Abstract
Predicting pathogen emergence and spillover risk requires understanding the determinants of a pathogens' host range and the traits involved in host competence. While host competence is often considered a fixed species-specific trait, it may be variable if pathogens diversify across hosts. Balancing selection can lead to maintenance of pathogen polymorphisms (multiple-niche-polymorphism; MNP). The causative agent of Lyme disease, Borrelia burgdorferi (Bb), provides a model to study the evolution of host adaptation, as some Bb strains defined by their outer surface protein C (ospC) genotype, are widespread in white-footed mice and others are associated with non-rodent vertebrates (e.g. birds). To identify the mechanisms underlying potential strain × host adaptation, we infected American robins and white-footed mice, with three Bb strains of different ospC genotypes. Bb burdens varied by strain in a host-dependent fashion, and strain persistence in hosts largely corresponded to Bb survival at early infection stages and with transmission to larvae (i.e. fitness). Early survival phenotypes are associated with cell adhesion, complement evasion and/or inflammatory and antibody-mediated removal of Bb, suggesting directional selective pressure for host adaptation and the potential role of MNP in maintaining OspC diversity. Our findings will guide future investigations to inform eco-evolutionary models of host adaptation for microparasites.
Keywords: Lyme disease, Borrelia, host adaptation, white-footed mouse, American robin
1. Introduction
Predicting the potential for pathogen spillover events to human and non-human hosts has become crucial with increasing frequency and magnitude of emergence events [1]. Critical traits involved in the potential for pathogen emergence are the range of hosts that a pathogen can infect (host range) and the competence of potential host species [2,3]. Host competence traits include the capacity to support initial infection establishment, development of infection over time, and pathogen transmission, while the pathogen's host range trait is defined as the breadth of species for which a pathogen can achieve these three criteria [4]. Given the importance to human health, recent reviews and meta-analyses have sought to identify host- and pathogen-derived traits contributing to potential zoonotic emergence. Host-associated traits included life-history traits [5–7], relationships among hosts [8], species interaction dynamics (e.g. domestication) [9–11] and phylogenetic relationships [12,13]. Analyses of pathogen traits suggest that host range, adaptive potential, and mechanisms of infection are key predictors of zoonoses [2,14,15]. However, with a focus on observed patterns of host–pathogen association, these studies offer limited insights into the underlying molecular mechanisms that determine host competence and potentially shape pathogen adaptation.
A challenge in characterizing host competence is that it is a composite trait resulting from multiple processes encompassing host–pathogen interactions, any of which exhibits a broad range of variation within species [1,16,17]. Intraspecific variability in host competence has often been linked to host-specific traits such as local adaptation [16], phenotypic plasticity [1], development [18], sex [19] or environmentally dependent physiological condition (e.g. resource availability, stressors) [20,21]. However, there is limited understanding of how competence varies depending on the host genotype–pathogen genotype interaction [16]. In fact, pathogen polymorphisms within host populations (or ‘niches'), have been proposed to maintain multiple phenotypes within a pathogen and drive variable outcomes in host competence through a balancing selection mechanism, known as ‘multiple niche polymorphism (MNP)' [22,23]. MNP is predicted when the environment (hosts) is heterogeneous, and no single pathogen genotype reaches the highest fitness in all hosts [24,25]. Thus, MNP assumes selective advantages for pathogen strains to adapt to certain hosts, reducing their host range compared to more generalist strains [26]. These selective advantages may be manifested as increased probability of initial establishment, faster development of infection (i.e. growth rates), or increased transmissibility [3,27].
Borrelia burgdorferi sensu stricto (Bb), the Lyme disease agent in North America, is an ideal system to study the evolution of host–pathogen interactions because it is one of the few zoonotic pathogens for which molecular mechanisms for host adaptation have been proposed [22,28,29]. Bb is transmitted by generalist tick vector, Ixodes spp., driving unique selective pressures across many host species, which exhibit strong variability in overall competence [28,30]. MNP is proposed as a mechanism maintaining extensive Bb strain diversity, particularly driving the polymorphism in a Bb gene, ospC [22]. Evidence of MNP is based on laboratory studies documenting fitness variation among Bb strains across hosts, as well as observations of the associations between hosts and genetic markers in nature [22,31–36]. Determining the extent of intraspecific variation in pathogen fitness and evaluating molecular mechanisms is pivotal for characterizing the potential selective pressures mediating host–pathogen interactions and influencing human disease risk.
To understand the role of intraspecific Bb variation in host competence and assess the plausibility of MNP as a driver of Bb diversification, we experimentally evaluated the interaction between two reservoir vertebrate hosts and three genotypically distinct Bb strains. Hosts were rodents (white-footed mice, Peromyscus leucopus) and birds (American robins, Turdus migratorius) that are competent for Bb and frequently infected in nature [5]. We aimed to address the following questions: (i) do Bb strains with distinct ospC genotypes exhibit variation in fitness (transmission to ticks) across two representative host species? (ii) are strains with higher fitness in one host (‘host adapted'), less fit in the other host compared to strains with similar fitness across hosts (‘generalists')? That is, are there trade-offs involved in host adaptation? and (iii) which cellular and immunological mechanisms are involved in initial strain survival and persistence and are these consistent with differences in fitness, resulting in divergent strain × host phenotypes?
2. Results
(a) . Bb strains vary in their fitness across hosts
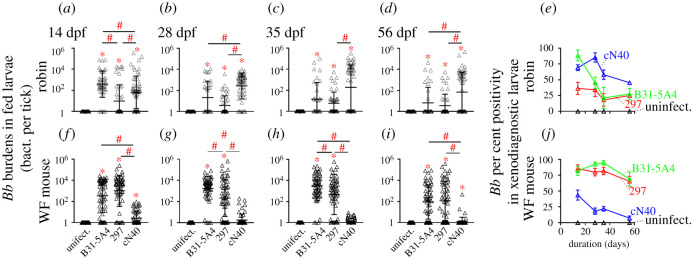
We compared fitness between Bb genotypes commonly found in wild-caught rodents (ospC type A, K) [14,27,28] or rarely found in rodents but infectious in birds (ospC type E) [29]. These strains included tick-isolated B31-5A4 (ospC type A) and cN40 (ospC type E), or human patient-isolated 297 (ospC type K) (electronic supplementary material, table S1) and displayed similar growth rates in culture in vitro (electronic supplementary material, figure S1). We generated Ixodes scapularis nymphs carrying each of these strains and permitted these nymphs to feed on robins and white-footed mice. We found indistinguishable bacterial burdens between the three strains in flat and engorged nymphs (electronic supplementary material, figure S2 and table S2). We then compared the ability of B31-5A4, 297 and cN40 to be transmitted from these Bb-infected robins and mice to naive ticks by determining the Bb burdens in xenodiagnostic I. scapularis larvae at various time points. We found significantly higher proportions of robin-fed cN40-positive larvae than 297-positive larvae, when all time points were combined (electronic supplementary material, table S3–S4). Additionally, the proportion of cN40-positive larvae was significantly lower at 14 days post feeding (dpf), but significantly higher at 28 dpf, than B31-5A4-positive larvae (electronic supplementary material, table S3–S4). We also found significantly greater Bb loads in the larvae that fed on cN40-infected robins relative to 297-infected robins throughout the experiments. The bacterial burdens in larvae from cN40-infected robins were significantly lower at 14 dpf but significantly higher at 28 and 56 dpf than larvae from B31-5A4-infected robins (figure 1a–d; electronic supplementary material, table S2). Although the percent positivity of larvae varied over time and among different strains (figure 1e), the percentage of positive larvae at 56 dpf (45, 27, 25% for larvae that fed on cN40-, B31-5A4-, 297-infected robins, respectively) suggest that cN40 is more fit in robins than B31-5A4 and 297.
