Abstract
BACKGROUND
Clinical trials of the KRAS inhibitors adagrasib and sotorasib have shown promising activity in cancers harboring KRAS glycine-to-cysteine amino acid substitutions at codon 12 (KRASG12C). The mechanisms of acquired resistance to these therapies are currently unknown.
METHODS
Among patients with KRASG12C-mutant cancers treated with adagrasib monotherapy, we performed genomic and histologic analyses that compared pretreatment samples with those obtained after the development of resistance. Cell-based experiments were conducted to study mutations that confer resistance to KRASG12C inhibitors.
RESULTS
A total of 38 patients were included in this study: 27 with non–small-cell lung cancer, 10 with colorectal cancer, and 1 with appendiceal cancer. Putative mechanisms of resistance to adagrasib were detected in 17 patients (45% of the cohort), of whom 7 (18% of the cohort) had multiple coincident mechanisms. Acquired KRAS alterations included G12D/R/V/W, G13D, Q61H, R68S, H95D/Q/R, Y96C, and high-level amplification of the KRASG12C allele. Acquired bypass mechanisms of resistance included MET amplification; activating mutations in NRAS, BRAF, MAP2K1, and RET; oncogenic fusions involving ALK, RET, BRAF, RAF1, and FGFR3; and loss-of-function mutations in NF1 and PTEN. In two of nine patients with lung adenocarcinoma for whom paired tissue-biopsy samples were available, histologic transformation to squamous-cell carcinoma was observed without identification of any other resistance mechanisms. Using an in vitro deep mutational scanning screen, we systematically defined the landscape of KRAS mutations that confer resistance to KRASG12C inhibitors.
CONCLUSIONS
Diverse genomic and histologic mechanisms impart resistance to covalent KRASG12C inhibitors, and new therapeutic strategies are required to delay and overcome this drug resistance in patients with cancer. (Funded by Mirati Therapeutics and others; ClinicalTrials.gov number, NCT03785249.)
KRAS IS ONE OF THE MOST COMMONLY mutated oncogenes in cancer. The KRAS protein normally functions as a molecular switch that cycles between an active, guanosine-5′-triphosphate (GTP)–bound state and an inactive, guanosine-5′-diphosphate (GDP)–bound form. Oncogenic mutations in KRAS typically occur at hotspots in the protein (e.g., codons 12, 13, and 61), increasing the steady-state levels of KRAS proteins in the GTP-bound state that are capable of driving protumorigenic signaling through downstream effector pathways, such as the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) pathways. Therapeutic targeting of mutant KRAS has proven challenging because of its high affinity for nucleotide and the lack of tractable binding pockets for small-molecule inhibitors.1,2 In the past several years, compounds have been identified that bind covalently to cysteine 12 within the switch II pocket of the GDP-bound form of the KRASG12C protein, preventing oncogenic signaling and causing potent tumor regressions in preclinical models.3-7 KRASG12C mutations occur in approximately 13% of lung adenocarcinomas and approximately 3% of colorectal adenocarcinomas and less commonly in cancers of the uterus, pancreas, breast, bladder, cervix, and ovaries.8,9
In early-phase clinical trials, two potent, selective, and irreversible small-molecule KRASG12C inhibitors have shown promising results in non–small-cell lung cancer (NSCLC) and more modest efficacy in colorectal cancer: adagrasib (MRTX849) and sotorasib (AMG510).6,7,10 An ongoing phase 1–2 study of adagrasib that involves patients with KRASG12C-mutant cancer has shown an objective response rate of 45% among patients with NSCLC11,12 and 17% among those with colorectal cancer.13 Sotorasib was associated with an objective response rate of 37.1% and a median progression-free survival of 6.8 months among patients with NSCLC14 and a response rate of 7.1% among those with colorectal cancer.10 Despite the clinical benefit that was observed for many patients treated with KRASG12C inhibitors, acquired resistance to single-agent therapy eventually occurred in most patients. Little is currently known about the clinical mechanisms of resistance to KRASG12C inhibition.6,7,15-17
Among patients with KRASG12C-mutant cancers treated with adagrasib monotherapy in the KRYSTAL-1 trial, we performed histologic and genomic analyses of samples obtained at the time of disease progression to identify mechanisms of resistance to adagrasib. Moreover, we used a deep mutational scanning screen with a library of KRASG12C missense variants to systematically define possible second-site mutations that confer resistance to KRASG12C inhibition.
METHODS
PATIENTS
Patients with KRASG12C-mutant cancers who had disease progression while receiving adagrasib monotherapy were included in this study. Acquired resistance to therapy was defined as stable disease for at least 12 weeks or a partial or complete response followed by disease progression according to the Response Evaluation Criteria in Solid Tumors, version 1.1. Institutional review board–approved correlative studies were performed at participating institutions before adagrasib treatment and at the time of resistance to adagrasib.
ANALYSIS OF RESISTANCE MECHANISMS
Histologic features of available tissue-biopsy samples that were obtained at the time of acquired resistance to adagrasib were compared with those of samples obtained before adagrasib treatment. Next-generation sequencing was performed on tissue samples or circulating tumor DNA (ctDNA) obtained at the time of adagrasib resistance, and the results were compared with available results of tissue or plasma sequencing before adagrasib treatment (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Genomic alterations were represented in relation to the wild-type reference coding sequence and according to the variant allele frequency of each specific mutation.
IN VITRO VALIDATION OF DRUG RESISTANCE
Individual resistance mutations were generated by site-directed mutagenesis and transduced into Ba/F3 cells.18 Ba/F3 cells that expressed the mutant or control KRASG12C alleles were treated with adagrasib or sotorasib across the dose range, and the area under the dose–response curve was calculated for comparison of dose response across alleles. Immunoblotting was performed to assay biomarker response to KRASG12C inhibition. The methods of validation are detailed in the Supplementary Appendix.
