Abstract
Drug resistance is a major problem to overcome in the treatment of cancer; therefore, identifying therapeutic targets for drug resistance is a point of focus in the field of cancer research. Long non-coding RNAs (lncRNAs) and microRNAs (miRs) not only affect gene expression regulation during cell proliferation, but also have several potential roles in the drug resistance of malignant tumors. Reverse transcription-quantitative PCR was used to detect the expression levels of DDX11 antisense RNA 1 (DDX11-AS1) and miR-497 in MCF-7 and MDA-MB-231 cells. Cell transfection techniques were used to interfere with the expression levels of DDX11-AS1 and miR-497. Cell Counting Kit-8 and MTT assays were used to detect cell viability. A colony formation assay was used to detect cell proliferation. Wound-healing and Transwell assays were performed to measure the levels of cell migration and invasion. Western blotting was used to analyze the expression levels of migration-associated proteins, and immunofluorescence and western blotting were used to determine the expression levels of the epithelial-mesenchymal transition-related proteins E-cadherin and N-cadherin, respectively. A luciferase reporter gene assay was used to verify the targeted binding of DDX11-AS1 and miR-497. The present study demonstrated that the expression levels of lncRNA DDX11-AS1 were markedly increased in paclitaxel (PTX)-resistant breast cancer cell lines. By contrast, knockdown of DDX11-AS1 expression inhibited PTX resistance of breast cancer cells, and suppressed the proliferation, invasion and migration of breast cancer cells, which was achieved via upregulation of miR-497 expression. In conclusion, knockdown of lncRNA DDX11-AS1 could inhibit the proliferation, migration and PTX resistance of breast cancer cells by upregulating miR-497 expression.
Keywords: breast cancer, long non-coding RNA DDX11 antisense RNA 1, proliferation, migration, paclitaxel resistance, microRNA-497
Introduction
Breast cancer is the most common type of malignant tumor in women. At present, advanced breast cancer can be treated with different drug approaches, including chemotherapy drugs (paclitaxel and anthracyclines), targeted therapy (trastuzumab) and immunotherapy using immune checkpoint inhibitors (1–4). However, multi-course chemotherapy can induce resistance of tumor cells to chemotherapy drugs, leading to treatment failure and tumor progression, which seriously affects the quality of life and long-term survival of patients (5,6).
Research on the function and mechanism of non-coding RNA (ncRNA) has garnered attention in the field of cancer (7). ncRNAs, including microRNAs (miRNAs/miRs) and long ncRNAs (lncRNAs), are involved in different levels of gene expression, including chromatin structure, epigenetic memory, transcription, RNA splicing and translation (8). It has been reported that lncRNAs, as competing endogenous RNAs (ceRNAs), adsorb miRNAs through sponge action, and subsequently regulate the expression of downstream genes at the translational level, thus serving an important regulatory role in the occurrence, development and prognosis of cancer (9). Previous studies have revealed that lncRNAs function as ceRNAs in breast cancer cells, and affect invasion, metastasis, cell proliferation and epithelial-mesenchymal transition (EMT) of breast cancer (10–12). These findings have improved the knowledge of the molecular mechanisms underlying the occurrence and development of breast cancer.
The expression levels of lncRNA DDX11 antisense RNA 1 (DDX11-AS1) in colorectal cancer have been revealed to be positively associated with lymphatic metastasis and TNM stage, and to have a marked effect on the overall survival of patients with colorectal cancer. Conversely, knockdown of DDX11-AS1 has been shown to inhibit the proliferation, migration and invasion of colorectal cancer cells, and to stimulate cell apoptosis (13). A previous study demonstrated that DDX11-AS1 is highly expressed in esophageal cancer tissues, and knockdown of DDX11-AS1 can inhibit DNA topoisomerase IIα transcription by inhibiting TATA-box binding protein-associated factor 1, thus reducing the sensitivity of esophageal cancer cells to paclitaxel (PTX) and inhibiting tumor growth. Therefore, DDX11-AS1 knockdown may be a promising therapeutic strategy for esophageal cancer (14). However, to the best of our knowledge, the effect of lncRNA DDX11-AS1 on proliferation, migration and PTX resistance in breast cancer has yet to be determined.
