Abstract
Females and males often exhibit different survival in nature, and it has been hypothesized that sex chromosomes may play a role in driving differential survival rates. For instance, the Y chromosome in mammals and the W chromosome in birds are often degenerated, with reduced numbers of genes, and loss of the Y chromosome in old men is associated with shorter life expectancy. However, mosaic loss of sex chromosomes has not been investigated in any non-human species. Here, we tested whether mosaic loss of the W chromosome (LOW) occurs with ageing in wild birds as a natural consequence of cellular senescence. Using loci-specific PCR and a target sequencing approach we estimated LOW in both young and adult individuals of two long-lived bird species and showed that the copy number of W chromosomes remains constant across age groups. Our results suggest that LOW is not a consequence of cellular ageing in birds. We concluded that the inheritance of the W chromosome in birds, unlike the Y chromosome in mammals, is more stable.
Keywords: mosaic loss of chromosome W, Sula nebouxii, Fregata magnificens, ageing
1. Introduction
Many sex chromosomes in amniote species originated greater than 50 Myr [1–5] following the emergence of genes that acted as regulators of gonadal development [6]. Mammals show X/Y sex chromosomes, whereas birds have Z/W sex chromosomes. During evolution, Y and W chromosomes underwent recombination arrests to preserve the sex-determining loci, a process that is often associated with the accumulation of repetitive DNA and massive genetic loss due to large-scale deletions [7–9].
Recently, it has been shown that sex-specific survival is more strongly associated with the type of sex chromosome system (X/Y or Z/W) than with typical ecological factors [10]. In general, the sex that carries the sex-limited chromosome (Y/W) dies earlier in vertebrates and invertebrates [11]. In birds, specifically, males live longer than females [12], a pattern likely caused by still unknown genetic factors linked to the W chromosome that may affect female survival. One hypothetical cause is particularly appealing: mosaic loss of the sex-limited chromosome during ageing. It has been noticed that mosaic loss of chromosome Y (LOY) in blood cells of aged men is strongly associated with reduced life expectancy [13–16]. We recently showed that LOY is likely shared across mammals [17], but its presence in species with other sex chromosomes is still unresolved.
It has long been debated whether senescence in birds is analogous to that in mammals [18] because birds do not show clear external signs of ageing. However, both taxa evolved endothermy, and higher body temperatures appear to foster cellular senescence [19–21]. Seabirds are among the birds with the longest longevity [22]; for example, the blue-footed booby (Sula nebouxii) can live up to 22 years [23,24], and the magnificent frigatebird (Fregata magnificens) up to 30 years [25]. Adult populations of the blue-footed booby are slightly male-biased and those of F. magnificens are strongly male-biased (male/female ratios of greater than 1 and greater than 2, respectively; electronic supplementary material, figure S1) [26–28]. We studied a wild population of S. nebouxii off the Pacific coast of México that has been monitored over the past three decades and for which we know the exact age of individuals [29,30]. We also analysed data from nestlings and adults of the magnificent Frigatebird from a wild population in Baja California Sur, México. In this work, we tested whether blood cells in long-lived birds evolved age-related mosaic loss of W chromosome (LOW) as a natural consequence of cellular senescence. Based on these sex ratios, we also tested whether LOW could be associated with differential female survival (i.e. we expected higher LOW in F. magnificens).
2. Material and methods
(a) . Study site and sample collection
Blood samples were used to obtain genomic DNA. Blood samples were obtained in the booby colony of Isla Isabel (21° 52′ N, 105°54′ W), México, where monitoring of birds has been carried out annually since 1989 [29,30]; we sampled 61 females: 13 nestlings–fledglings (0–1 year), 19 young adults (2–7 years), 10 middle-aged adults (8–11 years) and 19 old adults (12–18 years). For the magnificent frigatebird, blood samples were obtained from a population on Isla Espiritu Santo, in Baja California Sur, México. Individuals in this population have been monitored for the past four years; we sampled 41 females: 12 nestlings of 1 month old and 29 adult females of 6–30 years of age (with a likely average of approx. 14 years of age according to the species' population structure [25]). For both species, 0.5 ml of blood was stored in 1 ml of DNA/RNA shield buffer by Zymo Research (cat. no. R1200–125) supplied with 0.3 ml of heparin. Permission for fieldwork and sampling was granted by the Secretaría del Medioambiente y Recursos Naturales (SEMARNAT; permit nos. SGPA/DGVS/08333/10, SGPA/DGVS/05216/20 and SGPA/DGVS/03619/21).
