Abstract
Using a strategy to clone large genomic sequences containing repetitive elements from the infectious yeast Candida dubliniensis, the three unrelated sequences Cd1, Cd24, and Cd25, with respective molecular sizes of 15,500, 10,000, and 16,000 bp, were cloned and analyzed for their efficacy as DNA fingerprinting probes. Each generated a complex Southern blot hybridization pattern with endonuclease-digested genomic DNA. Cd1 generated an extremely variable pattern that contained all of the bands of the pattern generated by the repeat element RPS of Candida albicans. We demonstrated that Cd1 does not contain RPS but does contain a repeat element associated with RPS throughout the C. dubliniensis genome. The Cd1 pattern was the least stable over time both in vitro and in vivo and for that reason proved most effective in assessing microevolution. Cd24, which did not exhibit microevolution in vitro, was highly variable in vivo, suggesting in vivo-dependent microevolution. Cd25 was deemed the best probe for broad epidemiological studies, since it was the most stable over time, was the only truly C. dubliniensis-specific probe of the three, generated the most complex pattern, was distributed throughout all C. dubliniensis chromosomes, and separated a worldwide collection of 57 C. dubliniensis isolates into two distinct groups. The presence of a species-specific repetitive element in Cd25 adds weight to the already substantial evidence that C. dubliniensis represents a bona fide species.
Candida albicans is the most common yeast species carried as a commensal by healthy individuals (42) and the most common species isolated from yeast infections (21, 44). The next most common species include Candida tropicalis, Candida glabrata, Candida parapsilosis, and Candida krusei (9, 25). All of these species are readily distinguishable from C. albicans both phenotypically and genotypically. However, over the years there have been an ever-increasing number of reports of atypical strains of C. albicans that have been distinguished primarily by unusual fingerprint patterns (3, 18, 19, 22, 27, 45). Based upon phenotypic and genotypic differences, Coleman, Sullivan, and coworkers in Dublin, Ireland, were the first to argue that a majority of these atypical C. albicans isolates represented a separate species, which they named Candida dubliniensis (46).
Because strains of the putative species C. dubliniensis exhibited the two major phenotypic characteristics used to identify C. albicans, chlamydospore and true hypha formation (46), and because they hybridized with putative C. albicans-specific fingerprinting probes (3, 22, 45, 46), there has been some hesitation in considering them members of a bona fide species separate from C. albicans. However, the fact that several DNA typing methods, including species-specific probes (3, 22, 45, 46), multilocus enzyme electrophoresis (3, 12, 27, 47), randomly amplified polymorphic DNA (45, 46), electrophoretic karyotyping (46), oligonucleotide fingerprinting probes (45, 46), microsatellite DNA analysis (20), and restriction fragment length polymorphism without probes (46), discriminated these strains from typical C. albicans strains suggested that they indeed represent a separate species. For this reason, several methods based on phenotype have recently been developed to distinguish isolates of C. dubliniensis from isolates of C. albicans, including growth at 42°C (7, 33, 46, 47), β-glucosidase activity (3, 33, 47), colony color on CHROMagar (7, 33), and fluorescence after growth on methyl blue-Sabouraud agar (33). However, none of these methods appear to definitively identify all C. dubliniensis isolates.
If C. dubliniensis is a bona fide species, it should contain species-specific repetitive elements dispersed throughout the genome, as has been demonstrated for C. albicans (28, 31, 40), C. tropicalis (11, 41), C. glabrata (15), C. krusei (4) and C. parapsilosis (7a). Demonstration of such species-specific sequences in the C. dubliniensis genome would not only reinforce the arguments by Sullivan and Coleman (48) that it is a bona fide species but also provide the basis for a DNA fingerprinting system (39). We therefore screened for and cloned three DNA fingerprinting probes that included moderately repetitive elements. One of them, Cd25, not only proved to be effective in broad epidemiological studies but also contained one or more C. dubliniensis-specific sequences dispersed throughout the genome, supporting the argument that C. dubliniensis is a distinct species.
MATERIALS AND METHODS
Strains and growth conditions.
The C. dubliniensis strains used in this study are listed in Table 1. Each isolate was stored at room temperature on a YPD agar slant (2% glucose, 2% Bacto Peptone, 1% yeast extract, 2% agar) in a capped tube. For DNA extraction, the cells were transferred to an Erlenmeyer flask containing YPD medium, grown for 24 h at 25°C, and harvested. To assess the stability of Southern blot hybridization patterns over many generations, cells from a single clone were inoculated into YPD medium at an initial concentration of 5 × 104 per ml and grown to stationary phase (∼24 h at 37°C). Stationary-phase cells were diluted in fresh medium, and the process was repeated. After 200 generations, the cells were harvested and plated on agar at low density and cells from nine colonies of each strain were removed and stored on YPD slants in capped tubes for further analysis.
TABLE 1.
History of C. dubliniensis isolates used in the characterization of the cloned fingerprinting probes
| Patient | Isolate | Date isolated (mo-day-yr) | Body location (pathologya) | Place of collectionb | Patient | Isolate | Date isolated (mo-day-yr) | Body location (pathology) | Place of collectionb | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P30 | 10-26-93 | Oral (HIV+) | Lausanne, Switzerland | 30 | ANSA3 | 94–95 | Oral (HIV+) | UHA, Antwerp, Belgium | |
| P5 | 11-23-92 | 31 | ANSA5 | 95–95 | Oral (HIV+) | UHA, Antwerp, Belgium | ||||
| 2 | P14 | 03-12-93 | Oral (HIV+) | Lausanne, Switzerland | 32 | ANSA9 | 94–95 | Oral (HIV+) | UHA, Antwerp, Belgium | |
| Co4 | 09-26-93 | 33 | ANSA26 | 94–95 | Oral (HIV+) | UHA, Antwerp, Belgium | ||||
| 3 | Co5 | 09-29-93 | Oral (HIV+) | Lausanne, Switzerland | 34 | ANSA27 | 94–95 | Oral (HIV+) | UHA, Antwerp, Belgium | |
| 4 | d959 | 1986 | Oral (DA) | Valencia, Spain | 35 | ANSA28 | 94–95 | Oral (HIV+) | UHA, Antwerp, Belgium | |
| d961 | 1986 | 36 | d930936 | 1993 | Oral (HIV+) | JWU, Frankfurt, Germany | ||||
| 5 | d1046 | 1986 | Oral (DA) | Valencia, Spain | 37 | d930953 | 1993 | Oral (HIV+) | JWU, Frankfurt, Germany | |
| 6 | d1003 | 1986 | Oral (DA) | Valencia, Spain | 38 | d931021 | 1993 | Oral (HIV+) | JWU, Frankfurt, Germany | |