It has been reported that miR-497 expression is markedly downregulated in breast cancer tissue samples and cell lines (15). In other types of cancer, miR-497 has a tumor-suppressive effect (15,16). Furthermore, previous studies have shown that miR-497 may serve as a biomarker for cancer prognosis (17,18). Notably, the expression levels of miR-497 have been shown to be associated with the chemoresistance phenotype of ovarian cancer, and the overexpression of miR-497 may induce sensitivity of resistant ovarian tumors to cisplatin treatment (19).
In the present study, the expression levels of lncRNA DDX11-AS1 in breast cancer were detected, and its effects on the proliferation and migration of breast cancer cells, as well as PTX resistance, were assessed. Additionally, the present study discussed the mechanism underlying these effects, in order to provide a theoretical basis for targeted therapy of chemotherapy resistance in breast cancer.
Materials and methods
Cell culture
MCF-10A human normal mammary cells, and MCF-7 and MDA-MB-231 human breast cancer cell lines were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. The cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. The PTX-resistant MCF-7/PTX and MDA-MB-231/PTX cell lines were generated by an intermittent and stepwise method (20). Briefly, the parental MCF-7 and MDA-MB-231 cells were initially incubated with PTX [Beyotime Institute of Biotechnology; the concentration of PTX used was the IC50: 0.46 µmol/l in MCF-7 cells; 0.53 µmol/l in MDA-MB-231 cells, as determined using a Cell Counting Kit-8 asay (CCK-8)] under the same conditions as cell culture for 4 days at 37°C, and then cultured in drug-free medium for 3–4 days until normal proliferative ability was recovered. PTX treatment was performed six times during the induction period and the PTX-resistant clones were then harvested. The MCF-7/PTX and MDA-MB-231/PTX cell lines were subsequently cultured in complete medium without PTX.
Datebase
The LncBase website (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex) was used to indicate the binding site between lncRNA DDX11-AS1 and miR-497.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cultured cells using RNAzol® RT (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol. The concentration and purity of RNA were measured using a Nanodrop 2000 (Nanodrop; Thermo Fisher Scientific, Inc.). The RevertAid First Strand cDNA Synthesis kit (cat. no. K1622; Thermo Fisher Scientific, Inc.) was used to synthesize cDNA according to the manufacturer's protocol. The amplification conditions were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 60 sec, according to the manufacturer's protocol, and then amplified in triplicate via qPCR (cobas Z 480 system; Roche Diagnostics) using SYBR Green (final reaction volume, 20 µl; Roche Diagnostics GmbH), according to the manufacturer's protocol. The primer sequences used for qPCR were obtained from GenScript. U6 was used as a reference gene for normalization of DDX11-AS1 and miR-497. GAPDH was used as a reference gene for normalization. The primer sequences used for qPCR were as follows: DDX11-AS1 forward, 5′-CTGGCTACTCTTCCTCCTGG-3′ and reverse, 5′-CAGAGGACATGTGGGAGGTT-3′; miR-497 forward, 5′-GTGCAGGGTCCGAGGT-3′ and reverse, 5′-TAGCCTGCAGCACACTGTGGT-3′; and U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. Fold-changes in lncRNA expression and miR-497 were calculated using the 2−∆∆Cq method (21).
Cell transfection
MCF-7, MDA-MB-231, MCF-7/PTX and MDA-MB-231/PTX cells (5×105 cells/well) were seeded into 12-well plates, and then transfected with short hairpin RNAs (shRNAs) contained in a vector targeting DDX11-AS1 (shRNA-DDX11-AS1-1 and shRNA-DDX11-AS1-2) and negative control (shRNA-NC), miR-497 inhibitors (miR-497 inhibitor-1 and miR-497 inhibitor-2, cat. no. miR20004768-1-5; Guangzhou RiboBio Co., Ltd.) and scrambled negative control (NC) sequences all at a concentration of 20 nM (Shanghai GenePharma Co., Ltd.) for 6 h at 37°C. Cell transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RT-qPCR was used to detect the transfection efficiency at 48 h after transfection. Cells were divided into control, shRNA-DDX11-AS1, shRNA-DDX11-AS1 + inhibitor-NC and shRNA-DDX11-AS1 + miR-497 inhibitor groups.