DEEP MUTATIONAL SCANNING
Deep mutational scanning is an approach that uses massively parallel sequencing to simultaneously measure the functional effect of many variants in a protein in a single experiment.19,20 We designed and synthesized a lentiviral variant library that expressed nearly all possible distinct alleles with a single amino acid substitution in the KRASG12C backbone allele. We performed a positive-selection deep mutational scanning screen for mutations in KRASG12C that confer resistance to KRASG12C inhibition in Ba/F3 cells cultured for 7 days in the absence of interleukin-3 and in the presence of one of two KRASG12C inhibitors, MRTX1257 (a compound highly related to adagrasib)7 or sotorasib. Relative allele abundance at the screen end point was evaluated, and z scores were calculated to define resistance mutations as those mutant alleles that permitted cell proliferation in the context of KRASG12C inhibitor treatment (see the Supplementary Appendix).
RESULTS
MECHANISMS OF ACQUIRED RESISTANCE TO ADAGRASIB
A total of 38 patients treated with adagrasib who initially had stable disease for at least 12 weeks or an objective response to therapy followed by subsequent disease progression were included in this study: 27 patients had NSCLC, 10 had colorectal cancer, and 1 had appendiceal cancer. At the time of acquired resistance to adagrasib, tissue was available for analysis for 10 patients, and ctDNA was available for 32 patients; both tissue and ctDNA were obtained from 4 of these patients. In 32 of the 38 patients (84%), the original KRASG12C mutation was identified at the time of adagrasib resistance. For the six instances in which KRASG12C mutation was not observed at the time of resistance, only plasma was available for analysis, and the lack of detection of this allele may reflect insufficient tumor shedding of ctDNA in those patients.
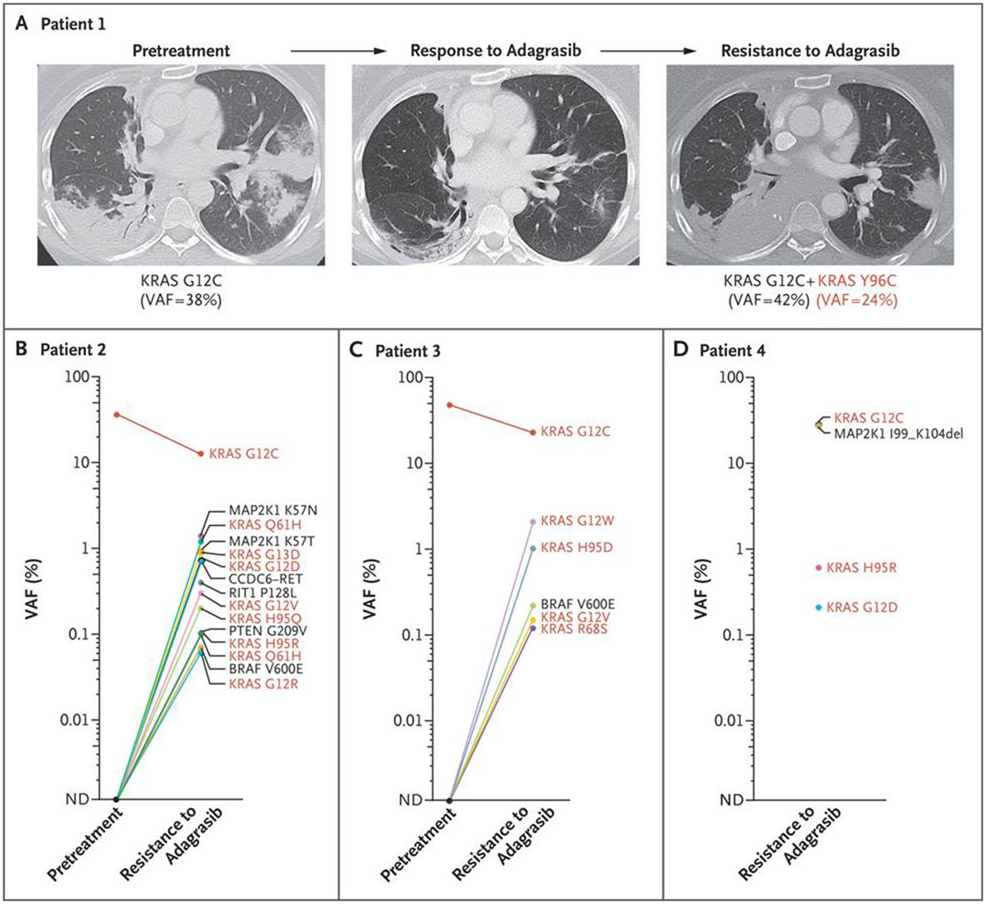
Putative mechanisms of resistance to adagrasib were identified in 17 of 38 patients (45%). Novel acquired secondary KRAS mutations within the adagrasib-binding pocket were found in 4 patients, including a Y96C mutation in 1 patient with NSCLC (Fig. 1A and Fig. S1), H95Q and H95R mutations in 1 patient with colorectal cancer (Fig. 1B), R68S and H95D mutations in 1 patient with NSCLC (Fig. 1C), and an H95R mutation in another patient with colorectal cancer (Fig. 1D). In addition, activating mutations in KRAS, such as G12D, G12V, and G13D (each occurring in trans on a separate KRAS allele than the G12C allele) as well as G12R and Q61H (status with respect to cis or trans position unable to be assessed), were identified in several patients (Figs. 1B through 1D and 2A and 2B and Fig. S2). In 1 patient, the G12C-mutant codon 12 (c.34G→T) apparently acquired an additional mutation (c.36T→G), leading to a mutation from cysteine to tryptophan (c.34_36 GGT→TGT→TGG, ultimately noted as a G12W mutation) (Fig. 1C and Fig. S2B), which conferred resistance to adagrasib.
Figure 1. Resistance to Adagrasib Conferred by Acquired KRAS Mutations.
