Abstract
The increasing threat to global health posed by antibiotic resistance remains a serious concern. This troublesome scenario has steered a need for the discovery and evaluation of novel antibacterial agents. Natural products are the main sources of antimicrobials used in clinical practice, serving as a rich reservoir for the discovery of new antibiotics. Pharmaceutical phenolics especially xanthones widely exist in the plant kingdom, and are important plant metabolites. They possess versatile biological activities, including antiviral, antibacterial, neurotrophic, and anticancer. In the present study, we focus on the antibacterial activities of phytoxanthones and summarize their structures and sources, categories and drug-likeness evaluations, and antibacterial activities. A total of 226 different plant xanthones are identified through the NETs screening, and most of them are distributed in Clusiaceae family. These phytoxanthones are divided into four groups according to the intrinsic structural properties, including the most common simple xanthones and the majority of biprenylated ones. Moreover, their physicochemical parameters are calculated and the structure–activity relationships are discussed as well. These results indicate that the biprenylated xanthone derivatives may be promising antibacterial candidates and that the natural products of plants may be a poorly understood repository for the discovery of novel antibacterial agents.
This review outlines the structures, drug-likeness evaluations and antibacterial activities of plant-derived xanthones. It reveals that natural products isolated from plants may be promising antibacterial candidates.
1. Introduction
Antibiotics are undoubtedly one of the most influential discoveries in medicine.1 The discovery and introduction of antibiotics, starting with penicillin, has revolutionized the treatment of bacterial infections and greatly reduced the morbidity and mortality of human beings caused by infectious diseases.2 Nonetheless, antibiotics are a double-edged sword. The excessive and improper application of antibiotics has also led to the increased number of pathogenic bacteria that can resist antibacterial treatments especially multidrug resistant (MDR) bacteria,3,4 resulting in that the clinicians have almost no choices and posing a global threat to public health.5 For instance, methicillin-resistant Staphylococcus aureus (MRSA) has become one of the most frequently reported nosocomial pathogens worldwide, and is responsible for more than 11 000 deaths annually in the USA alone.6,7 Therefore, there is an urgent need for new high-efficiency antibacterial agents and alternative strategies to fill the gaps in antibiotic discovery and development.
Natural products (NPs) are a large family of various chemical entities, with diverse biological activities and wide uses, especially in clinics and agriculture.8–10 Many of the examples, such as tetracyclines, aminoglycosides, β-lactams, and polypeptides, represent FDA-approved NPs or NP-inspired synthetic or semi-synthetic derivatives originally obtained from bacterial and fungal sources are currently in clinical trials as antibacterial agents. Compared to synthetic chemotherapeutic drugs and other potential methods, natural antibacterial agents have advantages in obtainability, structural diversity, high efficacy and unique mode of action.11 The processes in biotechnology accelerate the speed of discovering new antimicrobials, but the repeated discovery of known antibiotics has an adverse impact on the screening of new antibiotics. Hence, it is important to search for new sources, mainly natural origins, to advance the discovery of antimicrobial agents.
Plants are interesting sources of antimicrobial leads, because they can resist pathogens through various mechanisms, including producing secondary metabolites. Phenolics are the most widely distributed metabolites, ubiquitously present in the plant kingdom. Xanthones (9H-xanthen-9-one) comprise a family of O-heterocyclic symmetrical compounds with a dibenzo-γ-pyrone scaffold (Fig. 1a)12 and can be found in plants, fungi and lichen.13,14 A great variety of xanthones with volatile patterns of substitutions have been isolated. Since their discovery, xanthones display a wide range of biological and pharmacological activities, including antitumor,15,16 antioxidant,17,18 anti-inflammatory,19 antimicrobial,20–22 and antiviral.16,23 To explore the potential compounds with antibacterial activities, xanthone derivatives derived from plants reported in recent years are collected in the present review.
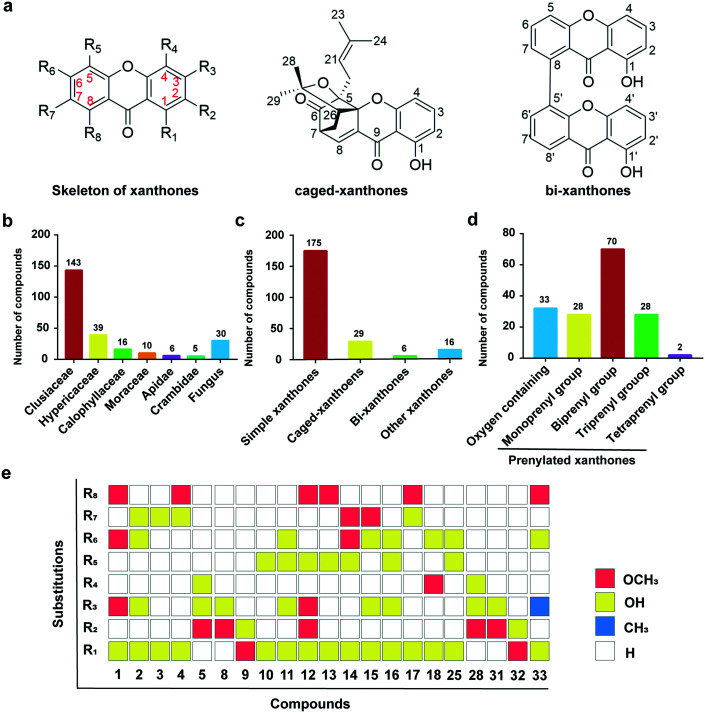
Fig. 1. The sources and classifications of xanthone derivatives. a, Chemical structures of the skeleton of simple xanthones, caged xanthones, and bi-xanthones. b, Various plant sources are classified according to the family categories of xanthones distribution. c and d, The proportions of xanthones and simple xanthones with prenylated modifications. e, The distribution of substituents of xanthone derivatives containing methoxy and hydroxyl groups.
In this study, the plant-derived antibacterial xanthones with emphasis on minimum inhibitory concentration (MIC), from 2000 to October 2021 have been reviewed. The MIC, commonly used as an index of antimicrobial efficacy, here refers to the lowest concentration of an antimicrobial agent that inhibits the visible growth of a microorganism in vitro, rather than the size of the bacteriostatic circle. The literature survey on this topic was conducted in the following databases: PubMed, Web of Science, and SciFinder, with specific keywords: plant, antimicrobial activity/antibacterial activity, and xanthones, covering the period from 2000 to October 2021. We finally obtained 226 botanical xanthones with a total of 1120 MIC values reported in Table S1.† It provides the names, structures, origin of plant species, and the antibacterial activities with MIC values and bacterial species. The names and structures of all compounds were checked for accuracy with the SciFinder database.
