Summary
The spacing of nodes of Ranvier crucially affects conduction properties along myelinated axons. It is assumed that node position is primarily driven by growing myelin sheaths. Here, we reveal an additional mechanism of node positioning that is driven by the axon. Through longitudinal live imaging of node formation dynamics in the zebrafish central nervous system, we show that stable clusters of the cell adhesion molecule neurofascin a can accumulate at specific sites along axons prior to myelination. While some of these clusters are pushed into future node position by extending myelin sheaths, others are not and thus prefigure the position of where a mature node forms. Animals that lack full-length neurofascin a show increased internodal distances and less regular nodal spacing along single axons. Together, our data reveal the existence of an axonal mechanism to position nodes of Ranvier that does not depend on regulation by myelin sheath growth.
Keywords: myelination, axon-oligodendrocyte interactions, node of Ranvier, central nervous system, zebrafish
Graphical abstract
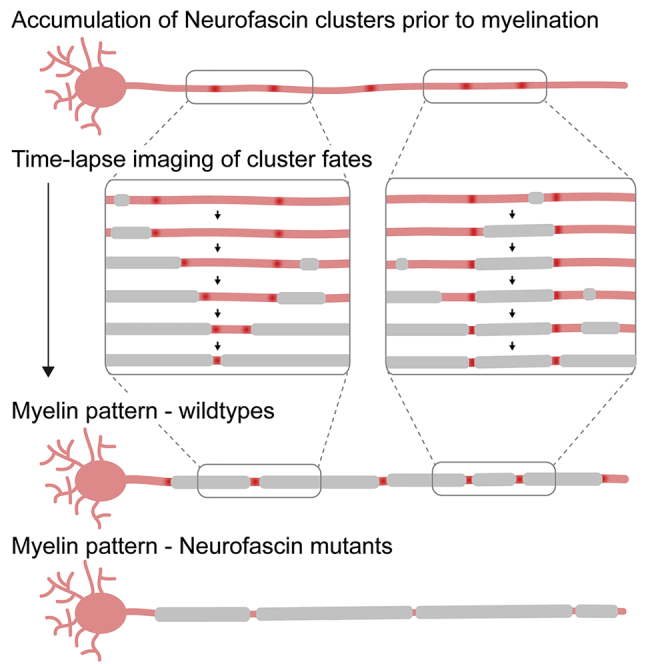
Highlights
-
•
Stable clusters of neuronal neurofascin can form along axons prior to myelination
-
•
Some clusters are pushed in place by developing myelin sheaths
-
•
Other clusters restrict sheath growth and thus prefigure node position
-
•
Animals lacking axonal neurofascin have longer and less regular internodal spacing
How nodes of Ranvier get positioned to pattern myelinated axons is unclear. Vagionitis et al. reveal that stable clusters of axonal neurofascin can prefigure future node position, thus revealing a neuronal mechanism to determine node position, which acts in addition to their regulation by myelin sheath growth.
Introduction
The myelination of axons is crucial for fast propagation of action potentials. Myelin restricts the localization of voltage-gated ion channels to discrete axonal subdomains around the nodes of Ranvier (Rasband and Peles, 2020; Sherman and Brophy, 2005). Not all axons are myelinated equally along their length but can instead show variable and highly organized internodal distances, with implications for conduction speed and circuit function (Brill et al., 1977; Ford et al., 2015; Waxman, 1997; Xin and Chan, 2020). Although we have a detailed understanding of the molecules that organize node of Ranvier architecture in general (Rasband and Peles, 2020), the mechanisms that determine where nodes are positioned along the length of individual myelinated axons are not understood.
It is known that myelin sheath length determines node position and thus the distance between consecutive nodes (the internodal spacing). This is due to the fact that a node is characterized as being localized to the gap between two adjacent myelin sheaths, and because the paranodal junctions act as molecular sieves that restrict the localization of ion channels and adhesion molecules to discrete microdomains around the node (Pedraza et al., 2001). Furthermore, it has been shown that developing myelin sheaths grow to different lengths in response to neural activity (Baraban et al., 2017; Krasnow et al., 2017). Therefore, the prevailing view is that sheaths push all nodes in position and that nodal spacing is thus determined by regulation of myelin growth. However, during the myelination of a single axon along its length, individual sheaths are added at different points in time and by different oligodendrocytes. It remains unclear how specific myelination patterns, such as even spacing along the axon, or their progressive shortening toward the synapse, can be achieved by only controlling myelin growth.
One simple alternative mechanism besides oligodendrocyte control of node position would be that the axon itself provides cues that restrict myelin sheath growth. One axonal feature that inherently limits sheath growth and determines node position is their branching pattern. However, we have previously shown that extension of developing myelin sheaths often occurs asymmetrically, even in the absence of obvious physical barriers, such as axon branches or adjacent myelin sheaths that could restrict lateral myelin growth (Auer et al., 2018). These observations suggest that other axonal cues might restrict sheath growth beyond certain points and therefore determine the position of where a node will form. It is known that neurons can form pre-nodal clusters along unmyelinated axons in vitro and in vivo (Freeman et al., 2015; Kaplan et al., 1997). However, if the localization of such pre-myelinating node clusters is a determinant of node of Ranvier position along axons in vivo is not known.
Here, we investigated node of Ranvier positioning along single axons using in vivo live cell imaging in the zebrafish spinal cord. We show that the neuronal cell adhesion molecule neurofascin a (Nfasca) accumulates along single axons prior to myelination. These clusters were either pushed in place by developing myelin sheaths, or appear to restrict sheath extension and were therefore predictive for future node position. Animals that lack neuronal Nfasca showed increased internodal distances and less regular nodal spacing along single axons. Therefore, we reveal the existence of an axonal mechanism of node positioning, which acts in addition to and independently of regulation of myelin sheath length.
Results
Localization dynamics of node of Ranvier reporter constructs during myelination of single axons
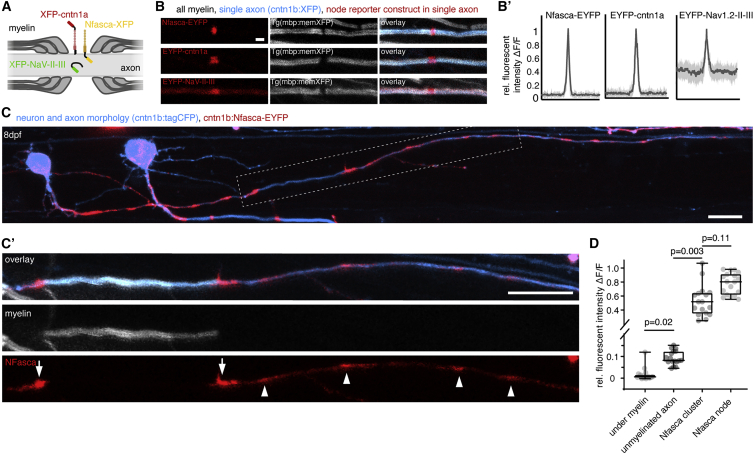
We studied node of Ranvier positioning along axons of commissural primary ascending (CoPA) and circumferential descending (CiD) neurons of larval zebrafish, which are both glutamatergic spinal interneurons with frequently myelinated axons (Higashijima et al., 2004; Koudelka et al., 2016). Nodes were labeled using three transgenic live-cell reporters that localize by different mechanisms: the GPI-anchored adhesion molecule contactin1a with EYFP fused to its N terminus (EYFP-cntn1a); the intracellular ankyrin G targeting motif of zebrafish NaV1.2 (EYFP-NaV-II-III); and neuronal neurofascin (Nfasca) as a transmembrane protein with a C-terminal fusion of EYFP (Nfasca-EYFP) (Figure 1A) (Auer et al., 2018; Garrido et al., 2003; Koudelka et al., 2016). Co-expression of these constructs in single neurons alongside a cytoplasmic fluorescent protein to outline axon morphology (cntn1b:XFP) in animals that have all myelin labeled (Tg(mbp:memXFP)) showed that all three nodal reporter constructs specifically accumulate in the gap between two adjacent myelin sheaths (Figure 1B). EYFP-cntn1a and Nfasca-EYFP fluorescence was completely excluded from myelinated axon parts (in agreement with previous observations) (Auer et al., 2018; Koudelka et al., 2016), whereas EYFP-NaV-II-III retained some fluorescence within/underneath ensheathed axon stretches (Figures 1B and 1B′). In contrast to the highly organized localization of our reporters along myelinated and nodal regions, the distribution of all three live-cell reporters was rather diffuse along unmyelinated stretches of partially myelinated axons (Figures 1C and S1A–S1C). However, we did regularly observe accumulations of Nfasca-EYFP fluorescence along unmyelinated stretches that resembled the accumulations at nodes and heminodes (Figures 1C′ and S1A). No signs of oligodendrocyte ensheathment could be detected in the vicinity of these Nfasca-EYFP accumulations, nor was diffuse Nfasca-EYFP fluorescence excluded from the respective axon stretches, as it would be the case upon myelin ensheathment (Figures 1D and S1D). Nfasca-EYFP clusters were about five times brighter than unmyelinated regions and of comparable, yet slightly lower, fluorescence intensity than nodes (relative Nfasca fluorescence: 0.1 ± 0.03 unmyelinated versus 0.5 ± 0.2 at cluster versus 0.8 ± 0.2 at node, n = 7/7 axons/animals, p = 0.003 unmyelinated versus cluster, p = 0.11 cluster versus node [Kruskal-Wallis test with multiple comparisons]; Figure 1D). We did not detect any cluster accumulations using cntn1a and NaV-II-III reporter constructs (Figures S1B and S1C). Pre-nodal clusters have previously been reported in different systems, including cell cultures and in rodents in vivo (Freeman et al., 2015; Kaplan et al., 1997). To confirm that also zebrafish axons can endogenously form such pre-nodal clusters, we carried out whole-mount immunohistochemistry for neurofascin and large-caliber axons in a transgenic myelin reporter line. Using this assay, we were able to identify nodes, heminodes, and Nfasca clusters along unmyelinated axon stretches, just as we did using the Nfasca-EYFP reporter (Figure S1E). Thus, we conclude that Nfasca-EYFP accumulations represent pre-nodal clusters as previously reported in other models.