Serial axial computed tomographic scans are shown for a patient with KRASG12C-mutant lung adenocarcinoma (Panel A) who had a partial response to adagrasib followed by disease progression. A lung tumor biopsy at the time of acquired resistance showed an acquired KRAS Y96C mutation. A patient with colorectal cancer (Panel B) showed multiple mechanisms of adagrasib resistance, including mutations in KRAS H95Q, H95R, G12R/D/V, G13D, and Q61H (listed twice owing to distinct c.183A→C and c.183A→T mutations leading to the same amino acid substitution); MAP2K1 K57N/T mutations; and a CCDC6-RET fusion. Pretreatment sequencing of RIT1 and PTEN was not performed. In this patient, the pretreatment variant allele frequency (VAF) for KRASG12C was from tissue, whereas the VAF at resistance was from circulating tumor DNA. A patient with lung adenocarcinoma (Panel C) was found to have KRAS R68S, H95D, and G12V/W along with BRAF V600E mutations at the time of acquired resistance to adagrasib. A patient with colorectal cancer (Panel D) showed KRAS H95R and G12D mutations as well as an activating MAP2K1 deletion. In Panels B through D, red coloring indicates KRAS alleles. ND denotes not detected.
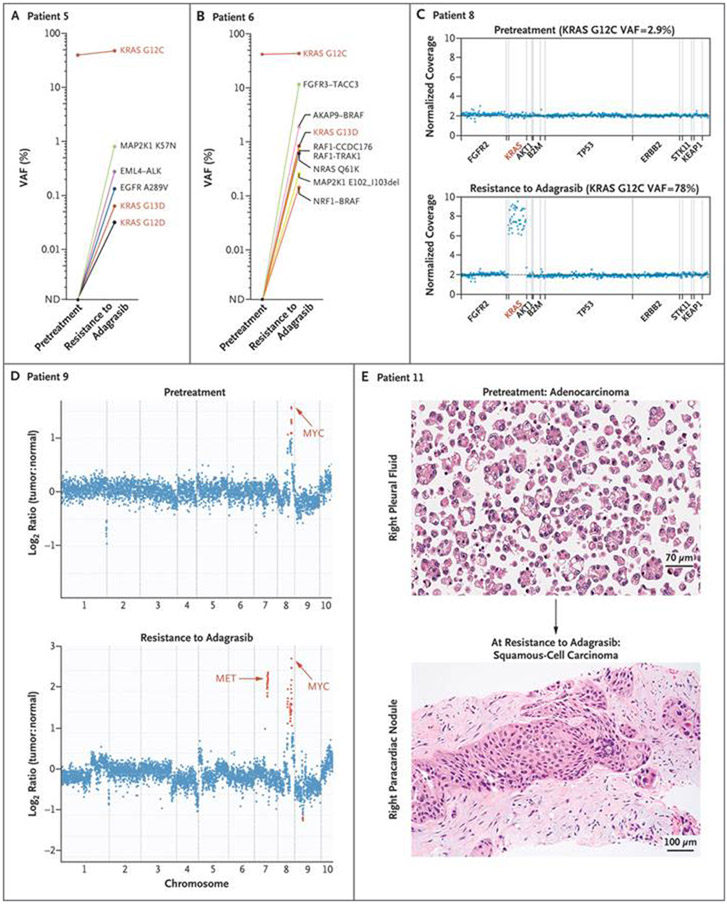
Figure 2. Genetic and Nongenetic Mechanisms of Resistance to Adagrasib.
A patient with colorectal cancer (Panel A) was found to have multiple acquired mutations in KRAS, EGFR, and MAP2K1 and an EML4-ALK rearrangement. Multiple oncogenic fusions (Panel B) involving FGFR3, BRAF, and RAF1, along with activating mutations of KRAS, NRAS, and MAP2K1, were detected in a patient with colorectal cancer. A patient with colorectal cancer (Panel C) was observed to have focal amplification of the KRASG12C allele at the time of adagrasib resistance. In Panels A through C, red coloring indicates KRAS alleles. Acquired focal MET amplification (Panel D) is shown for a patient with lung adenocarcinoma. Focal MYC amplification was also observed in both the pretreatment and postresistance biopsies. The y axis shows the log2 ratio of copy number for tumor to paired normal tissue. Histologic transformation from lung adenocarcinoma to squamous-cell carcinoma (Panel E) was observed at the time of adagrasib resistance in a patient for whom a genetic mechanism of resistance could not be identified.
Pathogenic mutations in other receptor tyrosine kinase (RTK)–RAS–MAPK pathway members were also detected in patients with acquired resistance to adagrasib, including NRAS (Q61K in one patient), BRAF (V600E in two patients), MAP2K1/MEK1 (K57T in one patient, K57N in two patients, an I99_K104 deletion in one patient, and an E102_I103 deletion in two patients), and EGFR (A289V in one patient) (Figs. 1B through 1D and 2A and 2B). Oncogenic gene rearrangements emerged after treatment with adagrasib in three patients with colorectal cancer, including CCDC6-RET (Fig. 1B) and EML4-ALK (Fig. 2A). Numerous oncogenic gene rearrangements were detected in a single patient with colorectal cancer (Patient 6), including distinct fusions involving RAF1, BRAF, and FGFR3 (Fig. 2B and Fig. S3). In several patients, multiple mechanisms of resistance were identified simultaneously through ctDNA analysis (Figs. 1B through 1D and 2A and 2B).
In two patients, high-level focal amplifications of the KRASG12C allele were identified in the adagrasib-resistant samples with no other identifiable resistance mechanisms (Fig. 2C and Fig. S4). Acquired MET amplification was the only potential genomic mechanism of adagrasib resistance identified in two other patients (Fig. 2D and Fig. S5). Lastly, in 2 of 10 patients (9 with NSCLC and 1 with colorectal cancer) from whom a repeat tissue-biopsy sample was obtained at the time of disease progression during adagrasib treatment, histologic transformation from adenocarcinoma to squamous-cell carcinoma was observed without any identifiable genomic mechanisms of resistance (Fig. 2E and Fig. S6).