2. Sources and structures of antibacterial xanthone derivatives
According to plant sources, these compounds were isolated from at least 6 plant families, 10 plant genera and 27 different plant species, and also contained endophytic fungus (Fig. 1b). In detail, these xanthones have a wide distribution in Clusiaceae with 143 metabolites in 16 genera accounting for 57.4%, followed by Hypericaceae with 39 compounds in three genera (15.7%) and Calophyllaceae with 16 structures in two plants of Calophyllum inophyllum and Kielmeyera variabilis (6.4%) and other plants (4.9%) or endophytic fungus (20.5%). These results are consistent with the estimate reported in the literature that 80% xanthone derivatives are isolated from the Clusiaceae family.24–28
Xanthones can be modified by oxidation and prenylation on the rings, to increase the lipophilicity of the backbone compounds and biological activities. All compounds share a dibenzo-γ-pyrone skeleton and are categorized into simple xanthones (1–175), caged xanthones (176–204), bi-xanthones (205–210), and other derivatives (211–226) based on their chemical structural features.
2.1. Simple xanthones
Simple xanthones are a group of compounds that retain the original core of dibenzo-γ-pyrone, but are fused by simple substituents such as methoxy, hydroxyl and isoprenyl groups, accounting for 77.4% of the 226 xanthones (Fig. 1c). The structural types of isoprenyl moiety mainly contain five side chains: isopentenyl, hydroxy-isopentenyl, pyran ring isopentenyl, methyl furan ring isopentenyl and lavender,29 remarkably increasing the structural diversity of xanthones. The simple xanthones can be roughly divided into two categories: oxygen-containing group (18.9%) and prenylated group (81.1%).
Analysing the substituents of compounds 1–33 of the oxygen-containing group (Fig. 1e), we found that hydroxyl represents the majority, particularly at the 1-position of the xanthone skeleton, contributing to the structural stability due to the hydroxyl–carbonyl hydrogen bonding. In addition, some hydroxyl groups are methylated into methoxy groups to increase the lipophilicity of the compounds and further enhance their interactions with cell membrane. According to the number of isopentenyl moieties, the prenylated xanthones can be further classified into four categories, in which the prenylation with two groups makes up the majority, accounting for about 43.5%, while monoprenyl and triprenyl groups account for 17.4%, and tetraprenyl group accounts for 1.2%.
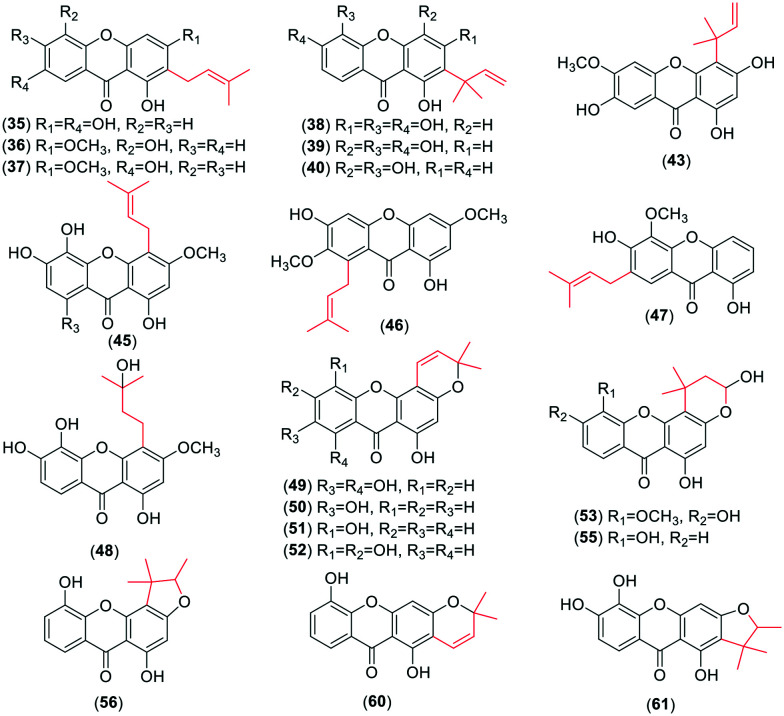
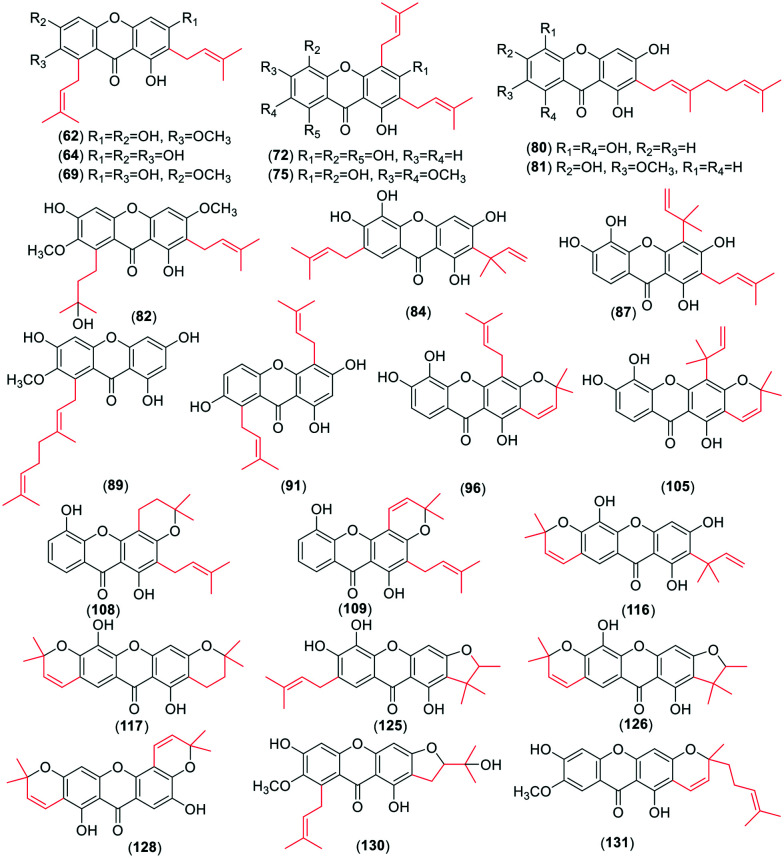
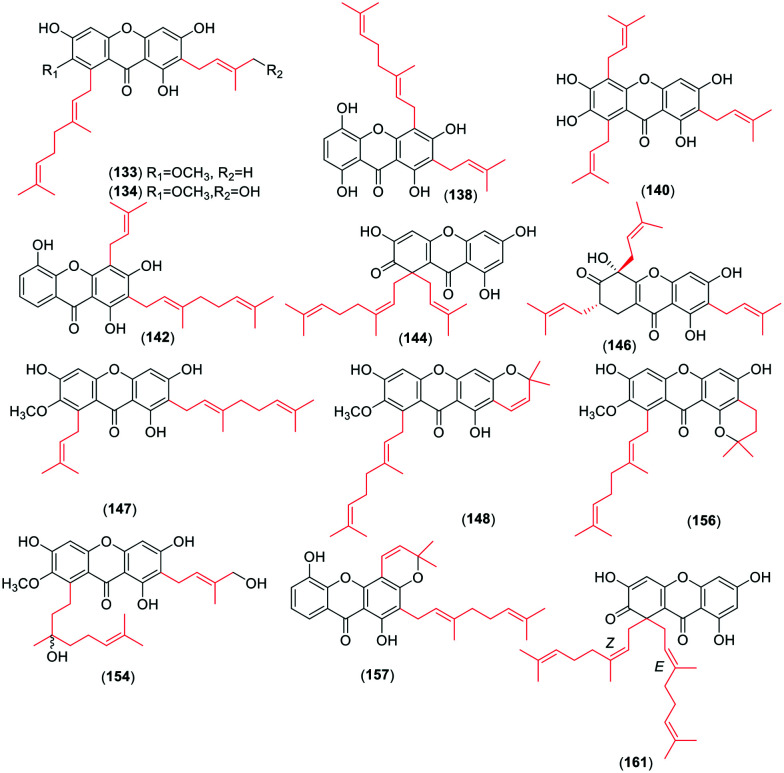
For the monoprenyl group xanthones (Fig. 2), 15 compounds (34–48) bear the intact C5-isopentenyl moiety and 13 compounds (49–61) of them are modified as five- or six-membered rings in this review. The formation of the rings is a hydroxyl group at the C-3 position and an isoprenyl moiety at their ortho-position. With respect to the dominant group of the biprenyl group, the xanthones with two intact C5 isoprenyl moieties are a few more than those with cyclization. There are 31 xanthones (62–92) (Fig. 3) with two intact C5 units. Four of them carry both isoprenyl moieties at the same side, and 27 at each side of the xanthone skeleton. 39 xanthones (93–131) with two modified C5-isopentenyl moieties are described in this review. 33 of them carry an intact C5 unit and a derived chromene formed by the other isoprenyl moiety with its ortho-hydroxyl group, and six (126–131) with two chromanes or chromenes. Similarly, 16 xanthone derivatives (132–147) (Fig. 4) sustain three intact C5-prenyl moieties, while 12 (148–161) are modified. Among compounds 132–147, 12 are regularly geranylated (C10-prenylated) derivatives. As for the tetraprenyl group xanthones, only one pair of isomers (160–161) is reported,30 and both of them carry two geranyl moieties. In summary, the diverse structures of isopentenyl substituents and the substitution sites of the xanthone skeleton contribute to the large number of the isoprenylated compounds.