CCK-8 assay
Cell viability was measured using a CCK-8 assay (Dojindo Molecular Technologies, Inc.), according to the manufacturer's instructions. Cells were seeded into 96-well plates, and trypsinized to prepare a single cell suspension with a density of 2×103 cells/ml. Following incubation at 37°C for 48 h, 10 µl CCK-8 solution was added to each well, and cells were further incubated for 2 h at 37°C. The absorbance was measured at 450 nm using a microplate reader (BioTek Instruments, Inc.). The Y value for 50% suppression (IC50) was calculated according to a linear formula (22).
MTT assay
Cell proliferation was determined using an MTT assay. The treated cells were plated into 96-well plates (5×103 cells/well) and incubated with 0.5 mg/ml MTT (Sigma-Aldrich; Merck KGaA) for 3 h at 37°C. Subsequently, formazan was dissolved with 600 µl DMSO for 15 min. The optical density was measured using an enzyme-linked detector at a wavelength of 490 nm.
Colony formation assay
Cells were seeded into six-well plates at a density of 200 cells/well and cultured at 37°C for 21 days until visible colonies appeared. Subsequently, cells were washed with PBS three times, fixed with methanol for 15 min at room temperature and stained with 0.1% (w/v) crystal violet. Megascopic cell colonies (>10 cells) were counted using Image-Pro Plus 5.0 software (Media Cybernetics, Inc.).
Wound-healing assay
Cell migration was determined using a wound-healing assay. Briefly, transfected cells were plated in 12-well plates at a density of 1×105 cells/well. Once cells reached 80% confluence, the medium was replaced with serum-free DMEM and cells were incubated at 37°C overnight before initiating the experiment. Subsequently, a wound was created on the surface of the cell monolayer using a 200-µl pipette tip. The cells were then rinsed twice with serum-free medium in order to remove free-floating cells and debris. An inverted light microscope (magnification, ×20; BX51; Olympus Corporation) was used to monitor cells at the edges of the scratch. The percentage of wound closure was determined according to the following equation: [(Ai-At)/Ai] ×100, where Ai represents the initial area of the wound at 0 h and At represents the area of the wound after 24 h.
Transwell assay
The 24-well Transwell chambers (pore size, 0.1 µm; Costar; Corning, Inc.) precoated with 50 µl Matrigel (BD Biosciences) at 37°C for 30 min were used to detect cell invasion. Prior to the experiments, the upper and lower chambers were filled with serum-free DMEM and with DMEM supplemented with 10% FBS, respectively. Cells (1.0×105 cells/well) were seeded into the upper chamber and were allowed to invade through the membrane for 48 h at 37°C in an atmosphere containing 5% CO2. Finally, the cells on the upper surface of the membranes (non-invaded cells) were removed and the Transwell filters were fixed with 10% cold methanol at room temperature for 15 min prior to staining with crystal violet (0.1% in 20% ethanol) for 30 min at room temperature. Invaded cells were directly counted using an Olympus light microscope (magnification, ×100; Olympus BX51; Olympus Corporation) and the average cell counts in five random fields were calculated.
Western blotting
Total protein was extracted from cells with RIPA lysis buffer (Thermo Fisher Scientific, Inc.). Total protein was quantified using a BCA protein assay kit (Bio-Rad Laboratories, Inc.). Equal amounts (30 µg) of protein samples were separated by SDS-PAGE on 10% gels and then electrophoretically transferred onto PVDF membranes (EMD Millipore). The PVDF membranes were incubated with primary antibodies overnight at 4°C after blocking with 10% non-fat milk for 1 h at room temperature. Subsequently, the PVDF membranes were incubated with HRP-conjugated secondary antibody (1:5,000; cat. no. ab150077; Abcam) at room temperature for 2 h. Protein signals were detected using a chemiluminescence detection kit (Amersham; Cytiva) and a semi-quantitative analysis was conducted using ImageJ software (version 1.8.0; National Institutes of Health). The following primary antibodies were purchased from Abcam: Anti-matrix metalloproteinase (MMP)2 (1:1,000; cat. no. ab92536), anti-MMP9 (1:1,000; cat. no. ab76003), anti-N-cadherin (1:1,000; cat. no. ab76011) and anti-GAPDH (dilution, 1:1,000; cat. no. ab181602) antibodies.