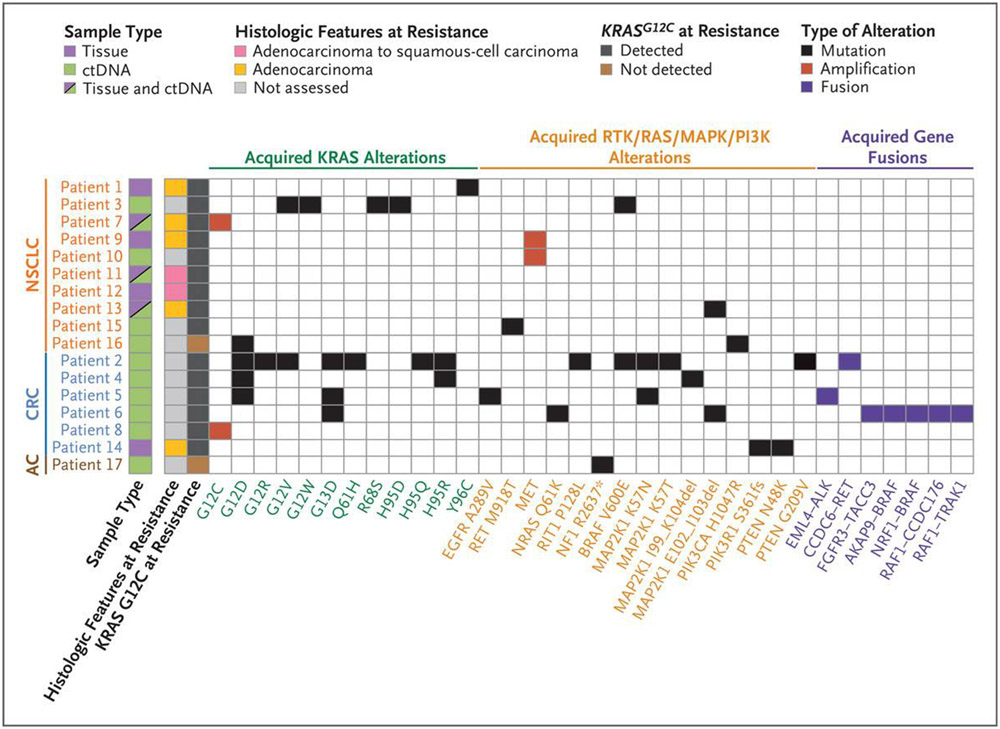
Observed mechanisms of resistance to adagrasib are summarized in Figure 3. Among the 17 of 38 patients with putative resistance mechanisms identified, 9 (53%) had at least one acquired KRAS mutation or amplification. Furthermore, putative resistance mechanisms that did not directly involve KRAS but were related to the RAS signaling pathway were observed in 12 of the 17 patients (71%). In addition, 7 of the 17 patients (41%) had more than one concurrent potential resistance mechanism identified. In our cohort, multiple resistance mechanisms appeared to be more common in colorectal cancer; among patients with available ctDNA sequencing, 4 of 5 patients with colorectal cancer and 2 of 7 patients with NSCLC had multiple resistance alterations, although this difference was not significant.
Figure 3. Summary of Putative Mechanisms of Acquired Resistance to Adagrasib Treatment.
Of 38 patients with KRASG12C-mutant cancers, 17 had at least one putative resistance mechanism. A summary of the genomic and histologic mechanisms of resistance among these 17 patients is shown in the comutation plot, with each row indicating a patient and each column indicating a specific acquired alteration. Seven patients had more than one putative resistance mechanism identified. AC denotes appendiceal cancer, CRC colorectal cancer, ctDNA circulating tumor DNA, and NSCLC non–small-cell lung cancer.
ROLE OF DRUG-BINDING SITE MUTATIONS
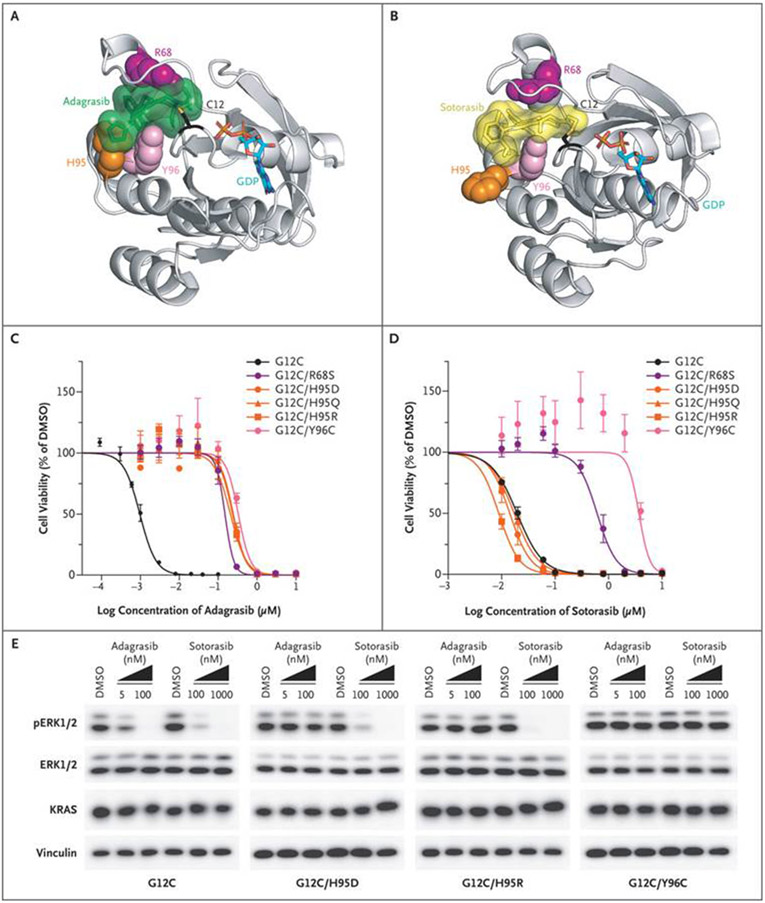
Given the required covalent mode of adagrasib binding to the cysteine at codon 12 in the KRASG12C protein, the observed KRAS G12D/R/V/W mutations would not be expected to bind adagrasib or to be inhibited by the drug. We also identified the KRAS R68S, H95D/Q/R, and Y96C mutations within the switch II pocket where both adagrasib and sotorasib bind (Fig. 4A and 4B). Analysis of these residues in the x-ray crystallographic structure of adagrasib bound to KRASG12C (6UT0)21 revealed that mutations at these positions could result in disruption of noncovalent binding interactions of the drug (Fig. 4A and Figs. S7 through S9). By comparison, mapping of these same residues onto the x-ray crystallographic structure of sotorasib bound to the KRASG12C protein (6OIM)6 suggested strong interactions of R68 and Y96 with sotorasib but probably weaker interactions with the H95 residue, although additional studies would be needed to quantify the strength of these interactions (Fig. 4B).