Figure 1.
Bb displayed strain-specific, robin- or white-footed (WF) mouse-dependent spirochete transmission to larvae. Nymphs carrying Bb strains B31-5A4, 297, cN40 or naive nymphs (uninfect.) fed to repletion on robins or WF mice. (a–d, f–i) Data represent the geometric mean ± geometric s.d. of Bb burdens in xenodiagnostic larvae from indicated animals. Significance (p ≤ 0.05) from the uninfected group (*) or between groups (#) is indicated. (e and j) Data represent the geometric mean ± s.e.m. of Bb per cent positivity in the xenodiagnostic larvae derived from each animal. (Online version in colour.)
In white-footed mice, the proportion of cN40-positive larvae was significantly lower than B31-5A4- or 297-positive larvae when the results from all time points were combined (electronic supplementary material, tables S3–S4). At 28 and 35 dpf, there were significantly lower bacterial burdens in 297-positive than B31-5A4-positive larvae (figure 1f–i; electronic supplementary material, table S2). We also found Bb burdens in cN40-positive larvae were significantly lower than those from B31-5A4- or 297-positive larvae throughout the experiments (figure 1f–i; electronic supplementary material, tables S2–S4). While the percent positivity differed in the larvae from B31-5A4-, 297- or cN40-infected mice at different time points and among different strains (figure 1j), the percentage of positive larvae at 56 dpf (7, 69, 65% for larvae that fed cN40-, B31-5A4-, 297-infected mice, respectively) suggest cN40 is less fit in mice than B31-5A4 or 297.
(b) . The fitness of Bb strains corresponds with their ability of initial survival and persistence
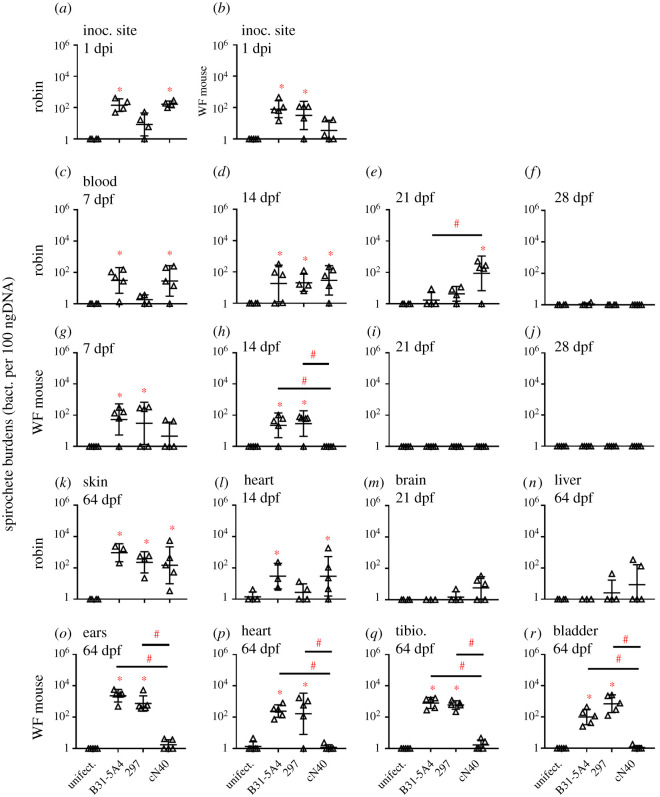
Because tick-transmitted bacterial loads can vary [37], we confirmed the above data and also evaluated early injection site survival of Bb by intradermally injecting robins and white-footed mice with each strain (104 spirochetes per individual). At 1 day post injection (dpi), B31-5A4- or cN40-infected robins showed significantly greater burdens at the injection sites than uninfected robins, whereas B31-5A4- or 297-infected mice had significantly higher burdens at these sites than uninfected mice (figure 2a,b; electronic supplementary material, table S2). These findings indicate that B31-5A4, 297 and cN40 differ in their initial survival in robins and white-footed mice, corresponding with their fitness.
Figure 2.
Bb strains varied in their ability for early colonization, bacteremia induction and persistence at tissues in robins and white-footed (WF) mice. (a-b) Robins (four per group) and WF mice (five per group) were intradermally inoculated with indicated Bb strains or BSK-II medium (uninfect.). The inoculation site of skin (inoc. site) was collected at 1 day post injection (dpi) to determine bacterial burdens by quantitative polymerase chain reaction (qPCR). (c-r) The bacterial loads in blood and tissues at indicated time points post nymph feeding (dpf) determined by qPCR. Data represent geometric mean ± geometric s.d. Significance (p ≤ 0.05) from the uninfected group (*) or between groups (#) is indicated.
Because of the correspondence of data trends during both needle and tick infections, we used a more physiologically relevant route of tick infections to evaluate the ability of these strains to survive in the bloodstream of American robins and white-footed mice. At 7 dpf, B31-5A4- and cN40-, but not 297-infected, robins developed significantly greater levels of bacteremia than uninfected robins, whereas robins infected with all strains displayed significantly greater bacteremia than uninfected robins at 14 dpf (figure 2c,d; electronic supplementary material, table S2). At 21 dpf, cN40-infected robins yielded significantly greater levels of bacteremia than uninfected robins, and bacteremia in cN40-infected robins was significantly greater than B31-5A4-infected robins (figure 2e; electronic supplementary material, table S2). At 28 dpf, robins infected with all strains displayed levels of bacteremia indistinguishable from uninfected robins (figure 2f; electronic supplementary material, table S2). Thus, cN40 induces longer-lasting bacteremia in robins. By contrast, the bacteremia levels from B31-5A4- or 297-, but not cN40-, infected white-footed mice were significantly greater than uninfected mice at 7 and 14 dpf, and B31-5A4- or 297-infected mice had significantly higher bacteremia levels than cN40-infected mice at 14 dpf (figure 2g,h; electronic supplementary material, table S2). None of these strains were detected in mouse blood at 21 and 28 dpf (figure 2i,j). Thus, cN40 is less capable than B31-5A4 and 297 at surviving in the white-footed mouse bloodstream.