| 7 | d950 | 1986 | Oral (DA) | Madrid, Spain | 39 | B71507 | 1992 | Oral (HIV+) | TCDH, Dublin, Ireland | |
| 8 | d952 | 1986 | Oral (DA) | Madrid, Spain | 40 | NCPF3949 | 88–94 | Oral (HIV+) | TCDH, Dublin, Ireland | |
| 9 | d895 | 06-12-85 | Sputum (Hem) | RFH, London, UK | 41 | B69819-1 | 1992 | Oral (HIV+) | HSL, Paris, France | |
| d872 | 06-18-85 | Fecal (Hem) | 42 | d1419-2 | 1993 | Oral (HIV+) | CMI, Montpellier, France | |||
| d879 | 06-19-85 | Oral (Hem) | 43 | d1558 | 1993 | Oral (HIV+) | CMI, Montpellier, France | |||
| 10 | d457 | 11-26-85 | Fecal (Hem) | RFH, London, UK | 44 | d941841 | 1994 | Oral (Hem) | UH, Nijmegen, Netherlands | |
| d251 | 11-26-85 | 45 | d930822 | 1993 | Oral (HIV+) | VAH, San Antonio, Tex. | ||||
| 11 | d456 | 1986 | (Hem) | RFH, London, UK | 46 | d109534 | 05-30-95 | (Cancer) | MH, Bakersfield, Calif. | |
| 12 | d681 | 12-04-86 | Oral (Hem) | RFH, London, UK | 47 | d1425638 | 12-19-95 | (Cancer) | BIMC, New York, N.Y. | |
| d710 | 04-14-86 | Oral (Hem) | 48 | d81217 | 1981 | Oral (NA) | BMC, Houston, Tex. | |||
| d675 | 04-22-86 | Fecal (Hem | 49 | d126423 | 05-23-95 | (Cancer) | UIHC, Iowa City, Iowa | |||
| d632 | 04-24-86 | Oral (Hem) | 50 | 10-25-94 | Oral (HIV+) | |||||
| 13 | d88029 | 1988 | Oral (HIV+) | ULMS, Leicester, UK | d166 | 04-02-95 | Asymptomatic | |||
| d90006 | 1990 | d167 | 04-02-95 | Asymptomatic | ||||||
| 14 | d89014 | 1989 | Oral (HIV+) | ULMS, Leicester, UK | d168 | 04-02-95 | Asymptomatic | |||
| d90013 | 1990 | Oral (HIV+) | d169 | 04-02-95 | Asymptomatic | |||||
| d90015 | 1990 | Oral (HIV+) | d170 | 04-02-95 | Asymptomatic | |||||
| d90033 | 1990 | Oral (HIV+) | d171 | 04-02-95 | Asymptomatic | |||||
| 15 | d78008 | 1978 | Oral (Healthy) | ULMS, Leicester, UK | d173 | 04-02-95 | Asymptomatic | UIHC, Iowa City, Iowa | ||
| 16 | d81060 | 1981 | Oral (Healthy) | ULMS, Leicester, UK | d174 | 06-21-95 | Asymptomatic | |||
| 17 | d88014 | 1988 | Oral (HIV+) | ULMS, Leicester, UK | d176 | 06-21-95 | Asymptomatic | |||
| 18 | d931111 | 1993 | Oral (HIV+) | CWHCT, London, UK | d178 | 10-04-95 | Oral thrush | |||
| 19 | d931113 | 1993 | Oral (HIV+) | CWHCT, London, UK | d180 | 10-04-95 | Oral thrush | |||
| 20 | d11 | 1985 | Oral (ICU) | Bangor, UK | d181 | 10-04-95 | Oral thrush | |||
| 21 | d514 | 1985 | Vaginal (ICU) | Bangor, UK | d182 | 10-04-95 | Oral thrush | |||
| 22 | d82006 | 1982 | Oral (Dental) | USDS, Sheffield, UK | d184 | 10-04-95 | Oral thrush | |||
| 23 | d73089 | 1973 | Oral (Cardiac) | LGI, Leeds, UK | 51 | d930664 | 1992 | Oral (HIV+) | LSP, Québec, Canada | |
| 24 | d75043 | 1975 | Oral (Diabetes) | LGI, Leeds, UK | 52 | d930666 | 1992 | Oral (HIV+) | LSP, Québec, Canada | |
| 25 | d75004 | 1975 | Oral (Diabetes) | LGI, Leeds, UK | 53 | M1 | 89–92 | Oral (HIV+) | FH, Victoria, Australia | |
| 26 | d920710 | 1992 | Oral (HIV+) | UCL, Brussels, Belgium | 54 | M2 | 89–92 | Oral (HIV+) | FH, Victoria, Australia | |
| 27 | d930713 | 1993 | Oral (HIV+) | UCL, Brussels, Belgium | 55 | M3 | 89–92 | Oral (HIV+) | FH, Victoria, Australia | |
| 28 | d932634 | 1994 | Oral (GU) | GC, Turnhout, Belgium | 56 | M4 | 89–92 | Oral (HIV+) | FH, Victoria, Australia | |
| 29 | d940613 | 1994 | Oral (GU) | GC, Antwerp, Belgium | 57 | M6 | 89–92 | Oral (HIV+) | FH, Victoria, Australia |
HIV+, HIV positive; DA, drug addict; Hem, hematology; ICU, intensive care unit; GU, gynecology unit; NA, not available.
RFH, Royal Free Hospital; ULMS, University of Leicester Medical School; CWHCT, Chelsea & Westminster Health Care Trust; USDS, University of Sheffield Dental School; LGI, Leeds General Infirmary; UCL, Université Catholique de Louvain; GC, Gynecology clinic; UHA, University Hospital of Antwerp; JWU, Johann Wolfgang Universität; TCDH, Trinity College Dental Hospital; HSL, Hôpital Saint Louis; CMI Clinique des Maladies Infectieuses; UH, University Hospital; VAH, VA Hospital; MH, Mercy Hospital; BIMC, Beth Israel Medical Center; BMC, Baylor Medical Center; UIHC, University of Iowa Hospital and Clinics; LSP, Laboratoire de Santé Publique; FH, Fairfield Hospital; UK, United Kingdom.
Cloning of genomic fragments containing moderately repetitive sequences.
Genomic DNA from C. dubliniensis d1558 (Table 1) was used to construct a λ phage genomic library as previously described for C. albicans (16, 28, 31, 40, 41). Briefly, the library was constructed in phage λ EMBL3 according to established protocols (29) with 9- to 23-kb fragments from a Sau3AI partial digest of genomic DNA size fractionated in a sucrose gradient (16). The library was amplified in Escherichia coli P2-392 and plated at a density of 10,000 plaques per 150-mm-diameter petri dish. Duplicate nitrocellulose filters were prepared from each plate (29) and incubated at 65°C for 20 min in a solution containing 1% bovine serum albumin, 7% sodium dodecyl sulfate (SDS), 0.5 M NaH2PO4 (pH 7.0), and 1 mM EDTA (6). One filter of each set (filter A) was hybridized overnight with a total of 106 cpm of random primer-labeled [32P]dCTP-Sau3AI-TaqI-digested genomic DNA of C. dubliniensis per ml. The second filter (filter B) was hybridized overnight with 106 cpm of random primer-labeled [32P]dCTP-ribosomal DNA of C. albicans per ml (43). The filters were washed first with a solution containing 5% SDS, 40 mM NaH2PO4, and 1 mM EDTA for 20 min and then with a solution containing 1% SDS, 40 mM NaH2PO4, and 1 mM EDTA for 20 min. The final filters were autoradiographed. Filter A was compared to filter B to eliminate plaques that contained ribosomal inserts. Clones that exhibited the more intense levels of hybridization expected of repeat sequences (16) and that did not hybridize to the ribosomal probe in filter B were selected, rescreened under the same conditions, and plaque purified (29). Since all clones selected as putative DNA fingerprinting probes in the first screen generated similar hybridization patterns with EcoRI-digested DNA from the same test strains of C. dubliniensis, they were deemed to represent the same family of repeats. To obtain additional unrelated clones from the library, a second screen was conducted under less stringent conditions. In the second screen, a representative sequence that had been selected in the first screen, Cd1, was used to probe a duplicate filter to exclude homologous clones. For the second screen, Cd1 was subcloned into the pGEM-3Zf(+) plasmid vector. Prehybridization (7 h at 65°C) and hybridization (overnight at 65°C) of the filters were conducted in a solution of 50 mM NaH2PO4 (pH 7.5), 50 mM EDTA, 0.9 M NaCl, 5% dextran sulfate, 150 μg of sheared denatured salmon sperm DNA per ml, and 0.3% SDS. The filters were washed at 45°C with a solution containing 0.3 M NaCl, 0.03 M sodium citrate (pH 7.0), and 0.2% SDS. In the second screen, clones hybridizing to Cd1 or ribosomal DNA were excluded.