Figure 4. Resistance to Adagrasib or Sotorasib Conferred by Acquired Missense Mutations in the KRASG12C Drug-Binding Site.
Putative second-site resistance mutations were mapped onto crystal structures of KRASG12C bound to adagrasib (Panel A) or sotorasib (Panel B). Shown are representative dose–response curves for Ba/F3 cells that were grown in the absence of interleukin-3, that expressed the indicated KRASG12C variants, and that were treated for 5 days with adagrasib (Panel C) or sotorasib (Panel D). The curves represent means ±SE from at least three independent experiments. Immunoblots (Panel E) are shown for extracellular signal-regulated kinase (ERK) phosphorylation levels (pERK1/2 T202/Y204) and KRAS total protein levels from Ba/F3 cells that expressed KRASG12C or three clinically observed resistance mutations: KRASG12C/H95D, KRASG12C/H95R, and KRASG12C/Y96C. An immunoblot for vinculin is shown as a loading control. Six-hour adagrasib or sotorasib treatment of cells that expressed sensitive but not insensitive alleles conferred suppression of pERK and upward band shift of KRAS due to covalent binding of the inhibitors. GDP denotes guanosine-5′-diphosphate, and DMSO dimethyl sulfoxide.
To investigate the effect of these switch II pocket mutations in KRASG12C on adagrasib and sotorasib sensitivity, we generated double-mutant alleles of KRASG12C with the clinically observed second-site mutations, including R68S, H95D, H95Q, H95R, and Y96C. We exogenously expressed these double-mutant alleles or the control KRASG12C allele in the Ba/F3 cell line, a well-established system for studying oncogene dependence (Fig. S10).18 Ba/F3 cells that express KRASG12C are highly sensitive to treatment with either adagrasib or sotorasib and show dose-dependent inhibition of downstream KRAS effector signaling (Fig. 4C through 4E). Expression of all clinically observed switch II pocket mutations (R68S, H95D, H95Q, H95R, and Y96C) conferred marked resistance to adagrasib in Ba/F3 cells (Fig. 4C). Similarly, expression of the G12C/R68S and G12C/Y96C double mutants mediated resistance to sotorasib; in contrast, the G12C/H95D, G12C/H95Q, and G12C/H95R mutants remained sensitive to sotorasib (Fig. 4D). Biochemical analysis of the response to adagrasib in Ba/F3 cells showed that the R68S, H95D/Q/R, and Y96C second-site mutations in KRASG12C blocked drug binding, as indicated by the absence of KRAS band shift mediated by covalent drug binding, and completely prevented the drug-mediated suppression of RAS–MAPK signaling, as measured by sustained phosphorylated extracellular signal-regulated kinase levels (Fig. 4E and Fig. S11). On the other hand, the R68S and Y96C but not the H95D/Q/R mutations mediated resistance to sotorasib in these biochemical assays.
Collectively, mutations that disrupt covalent or potentially noncovalent drug binding can account for clinical resistance to KRASG12C inhibition. Moreover, differential drug-binding mechanisms between adagrasib and sotorasib can lead to the emergence of drug-specific resistance mutations.
RESISTANCE MUTATIONS REVEALED BY DEEP MUTATIONAL SCANNING
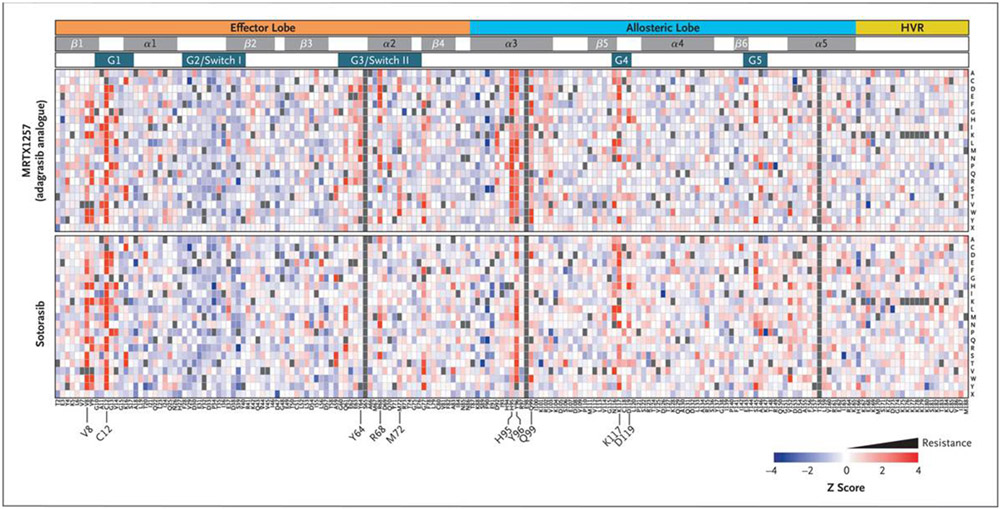
We sought to define all possible secondary mutations in KRASG12C that can confer resistance to KRASG12C inhibitors. Using a lentiviral library that encoded nearly all possible alleles with a single amino acid substitution within the KRASG12C backbone allele, we performed a positive-selection screen in the Ba/F3 cell line for mutations that cause resistance to two different KRASG12C inhibitors: sotorasib and MRTX1257, a compound highly related to adagrasib with an identical mode of binding (Figs. S12 through S14).7 Drug-resistance mutations were defined as those that enabled cell survival during treatment with a KRASG12C inhibitor (Fig. 5 and Table S2). Multiple mutations that were identified in the screen at codons 12, 68, 95, and 96 conferred strong resistance to MRTX1257. Additional strong MRTX1257-resistance mutations were detected at codons 8, 9, 64, 99, and 117. All clinically observed adagrasib-resistance mutations in KRASG12C scored as strong resistance mutations in the MRTX1257 screen (Fig. 5 and Fig. S15). In the sotorasib screen, numerous strong resistance mutations were observed at codons 8, 9, 12, 96, and 117.