We also evaluated the ability of B31-5A4, 297 and cN40 to colonize robin and white-footed mouse tissues at 64 dpf. Robins infected with all strains had significantly greater spirochete burdens in skin than uninfected robins (figure 2k; electronic supplementary material, table S2). Additionally, Bb burdens were significantly greater in the heart and brain of cN40-infected, and the heart of B31-5A4-infected, than uninfected robins (figure 2l,m; electronic supplementary material, table S2). The burdens in livers from robins infected with each strain were indistinguishable from uninfected robins (figure 2n; electronic supplementary material, table S2). These data showed persistent robin skin colonization of all tested strains, but the ability of each strain to colonize other tissues varied. In mice, the bacterial burdens in the outer ears (skin), joints, heart and bladder from B31-5A4 or 297 infections were significantly greater than uninfected or cN40-infected mice (figure 2o–r; electronic supplementary material, table S2). Thus, B31-5A4 and 297, but not cN40, persistently colonized these mice. Taken together, these results showed a positive correspondence of these Bb strains' initial survival and persistence in robins and white-footed mice.
(c) . Multiple cellular and immunological phenotypes correspond with Bb strain-specific fitness, providing mechanistic insights for Bb survival and persistence within a host
(i) . Cell adhesion: Bb strains differed in adhesion to robin and white-footed mouse fibroblasts
We incubated B31-5A4, 297 and cN40 with robin or white-footed mouse fibroblasts and compared the levels of bacterial attachment microscopically with a non-adherent Bb strain, B314 [38] (electronic supplementary material, figure S3). Compared to B314, we detected significantly greater attachment of B31-5A4 and cN40 (figure 3a; electronic supplementary material, table S2). We also observed significantly greater numbers of B31-5A4 and 297 attaching to white-footed mouse fibroblasts relative to B314, and 297 had greater attachment than cN40 (figure 3b; electronic supplementary material, table S2). These results showed differential cell adhesion of these Bb strains as a cellular mechanism that positively corresponds with these strains’ fitness in robins and white-footed mice.
Figure 3.
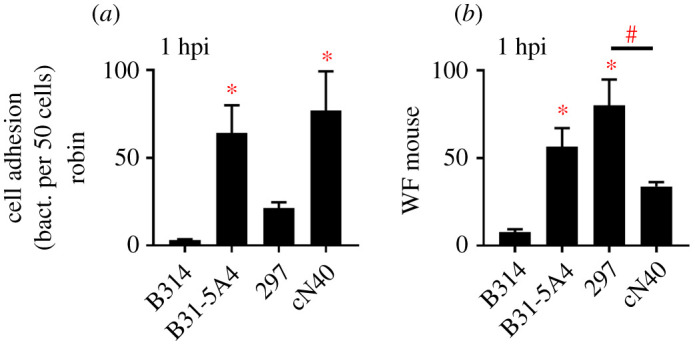
Bb strains differed in the levels of cell adhesion in a robin and/or white-footed (WF) mouse-specific manner. Spirochete strains (2 × 106 spirochetes) were incubated with fibroblasts (2 × 105 cells) from robins and WF mice, stained by anti-Bb antibodies and 4',6-diamidino-2-phenylindole (DAPI) and counted microscopically from four determinations at 1 h post incubation (hpi). Data represents mean ± s.d. Statistical significance (p ≤ 0.05) from B314-treated group (*) or between groups is indicated (#).
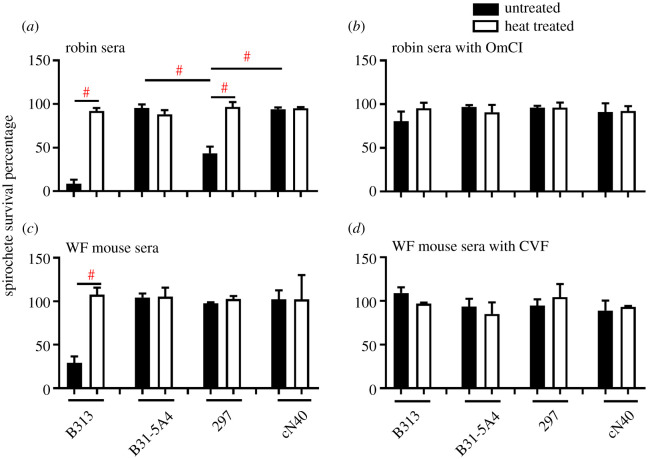
(ii) . Complement evasion: robin, but not white-footed mouse complement, differentiated between the Bb strains serum survivability
We first defined the capability of B31-5A4, 297, and cN40 to evade complement-mediated killing by evaluating the spirochete survival after incubating these strains with naive robin or white-footed mouse sera. More than 88.2% of the spirochetes of these strains survived in heat inactivated robin sera (rendered complement non-functional) [38], as did the high passage, non-infectious and serum sensitive control Bb strain, B313 (figure 4a). When incubated in normal robin sera, 8.6% of B313 survived, verifying the bactericidal activity of robin sera (figure 4a). 95.5% of B31-5A4 and 94.1% of cN40, but 43.6% of 297, survived normal robin sera (figure 4a). All strains survived indistinguishably (at least 80.6% survival) in robin sera treated with Ornithodoros moubata complement inhibitor (OmCI), which inactivates avian complement (figure 4b) [37], indicating that 297 is more vulnerable to robin complement-mediated killing than B31-5A4 and cN40. Similarly, at least 97.1% of all strains survived in heat-inactivated, white-footed mouse sera (figure 4c). Whereas only 29.1% of B313 survived normal white-footed mouse sera, at least 97% of all other strains survived in this sera (figure 4c). Additionally, at least 89.1% of all strains survived in white-footed mouse sera treated with cobra venom factor (CVF), which inactivates mammal complement (figure 4d) [37]. These results indicate that these strains evade killing by white-footed mouse complement at indistinguishable levels.
Figure 4.
Bb strains varied in their ability to survive in robin but not white-footed (WF) mouse sera in a complement-dependent fashion. Percent survival of indicated Bb strains is shown in uninfected robin sera in the (a) absence or (b) presence of OmCI, or uninfected WF mouse sera in the (c) absence or (d) presence of CVF. Heat-treated sera were also included. Data represent mean ± s.d. of per cent survival from three occasions. Significance (p ≤ 0.05) between groups is indicated (#).