Fig. 2. Representative chemical structures of xanthones with one intact or modified C5-isopentenyl moiety.
Fig. 3. Representative chemical structures of xanthones with two intact or modified C5-isopentenyl moieties and one or two modified C5-isopentenyl moieties.
Fig. 4. Representative chemical structures of xanthones with three or four intact or modified C5-isopentenyl moieties.
2.2. Caged xanthones
Caged xanthones are a kind of compounds, accounting for 12.8% of the 226 xanthones, which has an unique 4-oxatricyclo[4.3.1.0]dec-2-one backbone.22 Gambogic acid is the first reported caged xanthone, which is the main active component of gamboge.31 Gamboge is a brown-orange resin exuded from the G. hanburyi tree in southeast Asian, which is traditionally used as a colouring material and folk medicine due to its unique colour and broad spectrum of cytotoxic activity.31 The caged xanthones have been found to possess a wide range of pharmacological activities,25,30–32 particularly antibacterial property.25,32 Here, 29 caged xanthones (176–204) (ESI† Fig. S2) with the C5-isopentenyl moiety are summarized. All of them have antibacterial effects against Gram-positive bacteria, such as S. aureus. Simultaneously, these compounds were reported from G. scortechinii,31G. propinqua,32 and G. hanburyi in this review,33 which is consistent with previous reports.25,33–35 However, caged xanthones are rarely found outside the Garcinia genus.36
2.3. Bi-xanthones
Bi-xanthones are formed by xanthone dimerization, accounting for 2.6% of the 226 xanthones. Here, six xanthone dimers (205–210) with antibacterial activities are discussed. The dimerization of most compounds occurs at the ortho-position with two hydroxyl groups due to the low charge density of the hydroxyl group at ortho-position, making it easier for other xanthones to be introduced. The antibacterial activities of such compounds are better than other derivatives, with the lowest MIC of 3.12 μg ml−1.
2.4. Other xanthone derivatives
Other derivatives are a class of compounds containing the xanthone skeleton bound with polycyclic heteroatoms, accounting for 7.2% of the 226 xanthones. Most of them are distributed in the endophytes of plants, but are rarely found in plant metabolites. These compounds show promising antibacterial effects with abundant structural diversity.
3. Drug-likeness of xanthone derivative
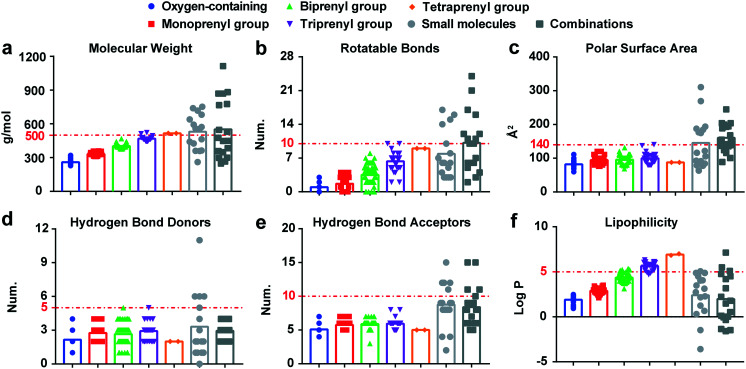
To get better understanding of the failure of candidates, there has been considerable interest in analysing the physicochemical properties of marked drugs, to steer the design of leads. The suitability of simple xanthone derivatives as drugs can be measured using well-established fundamental rules. These rules include the Lipinski rule-of-five,38 Veber rule,39 Lead like rule,40 Cmc like rule41 and Wdi like rule.42 We evaluated these compounds based on the Lipinski rule-of-five, the widely accepted procedure to assess the similarity of a compound to a drug substance. The Lipinski rule-of-five, implemented by Christopher Lipinski and his colleagues at Pfizer, can assess the physicochemical properties related to the oral bioavailability of drugs and advanced candidates.38 According to the Lipinski rule-of-five, bioactivity parameters predicted are molecule weight (≤500), log P (≤5), number of hydrogen bond acceptors (≤10), number of hydrogen bond donors (≤5), and number of rotatable bonds (≤10).43 Calculating such parameters is important for targeting the sweet spot of suitable pharmacodynamics and pharmacokinetics properties. Thus, the molecular descriptors of the simple xanthones were calculated using ADMETlab 2.0 (https://admetmesh.scbdd.com/service/evaluation/index) provided by the Xiangya School of Pharmaceutical Science, Central South University, China. For each category of the compounds, mean values are calculated. In comparison with FDA-approved antibacterial drugs, the physicochemical properties of simple xanthones are detailed as below.
