR detection sensitivity.
The sensitivity of the method for each target was tested by performing PCR on dilutions of purified M. ulcerans genomic DNA. IS2404 PCR could detect at least 0.1 genome equivalent (Fig. 4), which approaches the theoretical limit of detection for this target based on an estimated 50 copies per genome. IS2606 PCR was approximately 10-fold less sensitive, detecting 1 genome equivalent (Fig. 4), while the 16S ribosomal DNA (rDNA) genus-specific PCR detected 100 genomes (data not shown).
FIG. 4.
Sensitivity of PCR in detection of M. ulcerans genomes by amplification of IS2404 (lanes 1 to 4) and IS2606 (lanes 5 to 8), as determined by silver-stained polyacrylamide gel electrophoresis. Lanes 1 and 5, 10 genomes; lanes 2 and 6, 1 genome; lanes 3 and 7, 0.1 genome; lanes 4 and 8, 0 genomes; lane M, 100-bp ladder size 1arker (Gibco).
DISCUSSION
IS2404 has previously been reported as a highly repeated 1,109-bp AluI restriction fragment from M. ulcerans, present in at least 50 copies per genome and useful as a template for a specific and sensitive PCR-based diagnostic assay (25). The repetitive nature of the fragment was deduced after Southern blot hybridization of SacI-digested DNA resulted in more than 50 bands. These data suggested that the AluI restriction fragment was an internal segment of a repetitive element. In the present study, further analysis revealed that the entire element was 1,274 bp long and contained several features that suggested it was an IS element. However, while the putative transposase amino acid sequence shared similarity with three other bacterial elements, none of these were from mycobacteria (Fig. 2). The sequences were all from gram-negative bacteria, and the finding of a similar element in M. ulcerans probably indicates that this family of IS elements is widely distributed through several bacterial families.
IS2606 was discovered while screening an M. ulcerans clone library for sequences that may be specific for M. ulcerans. This revealed a 1,404-bp element with 12-bp inverted repeats and an open reading frame that could potentially encode a transposase of 445 amino acids. This element was repeated 30 to 40 times per genome (Fig. 1B). Significant DNA identity (80%) was found with IS1554 from M. tuberculosis, of which there is only a single copy, but with no other DNA sequences in the databases, including IS2404. However significant amino acid identity (60 to 80%) was found with six other bacterial IS elements (Fig. 2), suggesting that IS2606 belongs to a family of related elements spread through many different bacterial genera.
We have previously described a very sensitive PCR based on IS2404 that was capable of detecting two molecules of M. ulcerans DNA (25). We have now improved that detection limit to 0.1 molecule by redesigning the primers and amplification conditions and detecting the PCR products by silver-stained polyacrylamide gel electrophoresis. Detection of one molecule of M. ulcerans DNA was possible with IS2606 PCR under these conditions.
Specificity testing of the IS2404 and IS2606 PCR assays indicated that IS2404 appears specific for M. ulcerans while IS2606 was also present in M. lentiflavum (Fig. 3). Phylogenetic studies of M. lentiflavum and M. ulcerans, based on 16S rRNA comparisons, indicate the two species are not closely related (26). The carriage of IS2606 in M. lentiflavum was confirmed by sequence analysis of the PCR product. This revealed 98% identity between the 332-bp PCR products from M. lentiflavum and M. ulcerans (data not shown). We are currently attempting to design primers that will ensure PCR specificity for M. ulcerans based on the small regions of nucleotide differences between the elements in each species. The presence of IS2606 in a species apparently unrelated to M. ulcerans has interesting evolutionary implications and suggests horizontal transmission of the element to or from M. lentiflavum. This ability to cross species boundaries may make IS2606 a useful tool for molecular genetic analysis of some mycobacteria. The presence of IS2606 or IS2404 in another, perhaps as yet unidentified, species of mycobacteria is also possible. However the likelihood of another organism carrying both elements is probably very small. Based on the results of this validation we suggest that, in the absence of a culture isolate, concurrent detection of IS2404 and IS2606 in a sample can be used to provide convincing evidence of the presence of M. ulcerans.
Southern blot analysis to detect IS2404 and IS2606 demonstrated considerable RFLP among different strains (Fig. 1). Strain differences are most clearly evident with PvuII-digested genomic DNA probed with the IS2606 probe. Unfortunately the high copy numbers of both elements make the banding patterns very difficult to interpret and therefore limit the value of a Southern blotting method to type M. ulcerans isolates. However, it may be possible to capture these sequence polymorphisms by developing a simple, robust PCR typing scheme, following the approach used for M. tuberculosis (23). This scheme is based on PCR amplification between tandom copies of repeated sequences with outward-facing primers.
The results of this work should permit the development of improved clinical and environmental PCR assays which, when combined with the ability to rapidly type M. ulcerans strains, will be essential tools for understanding the ecology of this organism and the epidemiology of M. ulcerans disease.
ACKNOWLEDGMENTS
We are indebted to Françoise Portaels and David Dawson for provision of mycobacterial isolates.
This work was substantially supported by funding from the Government of Victoria through the Department of Human Services and also AWT Victoria.