(b) . DNA purification
Purified genomic DNA was required for the analyses and 150 µl of blood was used to purify DNA using the Blood DNA Isolation Mini kit from NORGEN BIOTEK CORP (cat. no. 46300/ 46380).
(c) . RNA purification and sequencing
We generated transcriptomic data to gather genetic information for the blue-footed booby. RNA was purified from blood using the RNAeasy QIAGEN kit. We generated strand-specific RNA-seq libraries, using the Illumina TruSeq Stranded mRNA Library protocol. Each library was sequenced on Illumina HiSeq 2500 platforms at the Macrogene facility in Korea (101 nucleotides, paired-end).
(d) . Assembly of W-linked transcripts in the blue-footed booby
To assemble W-linked sequences in the blue-footed booby we used a subtraction approach that compared male and female transcriptomic data; we used this method previously for other amniote species [1,4,31,32]. Briefly, we removed RNA-seq reads shared between males and females and then used Trinity (v. 2.0.2, k-mer of 25 bp) [33] to assemble a female-specific transcriptome.
(e) . Primer design
The PCR-based method required the design of W, Z and autosomal primers. We worked for the blue-footed booby with the male transcriptome assembly and for the magnificent frigatebird, we worked with a publicly available genome assembly (ASM1338994v1 [34]). We identified genes that could be autosomal or Z-linked by BLASTn [35] searches against orthologous genes on the chicken reference genome (https://www.ensembl.org/Gallus_gallus/Info/Index, v.98). We identified W-linked transcripts from the female-specific transcriptome assembly of the blue-footed booby. We designed primers that amplified around 550 base pairs of exonic sequences using the AmplifX software (v.2.0.7, https://inp.univ-amu.fr/en/amplifx-manage-test-and-design-your-primers-for-pcr). W-specific primers were required to show at least two mismatches with the Z gametologues to increase specificity. For PCR amplification we used the Phusion Flash High Fidelity from Thermo Fisher Scientific (cat. no. F548 L) with male and female genomic DNA. We confirmed the expected copy numbers in males and females using standard qPCR curves. We used four DNA dilutions: 0 ng/µl, 0.2 ng/µl, 2 ng/µl and 20 ng/µl and the PowerUp SYBR Green Master Mix from Thermo Fisher (cat. no. A25741). We chose NCK2 (autosomal), VCAN (Z-linked) and RICTOR (W-linked) for the blue-footed booby; and NCK2 (autosomal), DMRT1 (Z-linked) and APC1 (W-linked) for the magnificent frigatebird. We could not use the same Z/W genes in both species due to the lack of the corresponding sequences in the datasets. Primers are provided in electronic supplementary material, table S1.
(f) . Loci-specific PCR and target illumina sequencing
Loci-specific PCR for autosomal, Z and W markers were used as a proxy to quantify the coverage of the sex chromosomes. DNA samples were standardized to 10 ng/µl. We amplified the autosomal, Z-linked and W-linked loci in the same PCR reaction using the Phusion Flash High Fidelity from Thermo Fisher Scientific (cat. no. F548 L). PCR products were purified using Agencourt AMPure XP (cat. no. A63882). PCR products were multiplexed and sequenced in a NextSeq 500 Illumina machine (paired-end, 75 nucleotides long) at UNAM. The quality of the reads was verified using FastQC, and the remaining adaptors were removed with Trimmomatic (v. 036) [36]. Reads were aligned using bowtie2 (v. 2.3.4.1) [37] against the genome sequence of the magnificent frigatebird or the transcriptome assembly of the blue-footed booby. The W-linked gene APC1 was missing from the genomic sequence of the magnificent frigatebird and was assembled from the sequenced data; the Z gametologue was present in the genomic assembly, which allowed us to confirm the identity of W-specific reads. We then extracted the reads that mapped uniquely to the expected loci and obtained on average 821 199 reads (s.d.: ± 156 573) and 185 578 reads (s.d.: ± 22 995) for the magnificent frigatebird and the blue-footed booby, respectively (electronic supplementary material, table S2). To normalize coverage estimates, we first calculated the difference in coverage for the autosomal marker between individual samples and the median across samples, assuming the same autosomal copy number for all samples of the same species. We then used these values to correct individual W/Z coverages (see electronic supplementary material, table S2 for more details). The median value of nestling birds indicated a copy number of one chromosome. All statistical analyses were performed using the R package, standard libraries. Data were plotted using the R package, ‘ggplot2’ library (https://ggplot2.tidyverse.org).