(iii) . Cytokine induction: Bb induced strain-specific varying levels of robin and white-footed mouse pro-inflammatory cytokines during infection
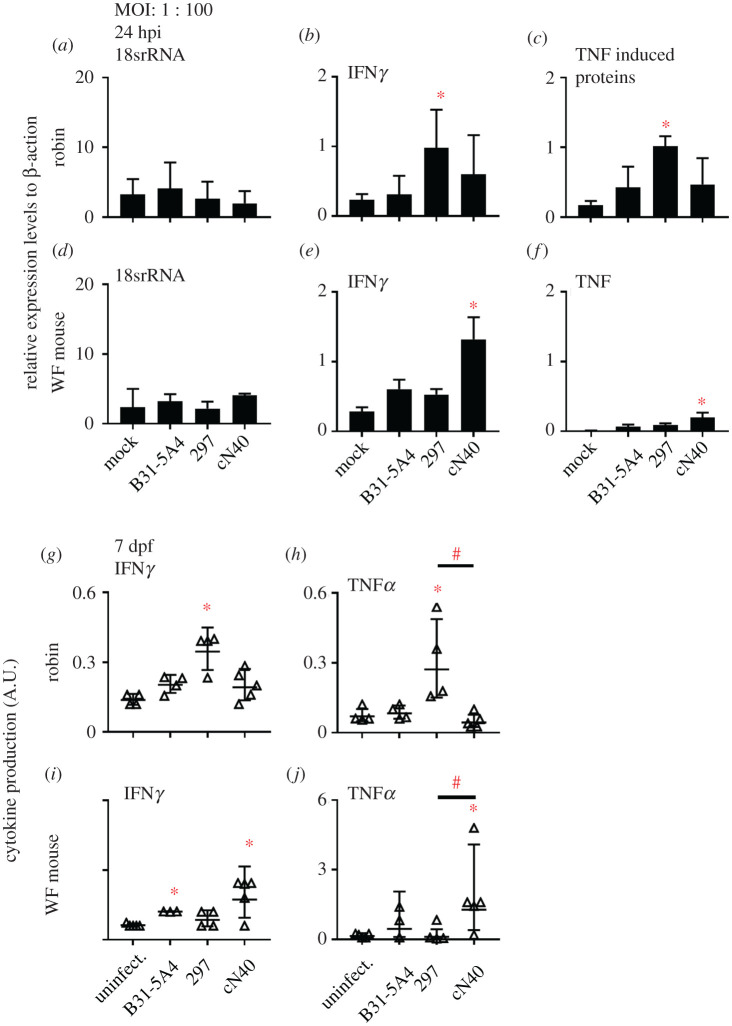
As an indicator of the overall immune response [39,40], we measured the ex vivo expression levels of the genes encoding pro-inflammatory cytokines, interferon gamma (IFNγ), tumour necrosis factor (TNF) or TNF-induced protein, after incubating robin or white-footed mouse fibroblasts with each Bb strain for 24 h. The expression levels of housekeeping genes (18S rRNA and β-actin) were included as controls. We found the expression levels for these genes in cells treated with all Bb strains were indistinguishable from mock-treated cells at 1 : 10 (fibroblasts : spirochetes) (electronic supplementary material, figure S4 and table S2). At a ratio of 1 : 100, 297-incubated robin cells had significantly greater expression of IFNγ and TNF-induced protein than mock-treated robin cells (figure 5a–c; electronic supplementary material, table S2). Conversely, cN40-treated mouse cells showed significantly greater expression of IFNγ and TNF than mock-treated cells (figure 5d–f).
Figure 5.
Bb strains triggered different levels of cytokine ex vivo and at early robin and white-footed (WF) mouse infection. (a–f) Fibroblasts (2 × 105 cells) from robins and WF mice were incubated with Bb strains (2 × 107 spirochetes) or cell medium (mock). The expression levels of indicated genes were determined at 24 h post incubation (hpi) by reverse transcription-qPCR (RT-qPCR) and normalized to expression of β-actin genes. (g–j) Sera were obtained 7 days post nymph feeding (dpf) to determine levels of cytokines using the enzyme linked immunosorbent assay (ELISA). Data represent the geometric mean ± geometric s.d. Significance (p ≤ 0.05) from the uninfected group (*) or between groups (#) is indicated. MOI, multiplicity of infection.
We also assessed the levels of the pro-inflammatory cytokine markers IFNγ and TNFα in vivo in robin and white-footed mouse sera at various times after tick infection. For 14 and 21 dpf, we found comparable levels of these cytokines in robins and mice, regardless of Bb strain (electronic supplementary material, figure S5 and table S2). At 7 dpf, 297-infected robins had significantly greater levels of IFNγ and TNFα than uninfected robins and greater levels of TNFɑ than cN40-infected robins, whereas cytokine levels in cN40 or B31-5A4-infected robins were indistinguishable from uninfected robins (figure 5g,h; electronic supplementary material, table S2). Conversely, cN40-infected mice displayed significantly greater levels of IFNγ and TNFα than uninfected mice and TNFɑ than 297-infected mice, while 297-infected mice showed indistinguishable cytokine levels from uninfected mice (figure 5i,j; electronic supplementary material, table S2). B31-5A4-infected mice had significantly greater levels of IFNγ at 7 dpf than uninfected mice (figure 5j; electronic supplementary material, table S2). These data indicate differences of pro-inflammatory cytokine induction ex vivo and in vivo by these Bb strains, consistent with the fitness of these strains in robins and white-footed mice.