3.1. Molecular weight (MW)
The average MWs of each type of simple xanthones are as follows: oxygen-containing = 259.50 g mol−1, monoprenyl = 331.69 g mol−1, biprenyl = 402.56 g mol−1, triprenyl = 468.13 g mol−1, and tetraprenyl = 516.67 g mol−1 (Fig. 5a). The MW of simple xanthones depends on the different substituents on the skeleton. For instance, with an increase in the number of isoprenyl groups, the mean MW of prenylated xanthones change in a MW of the isoprenyl group (69.07 g mol−1) dependent manner. Considering the MW of the Lipinski rule-of-five (≤500 g mol−1), it is apparent that most of the simple xanthones adhere to it except tetraprenyl xanthones. However, the mean MWs of small molecules and combinations approved as antibacterial agents by the FDA from 2015 until 2020 (ref. 37) contradict the Lipinski rule-of-five. In addition, it should be noted that antimicrobial drugs (544.40 g mol−1) are typically larger, compared with other therapeutics except cancer (620.05 g mol−1).44 Thus, the low MWs of simple xanthones display good drug-likeness properties.
Fig. 5. The physicochemical parameters of simple xanthone derivatives. a–f, Molecular weight, rotatable bonds, polar surface area, hydrogen bond donors, hydrogen bond acceptors, and lipophilicity of the simple xanthones, respectively. The small molecules in light gray (17 compounds) and the combinations in dark gray (18 compounds) are anti-infective agents approved by FDA from 2015 until June 2020. Among them, the small molecules contain cresemba, dakllnza, baxdela, xepi, benznidazole, solosec, prevymis, aemcolo, xerava, krintafel, moxidectin, zemdri, nuzyra, tpoxx, xofluza, pifeltro, fetroja, xenleta, egaten, pretomanid, artesunate, and the combinations contain elvitegravir, cobicistat, emtricitabine, tenofovir alafenanids, ceftazidime, avibactam, bictegravir, meropenem, velpatasvir, vaborbactam, sofosbuvir, elbasvir, pibrentasvir, glecaprevir, grazoprevir, voxilaprevir, lmipenem, relebactam, cilastatin.37.
3.2. Rotatable bonds (RBs) and polar surface area (PSA)
The number of rotatable bonds (RBs) is often used as a metric for molecular flexibility.39 Xanthones with a rigid heteroaromatic tricyclic core are considered as a privileged structure, and the number of RBs depends on the substituents, particularly the isoprenyl moieties. Therefore, tetraprenyl xanthones have higher numbers of nine RBs (Fig. 5b) owing to the substitutions of isoprenyl groups. Nevertheless, the compounds approved by FDA have complex RBs, with an average value of eight and ten. A total of ≤10 rotatable bonds are associated with good oral exposure. When combined with a PSA of ≤140 Å2, this criterion is considered sufficient to predict that the compound would exhibit a high probability of oral bioavailability in rats (≥20%).45 The PSA of simple xanthones is relatively concentrated (Fig. 5c), falling in the acceptable range of ≤140 Å2. Thus, the simple xanthone derivatives are potential candidates for the development of novel antimicrobial agents.
3.3. Hydrogen bond donors (HBDs) and hydrogen bond acceptors (HBAs)
The number of HBDs and HBAs characterize the polarity of compounds. They are expressed by the number of NH and OH bonds and N and O atoms, respectively.38 For simple xanthones, only the number of OH bonds and O atoms should be considered. Regarding the Lipinski's rule, the value is less than 5 for HBDs and less than 10 for HBAs. Antibacterial agents have the highest HBDs and HBAs values (2.4 and 8.7, respectively) in the distribution of drug characteristics, compared to other treatment fields.39 Moreover, almost all of simple xanthones mentioned here show lower HBDs and HBAs values than the compounds approved by the FDA from 2015 until June 2020 (Fig. 5d and e), following the Lipinski rule of five criteria. Such difference can be justified by the molecular formula and the classification of substitutes. Altogether, it indicates that simple xanthones should efficiently cross cell membranes.
3.4. Lipophilicity (log P)
Lipophilicity is expressed as a ratio of octanol solubility to water solubility. In almost all physicochemical properties related to absorption, lipophilicity occurs in some forms, such as ilog P, XLlog P, WLlog P, and MLlog P.46 Given the different forecasting methods resulting in the variant log P values, we used the mean value of log P to minimize potential errors. The mean log P values of simple xanthones increase linearly in a MW dependent manner (Fig. 5f). The acceptable limit of log P is less than 5, whereas the triprenyl and tetraprenyl xanthone derivatives do not fulfil such criterion. However, the mean log P of the FDA-approved antibacterial drugs are lower (mean values of 2.4 and 2.0) than that of prenylated xanthones (mean values ranging from 2.9 to 6.9). In addition, orally bioavailable anti-infective drugs tend to have lower log P values than other types of therapeutic drugs (mean log P of 1.56).44 Collectively, these results suggest that simple xanthones may have good biomembrane permeability.
4. Antibacterial activities of simple xanthones
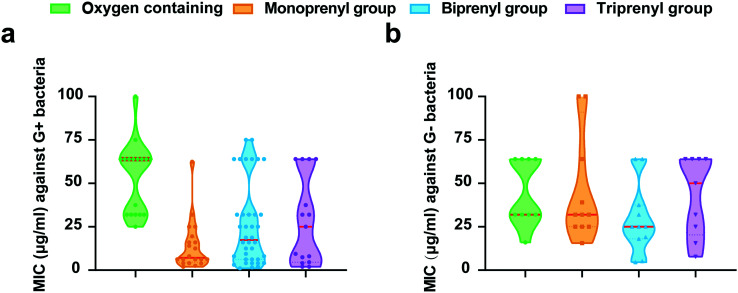
Subsequently, we used strict inclusion and exclusion criteria to narrow the scope of simple xanthones with significant antibacterial activities (MIC ≤ 100 μg ml−1). Xanthones show broad-spectrum antibacterial activities against diverse pathogenic bacteria (Fig. 6). The Gram-positive bacteria mainly include S. aureus and MRSA, while the Gram-negative bacteria include Escherichia coli and Pseudomonas aeruginosa. The structure–activity relationships (SAR) of these simple xanthones are discussed (Table 1).
Fig. 6. The MIC values of each type of simple xanthones against Gram-positive (a) and Gram-negative (b) bacteria.