Immunofluorescence
For staining of E-cadherin protein, the cells were fixed in 4% paraformaldehyde/PBS for 20 min and permeabilized with 0.1% Triton X-100/PBS for 3 min at room temperature. Subsequently, cells were blocked in 2% goat serum plus 1% BSA (both from Thermo Fisher Scientific, Inc.) in PBS for 30 min at room temperature, incubated with primary antibody at 4°C overnight, and then incubated with secondary antibody conjugated to Alexa Fluor 488 (cat. no. A32731; Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for 2 h. The nuclei were counterstained with DAPI. The primary antibody used was anti-E-cadherin (1:200; cat. no. ab40772; Abcam). Images were acquired using a light Leica DM5500B upright microscope (Leica Microsystems GmbH) with a Retiga SRV Cooled CCD camera (Teledyne Photometrics) and ImagePro Plus software (version 6.0; Media Cybernetics, Inc.).
Luciferase reporter gene assay
The interaction between DDX11-AS1 and miR-497 was predicted using LncBase (version 5.0; http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex). The interaction was validated using a dual-luciferase reporter system (cat. no. D0010; Beijing Solarbio Science & Technology Co., Ltd.). The luciferase vectors containing wild-type (WT) and mutant (MUT) binding sites of DDX11-AS1 were constructed using the pGL3 luciferase vector (Promega Corporation). miR-497 mimic and mimic negative control were co-transfected with 20 nM DDX11-AS1-WT or DDX11-AS1-Mut into MCF-10A cells (1.0×105 cells/well) using Lipofectamine 2000 reagent according to the manufacturer's protocol. After transfection for 48 h, the relative luciferase activity was measured using a microplate reader (BD Biosciences) and normalized to Renilla luciferase activity, which was measured using a Renilla luciferase activity kit (pRL-TK; Invitrogen; Thermo Fisher Scientific Inc.). The mimic sequences were as follows: Mimic negative control, sense UUCUCCGAACGUGUCACGUTT, antisense ACGUGACACGUUCGGAGAATT; miR-497 mimic, sense CAGCAGCACACUGUGGUUUGU, antisense AAACCACAGUGUGCUGCUGUU.
Statistical analysis
Data are presented as the mean ± standard deviation. SPSS v22.0 statistical software (IBM Corp.) was used for all statistical analyses. Comparisons among groups were analyzed by one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference. Each experiment was repeated three times.
Results
Expression levels of DDX11-AS1 in PTX-resistant breast cancer cell lines
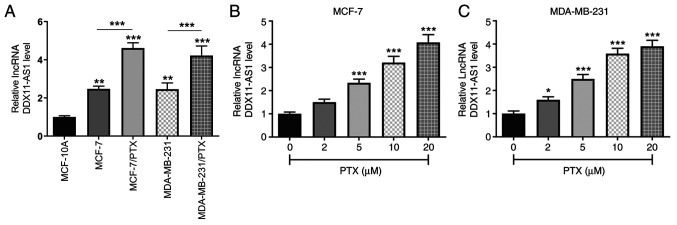
The expression levels of DDX11-AS1 in MCF-10A, MCF-7, MDA-MB-231, and PTX-resistant MCF-7/PTX and MDA-MB-231/PTX cell lines were detected by RT-qPCR (Fig. 1A). It was revealed that DDX11-AS1 expression was markedly increased in breast cancer cell lines compared with that in MCF-10A cells. Compared with those in MCF-7 cells, the expression levels of DDX11-AS1 in MCF-7/PTX cells were further increased, which was consistent with the expression levels detected in MDA-MB-231/PTX cells. Subsequently, the expression levels of DDX11-AS1 in MCF-7 and MDA-MB-231 cells treated with different concentrations of PTX were detected by RT-qPCR, and it was revealed that with increasing PTX concentration, the expression levels of DDX11-AS1 in breast cancer cells were increased (Fig. 1B and C).