(iv) . Antibodies: Bb strains differed in their ability to induce antibodies in a host- and infection stage-dependent manner
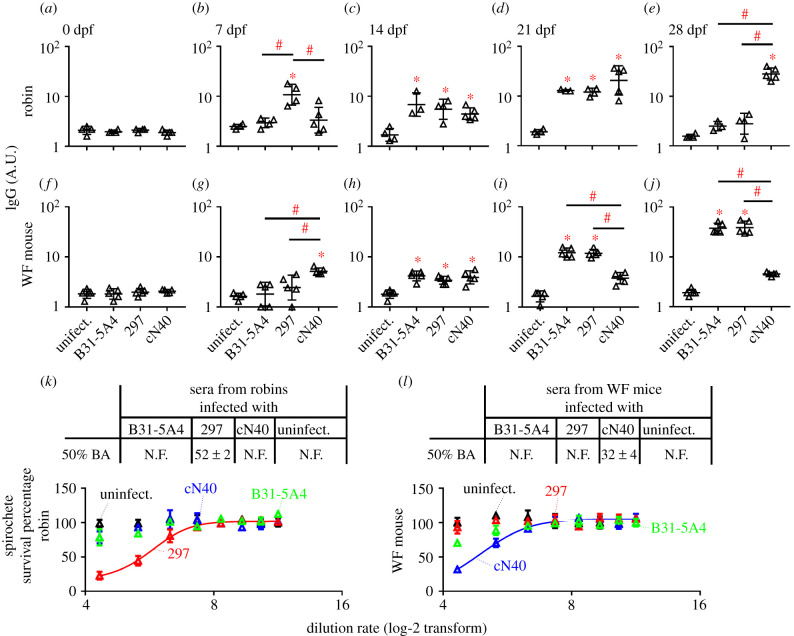
We quantified the levels of antibodies induced by infection of each Bb strain. To avoid strain-specific antibody recognition [41,42], we quantified the immunoglobulin G (IgG) titers in sera from infected robins and mice that recognized an equal ratio of mixed lysates of B31-5A4, 297 and cN40 (figure 6a–j). At 7 dpf, 297-infected robins generated higher anti-Bb IgG titers than uninfected or other strain-infected robins (figure 6b; electronic supplementary material, table S2). At 14 and 21 dpf, robins infected with any strain developed significantly greater IgG titers than uninfected robins (figure 6c,d; electronic supplementary material, table S2). By 28 dpf, cN40-infected robins generated significantly greater IgG titers than uninfected or other strain-infected robins (figure 6e; electronic supplementary material, table S2). In mice, cN40-infection induced significantly greater anti-Bb IgG titers than uninfected or other strain-infected mice at 7 dpf (figure 6g; electronic supplementary material, table S2). At 14 dpf, white-footed mice infected with any strain developed significantly greater anti-Bb IgG titers than uninfected mice (figure 6h). B31-5A4- or 297-infected white-footed mice generated significantly greater IgG titers than uninfected or cN40-infected mice at 21 and 28 dpf (figure 6i,j; electronic supplementary material, table S2). Thus, whereas Bb strains differed in their ability to induce antibodies in a host- and infection stage-dependent manner, the levels of anti-Bb antibodies in robins and white-footed mice at 7 dpf negatively corresponded to spirochete fitness in each of these hosts.
Figure 6.
Bb strains differed in their ability to induce anti-spirochete antibodies in robin and white-footed (WF) mouse infection. (a–j) Sera were obtained at indicated days post nymph feeding (dpf). The levels of IgG against an equal ratio of B31-5A4, 297 and cN40 lysates were determined by ELISA. Data represents the geometric mean ± geometric s.d. Significance (p ≤ 0.05) from the uninfected group (*) or between groups (#) are indicated. (k,l) Sera from infected robins and WF mice at 7 dpf, or naive nymphs (uninfect.). Data represent the mean ± s.e.m. of the survival percentage. 50% BA, the dilution rate that killed 50% of spirochetes, was obtained from curve-fitting. N.F., not fittable. (Online version in colour.)
In addition to whole cell lysates, we also compared IgG titers recognizing purified major Bb antigens in robins and white-footed mice infected with each Bb strain at 7 dpf and found similar negative correspondence between IgG titers and spirochete fitness (electronic supplementary material, figure S6 and table S2). These findings raised the hypothesis that 7 dpf antibodies are bactericidal. To avoid strain-specific bacterial killing, we incubated an equal ratio of B31-5A4, 297 and cN40 with different dilutions of sera from robins or white-footed mice infected with each strain at 7 dpf. We found B31-5A4- and cN40-infected robin sera did not kill spirochetes, but 297-derived sera eliminated 50% of spirochetes at the dilution rate of 1 : 52 (figure 6k). Conversely, cN40-infected white-footed mouse sera eliminated 50% of spirochetes at the dilution rate of 1 : 32 (figure 6l). These results correlate both titers and bactericidal activities of anti-Bb antibodies with the fitness of these strains at early stages of infection. Note, the lower IgG titers at late stages of infection (28 dpf) may reflect the lower fitness of the strains in their respective hosts, which warrants further investigation. However, our findings showed early stages-specific titers and bactericidal activities from the antibodies against B31-5A4-, 297- or cN40, consistent with the fitness of these strains in robins and white-footed mice.
3. Discussion
Our results provide evidence for strain-specific fitness variation across American robins and white-footed mice, two host species previously identified as highly competent for Bb. Based on significant differences in pathogen acquisition by xenodiagnostic larvae, strains 297 and cN40 appear to be white-footed mouse- and robin-adapted, respectively, and B31-5A4 is more generalist (particularly at early stages of infection). While we observed fitness reduction for host-adapted strains in their non-adapted hosts, our results suggest that Bb strains do not occupy strictly specialist or generalist phenotypes, as both strains 297 and cN40 achieved low levels of transmission in their non-adapted hosts.
Competing hypotheses exist for the fundamental drivers of Bb diversity, including MNP, suggesting that polymorphisms are maintained through differential fitness experienced by strains across different host environments. An alternative hypothesis, supported by parsimonious models of genome evolution, posits negative frequency-dependent selection (NFDS) to maintain high Bb diversity via adaptive immune responses that select against common genotypes in a host-independent manner [43]. Although the evolution and maintenance of Bb diversity has been the subject of much theoretical discussion [29,44–46], empirical and mechanistic approaches are uncommon. Previous evidence for host-adapted phenotypes among different ospC genotypes revealed weak associations from natural populations [22,47] and fitness variation for multiple Bb strains in a single species [29,41,48]. By demonstrating the presence of divergent host-adapted phenotypes among Bb strains across different host species, as well as the cellular and immunological mechanisms that influence host–pathogen interactions, this study provides novel evidence for the potential role of MNP in driving patterns of Bb diversity. This does not preclude the potential for NFDS to additionally structure Bb strain communities, and indeed synergistic effects of multiple evolutionary drivers have been suggested [46].
Our results indicate that intraspecific pathogen variability is associated with varying competence within and between hosts. While microparasite competence is defined as the capacity for hosts to permit establishment, development and transmission of infection, ‘realized host competence' for Bb has historically been measured as the percent of field-derived hosts, or individuals infected by field-derived nymphs, from which xenodiagnostic larvae can successfully acquire infection [30]. Our findings suggest that measures of competence may be influenced by the relative frequency of different host-adapted genotypes in the environment, as local Bb communities (measured by ospC types) appear to vary over time and space [33,47,49]. Thus, host overall competence would be an integrated measure of host genotype-specific competence.