Antibacterial activities of representative prenylated xanthones.
| Compounds | Names | Sources | Antibacterial activities (μg ml−1) | Ref. |
|---|---|---|---|---|
| One isopentenyl | ||||
| 39 | Subelliptenone F | G. subelliptica | MRSA (25), S. aureus (12.5), Escherichia coli (25) | 47 |
| 40 | 12-b-Dihydroxy-des-d-garcigerin | G. subelliptica | MRSA (3.13), S. aureus (6.25) | 15 |
| 50 | Nigrolineaxanthone F | G. nigrolineata | MRSA (2) | 34 |
| 55 | Pruniflorone N | Cratoxylum sumatranum | Micrococcus luteus (16), Bacillus cereus (32), S. epidermidis (16), Escherichia coli (32), Salmonella typhimurium (32), Pseudomonas aeruginosa (16) | 48 |
| Two isopentenyls | ||||
| 62 | α-Mangostin | G. cowa, G. staudtii, G. fusca, G. mangostana | Micrococcus luteus (1), S. aureus (3.13), S. epidermidis (1.56), Bacillus cereus (8), MRSA (6.25), Escherichia coli (12.5), Pseudomonas aeruginosa (12.5) | 30, 49–51 |
| 87 | Gerontoxanthone I | G. smeathmannii | Enterococci faecalis (3.13), Bacillus subtilis (2.5), S. aureus (1.1), MRSA (3.13), Micrococcus luteus (6.25), Pseudomonas aeruginosa (>50), Salmonella faecalis (4.6), Salmonella typhi (1.1) | 52 |
| 89 | Rubraxanthone | G. cowa, Allanblackia monticola, G. dioica | Bacillus cereus (2), Bacillus subtilis (1), Micrococcus luteus (2), MRSA (1.25), S. epidermidis (4), S. aureus (12), Escherichia coli (64), Salmonella typhimurium (64), Pseudomonas aeruginosa (64) | 15, 30, 47, 53, 54 |
| 91 | Gerontoxanthone H | Cudrania cochinchinensis | Enterococci faecalis (1.56), Bacillus subtilis (1.56), S. aureus (1.56), MRSA (1.56), Micrococcus lutes (1.56) | 55, 56 |
| 96 | Xanthone V1 | Cratoxylum formosum | Bacillus subtilis (1.1), S. aureus (1.1), Streptococcus faecalis (1.1), Salmonella typhi (1.1), Pseudomonas aeruginosa (9.3) | 52 |
| 116 | Gerontoxanthone B | Cudrania cochinchinensis | Bacillus subtilis (>25), S. aureus (25), Micrococcus luteus (12.5) | 56 |
| 128 | Nigrolineaxanthone I | G. nigrolineata | MRSA (4) | 34 |
| 130 | Mangostanin | Tetragonula laeviceps, G. cowa | Bacillus cereus (8), Listeria monocytogenes (0.78), Bacillus subtilis (2), Micrococcus luteus (12.5), S. aureus (12.5), S. epidermidis (25), Streptococcus pyogenes (3.13), MRSA (4), Escherichia coli (25) | 30, 57, 58 |
| 131 | Garciniacowone C | G. cowa | Bacillus cereus (32), Bacillus subtilis (64) | 30 |
| Three isopentenyls | ||||
| 133 | Cowanin | G. cowa, G. fusca | S. aureus (32), MRSA (2), Bacillus cereus (32), Bacillus subtilis (4), Micrococcus luteus (4), S. epidermidis (2), Salmonella typhimurium (64) | 49, 50, 53, 57, 59 |
| 140 | Garcinone E | G. mangostana | MRSA (25), S. aureus (12.5), Escherichia coli (25), Vibrio vulnificus (15.6), Vibrio rotiferianus (15.6), Vibrio campbellii (31.2) | 47, 60 |
| 144 | Garcinianone A | G. cowa | Bacillus cereus (4), Bacillus subtilis (2), S. aureus (16), MRSA (26), Escherichia coli (64), Salmonella typhimurium (64) | 30 |
| 146 | Garciniacowone | G. cowa | MRSA (2), S. aureus (2) | 49 |
| 157 | Formoxanthone B | Cratoxylum formosum | Bacillus subtilis (4.6), S. aureus (2.3), Streptococcus faecalis (18.7), Salmonella typhi (4.6), Pseudomonas aeruginosa (>50) | 52 |
| Four isopentenyls | ||||
| 161 | Garciniacowone B | G. cowa | Bacillus cereus (8), Bacillus subtilis (8), S. aureus (64), Escherichia coli (64), Salmonella typhimurium (64) | 30 |
4.1. Oxygen-containing xanthones
The oxygen-containing xanthones are a class of xanthones substituted by methoxy, hydroxyl and methyl groups. We focus on the oxygen-containing xanthones with particular inhibitory effects on either Gram-positive or Gram-negative bacteria. For instance, 1,5-dihydroxy-6,7-dimethoxyxanthone (14) has relatively good antibacterial activities against S. epidermidis and Bacillus cereus with a MIC of 16 μg ml−1 and 1,3,6-trihydroxy-7-methoxyxanthone (15) against Salmonella Typhimurium with a MIC of 4 μg ml−1 (ref. 48) probably due to their high log P (2.09 and 1.74, respectively). In addition, questin (33), a trioxygenated xanthone, exhibits antibacterial activity with the MIC value less than 50 μg ml−1 against both Gram-positive and Gram-negative bacteria.61 In general, there is almost no difference in the physicochemical parameters of all oxygen-containing xanthones in this review. Taken together, we deduce that simple substitutions of methoxy and hydroxyl groups have little effect on antibacterial activity.
4.2. Monoprenyl xanthones
The monoprenyl xanthones from natural plants are compounds containing one modified or unmodified isoprenyl group. Among them, we found that trioxygenated and tetraoxygenated xanthone skeletons exhibit better antimicrobial activities. For example, several compounds with a typical xanthone skeleton of 1,3,5-trioxygenated or 1,3,5,6-tetraoxygenated (53–56)48 show inhibition against either Gram-positive or Gram-negative bacteria. Compared to the cyclic compounds like pruniflorone N and pruniflorone M (55, 57),48 the antimicrobial activity is devoid when it is cyclized to a five-membered ring at C-3 and C-4 based on the SAR analysis. Moreover, the OH group at C-5 is replaced by the OCH3 group (53, 54),48 resulting in improved antibacterial activity. In addition, monoprenyl xanthones (36–40) share a typical trioxygenated or tetraoxygenated xanthone skeleton show promising antibacterial activities.47 Overall, we deduce that the trioxygenated and tetraoxygenated of the monoprenyl xanthones are essential for targeting bacteria.
4.3. Biprenyl xanthones
In this group, α-mangostin (62) is a typical compound substituted by two isoprene moieties with excellent antimicrobial activity. Previous studies reveal that it has direct inhibitory effect and synergism against diverse pathogenic S. aureus and MRSA within the MIC range of 0.5–1 μg ml−1.30,49,50 It has experimentally rapid bactericidal action and disrupts bacterial cytoplasmic membranes, enabling the ability to overcome drug resistance.21,51,62 Notably, a very recent mechanistic study demonstrates that α-mangostin not only displays rapid bactericidal activity against Gram-positive bacteria by binding to the bacterial membrane, leading to metabolic perturbation, but also restores the susceptibility of colistin against Gram-negative pathogens.51 The presence of a prenyl moiety and their modifications often results in increased lipophilicity and affinity to cell membranes facilitating biological consequences. To improve the antibacterial activity and selectivity of α-mangostin, a series of nonpeptidic xanthone-based peptidomimetics were designed and synthesized, and these molecules possess robust antibacterial properties and membrane selectivity with low toxicity.63 Altogether, these studies suggest that α-mangostin is an encouraging lead for the development of a new antibacterial drug. Furthermore, the antibacterial activity of four biprenyl xanthones (62–65) isolated from G. mangostana demonstrates a better inhibitory effect against Gram-positive bacteria.64 The SAR analysis shows that the 1,3,6,7-tetraoxygenated xanthone skeleton is more active against MRSA. In addition, the two isoprene groups can be cyclized with the adjacent oxygen atoms to form a ring among biprenyl xanthones. We find the cyclization of the C-2 isoprenyl group into a pyran ring with the C-3 hydroxyl group (87, 96) resulting in no effects on the antibacterial activities.52 However, it denotes that the antibacterial activity is lost when forming two ring structures.