Figure 1.
Localization of transgenic node of Ranvier reporters along unmyelinated and myelinated axon stretches
(A) Schematic of node of Ranvier reporter constructs used.
(B) Confocal images of nodal reporters along individually labeled myelinated axons in transgenic animals that have all myelin labeled. Scale bar, 2 μm. (B′) Fluorescence intensity traces of each reporter around the node. Bold lines represent mean, shaded areas SD; n = 3/3 (Nfasca), 4/3 (Cntn1a), and 5/4 (NaV-II-III) animals/technical replicates.
(C) Individually partially myelinated axon at 8 dpf co-expressing cntn1b:Nfasca-EYFP in a full transgenic myelin reporter background. (C′) Magnification of the boxed area in (C). Arrowheads indicate Nfasca-EYFP clusters along unmyelinated axon parts. Arrows point to heminodal Nfasca-EYFP. Scale bars, 10 μm.
(D) Fluorescence intensity quantifications at different regions. Median ± IQR (Kruskal-Wallis test with multiple comparisons); n = 7/7 animals/technical replicates.
See also Figure S1 for supporting information.
Neurofascin clusters remain stable along unmyelinated axons and do not associate with processes of oligodendrocyte precursor cells
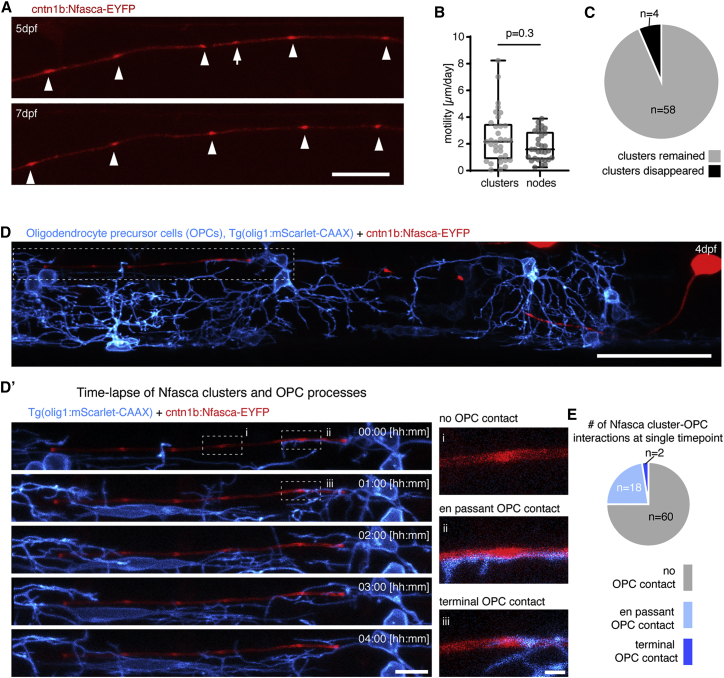
Having identified pre-nodal clusters in zebrafish, which is an excellent model for in vivo imaging over time, we sought to explore the dynamics and fates of these clusters to reveal their role for node of Ranvier positioning during the myelination of individual axons. Consecutive imaging of Nfasca-EYFP labeled single axons revealed that Nfasca clusters along unmyelinated axon stretches were largely stable and comparable to the low motility of nodes between myelin sheaths (2.5 ± 1.9 μm cluster movement versus 1.8 ± 1.1 μm node movement per day [n = 33/5 clusters/axons and 31/9 nodes/axons, p = 0.3; Mann-Whitney test]; Figures 2A and 2B). Only 6% (4/62) of all clusters analyzed disappeared over a 2-day period along unmyelinated axon domains (Figure 2C). It has been reported that processes of oligodendrocyte precursor cells (OPCs) can associate with nodes and pre-nodal clusters to varying extents (Biase et al., 2017; Serwanski et al., 2017; Thetiot et al., 2020). To test if OPC processes associate with Nfasca clusters where they could be held in position, we analyzed their co-localization in full transgenic olig1:mScarlet-CAAX, which have all OPC processes labeled. The vast majority of clusters analyzed had no associated OPC processes (60/80), 18/80 clusters had an OPC process crossing in the same imaging plane and only 2/80 clusters were associated with a terminal OPC process (n = 4 axons/animals; Figures 2D and 2E). Furthermore, cluster-OPC contacts were only transient and never lasted over the entire period of a 150 min time lapse (Figures 2D′ and S2). Therefore, we conclude that Nfasca clusters are stable over time and held in position by a mechanism that is independent of oligodendroglial contact.
Figure 2.
Clusters of neuronal neurofascin are stable over time and do not co-localize with processes of oligodendrocyte precursor cells
(A) Individual unmyelinated axon expressing Nfasca-EYFP at two different time points. Arrowheads depict stable clusters and the arrow points to a cluster that disappears between time points. Scale bar, 20 μm.
(B) Quantification of Nfasca cluster and node motility. Median ± IQR (Mann-Whitney test); n = 5/5 (cluster motility) and 9/6 (node motility) animals/technical replicates.
(C) Frequency of cluster disappearance.
(D) Axon expressing Nfasca-EYFP in a transgenic animal that has all OPCs labeled. Scale bar, 50 μm. (D′) (Left) Magnification of the boxed area in (D) showing OPC processes in relation to Nfasca-EYFP accumulations over time. Scale bar, 10 μm. (Right, i–iii) Examples of possible cluster-OPC process interactions. Scale bar, 2 μm.
(E) Quantification of the frequency of cluster-OPC process interactions; n = 4/2 animals/technical replicates.
See also Figure S2 for supporting information.