Theory suggests that more generalist parasites should experience fitness disadvantages compared to more host-adapted species [3]. However, our results did not indicate the presence of a fitness trade-off during the first two weeks of infection: B31-5A4 exhibited similar levels of Bb positive xenodiagnostic larvae and their bacterial burdens in both hosts at 14 dpf. Yet, we did observe a fitness cost for B31-5A4-infected robins during later stages of infection, where acquisition by ticks was significantly reduced compared to cN40 at all time points past 14 dpf. This shift in the infectivity profile [45], from ‘persistent' in white-footed mice to ‘rapidly cleared' in robins, can influence Bb fitness depending on the timing of tick feeding in nature (phenology). ‘Persistent' genotypes were found to be more frequent in the northeast US than the ‘rapidly cleared' genotypes, presumably owing to positive selection for the infection to persist from the nymphal abundance peak until they are acquired by the larvae that host-seek nearly two months later [50,51]. Future experiments using combinatorial Bb infections across different host species could reveal mechanisms that alter the fitness or infectivity profiles and improve our understanding of the evolutionary drivers of host adaptation and the elevated Bb strain diversity observed in nature.
Our results further support cellular and immunological mechanisms mediating the ability to ‘survive' at early stages and ‘persist' at late stages. These mechanisms include differential cell adhesion, cytokine induction, complement evasion and triggering of antibodies. During early stages of infection, each strain's ability to adhere to host fibroblasts positively corresponded to its overall fitness in that host, consistent with a role for Bb attachment to cells in permitting infection establishment [52]. Additionally, the tested cytokine markers (IFN, TNF) were selected as the documented strain-specific induction of those markers as representatives of overall immune responses [40]. Although these cytokines impact Lyme disease severity in humans, we did not notice any apparent similar symptoms in those reservoir animals (data not shown) [40]. Instead, we found that the extent of cytokine levels in vitro and in vivo corresponded with Bb fitness in reservoir hosts. These findings suggest cytokine marker-mediated signalling promotes Bb clearance, and host-adapted Bb strains avoid triggering this signalling in their adapted reservoir host. Although it is not clear if this is a result of active immune suppression or lack of stimulation, a lack of early immune responses in adapted reservoir hosts would presumably reduce selective pressures and fitness costs, increasing host permissiveness to Bb infection establishment and paving the road for persistence [53]. Moreover, the timing and intensity of antibody response also appeared to mediate variation in strain fitness across hosts, as both 297 and cN40 triggered antibody production earlier in their non-adapted hosts. Given the shift in the infection profile exhibited by B31-5A4 from persistent in mice to rapidly cleared in robins, the increased antibody response at 7 dpf by infected robins to Bb lipoproteins relative to mice is also striking. This suggests the capacity to delay host adaptive immune defences may be a key trait in determining infection persistence [53]. Finally, an important difference we observed among hosts is the role for complement-mediated clearance during early infection, which has been identified as a crucial bottleneck of transmission for Lyme borreliae [54]. While robin complement effectively reduced 297 survival, mouse complement did not have a similar effect on cN40 survival, consistent with the idea that some mechanisms limiting strain fitness are host-strain specific [54].
We acknowledge limitations of our findings, including that the in vitro cultivated fibroblasts do not necessarily reflect in vivo results and the chosen Bb strains and reservoir hosts do not recapitulate all the bacterial strains and hosts in nature. Thus, future studies should aim to increase both experimental hosts and strain diversity to better characterize these eco-evolutionary dynamics. Additionally, host-independent factors proposed to confer Bb adaptation to different hosts (e.g. host–tick species association), should also be considered [55]. Despite these limitations, our findings provide novel insights into the mechanisms structuring Bb diversity and provide a model for understanding host adaptation in a complex microparasite–host system. The patterns and mechanisms discussed here can guide future efforts to characterize host–pathogen interactions, reservoir competence dynamics, and the potential epidemiological impacts of intraspecific phenotypic variation.
4. Material and methods
(a) . Intradermal and tick infection
Animal infections, xenodiagnostic larval placement, and statistical analysis are detailed in the electronic supplementary material.
(b) . Quantitative (qPCR) and reverse transcription PCR (RT-qPCR)
Bb genomic equivalents were calculated through amplification of Bb 16SrRNA gene using primers detailed in the electronic supplementary material.
(c) . Adhesion assays
Fibroblasts were cultivated on 24-well plates and the levels of Bb attachment and statistical analysis are detailed in the electronic supplementary material.
(d) . Serum resistance, ELISA and borreliacidal assays
The uninfected animal sera or the sera from these animals at 7, 14, 21, or 28 dpf and statistical analysis are detailed in the electronic supplementary material.
Supplementary Material
Contributor Information
Yi-Pin Lin, Email: yi-pin.lin@health.ny.gov.
Maria A. Diuk-Wasser, Email: mad2256@columbia.edu.
Ethics
All experiments involving American robins and white-footed mice were performed in strict accordance with all provisions of the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals and the PHS Policy on Humane Care and Use of Laboratory Animals. Additionally, the protocol was approved by IACUC of Wadsworth Center, NYSDOH (no. 18–412 and 19–451) and Columbia University (no. AC-AAAY2450) and the City University in New York Advanced Science Research Center (no. 2016–20). To mistnet robins, personnel were approved on scientific collecting permits USFWS Collecting Permit MB035731 and NYSDEC Permit no. 1236.
Data accessibility
This article has no additional data.
Authors' contributions
Y.-P.L.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing—original draft, writing—review and editing; D.M.T.: conceptualization, data curation, formal analysis, investigation, methodology, resources, validation, writing—original draft, writing—review and editing; M.C.: data curation, investigation, methodology, writing—original draft, writing—review and editing; A.P.D.: data curation, investigation, methodology, resources, writing—review and editing; A.L.M.: data curation, formal analysis, investigation, methodology, writing—review and editing; A.D.H.: investigation, methodology, resources, writing—review and editing; A.J.D.: investigation, methodology, resources, writing—review and editing; J.L.S.: data curation, investigation, methodology, writing—review and editing; A.M.B.: resources, writing—review and editing; K.S.: conceptualization, writing—review and editing; A.D.D.: conceptualization, investigation, methodology, resources, supervision; L.D.K.: conceptualization, investigation, methodology, project administration, resources, supervision, writing—review and editing; S.-O.K.: writing—review and editing; M.A.D.-W.: conceptualization, funding acquisition, investigation, project administration, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
The funding information is described in the electronic supplementary material [56].