4.4. Triprenyl xanthones
With the increased number of isopentenyl substituents, the antibacterial activity of xanthones might increase.49 For example, triprenyl xanthone cowanin (133) and cowanol (134)49,50,57,59 show a better inhibitory effect on MRSA-SK1 than α-mangostin. Furthermore, garciniacowone (146)49 substituted with three independent isoprenyl moieties maintains antibacterial activity against MRSA (MIC = 2 μg ml−1). Interestingly, the cyclization of such isoprenyl groups always reduces the antibacterial activity of triprenyl xanthones, and may cause nonspecific cytotoxicity to host cells probably due to the intrinsic strong hydrophobicity. Hence, the antibacterial activity and safety of triprenyl xanthones should be systematically evaluated.
4.5. Tetraprenyl xanthones
There are few reports on tetraprenyl xanthones. We found only two isomers,30 and both show no difference on bacteria.
Altogether, our SAR analysis is consistent with the statistics of the MIC of simple xanthones (Fig. 6). We find that the median MIC value of monoprenyl xanthones is higher than that of biprenyl xanthones against Gram-positive bacteria (Fig. 6a). Interestingly, some biprenyl xanthones demonstrate impressive antibacterial activities. Moreover, the efficacy of biprenyl xanthones is better than other xanthones against Gram-negative bacteria (Fig. 6b), agreeing with the fact that proper hydrophobicity is the prerequisite for chemicals penetrating the hydrophobic barrier in Gram-negative bacteria.65 Overall, these results indicate that the biprenyl xanthone derivatives are promising broad-spectrum antibacterial candidates.
5. Drug-likeness and antibacterial activities of caged xanthones and bi-xanthones
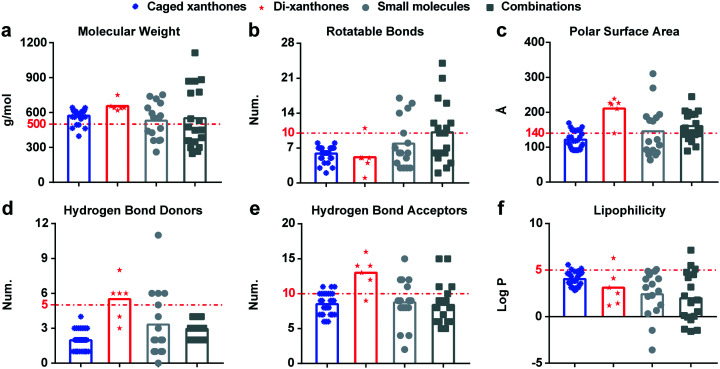
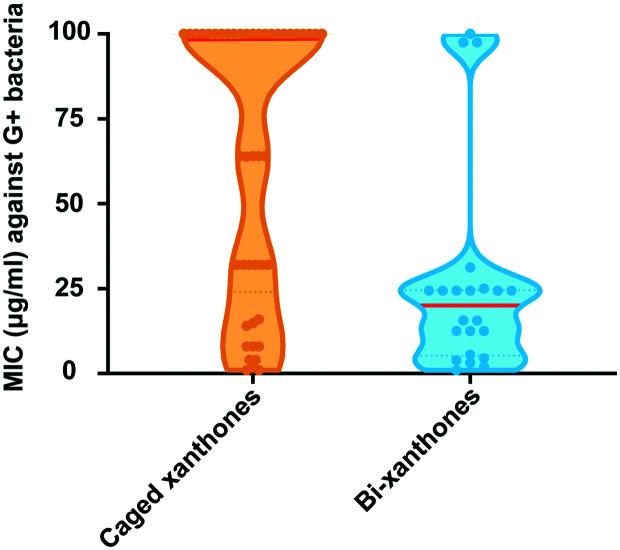
With deeper understanding of other xanthones, we discuss the drug-likeness and antibacterial activities of caged xanthones and bi-xanthones. The average MWs of caged xanthones and bi-xanthones are 572.94 g mol−1 and 656.28 g mol−1, respectively. Considering the MW of Lipinski rule-of-five (≤500 g mol−1), it is apparent that the two types of compounds do not fulfil such criterion. In addition, it should be noted that antimicrobial drugs (544.40 g mol−1) are typically larger, compared with other therapeutics except cancer (620.05 g mol−1). Thus, the caged xanthones are potential candidates for the development of novel antimicrobial agents. Caged xanthones have a higher number of RBs (5.9) owing to the substitutions. The PSA number of Caged xanthones is 121.32 Å2, falling in the acceptable range of ≤140 Å2. Combining the two parameters, the caged xanthones are indicated to have better oral exposure than bi-xanthones. Moreover, almost all of the caged xanthones show lower HBDs and HBAs values than bi-xanthones following the Lipinski rule-of-five criteria (Fig. 7d and e), suggesting the potential to cross membranes. In addition, lipophilicity relates to absorption. The mean log P of the two types of compounds adheres to the criteria (log P ≤ 5). Collectively, these results suggest that caged xanthones may be promising antibacterial candidates. Caged xanthones and bi-xanthones have a wide spectrum of bactericidal activities against Gram-positive and Gram-negative bacteria. There are few reports on the inhibition of Gram-negative bacteria, so we will not discussed here. We found that the median values of caged xanthones is lower than those of bi-xanthones against Gram-positive bacteria (Fig. 8), contributing to the small sample size. To evaluate the potential of caged xanthones and bi-xanthones as antibacterial agents, more attention should be paid to the biological activities.
Fig. 7. The physicochemical parameters of caged xanthones and bi-xanthones. a–f, Molecular weight, rotatable bonds, polar surface area, hydrogen bond donors, hydrogen bond acceptors, lipophilicity of caged xanthoens and bi-xanthones, respectively.
Fig. 8. The MIC values of caged xanthones and bi-xanthones against Gram-positive.
6. Conclusions
There is an urgent need for new antibacterial agents, and plants might be a huge source in the search for such compounds. We found that xanthones have good potential to become drug candidates, particularly those with one or two isoprenyl groups, inspired by the fact that many successful examples of antibacterial drugs from natural products do not accept the principle of Lipinski rule-of-five. The chemical diversity of xanthones provides abundant compounds to decipher the SAR particularly prenylation on antibacterial activity. However, these results advocate more in vivo investigations to further determine the efficacy and safety of promising candidates for clinical applications in the future. In conclusion, this review may also shed light on the discovery and development of other novel antibacterial drugs from numerous natural compounds.