Neurofascin clusters can prefigure node of Ranvier localization
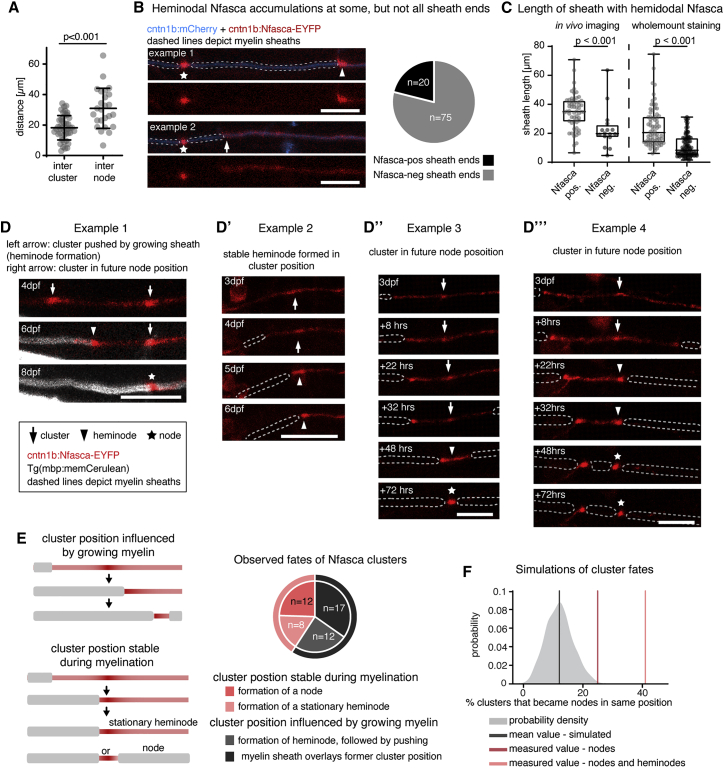
Because Nfasca clusters were stationary and interspersed on axon stretches that were not yet myelinated, we wondered how their positioning correlates with future node position. Indeed, Nfasca clusters appeared regularly spaced, but with overall shorter distancing than was seen for nodes (18.2 ± 8 μm intercluster distance versus 30.9 ± 13.2 μm internodal distance, n = 45/6/4 interclusters/axons/animals and 25/4/4 internodes/axons/animals, p < 0.001 [unpaired t test]; Figure 3A). It is known that components of nodes of Ranvier accumulate adjacent to the ends of myelin sheaths to form heminodes prior to forming nodes (Rasband and Peles, 2020). In our system, 79% of isolated myelin sheath ends that have no neighboring myelin showed a heminodal Nfasca accumulation (75/95 heminodes, n = 22/21 axons/animals) (Figure 3B). Interestingly, the remaining 21% of myelin sheaths that did not have a Nfasca-labeled heminode were overall shorter than the ones with heminodal Nfasca, both in live cell imaging using Nfasca-EYFP as well as in immunohistochemistry for endogenous Nfasca (live cell imaging: 35 ± 12 μm versus 23 ± 14 μm, n = 58/16 sheaths, p < 0.001 [Mann-Whitney test]; immunohistochemistry: 25 ± 14 μm versus 11 ± 8 μm, n = 72/90 sheaths, p < 0.001 [Mann-Whitney test]; Figures 3B, 3C, and S3A). Despite this overall correlation that longer sheaths have heminodal Nfasca, whereas shorter and presumably younger sheaths do not, we could not identify a strong linear correlation between sheath length and Nfasca intensity in a bulk analysis (n = 37/14 heminodes/axons [animals], Spearman r = 0.3546, two-tailed p value = 0.0313, α = 0.05). However, time-lapse imaging of individual axons did reveal a gradual increase of heminodal Nfasca over time (Figure S3B′).
Figure 3.
Clusters of neuronal neurofascin correlate with future node position and restrict myelin sheath extension
(A) Quantification of intercluster and internodal distances along single axons. Data are expressed as mean ± SD (unpaired t test); n = 4/3 animals/technical replicates.
(B) Example images of two individual myelinated areas (dashed lines) forming a node at their left side (asterisk), and the presence (top, arrowhead) and absence (bottom, arrow) of Nfasca-EYFP heminodal accumulation at their right sheath end. Pie chart shows frequency of observations; n = 5/3 animals/technical replicates. Scale bars, 10 μm.
(C) Length of Nfasca-EYFP positive and negative sheath ends. Data derived from both in vivo imaging (left) and imaging of whole-mount immunohistochemistry (right). Median ± IQR (Mann-Whitney test); n = 8/2 (live imaging) and 7/1 (histochemistry) animals/technical replicates.
(D–D‴) Individual axon expressing Nfasca-EYFP at different time points. Dashed lines indicate myelin sheaths. Arrows depict Nfasca clusters, arrowheads point to heminodes, and asterisks depict nodes of Ranvier. Scale bars, 10 μm.
(E) Possible cluster fates and quantification of frequencies observed by time lapse imaging; n = 5/3 animals/technical replicates.
(F) Modeling of cluster fates and predicted co-localization with nodes when random positioning is assumed. The shaded curve shows the probability distribution for the simulated values (gray line indicates the mean of simulated data). The light red and dark red lines indicate the quantified cluster fates analysis shown in (E).
See also Figure S3 for supporting information.
Shorter sheaths are an indication that they may have formed more recently and are therefore still growing. It is conceivable that nascent sheaths could initially form in between clusters, form a heminode when meeting a cluster, and continue to grow while pushing the cluster until apposing another sheath. In this scenario, sheath growth would determine cluster fate and future node position. Alternatively, developing myelin sheaths stop growing when they meet a cluster. In this scenario, cluster position would pre-determine node position. To investigate cluster fates, we followed single axons from unmyelinated through myelinated stages (Figure S3C). We saw that nascent sheaths could indeed encounter a Nfasca cluster and then continue to grow while pushing the cluster by forming a heminode, which ultimately fused with an adjacent myelin sheath to form a node (Figure 3D). In other cases, however, sheaths did not push the cluster further but instead stopped growing upon encountering a cluster, leading to the formation of either a stationary heminode, or of a node in this position (Figures 3D–3D‴). To determine how frequently Nfasca clusters prefigure node position, we analyzed timelines of single axons expressing Nfasca-EYFP during their myelination between 3 and 8 dpf. Of all 62 clusters analyzed (5/5 axons/animals), 9/62 (14.5%) remained clusters because the axon did not get myelinated and 4/62 (6.5%) clusters disappeared or merged with other clusters in the absence of any myelination. Among the remaining 49 Nfasca clusters, the position of 40.8% (20/49) was predictive of the location of a node (12/49) or a stationary heminode (8/49) by the end of analysis, while 59.2% (29/49) of clusters changed in position together with a growing myelin sheath (either by direct observation of heminode formation followed by pushing with growing myelin [12/49] or by a sheath ending up over former cluster position [17/49]) (Figure 3E). Likewise, a retrospective inspection on nodal origin showed that 55% of all nodes analyzed (18/33 nodes in 7 animals) had a Nfasca-EYFP cluster at the same position prior to any sign of myelination (Figure S3D). No indications for the site of future node positioning prior to myelination were detectable using the nodal reporters EYFP-cntn1a and EYFP-NaV-II-III (Figure S3D). To rule out that the observed co-localizations between cluster and node position occurred by chance, for example, because myelin sheaths just happened to stop extending in this position, we simulated how often clusters and nodes would randomly co-localize using the intercluster and internodal distances observed in vivo. Our modeling revealed no more than 12% ± 5% probability that a cluster would prefigure node position by chance in a prospective analysis of cluster fates, and no more than 20% ± 7% probability in a retrospective analysis of node origin (Figures 3F and S3E). In strong contrast, our measured values showed an at least 2-fold higher probability that clusters prefigure node position (24.5% [prospective] and 55% [retrospective], and even 40.8% [prospective] and 66.7% [retrospective] when also stationary heminodes were included, which ultimately convert into nodes as well). Together, these data show that the position of Nfasca clusters can correlate with future node of Ranvier position in many cases.