3. Results
(a) . The loci-specific PCR and target sequencing approach
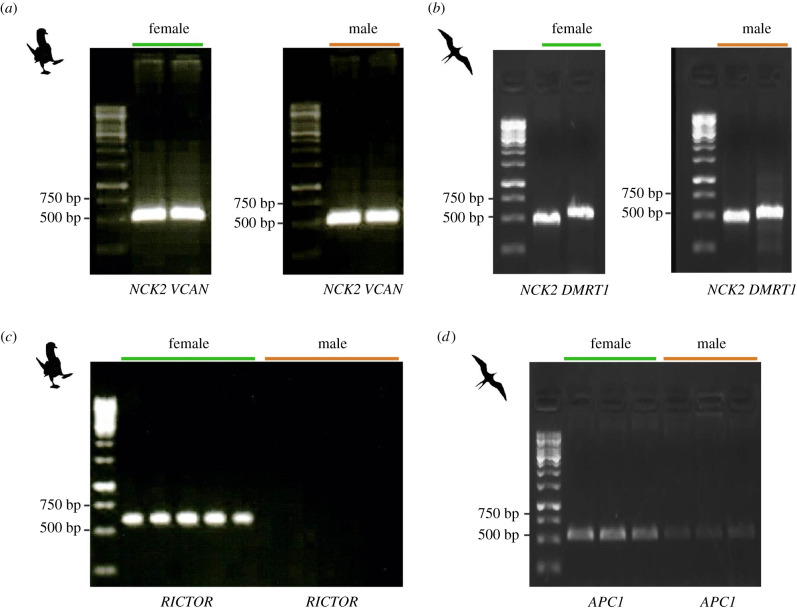
In humans, LOY is generally estimated using data from whole-genomes across age groups. Similar data, however, are lacking for birds. We developed a strategy to estimate LOW using as a proxy the combined amplification and target sequencing of three specific loci (an autosomal, a Z-linked and a W-linked). First, we confirmed that the primers showed the expected pattern: amplification of autosomal and Z loci in both sexes, and amplification of the W locus in females (figure 1).
Figure 1.
PCR products for the autosomal, Z-linked, and W-linked loci. (a) One per cent agarose gel showing a single band of approximately 550 bp for autosomal gene NCK2 and Z-linked gene VCAN in a female and a male of the blue-footed booby. (b) Same as (a) for autosomal gene NCK2 and Z-linked gene DMRT1 in a female and a male of the magnificent frigatebird. (c) The same as in (a) but for the W-linked gene RICTOR in five females and five males of the blue-footed boobies. (d) Same as in (a) but for the W-linked gene APC1 in three females and three males of the magnificent frigatebird; in this case, we observed minor cross-amplification of the Z gametologue in males.
(b) . The W chromosome is not lost during ageing in seabirds
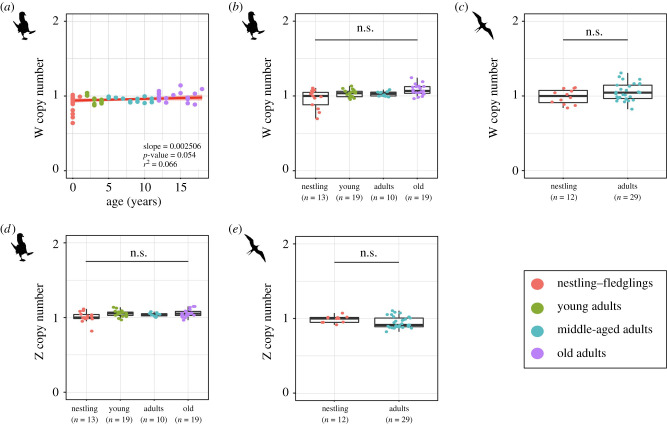
Sequencing data of Z and W markers were used as a proxy to estimate chromosomal copy numbers across age groups. For the blue-footed booby, we compared the autosomal-normalized coverage of the W-linked locus in 61 females distributed in four different age groups and found no statistically significant differences across age groups (figure 2a,b). We also analysed 41 females of the magnificent frigatebird from two different age groups and found that the autosomal-normalized coverage of the W-linked locus was not significantly different between nestlings and adult females (figure 2c).