Conflicts of interest
The authors confirm that this research article has no conflict of interest.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31772796, 21861142006), and Chinese Universities Scientific Fund (2021TC058).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1md00351h
References
- Matos de Opitz C. L. Sass P. Future Microbiol. 2020;15:703–708. doi: 10.2217/fmb-2020-0048. [DOI] [PubMed] [Google Scholar]
- He X. Yang Y. Guo Y. Lu S. Du Y. Li J. J. Zhang X. Leung N. L. C. Zhao Z. Niu G. Yang S. Weng Z. Kwok R. T. K. Lam J. W. Y. Xie G. Tang B. Z. J. Am. Chem. Soc. 2020;142:3959–3969. doi: 10.1021/jacs.9b12936. [DOI] [PubMed] [Google Scholar]
- Davies S. C. Fowler T. Watson J. Livermore D. M. Walker D. Lancet. 2013;381:1606–1609. doi: 10.1016/S0140-6736(13)60604-2. [DOI] [PubMed] [Google Scholar]
- Ghosh C. Sarkar P. Issa R. Haldar J. Trends Microbiol. 2019;27:323–338. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- Stoesser N. Mathers A. J. Moore C. E. Day N. P. Crook D. W. Lancet Infect. Dis. 2016;16:285–286. doi: 10.1016/S1473-3099(16)00010-4. [DOI] [PubMed] [Google Scholar]
- Vuong C. Yeh A. J. Cheung G. Y. Otto M. Expert Opin. Invest. Drugs. 2016;25:73–93. doi: 10.1517/13543784.2016.1109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olleik H. Yahiaoui S. Roulier B. Courvoisier-Dezord E. Perrier J. Peres B. Hijazi A. Baydoun E. Raymond J. Boumendjel A. Maresca M. Haudecoeur R. Eur. J. Med. Chem. 2019;165:133–141. doi: 10.1016/j.ejmech.2019.01.022. [DOI] [PubMed] [Google Scholar]
- Demain A. L. J. Ind. Microbiol. Biotechnol. 2014;41:185–201. doi: 10.1007/s10295-013-1325-z. [DOI] [PubMed] [Google Scholar]
- Giddings L. A. Newman D. J. J. Ind. Microbiol. Biotechnol. 2013;40:1181–1210. doi: 10.1007/s10295-013-1331-1. [DOI] [PubMed] [Google Scholar]
- Newman D. J. Cragg G. M. Snader K. M. Nat. Prod. Rep. 2000;17:215–234. doi: 10.1039/A902202C. [DOI] [PubMed] [Google Scholar]
- Newman D. J. Cragg G. M. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- Proenca C. Albuquerque H. M. Ribeiro D. Freitas M. Santos C. M. Silva A. M. Fernandes E. Eur. J. Med. Chem. 2016;115:381–392. doi: 10.1016/j.ejmech.2016.03.043. [DOI] [PubMed] [Google Scholar]
- Vieira L. M. Kijjoa A. Curr. Med. Chem. 2005;12:2413–2446. doi: 10.2174/092986705774370682. [DOI] [PubMed] [Google Scholar]
- El-Seedi H. R. El-Ghorab D. M. El-Barbary M. A. Zayed M. F. Goransson U. Larsson S. Verpoorte R. Curr. Med. Chem. 2009;16:2581–2626. doi: 10.2174/092986709788682056. [DOI] [PubMed] [Google Scholar]
- Mohamed G. A. Ibrahim S. R. Shaaban M. I. Ross S. A. Fitoterapia. 2014;98:215–221. doi: 10.1016/j.fitote.2014.08.014. [DOI] [PubMed] [Google Scholar]
- Reutrakul V. Anantachoke N. Pohmakotr M. Jaipetch T. Sophasan S. Yoosook C. Kasisit J. Napaswat C. F. Santisuk T. Tuchinda P. Planta Med. 2007;73:33–40. doi: 10.1055/s-2006-951748. [DOI] [PubMed] [Google Scholar]
- Luo C. T. Mao S. S. Liu F. L. Yang M. X. Chen H. Kurihara H. Li Y. Fitoterapia. 2013;91:140–147. doi: 10.1016/j.fitote.2013.08.021. [DOI] [PubMed] [Google Scholar]
- Jung H. A. Su B. N. Keller W. J. Mehta R. G. Kinghorn A. D. J. Agric. Food Chem. 2006;54:2077–2082. doi: 10.1021/jf052649z. [DOI] [PubMed] [Google Scholar]
- Chen L. G. Yang L. L. Wang C. C. Food Chem. Toxicol. 2008;46:688–693. doi: 10.1016/j.fct.2007.09.096. [DOI] [PubMed] [Google Scholar]
- Marona H. Szkaradek N. Karczewska E. Trojanowska D. Budak A. Bober P. Przepiorka W. Cegla M. Szneler E. Arch. Pharm. 2009;342:9–18. doi: 10.1002/ardp.200800089. [DOI] [PubMed] [Google Scholar]
- Yasunaka K. Abe F. Nagayama A. Okabe H. Lozada-Perez L. Lopez-Villafranco E. Muniz E. E. Aguilar A. Reyes-Chilpa R. J. Ethnopharmacol. 2005;97:293–299. doi: 10.1016/j.jep.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Nguemeving J. R. Azebaze A. G. Kuete V. Eric Carly N. N. Beng V. P. Meyer M. Blond A. Bodo B. Nkengfack A. E. Phytochemistry. 2006;67:1341–1346. doi: 10.1016/j.phytochem.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Groweiss A. Cardellina J. H. Boyd M. R. J. Nat. Prod. 2000;63:1537–1539. doi: 10.1021/np000175m. [DOI] [PubMed] [Google Scholar]
- Kureshi A. A. Dholakiya C. Hussain T. Mirgal A. Salvi S. P. Barua P. C. Talukdar M. Beena C. Kar A. Zachariah T. J. Kumari P. Dhanani T. Singh R. Manivel P. Kumar S. J. AOAC Int. 2019;102:1423–1434. doi: 10.5740/jaoacint.18-0335. [DOI] [PubMed] [Google Scholar]
- Han Q. B. Xu H. X. Curr. Med. Chem. 2009;16:3775–3796. doi: 10.2174/092986709789104993. [DOI] [PubMed] [Google Scholar]
- Raksat A. Maneerat W. Andersen R. J. Pyne S. G. Laphookhieo S. Fitoterapia. 2019;136:104175. doi: 10.1016/j.fitote.2019.104175. [DOI] [PubMed] [Google Scholar]
- Gao X. M. Yu T. Lai F. S. Zhou Y. Liu X. Qiao C. F. Song J. Z. Chen S. L. Luo K. Q. Xu H. X. Bioorg. Med. Chem. 2010;18:4957–4964. doi: 10.1016/j.bmc.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Xia Z. Zhang H. Xu D. Lao Y. Fu W. Tan H. Cao P. Yang L. Xu H. Molecules. 2015;20:11387–11399. doi: 10.3390/molecules200611387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. T. Zhu L. Yuan Y. Ling S. Xu J. W. Oxid. Med. Cell. Longevity. 2020;2020:2128513. doi: 10.1155/2020/2128513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriyatep T. Siridechakorn I. Maneerat W. Pansanit A. Ritthiwigrom T. Andersen R. J. Laphookhieo S. J. Nat. Prod. 2015;78:265–271. doi: 10.1021/np5008476. [DOI] [PubMed] [Google Scholar]