References
- 1.Gervasi SS, Civitello DJ, Kilvitis HJ, Martin LB. 2015. The context of host competence: a role for plasticity in host-parasite dynamics. Trends Parasitol. 31, 419-425. ( 10.1016/j.pt.2015.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolhouse ME, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238-244. ( 10.1016/j.tree.2005.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leggett HC, Buckling A, Long GH, Boots M. et al. 2013. Generalism and the evolution of parasite virulence. Trends Ecol. Evol. 28, 592-596. ( 10.1016/j.tree.2013.07.002) [DOI] [PubMed] [Google Scholar]
- 4.Stewart Merrill TE, Johnson PTJ. 2020. Towards a mechanistic understanding of competence: a missing link in diversity-disease research. Parasitology 147, 1159-1170. ( 10.1017/S0031182020000943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker DJ, Han BA. 2021. The macroecology and evolution of avian competence for Borrelia burgdorferi. Glob. Ecol. Biogeogr. 30, 710-724. ( 10.1111/geb.13256) [DOI] [Google Scholar]
- 6.Huang ZY, De Boer WF, Van Langevelde F, Olson V, Blackburn TM, Prins HHT. 2013. Species' life-history traits explain interspecific variation in reservoir competence: a possible mechanism underlying the dilution effect. PLoS ONE 8, e54341. ( 10.1371/journal.pone.0054341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker DJ, Downs CJ, Martin LB. 2019. Multi-scale drivers of immunological variation and consequences for infectious disease dynamics. Integr. Comp. Biol. 59, 1129-1137. ( 10.1093/icb/icz138) [DOI] [PubMed] [Google Scholar]
- 8.Wardeh M, Sharkey KJ, Baylis M. 2020. Integration of shared-pathogen networks and machine learning reveals the key aspects of zoonoses and predicts mammalian reservoirs. Proc. R. Soc. B 287, 20192882. ( 10.1098/rspb.2019.2882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells K, Morand S, Wardeh M, Baylis M. 2020. Distinct spread of DNA and RNA viruses among mammals amid prominent role of domestic species. Glob. Ecol. Biogeogr. 29, 470-481. ( 10.1111/geb.13045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caron A, Cappelle J, Cumming GS, De Garine-Wichatitsky M, Gaidet N. 2015. Bridge hosts, a missing link for disease ecology in multi-host systems. Vet. Res. 46, 83. ( 10.1186/s13567-015-0217-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borremans B, Faust C, Manlove KR, Sokolow SH, Lloyd-Smith JO. 2019. Cross-species pathogen spillover across ecosystem boundaries: mechanisms and theory. Phil. Trans. R. Soc. B 374, 20180344. ( 10.1098/rstb.2018.0344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mollentze N, Streicker DG. 2020. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proc. Natl Acad. Sci. USA 117, 9423-9430. ( 10.1073/pnas.1919176117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CK, Hitchens PL, Pandit PS, Rushmore J, Evans TS, Young CC, Doyle MM. 2020. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. R. Soc. B 287, 20192736. ( 10.1098/rspb.2019.2736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pulliam JR. 2008. Viral host jumps: moving toward a predictive framework. Ecohealth 5, 80-91. ( 10.1007/s10393-007-0149-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulliam JR, Dushoff J. 2009. Ability to replicate in the cytoplasm predicts zoonotic transmission of livestock viruses. J. Infect. Dis. 199, 565-568. ( 10.1086/596510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambrechts L, Chevillon C, Albright RG, Thaisomboonsuk B, Richardson JH, Jarman RG, Scott TW. 2009. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol. Biol. 9, 160. ( 10.1186/1471-2148-9-160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobey S. 2014. Pathogen evolution and the immunological niche. Ann. NY Acad. Sci. 1320, 1-15. ( 10.1111/nyas.12493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson PTJ, Kellermanns E, Bowerman J. 2011. Critical windows of disease risk: amphibian pathology driven by developmental changes in host resistance and tolerance. Funct. Ecol. 25, 726-734. ( 10.1111/j.1365-2435.2010.01830.x) [DOI] [Google Scholar]
- 19.Zuk M, McKean KA. 1996. Sex differences in parasite infections: patterns and processes. Int. J. Parasitol. 26, 1009-1023. ( 10.1016/S0020-7519(96)80001-4) [DOI] [PubMed] [Google Scholar]
- 20.Cressler CE, Nelson WA, Day T, Mccauley E. 2014. Disentangling the interaction among host resources, the immune system and pathogens. Ecol. Lett. 17, 284-293. ( 10.1111/ele.12229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gervasi SS, Burgan SC, Hofmeister E, Unnasch TR, Martin LB. 2017. Stress hormones predict a host superspreader phenotype in the West Nile virus system. Proc. R. Soc. B 284, 20171090. ( 10.1098/rspb.2017.1090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brisson D, Dykhuizen DE. 2004. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics 168, 713-722. ( 10.1534/genetics.104.028738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bown KJ, Lambin X, Ogden NH, Begon M, Telford G, Woldehiwet Z, Birtles RJ. 2009. Delineating Anaplasma phagocytophilum ecotypes in coexisting, discrete enzootic cycles. Emerg. Infect. Dis. 15, 1948-1954. ( 10.3201/eid1512.090178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsman A, Ahnesjö J, Caesar S, Karlsson M. 2008. A model of ecological and evolutionary consequences of color polymorphism. Ecology 89, 34-40. ( 10.1890/07-0572.1) [DOI] [PubMed] [Google Scholar]
- 25.Cohan FM, Koeppel AF. 2008. The origins of ecological diversity in prokaryotes. Curr. Biol. 18, R1024-R1034. ( 10.1016/j.cub.2008.09.014) [DOI] [PubMed] [Google Scholar]
- 26.Straub CS, Ives AR, Gratton C. 2011. Evidence for a trade-off between host-range breadth and host-use efficiency in aphid parasitoids. Am. Nat. 177, 389-395. ( 10.1086/658177) [DOI] [PubMed] [Google Scholar]