Figure 5. Landscape of On-Target Resistance Mutations to KRASG12C Inhibitors Revealed by Deep Mutational Scanning.
A heatmap depicts the results of a positive-selection screen in Ba/F3 cells for second-site missense mutations in KRASG12C that confer resistance to the KRASG12C inhibitors MRTX1257 (a close analogue of adagrasib) or sotorasib. Relative allele abundance is shown as a z score comparing the end point of the screen for each treatment group to the pretreatment allelic representation in the library, with red indicating enrichment of putative resistance variants. Columns represent the amino acid position within the KRASG12C allele, and each row represents 1 of 20 possible amino acids or the stop codon (X). The structural motifs within the KRAS protein are indicated in the tracks above the heatmap. Gray coloration indicates alleles that were not included in the screening library. HVR denotes hypervariable region.
Patterns of MRTX1257 and sotorasib resistance mutations were mapped onto existing structural representations of KRASG12C bound to adagrasib or sotorasib, respectively (Table S3), which revealed multiple resistance mutations surrounding the drug-binding pockets of each compound. A comparison of resistance patterns between MRTX1257 and sotorasib showed generally strong concordance, although several drug-specific resistance mutations were identified. In addition to differential resistance mutations observed at H95, a greater number of MRTX1257-specific resistance mutations were observed at codons 72 and 99, a finding that is probably related to differential drug-binding interactions between MRTX1257 and sotorasib with these residues (Fig. S16A through S16D). Although most second-site resistance mutations that were observed in the screen occurred at residues involved in drug binding, several mutations that were associated with resistance to both drugs were detected in amino acids located outside the drug-binding pockets of adagrasib and sotorasib, including known oncogenic mutations at codons 13, 59, 61, 117, and 146 that impede GTP hydrolysis or promote GDP-to-GTP nucleotide exchange (Fig. 5).
In addition to the previously validated R68S, H95D/Q/R, and Y96C resistance mutations (Fig. 4C through 4E), we validated the screening results for 17 additional mutations at residues that confer resistance to one or both KRASG12C inhibitors (Table S4). We evaluated adagrasib and sotorasib sensitivity in Ba/F3 cells that expressed each of these alleles (Figs. S17 and S18 and Tables S5 and S6). To enable comparison of drug resistance across different alleles and drugs, we defined a relative resistance score for each allele according to the degree of resistance conferred by that allele relative to the G12R mutant (Fig. S19). As expected, the Y96C drug-binding resistance mutation showed high-level resistance to both inhibitors, whereas the H95D, H95Q, and H95R mutations caused resistance to adagrasib but not to sotorasib. For both adagrasib and sotorasib, drug-resistance mutations at residues outside of the drug-binding pocket and known to be involved in enhanced nucleotide exchange, including G13D, A59S, K117N, and A146P, conferred more modest resistance than the G12R and Y96C drug-binding mutations.
In summary, deep mutational scanning has defined the landscape of KRAS mutations that cause resistance to KRASG12C inhibitors. These studies identified distinct mechanistic classes of resistance mutations as well as drug-specific resistance patterns.
DISCUSSION
We performed DNA sequencing of tumor-biopsy samples or ctDNA from 38 patients with acquired resistance to adagrasib and described putative mechanisms of resistance for 17 patients (45%) that can be classified into three main categories. The first category includes secondary mutations or amplifications in KRAS, the target of adagrasib; the second, alternative oncogenic alterations that activate the RTK–RAS signaling pathway but do not directly alter KRAS itself; and the third, histologic transformation from lung adenocarcinoma to squamous-cell carcinoma. In contrast to some targeted therapies for which dominant mechanisms of resistance have been observed, such as BCR-ABL mutations that cause resistance to imatinib in chronic myeloid leukemia,22 KRASG12C-mutant cancers appear to evolve diverse mechanisms of resistance to adagrasib. In the majority of patients for whom we identified a putative resistance mechanism, at least one of these mechanisms did not involve the KRAS gene itself (14 of 17 patients [82%]). Moreover, 7 of 17 patients (41%) had more than one concurrent potential resistance mechanism identified, which suggests that irreversible covalent inhibition of KRASG12C leads to strong selective pressures and convergent evolution of multiple distinct mechanisms of resistance.
Acquired mutations in KRAS were observed in 7 patients, most notably at codons 12, 13, 61, 68, 95, and 96. Five patients acquired codon 12 KRAS mutations that prevent adagrasib binding, and other secondary mutations in the switch II drug-binding pocket were observed in 4 patients (11% of the entire cohort) at the R68, H95, and Y96 positions. Although the Y96C mutation appeared to be a clonal resistance event, the R68S and H95D/Q/R secondary KRAS mutations were identified in ctDNA at relatively low variant allele frequency as compared with that of the KRASG12C allele and occurred in the presence of other concurrent resistance mutations, which suggests subclonality or heterogeneity of these mutations across each patient’s burden of disease. In recent work, a case was described of an acquired Y96D switch II pocket mutation identified at a low variant allele frequency in ctDNA at the time of acquired resistance to adagrasib.23 In our experimental studies, the R68S and Y96C mutations confer resistance to both adagrasib and sotorasib. In contrast, the H95D, H95Q, or H95R adagrasib-resistance mutations do not confer in vitro resistance to sotorasib. It is notable that the H95 adagrasib-specific resistance mutations occurred in a small number of patients (3 of 38), and each coevolved with multiple other alterations that would confer strong resistance to both adagrasib and sotorasib. Moreover, in contrast to EGFR-mutant lung cancer, in which discovery of the dominant T790M mutation as an on-target resistance mechanism for early-generation EGFR inhibitors led to the development of the mutant-selective inhibitor osimertinib, the diversity of KRAS mutations that have emerged in response to KRASG12C inhibition may pose an added challenge in the development of effective next-generation inhibitors.