Increased internodal distances in the absence of full-length neurofascin a
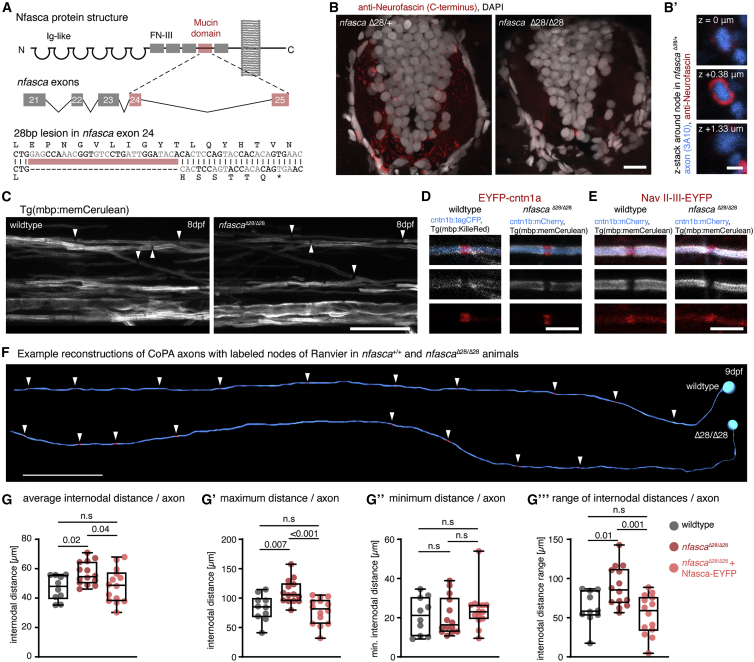
To test if Nfasca itself is important for regulating node positioning, we generated CRISPR mutants of nfasca. CRISPR-NfascaΔ28 mutants have a 28 bp deletion in the extracellular mucin domain (specific for the neuronal isoform of Nfasc186 in mammals), leading to a premature stop upstream of the transmembrane domain (Figures 4A, S4A, and S4B). We confirmed the absence of full-length Nfasca protein in homozygous mutants by immunostaining using an antibody against the intracellular C terminus of Nfasca (Figures 4B and 4B′). In contrast to neurofascin null mutant mice, which are lethal at early postnatal stages (Sherman et al., 2005), our zebrafish nfasca mutants are viable through adulthood and appear to breed normally (not shown). Nfasca mutants showed grossly normal myelination and even had discrete gaps between adjacent myelin sheaths that accumulated EYFP-cntn1a and EYFP-NaV-II-III reporters (Figures 4C–4E), consistent with the premise that myelination with an intact paranodal junction is sufficient to cluster nodal components in the absence of axonal Nfasc186 in rodents (Zonta et al., 2008). However, analysis of node position along single axons at 8–10 dpf using EYFP-cntn1a revealed an overall increase of internodal distances by about 18% in Nfasca mutants over wild types (47.1 ± 8.4 in wild types versus 56.7 ± 7.7 in nfascaΔ28/Δ28 in n = 10/14 axons in 9/14 animals, p = 0.02 [Brown-Forsythe ANOVA with multiple comparisons]; Figures 4F and 4G). Importantly, increased internodal distances observed in Nfasca mutants could be rescued to wild-type levels by re-expressing Nfasca-EYFP in individual axons of Nfasca mutants (56.7 ± 7.7 in nfascaΔ28/Δ28 versus 48.5 ± 11.5 in nfascaΔ28/Δ28 inj. Nfasca-EYFP n = 14/14 axons in 14/14 animals, p = 0.04 [Brown-Forsythe ANOVA with multiple comparisons]; Figure 4G). Expression of the nodal reporter Nfasca-EYFP and EYFP-cntn1a itself had no effect on internodal distances in wild types (47.1 ± 8.4 for EYFP-cntn1a versus 45.6 ± 10.9 for Nfasca-EYFP in n = 10/10 axons in 9/10 animals, p = 0.7 [unpaired t test]; Figure S4D). Closer analysis revealed that this increase in internodal distances seen in Nfasca mutants was caused by an increase in the maximum internode length per axon (83.5 ± 22.4 μm in wild type versus 110.6 ± 20.4 μm in nfascaΔ28/Δ28 versus 77.8 ± 22.4 μm in nfascaΔ28/Δ28 rescue; p = 0.007 [wild type versus nfascaΔ28/Δ28] and p < 0.001 [nfascaΔ28/Δ28 versus nfascaΔ28/Δ28 rescue; Brown-Forsythe ANOVA with multiple comparisons]; Figures 4G′ and S4C). Minimum internodal length did not change in either condition (21 ± 9.4 μm in wild type versus 20.8 ± 9.6 μm in nfascaΔ28/Δ28 versus 24 ± 10.1 μm nfascaΔ28/Δ28 rescue, p > 0.3 between all conditions [Kruskal-Wallis test with multiple comparisons]; Figures 4G″ and S4C). As a result, nodes in Nfasca mutants were less regularly spaced and showed an average 44% increase in the range of internodal distances along individual axons, which was again rescued upon re-expression of Nfasca-EYFP in nfascaΔ28/Δ28 (62.6 ± 21.5 μm in wild types versus 89.8 ± 25.7 μm in nfascaΔ28/Δ28 versus 53.8 ± 25.4 μm nfascaΔ28/Δ28 rescue, p = 0.01 [wild type versus nfascaΔ28/Δ28] and p = 0.001 [nfascaΔ28/Δ28 versus nfascaΔ28/Δ28 rescue; Brown-Forsythe ANOVA with multiple comparisons]; Figure 4G‴). Therefore, we conclude that axonal Nfasca plays a functional role in regulating the pattern of nodal spacing along individual axons, likely by restricting myelin sheath extension.
Figure 4.
Increased and less regular nodal spacing along individual axons in nfascaΔ28/Δ28 mutants
(A) Targeting strategy and genetic lesion in nfascaΔ28/Δ28 mutants.
(B) Immunohistochemistry against Nfasca on spinal cord cross-sections in adult heterozygous and homozygous nfascaΔ28/Δ28 animals. Scale bar, 10 μm. (B′) Cross-sectional views around a node of Ranvier at different z positions in a heterozygous nfascaΔ28/+ animal. Scale bar, 1 μm.
(C) Overviews of the spinal cord in full transgenic myelin reporter lines. Arrowheads highlight gaps between adjacent myelin sheaths. Scale bar, 20 μm.
(D and E) Close-up views showing single nodes of Ranvier marked with EYFP-cntn1a and EYFP-NaV-II-III in full transgenic myelin reporter lines. Scale bar, 5 μm.
(F) Reconstructions of two CoPA neurons and their node of Ranvier positions (arrowheads) from wild type and nfascaΔ28/Δ28 mutant animals. Scale bar, 50 μm.
(G) Average internodal distances along single axons per axon in wild type, nfascaΔ28/Δ28, and nfascaΔ28/Δ28 re-expressing Nfasca-EYFP in single axons. Median with IQR (Brown-Forsythe ANOVA with multiple comparisons); n = 9/4 (wild type), 10/8 (nfascaΔ28/Δ28), and 13/4 (nfascaΔ28/Δ28 rescue) animals/technical replicates.
(G′–G‴) Maximum (G′), minimum (G″), and range (G‴) of internodal distances per axon as quantified in (G). Median with IQR; Brown-Forsythe ANOVA with multiple comparisons for (G′ and G‴), Kruskal-Wallis test with multiple comparisons for (G″).
See also Figure S4 for supporting information.
Discussion
Our data provide evidence that the axon itself can play an instructive role in determining the position of its nodes of Ranvier. It is well established that formation and maintenance of node of Ranvier architecture requires intrinsic (linkage to axonal cytoskeleton) and extrinsic mechanisms (secreted glial ECM and paranodal adhesion) (Rasband and Peles, 2020; Susuki et al., 2013). It is also known that oligodendrocyte-conditioned medium is sufficient to induce sodium channel clustering in vitro, and it has been speculated that such clusters serve as pre-assembled immature nodes (Kaplan et al., 1997). However, the position where nodes form along an axon had remained essentially uninvestigated. Our data extend current knowledge by providing in vivo evidence that such pre-myelinating clusters can pre-pattern the axon and determine future node position by restricting sheath growth.
Axon-intrinsic mechanisms may be sufficient to target components of future nodes at specific sites along the length of axons through their linkage to the axon-cytoskeleton, as Nfasc186 does by directly binding to ankyrin G (Davis et al., 1993), although the traditional view has been that myelination precedes AnkG and Nfasc186 accumulation in the central nervous system (CNS) (Jenkins and Bennett, 2002; Rasband et al., 1999). Our finding that some clusters were stationary and thus prefigured node position, while other clusters were pushable by growing myelin sheaths, raises the possibility that stationary clusters may be anchored to the axon-cytoskeleton, whereas the moveable clusters are not (yet) and are thus more immature. However, we could not detect clustering of our NaVII-III live-cell reporter prior to myelination, which targets to nodes via its AnkG binding motif. Thus, it may be that AnkG does not localize to pre-myelinating Nfasca clusters. Alternatively, it is possible that the overall low signal-to-noise properties of our NaVII-III reporter do not allow us to detect potential early clustering of AnkG, which remains to be investigated in future studies.