Southern blot hybridization and computer-assisted analysis.
Southern blot hybridization was performed as previously described (11, 15, 16, 30, 32, 34, 40). Three μg of genomic DNA from each isolate was digested with EcoRI (4 U/μg of DNA) for 16 h at 37°C. The digested DNA was electrophoresed overnight at 45 V in a 0.65% agarose gel for Cd1 and a 0.8% agarose gel for the other clones. DNA was transferred to a nylon membrane by capillary blotting (17). Digested DNA of reference strain M6 (Table 1) was run in the far-right lane of each gel to facilitate computer-assisted analysis. The membrane was hybridized with a randomly primed 32P-labeled probe and autoradiographed as previously described (16, 32). Southern blots were then stripped of the initial radiolabeled probe by incubating them first in a solution of 0.4 M sodium hydroxide for 30 min at 45°C and then in a solution of 0.015 M NaCl, 0.0015 M sodium citrate, 0.2 M Tris-HCl (pH 7.5), and 0.1% (wt/vol) SDS for 15 min at room temperature.
To analyze gel patterns, the autoradiograms were digitized into the data file of the DENDRON software program version 2.0 (Solltech, Iowa City, Iowa) with a Scanjet IIcx flatbed scanner (Hewlett-Packard, Palo Alto, Calif.). Distortions in the gels were removed with the unwarping option of DENDRON, and the lanes and bands were automatically identified. Southern blot hybridization patterns were compared through a similarity coefficient (SAB) based on band position alone for every pair of patterns (isolates) according to the formula SAB = 2E/(2E + a + b), where E is the number of bands shared by strains A and B, a is the number of bands unique to A, and b is the number of bands unique to B. An SAB of 1.00 represented identical patterns, an SAB of 0.0 represented patterns with no correlate bands, and SABs ranging from 0.01 to 0.99 represented patterns with increasing proportions of bands at the same positions. Dendrograms based on SAB values were automatically generated by the DENDRON program based on the unweighted-pair group method (35).
CHEF electrophoresis.
Chromosomes were separated by contour-clamped homogeneous electric field (CHEF) electrophoresis. To generate spheroplasts, strains of C. dubliniensis and strain 3153A of C. albicans were grown overnight in YPD medium at 25°C. The cells were pelleted by centrifugation and washed twice with sterile water and once with 1 M sorbitol. The cells were then incubated for 30 min at room temperature in a solution containing 1 M sorbitol, 25 mM EDTA, and 50 mM dithiothreitol, harvested, washed with 1 M sorbitol, and resuspended at a concentration of 109 per ml in a solution containing 1 M sorbitol, 0.1 M sodium citrate (pH 5.8), 10 mM EDTA, and 0.4 mg of Zymolyase 100T (Seikagaku America, Rockville, Md.)/ml at 37°C for 30 min. The cells were then pelleted, washed twice with a solution containing 1 M sorbitol and 250 mM EDTA, and resuspended in that solution at a final density of 109 per ml. Agarose plugs were made by mixing in equal proportions the suspension of spheroplasts and a solution of 1% agarose (Low Melt Preparative Grade; Bio-Rad Laboratories, Hercules, Calif.) containing 10 mM Tris-HCl (pH 8.0) and 0.1 M EDTA. This mixture was poured into plug molds for the CHEF mapper (Bio-Rad Laboratories). The plugs were incubated in a solution containing 0.5 M EDTA, 1% Sarkosyl, and 5 mg of proteinase K/ml for 72 h at 37°C, washed five times with a solution containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA, and finally stored at 4°C. The plugs were loaded into wells of a 1% (wt/vol) agarose gel containing 22.5 mM Tris-HCl (pH 8.3), 22.5 mM boric acid, and 1 mM EDTA. The electrophoretic conditions providing the best separation of chromosomes were a multistate protocol run at 14°C at an angle of 120° and at a gradient of 4.5 V/cm. The pulse intervals were 120 s for the first 24 h and 240 s for the final 36 h. Following electrophoresis, the gel was stained with ethidium bromide, photographed, Southern blotted, and incubated with a particular DNA probe (29).
RESULTS
Cloning putative DNA fingerprinting probes.
A library was constructed in λEMBL3 from a Sau3AI partial digest of genomic DNA from C. dubliniensis d1558 (Table 1). Digestion products ranged between 9 and 23 kb. The library was plated on five dishes, generating approximately 10,000 plaques per dish. The library on each plate was transferred to duplicate nitrocellulose filters and hybridized in parallel with either radiolabeled C. dubliniensis genomic DNA or a radiolabeled ribosomal DNA probe of C. albicans. Under nonsaturating conditions, clones containing repeat sequences generated stronger signals than clones containing unique sequences. This difference was the basis of the screen (8, 11, 15, 16, 28, 31, 38). Twenty clones which hybridized intensively with genomic DNA, but not with the ribosomal probe, were selected for further analysis. The screen was repeated to verify that each clone contained a nonribosomal repeat sequence. The 20 clones were plaque purified, labeled, and used to probe Southern blots of EcoRI-digested DNA of three unrelated test strains of C. dubliniensis to assess the effectiveness of the clones as fingerprinting probes. Of the original 20 clones tested, 19 (95%) generated complex Southern blot hybridization patterns that varied among the three test strains. However, all of the 19 clones generated similar patterns for a particular strain, suggesting that the clones contained one or more common sequences. To assess the relatedness of the 19 clones, each was digested with SalI and EcoRI, electrophoresed in a 0.8% (wt/vol) agarose gel for 4 h, and stained with ethidium bromide. Seventeen different digestion patterns were generated, which suggested that 17 of the 19 related clones originated from different genomic sites or represented partial overlaps at the same sites. One representative of this family, Cd1, with an estimated size of 15,500 bp, was selected for further analysis.
To obtain additional, unrelated probes, a second screen was performed under the same conditions as the first, except that hybridization of the nitrocellulose membranes was performed at lower stringency. To exclude sequences homologous to Cd1, the Cd1 fragment was cloned into the pGEM-3Zf(+) plasmid vector and used to probe an additional filter in the screen. Of approximately 100,000 plaques, 100 displayed a signal of relatively high intensity and did not hybridize with either ribosomal DNA or the Cd1 plasmid. Of these, 26 were selected and analyzed. Fifteen generated complex polymorphic patterns when used to probe the three test strains of C. dubliniensis. Two general patterns were generated by the 15 probes, suggesting that two additional families of probes containing repetitive sequences were represented. One representative clone from each family, Cd24 and Cd25, were selected for further analysis. The estimated sizes of Cd24 and Cd25 were 10,000 and 16,000 bp, respectively.
Genetic variability displayed by the three selected probes.