Figure 1.
Expression of DDX11-AS1 in PTX-resistant breast cancer cell lines. (A) RT-qPCR assay was used to detect the expression levels of DDX11-AS1 in breast cancer cell lines. **P<0.01, ***P<0.001 vs. MCF-10A. RT-qPCR assay was used to detect the expression levels of DDX11-AS1 in (B) MCF-7 and (C) MDA-MB-231 cell lines. *P<0.05, ***P<0.001 vs. 0 µM PTX. DDX11-AS1, DDX11 antisense 1; lncRNA, long non-coding RNA; PTX, paclitaxel; RT-qPCR, reverse transcription-quantitative PCR.
Knockdown of DDX11-AS1 reduces the sensitivity of drug-resistant breast cancer cells to PTX
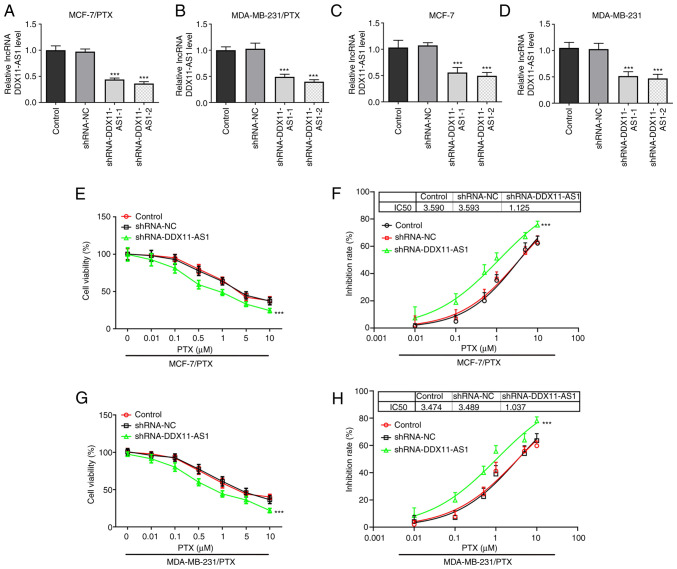
Cell transfection was used to knockdown DDX11-AS1 expression in two PTX-resistant cell lines, and MCF-7 and MDA-MB-231 cells. Cell transfection efficiency was detected by RT-qPCR (Fig. 2A-D). shRNA-DDX11-AS1-2 was selected for the subsequent experiments. The cells were divided into control, shRNA-NC and shRNA-DDX11-AS1 groups. Notably, a 3-fold reduction in PTX IC50 was observed in the shRNA-DDX11-AS1 groups compared with that in the shRNA-NC groups in MCF-7/PTX (Fig. 2E and F) and MDA-MB-231/PTX (Fig. 2G and H) cells.
Figure 2.
Knockdown of DDX11-AS1 enhances the sensitivity of drug-resistant breast cancer cells to PTX. Reverse transcription-quantitative PCR assay detected the expression levels of DDX11-AS1 in (A) MCF-7/PTX, (B) MDA-MB-231/PTX, (C) MCF-7 and (D) MDA-MB-231 cells after transfection. CCK-8 assay detected (E) cell viability and (F) cell inhibition rate in the MCF-7/PTX cell line. CCK-8 assay detected (G) cell viability and (H) cell inhibition rate in the MDA-MB-231/PTX cell line. ***P<0.001 vs. shRNA-NC. CCK-8, Cell Counting Kit-8; DDX11-AS1, DDX11 antisense 1; lncRNA, long non-coding RNA; NC, negative control; PTX, paclitaxel; shRNA, short hairpin RNA.