Our mutants that lack neuronal transmembrane Nfasca show overall increased internodal distances, suggesting that Nfasca is either important to form pre-nodal clusters in the first place, or to serve as a stop signal preventing myelin sheath extension beyond certain points. It is also possible that Nfasca affects axon-oligodendrocyte interactions in other ways that result in increased internodal distancing, which remains to be investigated. We can only speculate why our neuronal Nfasca mutants show no gross neurological defects, which is in contrast to mice that die at early postnatal stages, even upon re-expression of glial Nfasc155 (Sherman et al., 2005; Zonta et al., 2008). It may be that our mutants still express a functional extracellular portion of Nfasca that is dissociated from the axon membrane, as the stop codon is only just prior to the transmembrane domain. Furthermore, in zebrafish glial and neuronal neurofascin are encoded by two separate genes (nfasca and nfascb) (Klingseisen et al., 2019), which may together with the specifics of our CRISPR-NfascaΔ28 deletion alleviate the severe phenotypes seen in rodent nfasc mutants. Whether CRISPR-NfascaΔ28 affects axons in additional ways than presented here remains to be addressed in future studies.
The presence of axonal landmarks to determine the site of future node position by restricting myelin sheath growth provides a simple mechanism to pattern an axon along its length. It also provides a plausible explanation why myelin sheaths along partially myelinated axons are not longer than along fully myelinated axons (Auer et al., 2018; Hill et al., 2018; Hughes et al., 2018), and why developing myelin sheaths mostly grow asymmetrically, despite the absence of obvious physical barriers, such as adjacent myelin and axon collateral branches (Auer et al., 2018). It is likely that axonal clustering at future nodal sites is only one of several mechanisms to establish internodal distances along the axon. Observations by us and others argue that either multiple mechanisms are in place at the same time, and/or that not all axons are pattered by the same mechanisms. For example, it has been reported that only distinct subtypes of axons form clusters (Freeman et al., 2015). Similarly, it has been shown that neuronal activity only promotes myelination of specific axon types (Koudelka et al., 2016; Yang et al., 2020). Moreover, our work reveals that not all clusters convert into nodes, but that developing sheaths relocate cluster position and merge several clusters to single nodes, which is in line with another recent study showing that the number of pre-nodal clusters exceeds the number of nodes by about 2-fold (Thetiot et al., 2020). Together, these observations argue for the existence of two regulatory mechanisms to control node position; one that is axonal and independent of myelin, and another that involves regulation of myelin sheath length, which can intrinsically grow to variable lengths (Bechler et al., 2015), and in response to axonal signals (Baraban et al., 2017; Krasnow et al., 2017). Future studies will be required to elucidate to which degree these two mechanisms pattern different axon types, and whether one of both of these mechanisms can induce remodeling of internodal distances to adapt axon function.
Limitations of the study
This work provides insights into the fates of pre-nodal clusters and the mechanisms that determine node of Ranvier position along single axons. However, a few caveats remain. Most notably, it remains unclear how prevalent pre-nodal clusters are to prefigure node position in different types of CNS axons. Most live cell imaging data have been obtained from transgenic expression of an NFasca-EYFP fusion protein. Although we provide evidence for endogenous Nfasca cluster formation, and for the fact that overexpression of the NFasca-EYFP transgene itself does not affect internodal distances in wild types, we could not determine if this a general mechanism or rather one that is specific to some axons only. In addition, we provide no mechanistic insight into how Nfasca clusters might restrict myelin sheath growth, which remains to be addressed in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| chicken anti-GFP | Abcam plc. | RRID: AB_300798 |
| mouse IgG1 anti-3A10 | DSHB | RRID: AB_531874 |
| rabbit anti-Neurofascin | Abcam plc. | RRID: AB_2905463 |
| goat anti-mouse IgG, Alexa Fluor 633 | Thermo Fisher | RRID: AB_2535719 |
| goat anti-rabbit IgG, Alexa Fluor 555 | Thermo Fisher | RRID: AB_141784 |
| goat anti-chicken IgY, Alexa Fluor 488 | Thermo Fisher | RRID: AB_142924 |
| Experimental models: Organisms/strains | ||
| Tg(mbp:memCerulean)tum101Tg | Auer et al. (2018) | ZFIN: ZDB-ALT-180712-3 |
| Tg(mbp:KillerRed)tum103Tg | Auer et al. (2018) | ZFIN: ZDB-ALT-180712-5 |
| Tg(mbp:tagRFPt-CAAX)tum102Tg | Auer et al. (2018) | ZFIN: ZDB-ALT-180712-4 |
| Tg(cntn1b(3kb):KalTA4) | This paper | N/A |
| Tg(olig1:mScarlet-CAAX) | This paper | N/A |
| CRISPR-NfascaΔ28 | This paper | N/A |
| Oligonucleotides | ||
| For all oligonucleotides used, see Table S1 | this paper | N/A |
| Recombinant DNA | ||
| p5E_cntn1b(3kb) | this paper | N/A |
| pME_EYFPnostop_NaV-II-III | this paper | N/A |
| pME_sigpepEYFP_cntn1a | this paper | N/A |
| p5E_cntn1b(5kb) | Czopka et al. (2013) | N/A |
| pME_KalTA4GI | Almeida and Lyons (2015) | N/A |
| p5E_UAS(10x) | Kwan et al. (2007) | N/A |
| p3E_pA | Kwan et al. (2007) | N/A |
| pDestTol2_pA | Kwan et al. (2007) | N/A |
| pME_tagCFP | Auer et al. (2018) | N/A |
| pTol2_cntn1b(5kb):Nfasca-EYFP | Auer et al. (2018) | N/A |
| pTol2_cntn1b(5kb):EYFP-cntn1a | this paper | N/A |
| pTol2_cntn1b(5kb):EYFP-NaV-II-III | this paper | N/A |
| pTol2_UAS(10x):EYFP-cntn1a | this paper | N/A |
| pTol2_cntn1b(5kb):tagCFP | this paper | N/A |
| pTol2_cntn1b(5kb):mCherry | Czopka et al. (2013) | N/A |
| Software and algorithms | ||
| Fiji | http://fiji.sc/ | RRID: SCR_002285 |
| Imaris | Bitplane | RRID: SCR_007370 |
| Imaris FilamentTracer | Bitplane | RRID: SCR_007366 |
| Huygens Essential | Scientific Volume Imaging | RRID: SCR_014237 |
| Graphpad Prism 8 | Graphpad Software | RRID: SCR_015807 |
| Code for simulation of cluster-node correlation | This paper | ZENODO DOI: https://doi.org/10.5281/zenodo.5846391. |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact Tim Czopka (tim.czopka@ed.ac.uk), and will be fulfilled upon reasonable request.
Materials availability
All unique/stable reagents generated in this study are available from the lead contact without restriction.
Experimental model and subject details
Zebrafish lines and husbandry
The following existing zebrafish lines and strains were used in this study: Tg(mbp:memCerulean)tum101Tg (Auer et al., 2018), Tg(mbp:tagRFPt-CAAX)tum102Tg (Auer et al., 2018), Tg(mbp:KillerRed)tum103Tg (Auer et al., 2018). The following lines have been newly generated for this study: Tg(cntn1b(3kb):KalTA4), Tg(olig1:mScarlet-CAAX), Tg(mbp:mScarlet-CAAX), CRISPR-nfascaΔ28. All animals were kept at 28.5 degrees with a 14/10 hour light/dark cycle according to the local animal welfare regulations. Animals were used at embryonic and larval stages between 3dpf and 11dpf for live cell imaging, and as adults for histology and breeding. All experiments carried out with zebrafish at protected stages have been approved by the government of Upper Bavaria (animal protocols AZ55.2-1-54-2532-199-2015, ROB-55.2-2532.Vet_02-15-200, and ROB-55.2-2532.Vet_02-18-153 to T.C.).