We also performed deep mutational scanning screens of KRASG12C missense variants and systematically defined an atlas of KRAS mutations that confer resistance to multiple KRASG12C inhibitors. These studies defined two apparent major mutational mechanisms of resistance directly involving KRASG12C. As observed clinically, mutations within the drug-binding pocket also caused high-level resistance within our variant screening results and validation assays. Furthermore, we identified additional second-site resistance mutations at oncogenic hotspots that cause decreased GTP hydrolysis (e.g., G13D and Q61R) or enhanced GDP-to-GTP nucleotide exchange (e.g., G13D, A59S, and A146P). These resistance mutations probably increase the fraction of KRAS protein in the active GTP-bound form that does not bind to the drug.4 We have not yet observed these nucleotide-exchange resistance mutations in clinical samples, and whether they will cause resistance at clinically achievable concentrations of the drug remains to be determined.
Beyond KRAS mutations, numerous additional genomic alterations in the RTK–RAS signaling pathway were identified in 14 of 38 patients (37% of the entire cohort). In our cohort, multiple genomic alterations within the same patient, as well as acquired gene fusions, appeared to be more prevalent in colorectal cancer than in NSCLC. It is possible that underlying differences in genomic instability or DNA damage-response mechanisms between colorectal cancer and NSCLC at baseline or in response to KRAS inhibition may account for this observation. Whether the more modest activity of KRASG12C inhibitors in colorectal cancer is due to the earlier emergence of multiple acquired resistance mutations or to the presence of adaptive signaling pathways at baseline will require further investigation. The coevolution of multiple RTK–RAS–MAPK pathway alterations as mechanisms of adagrasib resistance strongly supports the rationale for ongoing clinical trials of adagrasib (ClinicalTrials.gov number, NCT04330664) or sotorasib (NCT04185883) in combination with inhibitors of RTKs or SHP2 (PTPN11) to address these adaptive resistance mechanisms.
In two patients with NSCLC for whom we were not able to identify a genomic resistance mechanism, we observed histologic transformation from adenocarcinoma to squamous-cell carcinoma, which suggests that nongenetic mechanisms of resistance to KRASG12C inhibition may occur, similar to those observed with other targeted therapies in lung cancer.24-26 In both patients, deep targeted panel sequencing was performed on the tissue sample at the time of resistance and showed sufficient sensitivity to detect the presence of multiple pretreatment baseline mutations in the sample, including the KRASG12C mutation. Furthermore, one of these patients (Patient 11) also had ctDNA sequencing at the time of resistance that identified the KRASG12C mutation as well as a TP53 alteration also present in the pretreatment sample. These data suggest that the tumor and ctDNA sequencing performed on these specimens did not miss a major known molecular driver of adagrasib resistance, but it remains possible that more extensive genomic sequencing of these samples could identify other acquired alterations that may have contributed to resistance. Moreover, investigation of the genetic or epigenetic features that drive the observed histologic conversion from adenocarcinoma to squamous-cell carcinoma may elucidate the mechanism by which this transformation causes resistance to KRASG12C inhibition.
The heterogeneous nature of the tumor and ctDNA sequencing platforms that were used to characterize acquired resistance is a limitation of this study. Future studies of resistance mechanisms to KRAS inhibitors will benefit from using consistent sequencing platforms, as well as analysis of matched pretreatment and postprogression biopsy samples to better define both genetic and nongenetic mechanisms of resistance. This study focused on acquired resistance mechanisms in patients treated with adagrasib and did not evaluate patients treated with sotorasib. Mechanisms of acquired resistance to sotorasib are currently unknown; thus, direct comparison between clinically observed adagrasib and sotorasib resistance mechanisms is not yet possible. That said, our deep mutational scanning experiments have highlighted common and contrasting patterns of KRAS mutations that confer resistance to different KRASG12C inhibitors and will provide a reference resource for oncologists to interpret additional KRAS mutations that emerge in the context of adagrasib and sotorasib therapy.
In aggregate, these data show that a diversity of on-target and off-target mechanisms can confer resistance to KRASG12C inhibitors and support the need for development of additional KRAS inhibitors with alternative modes of binding and different allele specificities. Moreover, development of effective combination therapy regimens will be required to fully combat resistance mechanisms that emerge during treatment with adagrasib or sotorasib.
Supplementary Material
Acknowledgments
Supported in part by Mirati Therapeutics and by grants from the Lustgarten Foundation (to Drs. Aguirre and Wolpin), the Dana–Farber Cancer Institute Hale Center for Pancreatic Cancer Research (to Drs. Aguirre and Wolpin), the Doris Duke Charitable Foundation (to Dr. Aguirre), the Pancreatic Cancer Action Network (to Drs. Aguirre and Wolpin), the National Cancer Institute (K08 CA218420-02, to Dr. Aguirre; P50 CA127003, to Drs. Aguirre and Wolpin; U01 CA210171, to Dr. Wolpin; and 1R01CA230745-01 and 1R01CA230267-01A1, to Dr. Lito), Stand Up to Cancer (to Dr. Wolpin), the Noble Effort Fund (to Dr. Wolpin), the Wexler Family Fund (to Dr. Wolpin), Promises for Purple (to Dr. Wolpin), the Bob Parsons Fund (to Dr. Wolpin), the Pew Charitable Trusts (to Dr. Lito), the Damon Runyon Cancer Research Foundation (to Dr. Lito), the Josie Robertson Investigator Program at Memorial Sloan Kettering Cancer Center (to Dr. Lito), the Mark Foundation for Cancer Research (19-029 MIA, to Dr. Jänne), and the American Cancer Society (CRP-17-111-01-CDD, to Dr. Jänne).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank Lesli Kiedrowski and Janice Patterson from Guardant Health for helpful discussions on genomic results.