Knockdown of DDX11-AS1 inhibits the proliferation of breast cancer cell lines
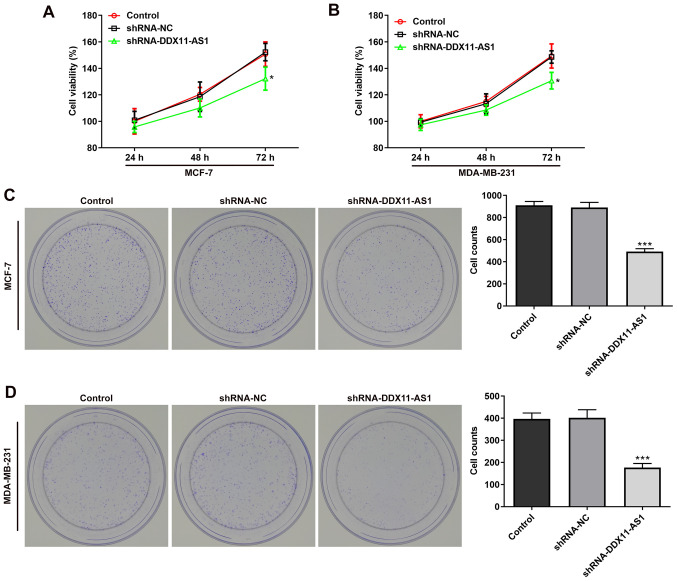
MTT (Fig. 3A and B) and colony formation (Fig. 3C and D) assays demonstrated that the viability of cells in the shRNA-DDX11-AS1 group was markedly decreased compared with that of parental cells in the shRNA-NC group. These results demonstrated that knockdown of DDX11-AS1 inhibited the proliferation of breast cancer cells.
Figure 3.
Knockdown of DDX11-AS1 inhibits the proliferation of breast cancer cells. MTT assay detected the viability of (A) MCF-7 and (B) MDA-MB-231 cells. Colony formation assay was used to detect the reproductive capacity of (C) MCF-7 and (D) MDA-MB-231. Clones were counted. *P<0.05, ***P<0.001 vs. shRNA-NC. DDX11-AS1, DDX11 antisense 1; NC, negative control; PTX, paclitaxel; shRNA, short hairpin RNA.
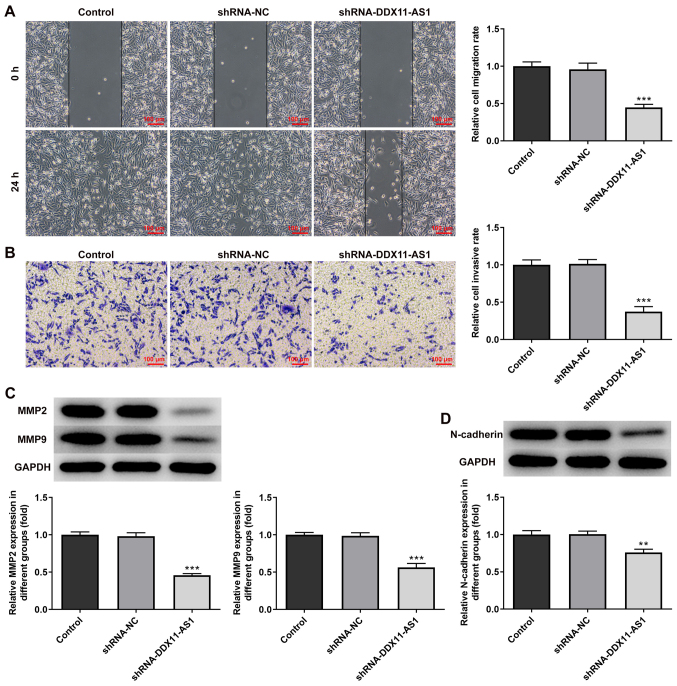
Knockdown of DDX11-AS1 inhibits the invasion and migration of breast cancer cells
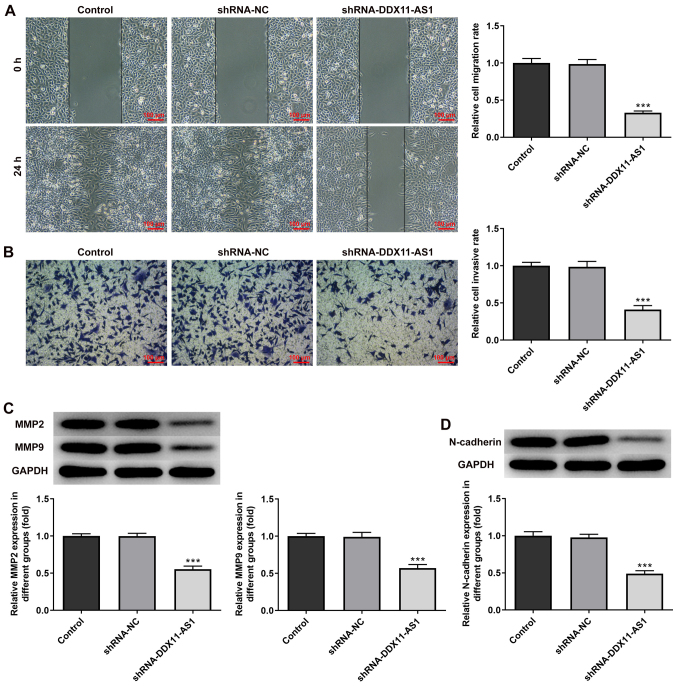
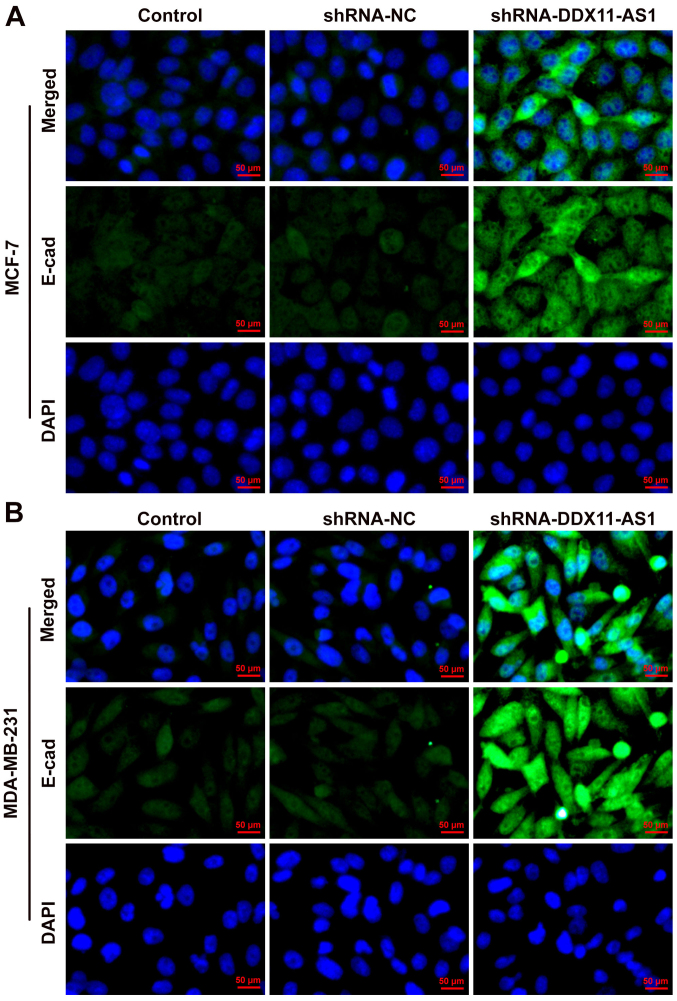
The present study used wound-healing and Transwell assays to detect the levels of cell migration and invasion. In MCF-7 cells, the results of the wound-healing assay revealed that compared with that of the shRNA-NC group, the cell migration ability of the shRNA-DDX11-AS1 group was markedly decreased (Fig. 4A). The results of the Transwell assay revealed that the invasion ability of cells in the shRNA-DDX11-AS1 group was markedly decreased compared with that of cells in the shRNA-NC group (Fig. 4B). Western blotting was used to detect the expression levels of migration-related proteins (MMP2 and MMP9); their trend was consistent with that of Transwell and wound-healing experiments (Fig. 4C). Subsequently, N-cadherin expression was detected by western blotting. The results revealed that N-cadherin expression was markedly decreased following DDX11-AS1 knockdown (Fig. 4D). In MDA-MB-231 cells, the effects of DDX11-AS1 knockdown on invasion and migration were consistent with those in MCF-7 cells (Fig. 5A-D). Subsequently, the expression of the EMT-related protein E-cadherin was detected by immunofluorescence; the results revealed that E-cadherin expression was markedly increased after DDX11-AS1 knockdown in MCF-7 and MDA-MB-231 cells (Fig. 6A and B).