Method details
Transgenesis constructs
The following existing expression constructs were used in this study: pTol2_cntn1b:Nfasca-EYFP (Auer et al., 2018), pTol2_cntn1b:mCherry (Czopka et al., 2013). The following expression constructs and entry clones were newly generated for this study, with the sequences of all primers used listed in Table S1: p5E_cntn1b(3kb), for which we cloned a 3kb fragment upstream of the start codon and until the second exon of the cntn1b gene (Ensembl(GRCz11): ENSDARG00000045685) from genomic DNA of AB zebrafish. The PCR product was recombined with pDONRP4-P1R using BP clonase (Invitrogen). pME_EYFP-NaV-II-III, for which a 1.3 kb fragment corresponding to the II-III region of Nav1.2 (Garrido et al., 2003) of the zebrafish scn1lab gene (between exons 16 and 18 of Ensembl(GRCz11): ENSDART00000151247.3) was commercially synthesized (BioCat). We then PCR amplified NaV-II-III from this donor plasmid using XhoI-NaV-II-III_F and XbaI-NaV-II-III_R primers and used these sites to clone it into pCS2+ containing EYFP with the stop codon removed between BamHI and EcoRI sites. The resulting plasmid pCS2+_EYFP-NaV-II-III was then used as template for a PCR reaction using attB1_YFP_F and attB2R-NavNaV-II-III_R primers, and recombination cloned into pDONR221 using BP clonase, creating pME_EYFP-NaV-II-III. To generate pME_EYFP-cntn1a, we first amplified 2.6 kb of the zebrafish contactin1a gene (Ensembl(GRCz11): ENSDART00000170348.2) lacking base pairs 1-135 (amino acids 1-45 coding for signal peptide for secretory pathway) from zebrafish cDNA using the primers EcoRI_cntn1a_F and XbaI_cntn1a_R and cloned into pCS2+ containing EYFP fused to the secretory pathway signal peptide of zebrafish cd59 (Ensembl(GRCz11): ENSDART00000126737.4) between BamHI and EcoRI sites to create pCS2+_sigpepEYFP-cntn1a. This plasmid was used as PCR template using the primers attB1_sigpepEYFP_F and attB2R_cntn1a_R and the PCR product was recombination cloned in pDONR221 using BP clonase to generate pME_EYFP-cntn1a. To generate pME_mScarlet-CAAX we used the commercially synthesized construct pUC57_attB1_mScarlet_attB2R (BioCat). This plasmid was then used as template for a PCR reaction using attB1_F and CAAX-mScarlet_R. The product of this PCR was used as a template for a subsequent PCR reaction with the primers attB1_F and AttB2R-CAAX-R and the product of this reaction was cloned into pDONR221 using BP clonase, creating pME_mScarlet-CAAX.
The expression constructs pTol2_cntn1b(3kb):KalTA4, pTol2_cntn1b(5kb):EYFP-cntn1a, pTol2_cntn1b(5kb):EYFP-NaV-II-III, pTol2_UAS(10x):EYFP-cntn1a, pTol2_cntn1b(5kb):tagCFP, pTol2_olig1:mScarlet-CAAX, and pTol2_mbp:mScarlet-CAAX were created via multisite LR recombination reactions using the newly generated entry clones described above, in addition to p5E_cntn1b(5kb) (Czopka et al., 2013), p5E_olig1 (Marisca et al., 2020), p5E_mbp (Almeida et al., 2011), pME:KalTA4 (Almeida and Lyons, 2015), pME:tagCFP (Auer et al., 2018) and p5E_UAS, p3E-pA, and pDestTol2_pA of the Tol2Kit (Kwan et al., 2007).
DNA microinjection and generation of transgenic lines
Zebrafish eggs were injected at the one cell stage using a pressure microinjector with 1nl of injection solution (20-30 ng/μl plasmid DNA, 25-60 ng/μl transposase mRNA and 1% phenol-red (Sigma Aldrich)). Injected animals were screened under a stereo-dissecting fluorescence microscope at 3-4 dpf for transgene expression and used for subsequent in vivo microscopy or were raised to adulthood. Injected adults were outcrossed to wild type animals and their progenies were screened for germline transmission to raise stable transgenic lines.
Generation of nfasca mutants using CRISPR/Cas9
Zebrafish CRISPR-NfascaΔ28 mutants were generated using CRISPR/Cas9 genome editing by injecting 12.5 ng/μl sgRNA and 300 ng/μl Cas9 encoding mRNA in AB wildtype embryos at the 1-cell stage. The sgRNA against exon 24 of the nfasca gene was designed using CHOPCHOP (Labun et al., 2016) and generated as described (Hruscha et al., 2013). F1 animals were sequenced for indel mutations and an individual with a 28 bp deletion 5' to the target site was selected to raise CRISPR-NfascaΔ28.
Mounting of zebrafish for live cell microscopy
For in vivo cell imaging, we anaesthetized zebrafish embryos and larvae using 0.2 mg/ml MS-222 (PHARMAQ, UK). Animals were mounted laterally in 1% ultrapure low melting point agarose (Invitrogen) on a grass coverslip as previously described in detail by us (Vagionitis and Czopka, 2018).
Immunohistochemistry
Zebrafish embryos or larvae were euthanised with an overdose of MS-222 (15 mM) and fixed overnight in 4% paraformaldehyde at 4°C. Following washes in phosphate buffer saline (PBS- 3 x 10 min), fish were incubated in incrementally increasing concentration of sucrose (10, 20 and 30%) until full submersion. Animals were mounted in TissueTek (OCT), sectioned with 14-17 μm thickness on a cryotome (Leica CM1850 UV) and subsequently stored at −20°C until further use. For immunofluorescence staining, sections were rehydrated using PBS and subsequently boiled in 10mM citrate buffer, pH = 6 for 10 min for antigen retrieval. After 30 min of cooling down in RT and subsequent PBS washes, the slides were permeabilised with PBS containing 0.1% Tween-20 (PBST) for 10-15 minutes. Following blocking in PBST containing 10% fetal calf serum (FCS), 3% normal goat serum (NGS) and 1% bovine serum albumin (BSA), slides were incubated with the primary antibodies in blocking solution overnight at 4°C, washed three times in PBST, and incubated with the secondary antibodies for 1h at room temperature. Finally, slides were washed two times with PBST, once with PBS and then mounted under a coverslip using Prolong Gold Antifade containing DAPI (Invitrogen).
Whole-mount immunohistochemistry
Zebrafish embryos or larvae were euthanised with an overdose of MS-222 (15 mM) and fixed in 2% paraformaldehyde and 1% trichloroacetic acid for 10 minutes at room temperature. Samples were washed in phosphate buffer saline supplemented with 0.1% Tween20 (PBST- 3 x 5 min). Subsequently, fish were incubated in distilled water for 5 minutes at room temperature. Animals were then permeabilized by being incubated for 10 minutes in acetone at −20°C. A second wash in distilled water was followed by a wash in PBST, both for 5 minutes at room temperature. Blocking was the same as described in the previous paragraph. Primary antibody incubation lasted for 68-72 hours at 4°C. This was followed by 3 washes of 1 hour each with PBST at 4°C. Then the samples were incubated with the secondary antibodies overnight at 4°C. Finally, the fish were again washed 3 times for at least 1 hour each at 4°C, followed by a final wash with PBS for 15 minutes at 4°C and embedding in 1% LMP agarose for confocal microscopy.
Microscopy and image acquisition
For imaging of embedded zebrafish embryos and larvae we used a confocal laser scanning microscope (Leica TCS SP8). Individual axons were identified and analysed along the whole length of the spinal cord (soma location somite level 5 to somite level 27). DAPI was excited with 405 nm wavelength, Cerulean and tagCFP with 448 and 458 nm, ALEXA488 with 488 nm, EYFP with 514 nm, tagRFPt, mCherry, and ALEXA555 with 552 and 561 nm, and ALEXA633 with 633 nm. During in vivo microscopy, confocal z-stacks were acquired as 12bit images with a pixel size ranging between 76 and 114 nm and a z-spacing of 1 μm using a Fluotar VISIR 25x/0.95 WATER immersion objective. Images are shown as lateral views of the zebrafish spinal cord with the anterior to the left and dorsal to the top of the image. Confocal z-stacks of zebrafish spinal cord cross-sections were acquired as 12bit images with a pixel size of 36 nm, z-spacing of 0.19 μm and pinhole set to 0.8AU using an HC PL APO CS2 63x/1.20 WATER immersion objective.
Quantification and statistical analysis
Reconstructions of confocal images
We used the FilamentTracer tool of Imaris 8.4.2 (Bitplane) to reconstruct the axon morphologies and node positioning. The cytoplasmic marker expressed by the axon was used for manual axon tracing while the nodal marker was automatically traced. The two traces were superimposed for visualisation of the node spacing along single axons.