Contributor Information
M.M. Awad, Dana–Farber Cancer Institute, Boston, Massachusetts
S. Liu, Dana–Farber Cancer Institute, Boston, and Broad Institute of MIT and Harvard, Cambridge, Massachusetts
I.I. Rybkin, Henry Ford Cancer Institute, Detroit
K.C. Arbour, Memorial Sloan Kettering Cancer Center, New York
J. Dilly, Dana–Farber Cancer Institute, Boston, Massachusetts
V.W. Zhu, Chao Family Comprehensive Cancer Center, University of California, Irvine, School of Medicine, Orange, California
M.L. Johnson, Sarah Cannon Research Institute, Tennessee Oncology/OneOncology, Nashville
R.S. Heist, Massachusetts General Hospital, Boston, Massachusetts
T. Patil, University of Colorado, Aurora
G.J. Riely, Memorial Sloan Kettering Cancer Center, New York
J.O. Jacobson, Dana–Farber Cancer Institute, Boston, Massachusetts
X. Yang, Broad Institute of MIT and Harvard, Cambridge, Massachusetts
N.S. Persky, Broad Institute of MIT and Harvard, Cambridge, Massachusetts
D.E. Root, Broad Institute of MIT and Harvard, Cambridge, Massachusetts
K.E. Lowder, Dana–Farber Cancer Institute, Boston, Massachusetts
H. Feng, Dana–Farber Cancer Institute, Boston, Massachusetts
S.S. Zhang, Chao Family Comprehensive Cancer Center, University of California, Irvine, School of Medicine, Orange, California
K.M. Haigis, Dana–Farber Cancer Institute, Boston, and Broad Institute of MIT and Harvard, Cambridge, Massachusetts
Y.P. Hung, Massachusetts General Hospital, Boston, Massachusetts
L.M. Sholl, Brigham and Women’s Hospital, Boston, Massachusetts
B.M. Wolpin, Dana–Farber Cancer Institute, Boston, Massachusetts
J. Wiese, Boundless Bio, La Jolla, California
J. Christiansen, Boundless Bio, La Jolla, California
J. Lee, Foundation Medicine, Cambridge, Massachusetts
A.B. Schrock, Foundation Medicine, Cambridge, Massachusetts
L.P. Lim, Resolution Bioscience, Kirkland, WA
K. Garg, Resolution Bioscience, Kirkland, WA
M. Li, Reso-lution Bioscience, Kirkland, WA
L.D. Engstrom, Mirati Therapeutics, San Diego, California
L. Waters, Mirati Therapeutics, San Diego, California
J.D. Lawson, Mirati Therapeutics, San Diego, California
P. Olson, Mirati Therapeutics, San Diego, California
P. Lito, Memorial Sloan Kettering Cancer Center, New York
S.-H.I. Ou, Chao Family Comprehensive Cancer Center, University of California, Irvine, School of Medicine, Orange, California
J.G. Christensen, Mirati Therapeutics, San Diego, California
P.A. Jänne, Dana–Farber Cancer Institute, Boston, Massachusetts
A.J. Aguirre, Dana–Farber Cancer Institute, Brigham and Women’s Hospital, Boston, and Broad Institute of MIT and Harvard, Cambridge, Massachusetts
REFERENCES
- 1.Papke B, Der CJ. Drugging RAS: know the enemy. Science 2017;355:1158–63. [DOI] [PubMed] [Google Scholar]
- 2.Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 2020;19:533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013;503:548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lito P, Solomon M, Li L-S, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016;351:604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janes MR, Zhang J, Li L-S, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 2018; 172(3):578.e17–589.e17. [DOI] [PubMed] [Google Scholar]
- 6.Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217–23. [DOI] [PubMed] [Google Scholar]
- 7.Hallin J, Engstrom LD, Hargis L, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D, Xue JY, Lito P. Targeting KRAS(G12C): from inhibitory mechanism to modulation of antitumor effects in patients. Cell 2020;183:850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRASG12C somatic mutations across race, sex, and cancer type. N Engl J Med 2021;384:185–7. [DOI] [PubMed] [Google Scholar]
- 10.Hong DS, Fakih MG, Strickler JH, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med 2020; 383:1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riely GJ, Ou S-HI, Rybkin I, et al. 99O_PR KRYSTAL-1: activity and preliminary pharmacodynamic (PD) analysis of adagrasib (MRTX849) in patients (Pts) with advanced non–small cell lung cancer (NSCLC) harboring KRASG12C mutation. J Thorac Oncol 2021;16:Suppl:S751–S752. [Google Scholar]
- 12.Janne PA, Rybkin I, Spira AI, et al. KRYSTAL-1: activity and safety of adagrasib (MRTX849) in advanced/metastatic non–small-cell lung cancer (NSCLC) harboring KRAS G12C mutation. Presented at the 32nd EORTC-NCI-AACR Symposium, virtual meeting, October 24–25, 2020:LBA3. abstract. [Google Scholar]
- 13.Johnson ML, Ou SI, Barve M, et al. KRYSTAL-1: activity and safety of adagrasib (MRTX849) in patients with colorectal cancer (CRC) and other solid tumors harboring a KRAS G12C mutation. Presented at the 32nd EORTC-NCI-AACR Symposium, virtual meeting, October 24–25, 2020:LBA4. abstract. [Google Scholar]
- 14.Skoulidis F, Li BT, Dy GK, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med 2021;384:2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan MB, Fece de la Cruz F, Phat S, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRASG12C inhibition. Clin Cancer Res 2020;26:1633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue JY, Zhao Y, Aronowitz J, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020;577:421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amodio V, Yaeger R, Arcella P, et al. EGFR blockade reverts resistance to KRASG12C inhibition in colorectal cancer. Cancer Discov 2020;10:1129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warmuth M, Kim S, Gu X, Xia G, Adrián F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol 2007;19:55–60. [DOI] [PubMed] [Google Scholar]
- 19.Starita LM, Fields S. Deep mutational scanning: a highly parallel method to measure the effects of mutation on protein function. Cold Spring Harb Protoc 2015;2015:711–4. [DOI] [PubMed] [Google Scholar]
- 20.Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods 2014;11:801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fell JB, Fischer JP, Baer BR, et al. Identification of the clinical development candidate MRTX849, a covalent KRASG12C inhibitor for the treatment of cancer. J Med Chem 2020;63:6679–93. [DOI] [PubMed] [Google Scholar]
- 22.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 2001;293:876–80. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka N, Lin JJ, Li C, et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov 2021. April 6 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou S, Han X, Ji H. Squamous transition of lung adenocarcinoma and drug resistance. Trends Cancer 2016;2:463–6. [DOI] [PubMed] [Google Scholar]
- 25.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 2020;26:2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.