Figure 2.
W and Z chromosome copy number estimates across age groups. (a) Dot plot of the estimated copy number of W chromosome relative to the age of females in the blue-footed booby. Nestling–fledglings are in red; young adults are in green; middle-aged adults are in blue; old adults are in purple. Significant differences, linear model: lm[W.coverage ∼ age], p < 0.05, excluding outliers. (b) Box plot of the estimated copy number of W chromosome in the four different age groups of the blue-footed booby: nestlings–fledglings (0–1 year), young adults (2–7 years), middle-aged adults (8–11 years) and old adults (12–18 years). N-values are indicated in parenthesis. Significant differences, Benjamin–Hochberg corrected Mann–Whitney U test, p < 0.05. (c) Box plot of the estimated copy number of W chromosome in the two different age groups of the magnificent frigatebird: nestlings (1 month old) and adults (6–30 years). N values are indicated in parenthesis. Significant differences, Mann–Whitney U test, p < 0.05. (d) Same as in (b) but for the estimated copy number of Z chromosome. (e) Same as in (c) but for the estimated copy number of Z chromosome.
We repeated the analyses using the autosomal-normalized coverage of the Z-specific loci in both species. Again, we did not find statistically significant differences between age groups (figure 2d–e).
4. Discussion
This is the first evaluation of the occurrence of mosaic loss of the W chromosome in birds. These species have 140 million years old Z/W chromosomes [2] that originated from a different pair of autosomes than the human X/Y system [38], where LOY was reported [15]. We found no signs of LOW during ageing of either species; accumulation of lower coverage values in older individuals, despite technical stochasticity from PCR amplification, would have indicated LOW. Cross-sectional study of wild populations of two long-lived seabirds allowed sampling across a wide age range, particularly in the booby where individuals have been monitored for over three decades, thus, providing the opportunity to explore the genetics of ageing in a bird species. Similar studies can be performed in other birds provided that proper data (DNA samples for individuals of known age and sex across multiple age groups) are available.
In vertebrates and insects, the sex that carries the Y/W chromosome dies earlier [10,11] and because LOY in humans has been correlated with the early death of men [13,15,39], the mosaic loss of sex chromosomes has been proposed as an important process shaping sex-specific survival rates across taxa [40]. Our results, however, are at odds with LOY/LOW reflecting a general process of cellular senescence associated with W chromosomes and/or the evolution of longer lifespans. Our work indicates that LOW does not influence sex-specific survival in seabirds. We could hypothesize that seabirds may be well-buffered against LOW and that alternative genetic or ecological forces are shaping male/female ratios.
We developed a PCR and target sequencing approach to estimate the coverage of W/Z chromosomes using data from an autosomal gene to standardize variations in sequencing depths across samples. This approach allowed us to analyse over 100 samples without the need to sequence whole-genomes. Although further work is needed to establish whether the PCR-based method can detect LOW at low frequencies, our results support the idea that the ploidy of the W chromosome remains constant across age groups, suggesting that the inheritance of this sex chromosome is stable in birds.
Aneuploidies involving the sex chromosomes are among the more frequent chromosomal aberrations in humans [41]. For example, one in 300 newborn babies is aneuploid, most commonly with a missing or additional sex chromosome [42]. By contrast, aneuploidies involving sex chromosomes in birds (ZO karyotype or triploids) are usually lethal at the embryonic stage [43]. Rare cases of adult females with ZZW triploidy have been reported in four species of birds [44–47]. And in chickens, for example, ZZW females develop as inter-sexes [46]. So, it appears that the lack of W chromosomes in birds may be more deleterious than the lack of Y chromosomes in mammals [48,49].