Fluorescence intensity measurements
Fluorescent intensities were measured using the x line tool in Fiji (distribution of ImageJ) with a line width adjusted to the thickness of each axon. For each axon, fluorescence intensity values were manually allocated to node, cluster and myelinated categories. One value for the background was obtained for each image and was subtracted. For the intensities of markers at the nodes of Ranvier, 1μm-long axon stretches were measured and averaged. Several nodes and myelinated areas were measured. The fluorescence intensity traces were normalised to the maximum value of the node and the peak was aligned. Average and standard deviation for all nodes were calculated and plotted.
For the partially myelinated axons, one node and one myelinated stretch were measured as well as the unmyelinated part, and all values were normalised to the maximum intensity of the brightest node on the axon of interest. ΔF/F was calculated and plotted.
For the swarmplot, background was subtracted. For comparing fluorescent intensities at different positions along the axon, a Kruskal-Wallis test with Dunn’s correction for multiple testing was used.
Node cluster correlation
Clusters were detected by visual impression, as obviously denser accumulations of nodal reporter construct compared to the diffused localisation along unmyelinated parts of the axon devoid of clusters. For the retrospective analysis, images were aligned to a landmark close to the node analysed. If more than one node was analysed on the same axon, the images were realigned for each node. The growth factor of the animal between two imaging sessions was determined as the % change in linear distance between 2 neuronal somata in the spinal cord. This factor was used to correct the aligned images for body growth. For retrospective analysis of node origin, a box with width equal to node width (1-2 μm) was drawn and superimposed to the aligned images from earlier timepoints. Only when a cluster or a heminode was found inside this border, it was counted as residing at the same position. For the prospective analysis of cluster fates, images were again aligned to a landmark close to the analysed cluster. Again, alignment was corrected for body growth. A box of 2-3 μm width was drawn around each cluster and it was used to assess the cluster fate over time. In these analyses, only clusters and nodes devoid of axon collateral branches were analysed, to exclude the effect of structural obstacles in the analysis.
Simulation of node cluster correlation
Cluster to node correlation was simulated using a custom written Matlab script available on https://github.com/Czopka-Lab/Vagionitis-Auer-et-al. The intercluster/-node distances were randomly generated from normal distributions with the observed means and SDs. Cluster and node width were set to 2 μm. The same number of clusters and nodes, respectively, were generated in our simulations as have been analysed from our measured data. The position of clusters/nodes on the simulated axons were calculated from the intercluster/-node distances by adding these, in addition to adding cluster/node width after each intercluster/-node distance. For the prospective analysis for each cluster, it was checked if there was a node at the same position. Therefore, the positions of the node and cluster were subtracted, and if the absolute difference was less than the node width it was counted as co-localisation. For the retrospective analysis, the same analysis was carried out but for each node it was checked if there was a cluster at the same position. These models give the mean percentage and SD of co-localisations occurring by chance, as well as the maximum observed values for both analyses.
Sheath length measurements
Individual isolated sheaths of partially myelinated axons were visually selected for being flanked by one or two heminodes. Sheath length was measured using the x line tool of ImageJ. Sheaths were then categorised based on the presence or absence of Nfasca accumulations at their ends.
Internodal and intercluster distance measurements
The distance along the axon between discrete nodal marker accumulations (clusters or nodes) was measured using the x line tool of ImageJ. The distance between the soma and the first node/cluster was not measured.
For the CRISPR-NfascaΔ28 analysis, internodal distance was measured along CoPA axons in the spinal cord of 8 and 9 dpf zebrafish. Average internodal distance was calculated for each animal and was directly compared to WT. Analysis was blind, as microscopy and analysis was completed prior to genotyping. Average, range, minimum and maximum internodal distance per individual animal was then plotted.
Node and cluster dynamics
The distance along individual axons between a node and a chosen landmark was measured over time. The measured distance was corrected for body growth by dividing by the respective growth factor, which was obtained by measuring the distance between two landmarks over time. The corrected internodal lengths were subtracted to calculate daily motility. For the node translocation plot, the initial position of the node was set to 0.
The directionality of movement was not analysed and all movements are expressed as absolute values.
Nfasca cluster and OPC process interactions
Single images of axons expressing Nfasca-EYFP and showing clusters were analyzed for the presence or not of an OPC process in contact with the clusters on the same 1 μm z-plane. Found contacts were categorized as en passant, when the OPC process seemed to pass by a Nfasca-EYFP cluster or terminal, when the OPC process tip was in contact with the observed cluster.
Statistics
Mean and standard deviation were always calculated using GraphPad Prism9 (GraphPad Software LCC) descriptive statistics tool. Normality tests were used to assess the Gaussian distribution of the data. Normally distributed data are expressed as mean with standard deviation (SD). Datasets that contain non-normally distributed data are expressed as median with interquartile range (IQR). Statistical testing was done appropriately according to the type of distribution of data. For simplicity and legibility, the main text always reports mean and standard deviation. Differences in the fluorescence intensity measurements were tested using the Kruskal-Wallis test with multiple comparisons, controlling for False Discovery Rate (FDR) with the two-stage step-up method of Benjamini, Krieger and Yekutieli. Differences in cluster and node motility were tested using the Mann-Whitney test, as was the difference between lengths of sheaths that showed Nfasca-positive or Nfasca-negative heminodes. Differences between intercluster and internode distance were tested using an unpaired t-test. Differences in average internodal distances between WT, Nfasca mutant and rescued Nfasca-EYFP expressing mutant were tested using Brown-Forsythe ANOVA with multiple comparisons. Differences in range of internodes measured per axon between WT, Nfasca mutant and rescued Nfasca-EYFP expressing mutant were tested using Brown-Forsythe ANOVA test with multiple comparisons, as were the differences in maximum measured values per axon. Differences in minimum of internodes measured per axon between WT, Nfasca mutants and rescue Nfasca-EYFP expressing mutants were tested using Kruskal-Wallis test with multiple comparisons. All multiple comparisons tests in the analyses of WT, Nfasca mutant and rescued Nfasca-EYFP expressing mutant were performed controlling for False Discovery Rate (FDR) with the two-stage step-up method of Benjamini, Krieger and Yekutieli.
Acknowledgments
We are grateful to Wenke Barkey for excellent technical assistance throughout this study. We thank Laura Fontenas from the Kucenas group (University of Virginia) for helpful advice with whole-mount immunohistochemistry and all members of the Czopka lab for critical input and discussion of the manuscript. This work was funded by the German Research Foundation DFG (ENP Cz226/1-1 and Cz226/1-2, SFB870 A14 #118803580, and EXC 2145 SyNergy #390857198 to T.C.), a Gertrud Reemtsma PhD student fellowship of the Max-Planck-Society to F.A., support from the TUM PhD program “Medical Life Science and Technology” to S.V., and a Medical Research Council project grant (MR/P006272/1) to R.G.A. and D.A.L.
Author contributions
Conceptualization, F.A., S.V., and T.C.; methodology, S.V., F.A., and R.G.A.; investigation, S.V., F.A., X.Y., and R.G.A.; formal analysis, S.V. and F.A.; visualization, S.V., F.A., and T.C.; supervision, D.A.L. and T.C.; writing – original draft, T.C.; funding acquisition, all authors.
Declaration of interests
The authors declare no competing interests.
Published: February 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110366.
Supplemental information
Data and code availability
-
-
Data reported in this paper will be shared by the lead contact upon reasonable request.
-
-
The code used for the simulation of node cluster correlation is available under https://doi.org/10.5281/zenodo.5846391.