REFERENCES
- 1.Alsop D G. The Bairnsdale ulcer. Aust N Z J Surg. 1972;41:317–319. [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Australian National Genome Information Service Website. 1996, copyright date. [Online.] http:\\mel1.angis.org.au. [18 August 1998, last date accessed.]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Barker D J. Epidemiology of Mycobacterium ulcerans infection. Trans R Soc Trop Med Hyg. 1973;67:43–50. doi: 10.1016/0035-9203(73)90317-9. [DOI] [PubMed] [Google Scholar]
- 5.Böddinghaus B, Rogall T, Flohr T, Blocker H, Bottger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buluwela L, Forster A, Boehm T, Rabbitts T H. A rapid procedure for colony screening using nylon filters. Nucleic Acids Res. 1989;17:452. doi: 10.1093/nar/17.1.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caetano-Anolles G, Gresshoff P M. Staining with silver. Promega Notes. 1994;45:13–18. [Google Scholar]
- 8.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeir K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 9.Feng D F, Doolittle R F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 10.Fyfe J A, Carrick C S, Davies J K. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a sigma 70 promoter during growth in vitro. J Bacteriol. 1995;177:3781–3787. doi: 10.1128/jb.177.13.3781-3787.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 12.Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. 1995;33:304–307. doi: 10.1128/jcm.33.2.304-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustafson C E, Chu S, Trust T J. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994;237:452–463. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- 14.Hermans P W, van Soolingen D, Dale J W, Schuitema A R, McAdam R A, Catty D, van Embden J D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen M A, Straus N. Effect of PCR conditions on the formation of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 1993;3:186–194. doi: 10.1101/gr.3.3.186. [DOI] [PubMed] [Google Scholar]
- 16.Mangiapan G, Vokurka M, Schouls L, Cadranel J, Lecossier D, van Embden J, Hance A J. Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J Clin Microbiol. 1996;34:1209–1215. doi: 10.1128/jcm.34.5.1209-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marston B J, Diallo M O, Horsburgh C R, Jr, Diomande I, Saki M Z, Kanga J M, Patrice G, Lipman H B, Ostroff S M, Good R C. Emergence of Buruli ulcer disease in the Daloa region of Cote d’Ivoire. Am J Trop Med Hyg. 1995;52:219–224. doi: 10.4269/ajtmh.1995.52.219. [DOI] [PubMed] [Google Scholar]
- 18.Picardeau M, Bull T J, Vincent V. Identification and characterization of IS-like elements in Mycobacterium gordonae. FEMS Microbiol Lett. 1997;154:95–102. doi: 10.1111/j.1574-6968.1997.tb12629.x. [DOI] [PubMed] [Google Scholar]
- 19.Portaels F. Epidemiology of mycobacterial diseases. Clin Dermatol. 1995;13:207–222. doi: 10.1016/0738-081x(95)00004-y. [DOI] [PubMed] [Google Scholar]
- 20.Portaels F, Agular J, Fissette K, Fonteyne P A, De Beenhouwer H, de Rijk P, Guedenon A, Lemans R, Steunou C, Zinsou C, Dumonceau J M, Meyers W M. Direct detection and identification of Mycobacterium ulcerans in clinical specimens by PCR and oligonucleotide-specific capture plate hybridization. J Clin Microbiol. 1997;35:1097–1100. doi: 10.1128/jcm.35.5.1097-1100.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulet S, Cole S T. Repeated DNA sequences in mycobacteria. Arch Microbiol. 1995;163:79–86. doi: 10.1007/BF00381780. [DOI] [PubMed] [Google Scholar]
- 22.Roberts B, Hirst R. Immunomagnetic separation and PCR for detection of Mycobacterium ulcerans. J Clin Microbiol. 1997;35:2709–2711. doi: 10.1128/jcm.35.10.2709-2711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross B C, Dwyer B. Rapid, simple method for typing isolates of Mycobacterium tuberculosis by using the polymerase chain reaction. J Clin Microbiol. 1993;31:329–334. doi: 10.1128/jcm.31.2.329-334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross B C, Johnson P D, Oppedisano F, Marino L, Sievers A, Stinear T, Hayman J A, Veitch M G, Robins-Browne R M. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl Environ Microbiol. 1997;63:4135–4138. doi: 10.1128/aem.63.10.4135-4138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross B C, Marino L, Oppedisano F, Edwards R, Robins-Browne R M, Johnson P D. Development of a PCR assay for rapid diagnosis of Mycobacterium ulcerans infection. J Clin Microbiol. 1997;35:1696–1700. doi: 10.1128/jcm.35.7.1696-1700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Springer B, Wu W K, Bodmer T, Haase G, Pfyffer G E, Kroppenstedt R M, Schroder K H, Emler S, Kilburn J O, Kirschner P, Telenti A, Coyle M B, Bottger E C. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol. 1996;34:1100–1107. doi: 10.1128/jcm.34.5.1100-1107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyler S D, Rozee K R, Johnson W M. Identification of IS1356, a new insertion sequence, and its association with IS402 in epidemic strains of Burkholderia cepacia infecting cystic fibrosis patients. J Clin Microbiol. 1996;34:1610–1616. doi: 10.1128/jcm.34.7.1610-1616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Soolingen D, Hermans P W, de Haas P E, van Embden J D. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol. 1992;30:1772–1777. doi: 10.1128/jcm.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vary P H, Andersen P R, Green E, Hermon-Taylor J, McFadden J J. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne’s disease. J Clin Microbiol. 1990;28:933–937. doi: 10.1128/jcm.28.5.933-937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veitch M G, Johnson P D, Flood P E, Leslie D E, Street A C, Hayman J A. A large localized outbreak of Mycobacterium ulcerans infection on a temperate southern Australian island. Epidemiol Infect. 1997;119:313–318. doi: 10.1017/s0950268897008273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C Y, Bond V C, Genco C A. Identification of a second endogenous Porphyromonas gingivalis insertion element. J Bacteriol. 1997;179:3808–3812. doi: 10.1128/jb.179.11.3808-3812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G C, Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheatcroft R, Laberge S. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991;173:2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S, Sandt C H, Feulner G, Vlazny D A, Gray J A, Hill C W. Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993;175:2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]