Acknowledgements
We thank the students that helped during fieldwork.
Contributor Information
Araxi O. Urrutia, Email: A.Urrutia@bath.ac.uk.
Diego Cortez, Email: dcortez@ccg.unam.mx.
Ethics
Permission for fieldwork and sampling was granted by the Secretaría del Medioambiente y Recursos Naturales (SEMARNAT; permit nos. SGPA/DGVS/08333/10, SGPA/DGVS/05216/20 and SGPA/DGVS/03619/21).
Data accessibility
RNA-seq data and PCR-based method and target Illumina sequencing data have been deposited to the NCBI-SRA database under BioProject PRJNA764264. Primers used for the PCR-based method can be found in electronic supplementary material, table S1. Raw coverage values and normalized estimates can be found in electronic supplementary material, table S2. Transcriptome for the blue-footed booby, sequence for APC1 (W-linked) used for the magnificent frigatebird, and code used for coverage and statistical analyses is available on Figshare https://figshare.com/articles/dataset/Trujillo_et_al_Lack_of_agerelated_mosaic_loss_of_W_chromosome_in_long-lived_birds/17086718.
Authors' contributions
N.T.: data curation, formal analysis, investigation, methodology, visualization, writing—original draft; M.M.: formal analysis, investigation, methodology, visualization, writing—review and editing; C.S.: conceptualization, data curation, methodology, resources, supervision, writing—review and editing; S.A.: data curation, formal analysis, investigation, resources, supervision, writing—review and editing; R.C.Y.: data curation, investigation, resources, writing—review and editing; Y.V.A.: data curation, investigation, methodology, resources, writing—review and editing; A.H.O.: data curation, methodology, writing—review and editing; C.R.: data curation, methodology, resources, writing—review and editing; T.S.: conceptualization, funding acquisition, project administration, resources, supervision, writing—review and editing; H.D.: conceptualization, project administration, resources, supervision, validation, writing—review and editing; A.O.U.: conceptualization, funding acquisition, project administration, resources, supervision, writing—original draft; D.C.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing—original draft. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by CONACYT Fronteras en la Ciencia (no. FC-2016/1682), CONACYT FORDECYT-PRONACES (no. 682142) and Royal Society Newton Advanced Fellowship (no. NA160564).
References
- 1.Cornejo-Paramo P, et al. 2020. Viviparous reptile regarded to have temperature-dependent sex determination has old XY chromosomes. Genome Biol. Evol. 12, 924-930. ( 10.1093/gbe/evaa104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grutzner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488-493. ( 10.1038/nature13151) [DOI] [PubMed] [Google Scholar]
- 3.Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. 2015. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 32, 1296-1309. ( 10.1093/molbev/msv023) [DOI] [PubMed] [Google Scholar]
- 4.Marin R, et al. 2017. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res. 27, 1974-1987. ( 10.1101/gr.223727.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rovatsos M, Rehak I, Velensky P, Kratochvil L. 2019. Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol. Biol. Evol. 36, 1113-1120. ( 10.1093/molbev/msz024) [DOI] [PubMed] [Google Scholar]
- 6.Bachtrog D, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12, e1001899. ( 10.1371/journal.pbio.1001899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113-124. ( 10.1038/nrg3366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponnikas S, Sigeman H, Abbott JK, Hansson B. 2018. Why do sex chromosomes stop recombining? Trends Genet. 34, 492-503. ( 10.1016/j.tig.2018.04.001) [DOI] [PubMed] [Google Scholar]
- 9.Wright AE, Dean R, Zimmer F, Mank JE. 2016. How to make a sex chromosome. Nat. Commun. 7, 12087. ( 10.1038/ncomms12087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pipoly I, Bokony V, Kirkpatrick M, Donald PF, Szekely T, Liker A. 2015. The genetic sex-determination system predicts adult sex ratios in tetrapods. Nature 527, 91-94. ( 10.1038/nature15380) [DOI] [PubMed] [Google Scholar]