- Johrer K. Cicek S. S. Cancers. 2021;13:2678. doi: 10.3390/cancers13112678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke H. Morrisey J. M. Qu S. Chantarasriwong O. Mather M. W. Theodorakis E. A. Vaidya A. B. Antimicrob. Agents Chemother. 2017;61:e01220. doi: 10.1128/AAC.01220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyakunvat P. Anantachoke N. Reutrakul V. Jiarpinitnun C. Bioorg. Med. Chem. Lett. 2016;26:2980–2983. doi: 10.1016/j.bmcl.2016.05.030. [DOI] [PubMed] [Google Scholar]
- Rukachaisirikul V. Tadpetch K. Watthanaphanit A. Saengsanae N. Phongpaichit S. J. Nat. Prod. 2005;68:1218–1221. doi: 10.1021/np058050a. [DOI] [PubMed] [Google Scholar]
- Alam S. Khan F. Sci. Rep. 2018;8:5524. doi: 10.1038/s41598-018-23768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q. B. Xu H. X. Curr. Med. Chem. 2009;16:3775–3796. doi: 10.2174/092986709789104993. [DOI] [PubMed] [Google Scholar]
- Bhutani P. Joshi G. Raja N. Bachhav N. Rajanna P. K. Bhutani H. Paul A. T. Kumar R. J. Med. Chem. 2021;64:2339–2381. doi: 10.1021/acs.jmedchem.0c01786. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A. Lombardo F. Dominy B. W. Feeney P. J. Adv. Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Veber D. F. Johnson S. R. Cheng H. Y. Smith B. R. Ward K. W. Kopple K. D. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Ghose A. K. Viswanadhan V. N. Wendoloski J. J. J. Comb. Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- Egan W. J. Merz K. M. Baldwin J. J. J. Med. Chem. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- Muegge I. Heald S. L. Brittelli D. J. Med. Chem. 2001;44:1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]
- Shen L. Yuan Y. Guo Y. Li M. Li C. Pu X. Front. Pharmacol. 2019;10:1310. doi: 10.3389/fphar.2019.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp D. Garavelas A. Campitelli M. J. Nat. Prod. 2015;78:1370–1382. doi: 10.1021/acs.jnatprod.5b00255. [DOI] [PubMed] [Google Scholar]
- Meanwell N. A. Chem. Res. Toxicol. 2011;24:1420–1456. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- Roque J. A. Barrett P. C. Cole H. D. Lifshits L. M. Shi G. Monro S. von Dohlen D. Kim S. Russo N. Deep G. Cameron C. G. Alberto M. E. McFarland S. A. Chem. Sci. 2020;11:9784–9806. doi: 10.1039/D0SC03008B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iinuma M. Tosa H. Tanaka T. Asai F. Kobayashi Y. Shimano R. Miyauchi K. J. Pharm. Pharmacol. 1996;48:861–865. doi: 10.1111/j.2042-7158.1996.tb03988.x. [DOI] [PubMed] [Google Scholar]
- Tantapakul C. Maneerat W. Sripisut T. Ritthiwigrom T. Andersen R. J. Cheng P. Cheenpracha S. Raksat A. Laphookhieo S. J. Agric. Food Chem. 2016;64:8755–8762. doi: 10.1021/acs.jafc.6b03643. [DOI] [PubMed] [Google Scholar]
- Siridechakorn I. Phakhodee W. Ritthiwigrom T. Promgool T. Deachathai S. Cheenpracha S. Prawat U. Laphookhieo S. Fitoterapia. 2012;83:1430–1434. doi: 10.1016/j.fitote.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Nontakham J. Charoenram N. Upamai W. Taweechotipatr M. Suksamrarn S. Arch. Pharmacal Res. 2014;37:972–977. doi: 10.1007/s12272-013-0266-4. [DOI] [PubMed] [Google Scholar]
- Song M. Liu Y. Li T. Liu X. Hao Z. Ding S. Panichayupakaranant P. Zhu K. Shen J. Adv. Sci. 2021;8:e2100749. doi: 10.1002/advs.202100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsri S. Karalai C. Ponglimanont C. Kanjana-opas A. Chantrapromma K. Phytochemistry. 2006;67:723–727. doi: 10.1016/j.phytochem.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Auranwiwat C. Trisuwan K. Saiai A. Pyne S. G. Ritthiwigrom T. Fitoterapia. 2014;98:179–183. doi: 10.1016/j.fitote.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Azebaze A. G. B. Meyer M. Bodo B. Nkengfack A. E. Phytochemistry. 2004;65:2561–2564. doi: 10.1016/j.phytochem.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Fukai T. Oku Y. Hou A. J. Yonekawa M. Terada S. Phytomedicine. 2005;12:510–513. doi: 10.1016/j.phymed.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Fukai T. Oku Y. Hou A. J. Yonekawa M. Terada S. Chem. Biodiversity. 2004;1:1385–1390. doi: 10.1002/cbdv.200490101. [DOI] [PubMed] [Google Scholar]
- Panthong K. Pongcharoen W. Phongpaichit S. Taylor W. C. Phytochemistry. 2006;67:999–1004. doi: 10.1016/j.phytochem.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Sanpa S. Popova M. Bankova V. Tunkasiri T. Eitssayeam S. Chantawannakul P. PLoS One. 2015;10:e0126886. doi: 10.1371/journal.pone.0126886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trisuwan K. Ritthiwigrom T. Arch. Pharmacal Res. 2012;35:1733–1738. doi: 10.1007/s12272-012-1004-z. [DOI] [PubMed] [Google Scholar]
- Wang W. Liao Y. Huang X. Tang C. Cai P. Nat. Prod. Res. 2018;32:1769–1774. doi: 10.1080/14786419.2017.1402315. [DOI] [PubMed] [Google Scholar]
- Guo L. Zhang F. Wang X. Chen H. Wang Q. Guo J. Cao X. Wang L. 3 Biotech. 2019;9:14. doi: 10.1007/s13205-018-1535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J. J. Qiu S. Zou H. Lakshminarayanan R. Li J. Zhou X. Tang C. Saraswathi P. Verma C. Tan D. T. Tan A. L. Liu S. Beuerman R. W. Biochim. Biophys. Acta. 2013;1828:834–844. doi: 10.1016/j.bbamem.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Lin S. Wade J. D. Liu S. Acc. Chem. Res. 2021;54:104–119. doi: 10.1021/acs.accounts.0c00550. [DOI] [PubMed] [Google Scholar]
- Boonnak N. Chantrapromma S. Sathirakul K. Kaewpiboon C. Bioorg. Med. Chem. Lett. 2020;30:127494. doi: 10.1016/j.bmcl.2020.127494. [DOI] [PubMed] [Google Scholar]
- Song M. Liu Y. Huang X. Ding S. Wang Y. Shen J. Zhu K. Nat. Microbiol. 2020;5:1040–1050. doi: 10.1038/s41564-020-0723-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.