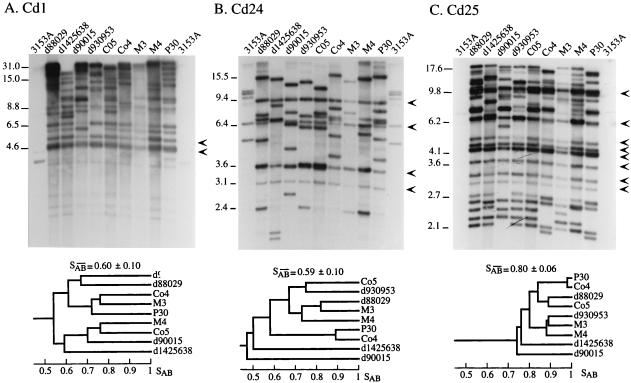
To test the effectiveness of the three cloned probes and cross-hybridization with C. albicans DNA, each clone was used to probe Southern blots of nine unrelated test isolates of C. dubliniensis and C. albicans 3153A (Fig. 1). The Cd1 probe generated between 9 and 15 bands ranging from 4.6 to 31 kb for each of the nine test strains (Fig. 1A). Cd1 generated significantly different patterns for all nine test strains, even the ones originating from patients treated in the same hospital (e.g., d88029 and d90015, and M3 and M4) (Fig. 1A). Cd1 generated two monomorphic bands (i.e., bands at the same molecular weight in the nine test strains) at 4.6 and 5.3 kb (Fig. 1A), as well as several moderately variable bands and several highly variable bands. In the dendrogram generated for Cd1 in Fig. 1A, the two pairs of isolates with the highest SABs (0.77 in both cases) were Co4 and M3, and Co5 and M4. The isolates in each pair were from different countries. The Cd1 probe generated three bands with EcoRI-digested DNA of C. albicans 3153A, a weak one at 31 kb and a weak one and a moderately intense one below 4.6 kb (Fig. 1A).
FIG. 1.
Southern blot hybridization patterns and resulting dendrograms of EcoRI-digested DNA of nine C. dubliniensis isolates and C. albicans 3153A probed with Cd1(A), Cd24 (B), and Cd25 (C). The DNA was electrophoresed in a 0.65% agarose gel, and the Southern blot was sequentially hybridized with the three probes. The arrowheads to the right of each gel represent prominent invariant (monomorphic) bands. Key molecular sizes are presented in kilobases to the left of each gel. The origins of these test isolates are presented in Table 1. A dendrogram (shown below each gel) for each set of hybridization patterns was generated from the similarity coefficient (SAB) computed for all possible pairs of the nine unrelated C. dubliniensis isolates. Since the M3 lane was underloaded, analyses of that pattern were performed on autoradiograms exposed for longer periods. The average SAB is indicated at the top of each dendrogram.
The Cd24 probe also generated significantly different patterns for all nine test strains (Fig. 1B). Cd24 generated four monomorphic bands, a few moderately variable bands, and several highly variable bands. In the dendrogram generated for Cd24 (Fig. 1B), the pair of isolates with the highest SAB (0.89) was P30 and Co4. The Cd24 probe generated five moderately intense bands and two very weak bands with DNA of C. albicans 3153A (Fig. 1B).
The Cd25 probe also generated significantly different patterns for all of the nine test strains (Fig. 1C). Cd25 generated the most complex pattern of the three tested probes. The patterns contained 10 monomorphic bands as well as several moderately variable bands and several highly variable bands. In the dendrogram generated for Cd25 (Fig. 1C), the pair of isolates with the highest SAB (0.98) was P30 and Co4, the same pair exhibiting the highest SAB in the Cd24 dendrogram (Fig. 1B). Cd25 was the only probe of the three with no significant hybridization to DNA of C. albicans 3153A (Fig. 1C).
The patterns generated by Cd1, Cd24, and Cd25 were distinctly different. None of the monomorphic or highly variable bands were common, supporting the conclusion that the three probes were unrelated.
The relationship of Cd1 and the C. albicans repetitive element RPS.
Two of the three probes, Cd1 and Cd24, hybridized with EcoRI-digested DNA of C. albicans 3153A (Fig. 1A and B). Since the C. albicans fingerprinting probes Ca3 (32) and 27A (31) have been demonstrated to cross-hybridize with C. dubliniensis DNA (3, 46, 47), we tested whether any of the three C. dubliniensis probes identified the same genomic fragments as Ca3 and 27A. The gel shown in Fig. 1 was reprobed with the C1 fragment of Ca3 (1, 14), which contains a portion of the C. albicans repetitive sequence RPS (13), with a cloned RPS element, RPS39 (27a), and with the probe 27A (31), which also contains an RPS element (27a). RPS39 has 99% homology with other published RPS sequences (27a). The probes C1, RPS39, and 27A all generated the same pattern for each of the nine C. dubliniensis test strains (Fig. 2B, C, and D, respectively). This pattern contained approximately 80% of the bands present in the respective Cd1 patterns (Fig. 2A).
FIG. 2.
Hybridization patterns of the nine test isolates of C. dubliniensis and C. albicans 3153A probed with the Cd1 probe (A), the C1 fragment of the C. albicans probe Ca3 (B) (1), the RPS39 element of C. albicans (C) (27a), and the C. albicans probe 27A (30). The origins of the test isolates are presented in Table 1. Molecular sizes are presented in kilobases to the left of each set of hybridization patterns.
While probes C1, RPS39, and 27A generated patterns of four to eight bands when hybridized to EcoRI-digested C. albicans DNA (Fig. 2A, B, and C, respectively), Cd1 generated only one moderately intense band and two very weak bands (Fig. 2A), none of which corresponded to the bands generated by C1, RPS39, and 27A. These results suggest that Cd1 contains a C. dubliniensis-specific sequence dispersed throughout the C. dubliniensis genome in close association with an RPS-like element but that the Cd1 probe itself does not contain an RPS sequence.
Cross-verification of the probes Cd1, Cd24, and Cd25.
Since the three C. dubliniensis probes represent unrelated genomic fragments, they can be considered unrelated fingerprinting methods and can, therefore, be used to cross-verify efficacy by cluster analysis (11, 15, 26, 39, 49). Cd25 generated the most complex Southern blot hybridization patterns and the highest average SAB when used as a probe to fingerprint the nine test isolates (Fig. 1A). It grouped isolates P30 and Co4 into the most related cluster, grouped M3 and M4 into a moderately related cluster, and identified d1425638 and d90015 as the most unrelated isolates in the collection. Cd24 also grouped isolates P30 and Co4 into the most related cluster, grouped M3 and M4 into a moderately related cluster, and identified d1425638 and d90015 as the most unrelated isolates in the collection. This was not the case for Cd1. Cd1 did not group P30 and Co4 into the most related cluster, did not group M3 and M4 in a moderately related cluster, and did not identify d1425638 and d90015 as the most unrelated isolates. These results demonstrate that relative parity exists between Cd25 and Cd24 as fingerprinting probes, but not between either of these probes and Cd1.
The distribution of sequences homologous to Cd1, Cd24, and Cd25 in the C. dubliniensis genome.
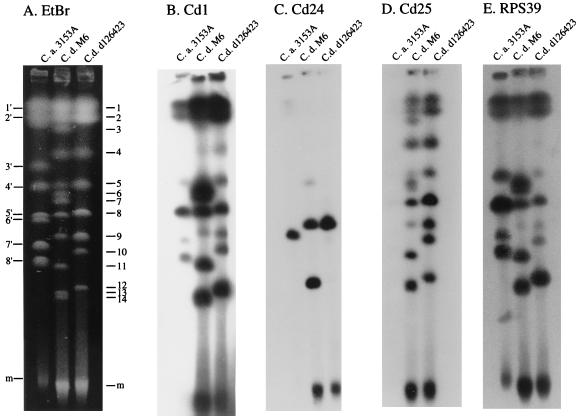
Twelve chromosomal bands and one minichromosomal band were separated by CHEF from C. dubliniensis M6, and eight chromosomal bands and one minichromosomal band were separated from C. dubliniensis d126423 (Fig. 3A). Eight chromosomal bands and one minichromosomal band were separated from C. albicans 3153A (Fig. 3A). Cd1 hybridized to all chromosomal and minichromosomal bands of both C. dubliniensis strains (Fig. 3B). The degree of hybridization varied substantially among chromosomal bands. Cd1 also hybridized to all C. albicans chromosomal bands except band 3 (Fig. 1B). RPS39 also hybridized to all chromosomal bands of the two C. dubliniensis strains except band 4 and hybridized to all C. albicans chromosomal bands except band three (Fig. 3E). Moreover, the relative differences between the Cd1 hybridization intensities of the C. dubliniensis bands (Fig. 3B) were the same when RPS39 was used as a probe (Fig. 3E). These results demonstrate that Cd1 sequences are dispersed throughout the C. dubliniensis genome and are associated in all or a majority of cases with RPS sequences.