- 27.Visher E, Boots M. 2020. The problem of mediocre generalists: population genetics and eco-evolutionary perspectives on host breadth evolution in pathogens. Proc. R. Soc. B 287, 20201230. ( 10.1098/rspb.2020.1230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Keeffe KR, Oppler ZJ, Brisson D. 2020. Evolutionary ecology of Lyme Borrelia. Infect. Genet. Evol. 85, 104570. ( 10.1016/j.meegid.2020.104570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanincova K, Ogden NH, Diuk-Wasser M, Pappas CJ, Iyer R, Fish D, Schwartz I, Kurtenbach K. 2008. Fitness variation of Borrelia burgdorferi sensu stricto strains in mice. Appl. Environ. Microbiol. 74, 153-157. ( 10.1128/AEM.01567-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. 2003. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc. Natl Acad. Sci. USA 100, 567-571. ( 10.1073/pnas.0233733100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerar T, Strle F, Stupica D, Ruzic-Sabljic E, Mchugh G, Steere AC, Strle K. 2016. Differences in genotype, clinical features, and inflammatory potential of Borrelia burgdorferi sensu stricto strains from Europe and the United States. Emerg. Infect. Dis. 22, 818-827. ( 10.3201/eid2205.151806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang I-N, Wormser GP, Schriefer ME, Luft BJ. 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 67, 3518-3524. ( 10.1128/IAI.67.7.3518-3524.1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanincova K, Mukherjee P, Ogden NH, Margos G, Wormser GP, Reed KD, Meece JK, Vandermause MF, Schwartz I. 2013. Multilocus sequence typing of Borrelia burgdorferi suggests existence of lineages with differential pathogenic properties in humans. PLoS ONE 8, e73066. ( 10.1371/journal.pone.0073066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di L, et al. 2018. Genotyping and quantifying lyme pathogen strains by deep sequencing of the outer surface protein C (ospC) locus. J. Clin. Microbiol. 56, e00940-18. ( 10.1128/JCM.00940-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbour AG, Travinsky B. 2010. Evolution and distribution of the ospC gene, a transferable serotype determinant of Borrelia burgdorferi. mBio 1, e00153-10. ( 10.1128/mBio.00153-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.States SL, Brinkerhoff RJ, Carpi G, Steeves TK, Folsom-O'Keefe C, DeVeaux M, Diuk-Wasser MA. 2014. Lyme disease risk not amplified in a species-poor vertebrate community: similar Borrelia burgdorferi tick infection prevalence and OspC genotype frequencies. Infect. Genet. Evol. 27, 566-575. ( 10.1016/j.meegid.2014.04.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart TM, et al. 2021. Host tropism determination by convergent evolution of immunological evasion in the Lyme disease system. PLoS Pathog. 17, e1009801. ( 10.1371/journal.ppat.1009801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadziene A, Wilske B, Ferdows MS, Barbour AG. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61, 2192-2195. ( 10.1128/iai.61.5.2192-2195.1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petzke MM, Iyer R, Love AC, Spieler Z, Brooks A, Schwartz I. 2016. Borrelia burgdorferi induces a type I interferon response during early stages of disseminated infection in mice. BMC Microbiol. 16, 29. ( 10.1186/s12866-016-0644-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strle K, Jones KL, Drouin EE, Li X, Steere AC. 2011. Borrelia burgdorferi RST1 (OspC type A) genotype is associated with greater inflammation and more severe Lyme disease. Am. J. Pathol. 178, 2726-2739. ( 10.1016/j.ajpath.2011.02.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baum E, Hue F, Barbour AG. 2012. Experimental infections of the reservoir species Peromyscus leucopus with diverse strains of Borrelia burgdorferi, a Lyme disease agent. mBio 3, e00434-12. ( 10.1128/mBio.00434-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanova L, Christova I, Neves V, Aroso M, Meirelles L, Brisson D, Gomes-Solecki M. 2009. Comprehensive seroprofiling of sixteen B. burgdorferi OspC: implications for Lyme disease diagnostics design. Clin. Immunol. 132, 393-400. ( 10.1016/j.clim.2009.05.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haven J, et al. 2011. Pervasive recombination and sympatric genome diversification driven by frequency-dependent selection in Borrelia burgdorferi, the Lyme disease bacterium. Genetics 189, 951-966. ( 10.1534/genetics.111.130773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brisson D, Drecktrah D, Eggers CH, Samuels DS. 2012. Genetics of Borrelia burgdorferi. Annu. Rev. Genet. 46, 515-536. ( 10.1146/annurev-genet-011112-112140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haven J, Magori K, Park AW. 2012. Ecological and inhost factors promoting distinct parasite life-history strategies in Lyme borreliosis. Epidemics 4, 152-157. ( 10.1016/j.epidem.2012.07.001) [DOI] [PubMed] [Google Scholar]
- 46.Qiu WG, Martin CL. 2014. Evolutionary genomics of Borrelia burgdorferi sensu lato: findings, hypotheses, and the rise of hybrids. Infect. Genet. Evol. 27, 576-593. ( 10.1016/j.meegid.2014.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mechai S, Margos G, Feil EJ, Barairo N, Lindsay LR, Michel P, Ogden NH. 2016. Evidence for host-genotype associations of Borrelia burgdorferi sensu stricto. PLoS ONE 11, e0149345. ( 10.1371/journal.pone.0149345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vuong HB, Canham CD, Fonseca DM, Brisson D, Morin PJ, Smouse PE, Ostfeld RS. 2014. Occurrence and transmission efficiencies of Borrelia burgdorferi ospC types in avian and mammalian wildlife. Infect. Genet. Evol. 27, 594-600. ( 10.1016/j.meegid.2013.12.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter KS, Carpi G, Caccone A, Diuk-Wasser MA. 2017. Genomic insights into the ancient spread of Lyme disease across North America. Nat. Ecol. Evol. 1, 1569-1576. ( 10.1038/s41559-017-0282-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatewood AG, et al. 2009. Climate and tick seasonality are predictors of Borrelia burgdorferi genotype distribution. Appl. Environ. Microbiol. 75, 2476-2483. ( 10.1128/AEM.02633-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levi T, Keesing F, Oggenfuss K, Ostfeld RS. 2015. Accelerated phenology of blacklegged ticks under climate warming. Phil. Trans. R. Soc. B 370, 1665. ( 10.1098/rstb.2013.0556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coburn J, Leong J, Chaconas G. 2013. Illuminating the roles of the Borrelia burgdorferi adhesins. Trends Microbiol. 21, 372-379. ( 10.1016/j.tim.2013.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tracy KE, Baumgarth N. 2017. Borrelia burgdorferi manipulates innate and adaptive immunity to establish persistence in rodent reservoir hosts. Front. Immunol. 8, 116. ( 10.3389/fimmu.2017.00116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin YP, Diuk-Wasser MA, Stevenson B, Kraiczy P. 2020. Complement evasion contributes to Lyme borreliae-host associations. Trends Parasitol. 36, 634-645. ( 10.1016/j.pt.2020.04.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margos G, Becker NS, Fingerle V, Sing A, Ramos JA, Carvalho IL, Norte AC. et al. 2019. Core genome phylogenetic analysis of the avian associated Borrelia turdi indicates a close relationship to Borrelia garinii. Mol. Phylogenet. Evol. 131, 93-98. ( 10.1016/j.ympev.2018.10.044) [DOI] [PubMed] [Google Scholar]
- 56.Lin Y-P, et al. 2022. Cellular and immunological mechanisms influence host-adapted phenotypes in a vector-borne microparasite. Figshare. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.