- 11.Xirocostas ZA, Everingham SE, Moles AT. 2020. The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life. Biol. Lett. 16, 20190867. ( 10.1098/rsbl.2019.0867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liker A, Szekely T. 2005. Mortality costs of sexual selection and parental care in natural populations of birds. Evolution 59, 890-897. [PubMed] [Google Scholar]
- 13.Forsberg LA. 2017. Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men. Hum. Genet. 136, 657-663. ( 10.1007/s00439-017-1799-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg LA, et al. 2019. Mosaic loss of chromosome Y in leukocytes matters. Nat. Genet. 51, 4-7. ( 10.1038/s41588-018-0267-9) [DOI] [PubMed] [Google Scholar]
- 15.Forsberg LA, et al. 2014. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 46, 624-628. ( 10.1038/ng.2966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright DJ, et al. 2017. Genetic variants associated with mosaic Y chromosome loss highlight cell cycle genes and overlap with cancer susceptibility. Nat. Genet. 49, 674-679. ( 10.1038/ng.3821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orta AH, et al. 2021. Rats exhibit age-related mosaic loss of chromosome Y. Commun. Biol. 4, 1418. ( 10.1038/s42003-021-02936-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ricklefs RE. 2010. Insights from comparative analyses of aging in birds and mammals. Aging Cell 9, 273-284. ( 10.1111/j.1474-9726.2009.00542.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conti B, et al. 2006. Transgenic mice with a reduced core body temperature have an increased life span. Science 314, 825-828. ( 10.1126/science.1132191) [DOI] [PubMed] [Google Scholar]
- 20.Flouris AD, Piantoni C. 2015. Links between thermoregulation and aging in endotherms and ectotherms. Temperature (Austin) 2, 73-85. ( 10.4161/23328940.2014.989793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keil G, Cummings E, de Magalhaes JP. 2015. Being cool: how body temperature influences ageing and longevity. Biogerontology 16, 383-397. ( 10.1007/s10522-015-9571-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber EA, Burger J. 2002. Biology of marine birds. Boca Raton, FL: CRC press. [Google Scholar]
- 23.Terres J. 1980. The Audubon Society encyclopedia of North American birds. New York, NY: Knopf. [Google Scholar]
- 24.Young RC, Kitaysky AS, Drummond HM. 2021. Telomere lengths correlate with fitness but assortative mating by telomeres confers no benefit to fledgling recruitment. Sci. Rep. 11, 5463. ( 10.1038/s41598-021-85068-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diamond AW, Schreiber EA. 2002. Magnificent frigatebird (Fregata magnificens), version 1.0. In Birds of the world (eds AF Poole, FB Gill). Ithaca, NY: Cornell Lab of Ornithology. ( 10.2173/bow.magfri.01) [DOI] [Google Scholar]
- 26.Drummond H, Osorno JL, Torres R, Chavelas CG, Larios HM. 1991. Sexual size dimorphism and sibling competition: implications for avian sex ratios. Am. Nat. 138, 623-641. ( 10.1086/285238) [DOI] [Google Scholar]
- 27.Torres R, Drummond H. 1999. Variably male-biased sex ratio in a marine bird with females larger than males. Oecologia 118, 16-22. ( 10.1007/s004420050698) [DOI] [PubMed] [Google Scholar]
- 28.Dearborn DC, Anders AD, Parker PG. 2001. Sexual dimorphism, extrapair fertilizations, and operational sex ratio in great frigatebirds (Fregata minor). Behav. Ecol. 12, 746-752. ( 10.1093/beheco/12.6.746) [DOI] [Google Scholar]
- 29.Ancona S, Drummond H, Rodríguez C, Zúñiga-Vega JJ. 2016. Long-term population dynamics reveal that survival and recruitment of tropical boobies improve after a hurricane. J. Avian Biol. 48, 320-332. [Google Scholar]