FIG. 3.
Hybridization of CHEF-separated chromosomes of C. dubliniensis (C.d.) and C. albicans (C.a.) with the C. dubliniensis probes Cd1, Cd24, and Cd25 and the C. albicans repeat element RPS39. Chromosomes of C. albicans 3153A and C. dubliniensis M6 and d126423 were separated by CHEF, and the gel was stained with ethidium bromide (EtBr) (A). The gels were then Southern blotted and probed with Cd1(B), Cd24 (C), Cd25 (D), and RPS39 (E). C. albicans bands are numbered to the left of the EtBr-stained image, and C. dubliniensis bands are numbered to the right of the EtBr-stained image. m, minichromosomal band.
Cd24, on the other hand, hybridized to only two chromosomal bands and the minichromosomal band of C. dubliniensis M6 and to only one chromosomal band and the minichromosomal band of C. dubliniensis d126423 (Fig. 3C). Cd24 also hybridized to one chromosomal band of C. albicans 3153A (Fig. 3C). Therefore, although Cd24 generates a complex Southern blot hybridization pattern with EcoRI-digested C. dubliniensis DNA, it is distributed on only one to two chromosomes.
Cd25 hybridized to all chromosomal bands and the minichromosomal band of the two C. dubliniensis strains (Fig. 3D). Cd25, therefore, is distributed on all C. dubliniensis chromosomes. It hybridized to no C. albicans chromosomal bands (Fig. 3D), supporting the conclusion that Cd25 is the only C. dubliniensis-specific probe of the three clones analyzed.
Species specificity of Cd1, Cd24, and Cd25.
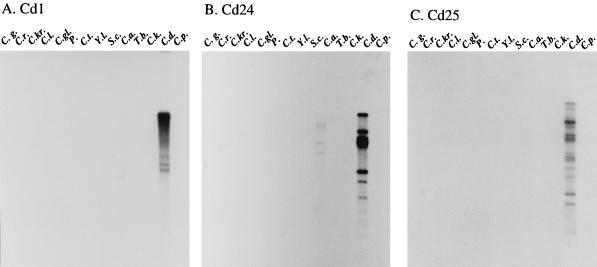
Each of the three clones was used to probe Southern blots of EcoRI-digested DNA of 14 related yeast species (Fig. 4). Cd1 hybridized strongly with C. dubliniensis DNA, very weakly with C. albicans DNA, and not at all with the DNA of the remaining 12 species (Fig. 4A). Cd24 hybridized strongly with C. dubliniensis DNA, weakly with C. albicans DNA, and not at all with DNA of the remaining 12 species (Fig. 4B). Cd25 hybridized strongly with C. dubliniensis DNA but not at all with DNA of the remaining 13 species (Fig. 4C). When the C. albicans repeat element RPS39 was used to probe the same blot, it hybridized strongly with C. dubliniensis and C. albicans DNA, but not at all with DNA from the remaining 12 species (data not shown). These results demonstrate that Cd1 and Cd24 are C. dubliniensis and C. albicans specific, and that Cd25 is C. dubliniensis specific.
FIG. 4.
Species specificity of the three C. dubliniensis probes. EcoRI-digested DNA of 14 different yeast species was sequentially probed with Cd1 (A), Cd24 (B), Cd25 (C), and the C. albicans repeat element RPS39 (D). Lanes: C.g., Candida guillermondii; C.r., Candida rugosa; C.kr., C. krusei; C.l., Candida lusitaniae; C.gl., C. glabrata; P., Pichia sp.; C.t., C. tropicalis; Y.l., Yarrowia lipolytica; S.c., Saccharomyces cerevisiae; C.a., C. albicans; T.b., Trichosporon beigelii; C.k., Candida kefyr; C.d., C. dubliniensis; C.p., C. parapsilosis.
Stability of the DNA fingerprint patterns generated by Cd1, Cd24, and Cd25.
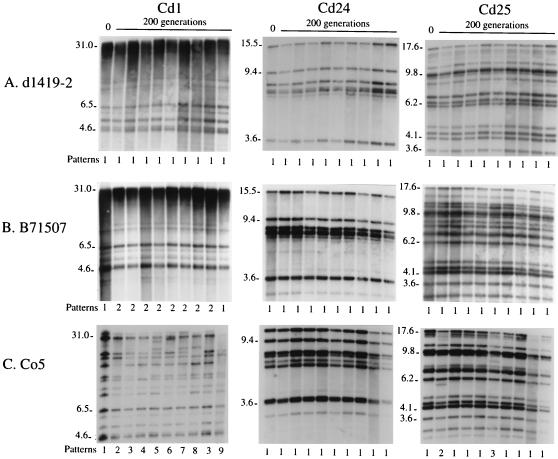
The stability of the patterns generated by the three C. dubliniensis probes was first tested in vitro for three unrelated strains grown from single colonies for 200 generations. Cells from each of the final growth cultures were plated on agar, and cells from nine colonies of each strain as well as from each original clone were analyzed by Southern blot hybridization with the probes Cd1, Cd24, and Cd25. Stability proved to be both strain and probe related. The nine clones of strain d1419-2 obtained after 200 generations exhibited no differences from each other or from the original clone when fingerprinted with any of the three probes (Fig. 5A). The nine clones of strain B71507 exhibited no differences from each other or from the original clone with probes Cd24 or Cd25 but displayed two patterns with probe Cd1 (patterns 1 and 2), one of which (pattern 1) was the same as that of the original clone (Fig. 5B). Pattern 1 differed from pattern 2 by an 8.8-kb band present in the latter but not in the former. The nine clones of strain Co5 exhibited no differences from each other or from the original clone with probe Cd24. However, the nine clones displayed three patterns (patterns 1, 2, and 3) when probed with Cd25 (Fig. 5C). One of the patterns (pattern 1) was the same as that of the original clone (Fig. 5C). The nine clones of strain Co5 exhibited eight different patterns (patterns 2 through 9) when probed with Cd1. All of these patterns differed from that of the original clone (pattern 1 [Fig. 5C]). These results demonstrate that Cd24 provides the most stable pattern, Cd25 provides the second most stable pattern, and Cd1 provides the least stable pattern over time in vitro. These results also demonstrate that strain d1419-2 has the most stable genotype and strain Co5 has the least stable genotype in vitro.
FIG. 5.
In vitro analysis of the stability of the patterns generated by C. dubliniensis probes Cd1, Cd24, and Cd25. Southern blots of EcoRI-digested DNA from clones of C. dubliniensis d1419-2 (A), B71507 (B), and Co5 (C), at zero hours (0) and after 200 generations, were probed with Cd1, Cd24, and Cd25. Nine individual clones of each strain were selected randomly for analysis at 200 generations. Variant patterns are numbered at the bottom of each blot. Key molecular sizes in kilobases are presented to the left of each blot.