-
-
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Almeida R.G., Lyons D.A. Intersectional gene expression in zebrafish using the split KalTA4 system. Zebrafish. 2015;12:377–386. doi: 10.1089/zeb.2015.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida R.G., Czopka T., ffrench-Constant C., Lyons D.A. Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development. 2011;138:4443–4450. doi: 10.1242/dev.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer F., Vagionitis S., Czopka T. Evidence for myelin sheath remodeling in the CNS revealed by in vivo imaging. Curr. Biol. 2018;28:549–559.e3. doi: 10.1016/j.cub.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Baraban M., Koudelka S., Lyons D.A. Ca2+ activity signatures of myelin sheath formation and growth in vivo. Nat. Neurosci. 2017;19:1–23. doi: 10.1038/s41593-017-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechler M.E., Byrne L., ffrench-Constant C. CNS myelin sheath lengths are an intrinsic property of oligodendrocytes. Curr. Biol. 2015;25:2411–2416. doi: 10.1016/j.cub.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biase L.M.D., Pucak M.L., Kang S.H., Rodriguez S.N., Bergles D.E. Sparse interaction between oligodendrocyte precursor cells (NG2+ cells) and nodes of Ranvier in the central nervous system. biorxiv. 2017 doi: 10.1101/185801. [DOI] [Google Scholar]
- Brill M.H., Waxman S.G., Moore J.W., Joyner R.W. Conduction velocity and spike configuration in myelinated fibres: computed dependence on internode distance. J. Neurol. Neurosurg. Psychiatry. 1977;40:769–774. doi: 10.1136/jnnp.40.8.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka T., ffrench-Constant C., Lyons D.A. Individual oligodendrocytes have only a few hours in which to generate new myelin sheaths in vivo. Dev. Cell. 2013;25:599–609. doi: 10.1016/j.devcel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.Q., McLaughlin T., Bennett V. Ankyrin-binding proteins related to nervous system cell adhesion molecules: candidates to provide transmembrane and intercellular connections in adult brain. JCB. 1993;121:121–133. doi: 10.1083/jcb.121.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.C., Alexandrova O., Cossell L., Stange-Marten A., Sinclair J., Kopp-Scheinpflug C., Pecka M., Attwell D., Grothe B. Tuning of Ranvier node and internode properties in myelinated axons to adjust action potential timing. Nat. Commun. 2015;6:8073. doi: 10.1038/ncomms9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S.A., Desmazieres A., Simonnet J., Gatta M., Pfeiffer F., Aigrot M.-S., Rappeneau Q., Guerreiro S., Michel P.P., Yanagawa Y., et al. Acceleration of conduction velocity linked to clustering of nodal components precedes myelination. Proc. Natl. Acad. Sci. U S A. 2015;112:E321–E328. doi: 10.1073/pnas.1419099112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido J.J., Giraud P., Carlier E., Fernandes F., Moussif A., Fache M.-P., Debanne D., Dargent B. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science. 2003;300:2091–2094. doi: 10.1126/science.1085167. [DOI] [PubMed] [Google Scholar]
- Higashijima S.-I., Schaefer M., Fetcho J.R. Neurotransmitter properties of spinal interneurons in embryonic and larval zebrafish. J. Comp. Neurol. 2004;480:19–37. doi: 10.1002/cne.20279. [DOI] [PubMed] [Google Scholar]
- Hill R.A., Li A.M., Grutzendler J. Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat. Neurosci. 2018;21:683–695. doi: 10.1038/s41593-018-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Hughes E.G., Orthmann-Murphy J.L., Langseth A.J., Bergles D.E. Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nat. Neurosci. 2018;21:696–706. doi: 10.1038/s41593-018-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins S.M., Bennett V. Developing nodes of Ranvier are defined by ankyrin-G clustering and are independent of paranodal axoglial adhesion. Proc. Natl. Acad. Sci. U S A. 2002;99:2303–2308. doi: 10.1073/pnas.042601799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.R., Meyer-Franke A., Lambert S., Bennett V., Duncan I.D., Levinson S.R., Barres B.A. Induction of sodium channel clustering by oligodendrocytes. Nature. 1997;386:724–728. doi: 10.1038/386724a0. [DOI] [PubMed] [Google Scholar]
- Klingseisen A., Ristoiu A.-M., Kegel L., Sherman D.L., Rubio-Brotons M., Almeida R.G., Koudelka S., Benito-Kwiecinski S.K., Poole R.J., Brophy P.J., et al. Oligodendrocyte Neurofascin independently regulates both myelin targeting and sheath growth in the CNS. Dev. Cell. 2019;51:730–744.e6. doi: 10.1016/j.devcel.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka S., Voas M.G., Almeida R.G., Baraban M., Soetaert J., Meyer M.P., Talbot W.S., Lyons D.A. Individual neuronal subtypes exhibit diversity in CNS myelination mediated by synaptic vesicle release. Curr. Biol. 2016;26:1447–1455. doi: 10.1016/j.cub.2016.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow A.M., Ford M.C., Valdivia L.E., Wilson S.W., Attwell D. Regulation of developing myelin sheath elongation by oligodendrocyte calcium transients in vivo. Nat. Neurosci. 2017;93:1–28. doi: 10.1038/s41593-017-0031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.-B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisca R., Hoche T., Agirre E., Hoodless L.J., Barkey W., Auer F., Castelo-Branco G., Czopka T. Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci. 2020;23:363–374. doi: 10.1038/s41593-019-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza L., Huang J.K., Colman D.R. Organizing principles of the axoglial apparatus. Neuron. 2001;30:335–344. doi: 10.1016/s0896-6273(01)00306-3. [DOI] [PubMed] [Google Scholar]
- Rasband M.N., Peles E. Mechanisms of node of Ranvier assembly. Nat. Rev. Neurosci. 2020;22:7–20. doi: 10.1038/s41583-020-00406-8. [DOI] [PubMed] [Google Scholar]
- Rasband M.N., Peles E., Trimmer J.S., Levinson S.R., Lux S.E., Shrager P. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J. Neurosci. 1999;19:7516–7528. doi: 10.1523/JNEUROSCI.19-17-07516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwanski D.R., Jukkola P., Nishiyama A. Heterogeneity of astrocyte and NG2 cell insertion at the node of ranvier. J. Comp. Neurol. 2017;525:535–552. doi: 10.1002/cne.24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman D.L., Brophy P.J. Mechanisms of axon ensheathment and myelin growth. Nat. Rev. Neurosci. 2005;6:683–690. doi: 10.1038/nrn1743. [DOI] [PubMed] [Google Scholar]
- Sherman D.L., Tait S., Melrose S., Johnson R., Zonta B., Court F.A., Macklin W.B., Meek S., Smith A.J., Cottrell D.F., et al. Neurofascins are required to establish axonal domains for saltatory conduction. Neuron. 2005;48:737–742. doi: 10.1016/j.neuron.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Susuki K., Chang K.-J., Zollinger D.R., Liu Y., Ogawa Y., Eshed-Eisenbach Y., Dours-Zimmermann M.T., Oses-Prieto J.A., Burlingame A.L., Seidenbecher C.I., et al. Three mechanisms assemble central nervous system nodes of Ranvier. Neuron. 2013;78:469–482. doi: 10.1016/j.neuron.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thetiot M., Freeman S.A., Roux T., Dubessy A.L., Aigrot M.-S., Rappeneau Q., Lejeune F.-X., Tailleur J., Sol-Foulon N., Lubetzki C., et al. An alternative mechanism of early nodal clustering and myelination onset in GABAergic neurons of the central nervous system. Glia. 2020;68:1891–1909. doi: 10.1002/glia.23812. [DOI] [PubMed] [Google Scholar]
- Vagionitis S., Czopka T. Visualization and time-lapse microscopy of myelinating glia in vivo in zebrafish. Methods Mol. Biol. 2018;1791:25–35. doi: 10.1007/978-1-4939-7862-5_3. [DOI] [PubMed] [Google Scholar]
- Waxman S.G. Axon-glia interactions: building a smart nerve fiber. Curr. Biol. 1997;7:R406–R410. doi: 10.1016/s0960-9822(06)00203-x. [DOI] [PubMed] [Google Scholar]
- Xin W., Chan J.R. Myelin plasticity: sculpting circuits in learning and memory. Nat. Rev. Neurosci. 2020;21:682–694. doi: 10.1038/s41583-020-00379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.M., Michel K., Jokhi V., Nedivi E., Arlotta P. Neuron class-specific responses govern adaptive myelin remodeling in the neocortex. Science. 2020;370:eabd2109. doi: 10.1126/science.abd2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonta B., Tait S., Melrose S., Anderson H., Harroch S., Higginson J., Sherman D.L., Brophy P.J. Glial and neuronal isoforms of Neurofascin have distinct roles in the assembly of nodes of Ranvier in the central nervous system. J. Cell Biol. 2008;181:1169–1177. doi: 10.1083/jcb.200712154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
-
Data reported in this paper will be shared by the lead contact upon reasonable request.
-
-
The code used for the simulation of node cluster correlation is available under https://doi.org/10.5281/zenodo.5846391.
-
-
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.