- 30.Torres R, Drummond H, Velando A. 2011. Parental age and lifespan influence offspring recruitment: a long-term study in a seabird. PLoS ONE 6, e27245. ( 10.1371/journal.pone.0027245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acosta A, Suarez-Varon G, Rodriguez-Miranda LA, Lira-Noriega A, Aguilar-Gomez D, Gutierrez-Mariscal M, Hernandez-Gallegos O, Mendez-de-la-Cruz F, Cortez D. 2019. Corytophanids replaced the pleurodont XY system with a new pair of XY chromosomes. Genome Biol. Evol. 11, 2666-2677. ( 10.1093/gbe/evz196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Pacheco M, et al. 2020. Expression evolution of ancestral XY gametologs across all major groups of placental mammals. Genome Biol. Evol. 12, 2015-2028. ( 10.1093/gbe/evaa173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644-652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng S, et al. 2020. Dense sampling of bird diversity increases power of comparative genomics. Nature 587, 252-257. ( 10.1038/s41586-020-2873-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Meally D, Ezaz T, Georges A, Sarre SD, Graves JA. 2012. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 20, 7-19. ( 10.1007/s10577-011-9266-8) [DOI] [PubMed] [Google Scholar]
- 39.Loftfield E, Zhou W, Graubard BI, Yeager M, Chanock SJ, Freedman ND, Machiela MJ. 2018. Predictors of mosaic chromosome Y loss and associations with mortality in the UK Biobank. Sci. Rep. 8, 12316. ( 10.1038/s41598-018-30759-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marais GAB, Gaillard JM, Vieira C, Plotton I, Sanlaville D, Gueyffier F, Lemaitre JF. 2018. Sex gap in aging and longevity: can sex chromosomes play a role? Biol. Sex Differ. 9, 33. ( 10.1186/s13293-018-0181-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, Saker D, Shen J, Zaragoza M. 1996. Human aneuploidy: incidence, origin, and etiology. Environ. Mol. Mutagen 28, 167-175. () [DOI] [PubMed] [Google Scholar]
- 42.Hassold T, Hunt P. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280-291. ( 10.1038/35066065) [DOI] [PubMed] [Google Scholar]
- 43.Forstmeier W, Ellegren H. 2010. Trisomy and triploidy are sources of embryo mortality in the zebra finch. Proc. Biol. Sci. 277, 2655-2660. ( 10.1098/rspb.2010.0394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arit D, Bensch S, Hansson B, Hasselquist D, Westerdahl H. 2004. Observation of a ZZW female in a natural population: implications for avian sex determination. Proc. R. Soc. Lond. B 271(Suppl. 4), S249-S251. ( 10.1098/rsbl.2003.0155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kupper C, Augustin J, Edwards S, Szekely T, Kosztolanyi A, Burke T, Janes DE. 2012. Triploid plover female provides support for a role of the W chromosome in avian sex determination. Biol. Lett. 8, 787-789. ( 10.1098/rsbl.2012.0329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin M, Thorne MH, Martin IC, Sheldon BL, Jones RC. 1995. Development of the gonads in the triploid (ZZW and ZZZ) fowl, Gallus domesticus, and comparison with normal diploid males (ZZ) and females (ZW). Reprod. Fertil. Dev. 7, 1185-1197. ( 10.1071/rd9951185) [DOI] [PubMed] [Google Scholar]
- 47.Tiersch TR, Beck ML, Douglass M. 1991. ZZW autotriploidy in a blue-and-yellow macaw. Genetica 84, 209-212. ( 10.1007/bf00127249) [DOI] [Google Scholar]
- 48.Graves JA. 2003. Sex and death in birds: a model of dosage compensation that predicts lethality of sex chromosome aneuploids. Cytogenet. Genome Res. 101, 278-282. ( 10.1159/000074349) [DOI] [PubMed] [Google Scholar]
- 49.Smith CA, Roeszler KN, Hudson QJ, Sinclair AH. 2007. Avian sex determination: what, when and where? Cytogenet. Genome Res. 117, 165-173. ( 10.1159/000103177) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data and PCR-based method and target Illumina sequencing data have been deposited to the NCBI-SRA database under BioProject PRJNA764264. Primers used for the PCR-based method can be found in electronic supplementary material, table S1. Raw coverage values and normalized estimates can be found in electronic supplementary material, table S2. Transcriptome for the blue-footed booby, sequence for APC1 (W-linked) used for the magnificent frigatebird, and code used for coverage and statistical analyses is available on Figshare https://figshare.com/articles/dataset/Trujillo_et_al_Lack_of_agerelated_mosaic_loss_of_W_chromosome_in_long-lived_birds/17086718.