To examine the patterns generated by the three probes in vivo, 15 C. dubliniensis isolates were recovered from the oral cavity of a human immunodeficiency virus (HIV)-positive patient at the University of Iowa Hospitals and Clinics over a 12-month period (Table 1, patient 50). One isolate was obtained at time zero, seven isolates were obtained at 5 months, two isolates were obtained at 7 months, and five isolates were obtained at 12 months. The last isolates, collected at 12 months, were obtained when the patient presented with his first episode of oral thrush. All isolates were analyzed by Southern blot hybridization with probes Cd1 (Fig. 6A), Cd24 (Fig. 6B), and Cd25 (Fig. 6C). All three probes separated the collection of 15 isolates into two groups distinguishable by the general fingerprint patterns. The first, group a, was composed of 13 isolates, and the second, group b, was composed of 2 isolates. The separation into two groups is evident in the dendrograms generated from the hybridization patterns of each probe (Fig. 6). The node separating group a and b isolates occurred at SABs of 0.39, 0.27, and 0.75 for probes Cd1, Cd24, and Cd25, respectively (Fig. 6). These node SABs are all below the average SAB for unrelated isolates (Fig. 1). Probe Cd1 distinguished differences between the two b isolates, but probes Cd24 and Cd25 did not (Fig. 6). It should be noted that the time interval between collection of the two b isolates was 7 months (Fig. 6). Variability was observed in group a isolates with all three probes (Fig. 6). However, the least variability occurred in the Cd25 patterns. For the 13 isolates in group a, Cd1 distinguished 11 patterns, Cd24 distinguished 7 patterns, and Cd25 distinguished 3 patterns (Fig. 6, see the pattern analysis at the bottom of each gel). These results demonstrate that Cd25 provides the most stable pattern in vivo and in vitro, while Cd1 and Cd24, on the other hand, distinguish greater variability within a strain over time and are, therefore, superior for assessing microevolution.
FIG. 6.
The variability of probe-generated patterns in a population colonizing an HIV-positive individual over time. EcoRI-digested DNA from multiple isolates from patient 50 (Table 1) were probed with Cd1 (A), Cd24 (B), and Cd25 (C). The isolates were obtained at 0, 5, 7, and 12 months. The patient presented with oral thrush at 12 months. Variations in patterns are indicated at the bottom of each blot. This patient was infected by two different strains displaying the general genotypes a and b. Changes in the patterns are numbered in chronological order of occurrence. Molecular sizes are presented in kilobases to the left of the gels. On the right side of each hybridized Southern blot the representative dendrogram is generated. The average SAB is presented at the top of each dendrogram.
Analysis of a broad collection of C. dubliniensis isolates with the probe Cd25.
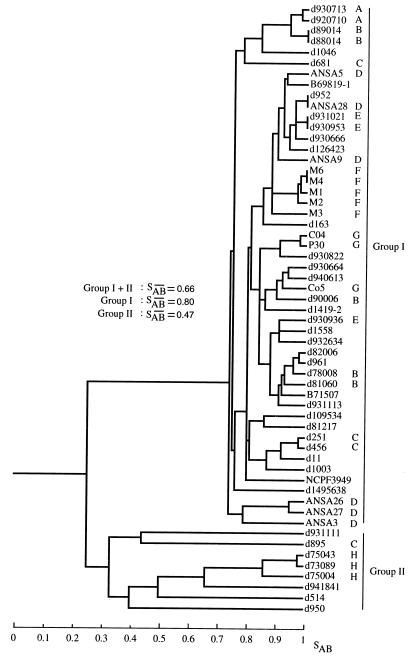
Because Cd25 was deemed the best probe for broad epidemiological studies, it was used to analyze the relatedness of 57 independent C. dubliniensis isolates collected in 11 countries (Table 1). Among the 57 isolates, Cd25 discriminated 53 different patterns (Fig. 7). Of the four pairs of identical isolates, three pairs consisted of isolates collected from different patients in the same hospitals, suggesting that each represented nosocomial strains endemic to the respective hospitals (23, 24). Furthermore, of 31 isolates collected from eight hospitals (Fig. 7, A through H), 67% grouped in clusters that contained only strains from a single hospital, suggesting the presence of nosocomial strains endemic to the respective hospitals. Only one pair of identical isolates in the entire collection, d952 and ANSA28 (Fig. 7), was derived from individuals in different hospitals and countries (Table 1).
FIG. 7.
Dendrograms generated from the similarity coefficients (SABs) computed for all pairs of 57 unrelated isolates collected worldwide and fingerprinted with Cd25. The origins of the isolates are presented in Table 1. The letters (A through H) to the right of the dendrogram indicate the hospitals of origin in cases where several isolates, each from a different individual, were collected from the same hospital: A, Brussels, Belgium; B, Leicester, United Kingdom; C, London, United Kingdom; D, Antwerp, Belgium; E, Frankfurt, Germany; F, Victoria, Australia; G, Lausanne, Switzerland; H, Leeds, United Kingdom. The two main clusters are delineated to the right of the dendrogram (groups I and II). The average SABs for each group, and for the total collection, are presented in the middle of the dendrogram.
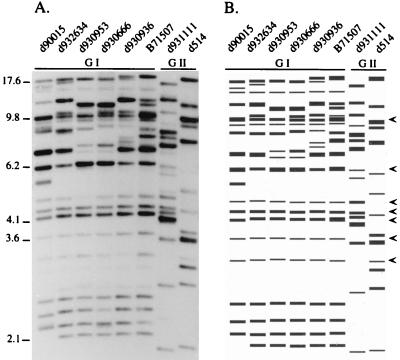
The isolates in this broad collection separated into two general groups, group I and group II (Fig. 7). The two groups were separated by a node at an SAB of 0.24 (Fig. 7). Group I, which contained 49 isolates, had an average SAB of 0.80. Group I isolates shared several common, or monomorphic, bands (Fig. 8). In contrast, Group II, which contained eight isolates, had a relatively low average SAB of 0.47 (Fig. 7). This low average SAB suggests a high level of diversity within the group. The difference between the two groups is evident in the representative patterns in Fig. 8. While group I patterns were similar, group II patterns differed markedly not only from group I patterns but also from each other. Group II isolates lacked, for the most part, the monomorphic bands described in the original comparison of the three probes (Fig. 1 and 5), which, by chance, was performed with only group I isolates.
FIG. 8.
Examples of Southern blot hybridization patterns of group I and group II isolates probed with Cd25. Hybridization patterns are presented in A, and a model generated by the DENDRON program is presented in B. Molecular sizes are presented in kilobases to the left of the hybridization pattern. The arrowheads to the right of the model indicate the prominent invariant bands shared by group I isolates but not by group II isolates.
Analysis of isolates from the same individuals with the probe Cd25.
In the worldwide collection analyzed in this study, multiple isolates were obtained from nine individuals. When these multiple isolates were added to the dendrogram in Fig. 7, in all cases but one the isolates from the same individual clustered at an SAB of 0.95 to 1.00 (data not shown).
DISCUSSION
At the phenotypic level of analysis, a number of traits are readily distinguishable between the majority of C. albicans and C. dubliniensis isolates (48). Perhaps the most definitive is the inability of C. dubliniensis isolates to express β-glucosidase (3, 33, 47). However, conclusive differences must be assessed at the genetic level before one would feel comfortable separating a set of atypical C. albicans isolates with common phenotypes into a bona fide new species, especially in view of the capacity of C. albicans and related species to change their phenotypes in so pleiotropic a fashion through reversible high-frequency phenotypic switching (36). The first clues that genetic differences also existed between typical and atypical C. albicans isolates came from reports that the latter exhibited diminished hybridization patterns with the related probes 27A (46, 47) and Ca3 (22) and that atypical strains could be readily distinguished from other species, including C. albicans, by restriction fragment length polymorphisms generated from HinfI-digested DNA (46), randomly amplified polymorphic DNA analysis (46), hybridization to oligonucleotides homologous to microsatellites (45, 46), PCR products of primers homologous to CARE-2 (18, 19), electrophoretic karyotypes (2, 46), multilocus enzyme electrophoresis (3, 12, 27, 47), and sequencing ribosomal DNA (46).
C. dubliniensis-specific repeat elements support its status as a species.
We have cloned and characterized complex DNA fingerprinting probes from C. albicans (28, 40), C. glabrata (15), C. tropicalis (11), and Aspergillus fumigatus (8) and have found in all cases that they contain repetitive, species-specific sequences. We hypothesized that the repetitive sequences harbored by these probes have evolved rapidly enough to be species specific and, therefore, to be good indicators of speciation. Here, we have cloned one complex probe, Cd25, that is homologous to sequences dispersed throughout the C. dubliniensis genome and produces no detectable signal when used to probe 13 related yeast species, including C. albicans. The evolution of this specific sequence in a subgroup of atypical C. albicans strains with common phenotypic traits supports the argument by Coleman and Sullivan (48) that C. dubliniensis represents a bona fide species. However, by the same argument, we have presented contradictory evidence that this subgroup of atypical C. albicans isolates also contains dispersed homologs of the C. albicans-specific RPS repetitive element (5, 10). These apparently contradictory results are accommodated by a relatively straightforward explanation. C. albicans and C. dubliniensis both may have evolved from a common ancestor that possessed RPS elements. After separation, C. dubliniensis acquired the repeat element in Cd25 that is dispersed throughout its genome. Therefore, C. albicans and C. dubliniensis are closely enough related to share a repetitive element (RPS) not found in any other Candida species but distant enough not to share the repetitive element in Cd25.
The potential of Cd1, Cd24, and Cd25 as C. dubliniensis DNA fingerprinting probes.
Although all three probes generated complex Southern blot hybridization patterns, they were not equal in their capacities to discriminate among isolates. Cd1 produced far more variable patterns between moderately related and unrelated isolates and did not achieve parity with the other two probes in a cluster analysis of nine test isolates. The pattern generated by Cd1 was also far less stable both in vitro and in vivo than those generated by the other two probes. Because of its decreased stability, Cd1 is less effective in clustering moderately related isolates and therefore will not perform well in broad epidemiological studies. On the other hand, Cd1 is superior to both Cd24 and Cd25 in discriminating microevolutionary changes in clonal populations. These characteristics are similar to those noted for the C. albicans probe 27A and the C1 fragment of Ca3, both of which are composed predominately of RPS sequences (27a). Curiously, Cd24 did not discriminate microevolution in the three clones grown for 200 generations in vitro but was an excellent indicator of microevolution in one clonal population of C. dubliniensis monitored over a 12-month period in an HIV-positive patient. These results suggest either that the infecting strain in the HIV-positive patient had gone through far more than 200 generations or that variations in the Cd24 pattern are induced by in vivo conditions.
Although Cd24 and Cd25 achieved parity in the cluster analysis, Cd24 generated the least complex pattern of the three probes and exhibited the highest level of hybridization with C. albicans DNA. Cd24 was also the least dispersed sequence in the C. dubliniensis genome. Cd25, therefore, was selected as the probe of choice for broad epidemiological studies. It generated the most stable pattern over time for clonal populations in vitro and in vivo, produced the most complex pattern, produced the pattern with the greatest number of monomorphic bands for group I isolates, and produced the pattern with the most highly resolved bands of the three probes. Cd25 was also the only C. dubliniensis-specific probe. However, as noted, Cd25 was the least effective of the three probes in discriminating microevolution within an infecting strain.
Analysis of a broad collection of C. dubliniensis isolates.
In an analysis by Southern blot hybridization with the Cd25 probe of 57 isolates collected in 11 countries, two groups were identified, group I and group II. Group I comprised 86% of all isolates and exhibited an average SAB of 0.80, which was relatively high given the complexity of the pattern generated by Cd25. Group II comprised 14% of all isolates and exhibited an average SAB of 0.47, which was relatively low. The lower SAB suggests that the isolates in group II are relatively unrelated. The greater degree of variability among group II isolates suggests either that they have a higher frequency of Cd25 sequence reorganization than group I isolates or that group I represents a younger and therefore more homogeneous subgroup of C. dubliniensis that has rapidly become predominant worldwide. The very low node value that separates groups I and II (SAB = 0.24) suggests reproductive isolation between the two groups.
Of the eight group II isolates, six (75%) were collected in the United Kingdom and the remaining two were collected in Spain and the Netherlands. Since the isolates from the United Kingdom comprised only 30% of the test collection, it would appear that group II isolates may be disproportionately concentrated in the United Kingdom. Further analysis of a larger number of C. dubliniensis isolates obtained worldwide is now being performed to verify the separation of C. dubliniensis into groups I and II described above.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants DE10758 and AI2392 from the National Institutes of Health.
We are indebted to F. Odds of Janssen Pharmaceutics; Patrick Boerlin of the Institute of Microbiology at Lausanne, Switzerland; M. McCullough of the School of Dental Science at Melbourne, Australia; and M. Pfaller of the University of Iowa for many of the isolates used in this study.
REFERENCES
- 1.Anderson J, Srikantha T, Morrow B, Miyasaki S, White T, Agabian N, Schmid J, Soll D R. Characterization and partial nucleotide sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J Clin Microbiol. 1993;31:1472–1480. doi: 10.1128/jcm.31.6.1472-1480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony R M, Midgley J, Sweet S P, Howell S A. Multiple strains of Candida albicans in the oral cavity of HIV-positive and HIV-negative patients. Microb Ecol Health Dis. 1995;8:23–30. [Google Scholar]
- 3.Boerlin P, Boerlin-Petzold F, Durussel C, Addo M, Pagani J L, Chave J P, Bille J. Cluster of oral atypical Candida albicans isolates in a group of human immunodeficiency virus-positive drug users. J Clin Microbiol. 1995;33:1129–1135. doi: 10.1128/jcm.33.5.1129-1135.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlotti A, Srikantha T, Schröppel K, Kvaal C, Villard J, Soll D R. A novel repeat sequence (CKRS-1) containing a tandemly repeated subelement (kre) accounts for differences between Candida krusei strains fingerprinted with the probe CkF1,2. Curr Genet. 1997;31:255–263. doi: 10.1007/s002940050203. [DOI] [PubMed] [Google Scholar]
- 5.Chindamporn A, Nakagawa Y, Nizuguchi I, Chibana H, Doi M, Tanaka K. Repetitive sequences (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology. 1998;144:849–857. doi: 10.1099/00221287-144-4-849. [DOI] [PubMed] [Google Scholar]
- 6.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman D C, Sullivan D J, Bennett D E, Moran G P, Barry H J, Shanley D B. Candidiasis: the emergence of a novel species Candida dubliniensis. AIDS. 1997;11:557–567. doi: 10.1097/00002030-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 7a.Enger, L., S. Joly, M. Pfaller, and D. R. Soll. Unpublished data.
- 8.Girardin H, Latge J-P, Srikantha T, Morrow B, Soll D R. Development of DNA probes for fingerprinting Aspergillus fumigatus. J Gen Microbiol. 1993;31:1547–1554. doi: 10.1128/jcm.31.6.1547-1554.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazen K C. New and emerging yeast pathogens. Clin Microbiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwaguchi S I, Homma M, Chibana H, Tanaka K. Isolation and characterization of a repeated sequence (RPS1) of Candida albicans. J Gen Microbiol. 1992;138:1893–1906. doi: 10.1099/00221287-138-9-1893. [DOI] [PubMed] [Google Scholar]
- 11.Joly S, Pujol C, Schröppel K, Soll D R. Development and verification of two species fingerprinting probes for Candida tropicalis amenable to computer analysis. J Clin Microbiol. 1996;34:3063–3071. doi: 10.1128/jcm.34.12.3063-3071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeGuennec R, Reynes J, Mallié M, Pujol C, Janbon F, Bastide J M. Fluconazole- and itraconazole-resistant Candida albicans strains from AIDS patients: multilocus enzyme electrophoresis analysis and antifungal susceptibilities. J Clin Microbiol. 1995;33:2732–2737. doi: 10.1128/jcm.33.10.2732-2737.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lockhart, S., J. J. Fritch, A. S. Meier, K. Schroeppel, T. Srikantha, R. Galask, and D. R. Soll. Colonizing populations of C. albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33:1501–1509. [DOI] [PMC free article] [PubMed]
- 14.Lockhart S R, Reed B D, Pierson C L, Soll D R. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J Clin Microbiol. 1996;34:767–777. doi: 10.1128/jcm.34.4.767-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart S R, Joly S, Pujol C, Sobel J D, Pfaller M A, Soll D R. Development and verification of fingerprinting probes for Candida glabrata. Microbiology. 1997;143:3733–3746. doi: 10.1099/00221287-143-12-3733. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart, S. R., and D. R. Soll. Generation of species-specific DNA probes and their applications for analysis of fungal populations. In D. Coleman and K. Haynes (ed.), Methods in molecular medicine series: medical mycology protocols, in press. Humana Press, Totowa, N.J.
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 18.McCullough M, Ross B, Reade P. Oral Candida albicans from patients infected with the human immunodeficiency virus and characterization of a genetically distinct subgroup of Candida albicans. Aust Dent J. 1995;40:91–97. doi: 10.1111/j.1834-7819.1995.tb03122.x. [DOI] [PubMed] [Google Scholar]
- 19.McCullough M, Ross B, Reade P. Characterization of genetically distinct subgroup of Candida albicans strains isolated from oral cavities of patients infected with human immunodeficiency virus. J Clin Microbiol. 1995;33:696–700. doi: 10.1128/jcm.33.3.696-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metzgar D, Field D, Haubrich R, Wills C. Sequence analysis of a compound coding-region microsatellite in Candida albicans resolves homoplasia and provides a high-resolution tool for genotyping. FEMS Immunol Med Microbiol. 1998;20:103–109. doi: 10.1111/j.1574-695X.1998.tb01116.x. [DOI] [PubMed] [Google Scholar]
- 21.Odds F C. Candida and candidosis: a review and bibliography. 2nd ed. London, United Kingdom: Balliere Tindall; 1988. [Google Scholar]
- 22.Odds F C, Schmid J, Soll D R. Epidemiology of Candida infection in AIDS. In: Vanden Bossche H, et al., editors. Mycoses in AIDS patients. New York, N.Y: Plenum Press; 1990. pp. 67–74. [Google Scholar]
- 23.Pfaller M A. Nosocomial candidiasis: emerging species, reservoirs and models of transmission. Clin Infect Dis. 1996;22(Suppl. 2):589–594. doi: 10.1093/clinids/22.supplement_2.s89. [DOI] [PubMed] [Google Scholar]
- 24.Pfaller M A, Lockhart S R, Pujol C, Swails-Wenger J A, Messer S A, Edmond M B, Jones R N, Wenzel R P, Soll D R. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J Clin Microbiol. 1998;36:1518–1529. doi: 10.1128/jcm.36.6.1518-1529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powderly W G. Mucosal candidiasis caused by non albicans species of Candida in HIV-positive patients. AIDS. 1992;6:604–605. [PubMed] [Google Scholar]
- 26.Pujol C, Joly S, Lockhart S R, Noel S, Tibayrenc M, Soll D R. Parity among the radomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pujol C, Renaud F, Mallié M, De Meeûs T, Bastide J M. Atypical strains of Candida albicans recovered from AIDS patients. J Med Vet Mycol. 1997;35:115–121. [PubMed] [Google Scholar]
- 27a.Pujol, C., S. Joly, T. Srikantha, S. Lockhart, and D. R. Soll. Unpublished data.
- 28.Sadhu C, McEachern M J, Rustchenko-Bulgac E P, Schmid J, Soll D R, Hicks J. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J Bacteriol. 1991;173:842–850. doi: 10.1128/jb.173.2.842-850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scherer S, Stevens D A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc Natl Acad Sci USA. 1988;85:1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid J, Voss E, Soll D R. Computer-assisted methods for assessing Candida albicans strain relatedness by Southern blot hybridization with repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoofs A, Odds F C, Colebunders R, Leven L, Goossens H. Use of specific isolation media for recognition and identification of Candida dubliniensis isolates from HIV-patients. Eur J Clin Microbiol Infect Dis. 1997;16:296–300. doi: 10.1007/BF01695634. [DOI] [PubMed] [Google Scholar]
- 34.Schröppel K, Rotman M, Galask R, Mac K, Soll D R. The evolution and replacement of C. albicans strains during recurrent vaginitis demonstrated by DNA fingerprinting. J Clin Microbiol. 1994;32:2646–2654. doi: 10.1128/jcm.32.11.2646-2654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sneath P, Sokal R. Numerical taxonomy. The principles and practice of numerical classification. San Francisco, Calif: W. H. Freeman; 1973. pp. 230–234. [Google Scholar]
- 36.Soll D R. High frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soll D R. DNA fingerprinting of Candida albicans. J Mycol Med. 1993;3:37–44. [Google Scholar]
- 38.Soll D R. Computer-assisted analysis of DNA fingerprinting. In: Tanaka K, Yamaguchi Y, Magee P T, editors. Genes and genomes in medically important fungi. Tokyo, Japan: Foundation for Advancement of International Science; 1996. pp. 97–108. [Google Scholar]
- 39.Soll, D. R. Unpublished data.
- 40.Soll D R, Langtimm C J, McDowell J, Hicks J, Rao T V G. High frequency switching in Candida strains isolated from vaginitis patients. J Clin Microbiol. 1987;25:1611–1622. doi: 10.1128/jcm.25.9.1611-1622.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soll D R, Staebell M, Langtimm C J, Pfaller M, Hicks J, Rao T V G. Multiple Candida strains in the course of a single systemic infection. J Clin Microbiol. 1988;26:1448–1459. doi: 10.1128/jcm.26.8.1448-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soll D R, Galask R, Schmid J, Hanna C, Mac K, Morrow B. Genetic dissimilarity of commensal strains carried in different anatomical locations of the same healthy women. J Clin Microbiol. 1991;29:1702–1710. doi: 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srikantha T, Guttell R, Morrow B, Soll D R. Partial nucleotide sequence of a single ribosomal coding region and secondary structure of the large subunit 25S rRNA of Candida albicans. Curr Genet. 1994;26:321–328. doi: 10.1007/BF00310496. [DOI] [PubMed] [Google Scholar]
- 44.Stevens D A. Fungal infections in AIDS patients. Br J Clin Pract. 1990;44(Suppl. 7):11–22. [PubMed] [Google Scholar]
- 45.Sullivan D, Bennett D, Henman M, Harwood P, Flint S, Mulcahy F, Shanley D, Coleman D. Oligonucleotide fingerprinting of isolates of Candida species other than C. albicans and of atypical Candida species from human immunodeficiency virus-positive and AIDS patients. J Clin Microbiol. 1993;31:2124–2133. doi: 10.1128/jcm.31.8.2124-2133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan D, Westerneng T, Haynes K, Bennett D, Coleman D. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan D, Haynes K, Bille J, Boerlin P, Rodero L, Lloyd S, Henman M, Coleman D. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997;35:960–964. doi: 10.1128/jcm.35.4.960-964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. . (Minireview.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tibayrenc M, Neubauer K, Barnabé C, Guerrini D, Skarecky D, Ayala F J. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]