Figure 4.
Knockdown of DDX11-AS1 inhibits invasion and migration of MCF-7 cells. (A) Wound-healing assay was used to detect cell migration. (B) Transwell assay was used to detect cell invasion. Western blotting detected the protein expression levels of (C) MMP2 and MMP9, and (D) N-cadherin. ***P<0.001 vs. shRNA-NC. DDX11-AS1, DDX11 antisense 1; MMP, matrix metalloproteinase; NC, negative control; PTX, paclitaxel; shRNA, short hairpin RNA.
Figure 5.
Knockdown of DDX11-AS1 inhibits invasion and migration of MDA-MB-231 cells. (A) Wound-healing assay was used to detect cell migration. (B) Transwell assay was used to detect cell invasion. Western blotting detected the protein expression levels of (C) MMP2 and MMP9, and (D) N-cadherin. **P<0.01, ***P<0.001 vs. shRNA-NC. DDX11-AS1, DDX11 antisense 1; MMP, matrix metalloproteinase; NC, negative control; PTX, paclitaxel; shRNA, short hairpin RNA.
Figure 6.
Expression of E-cadherin in breast cancer cell lines. Immunofluorescence was used to detect the expression of E-cadherin in (A) MCF-7 and (B) MDA-MB-231 cells. DDX11-AS1, DDX11 antisense 1; NC, negative control; shRNA, short hairpin RNA.
Knockdown of DDX11-AS1 upregulates the expression levels of miR-497
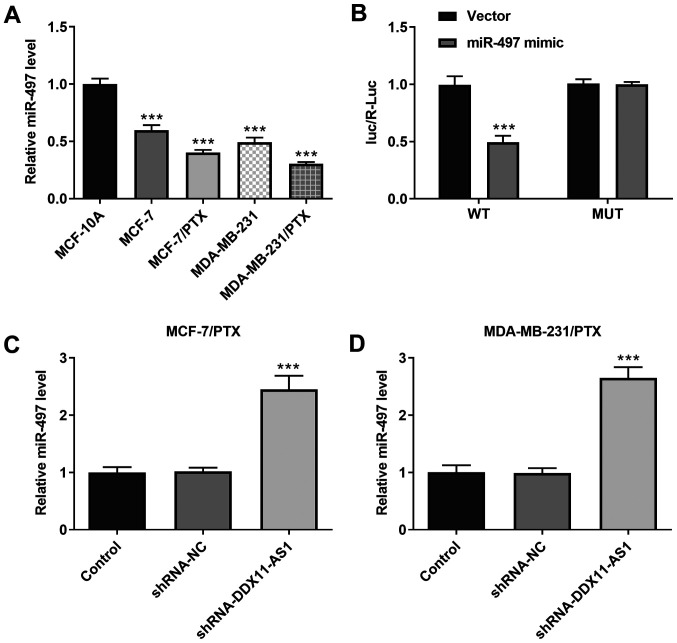
It was demonstrated that the expression levels of miR-497 in PTX-resistant cell lines were markedly decreased (Fig. 7A). In addition, the targeted binding of DDX11-AS1 and miR-497 was verified using a luciferase reporter gene assay (Fig. 7B). Furthermore, the expression levels of miR-497 in PTX-resistant cell lines were markedly increased following DDX11-AS1 interference (Fig. 7C and D). These results suggested that knockdown of DDX11-AS1 may upregulate miR-497 expression.
Figure 7.
Knockdown of DDX11-AS1 upregulates the expression levels of miR-497. (A) RT-qPCR assay was used to detect the expression levels of miR-497 in breast cancer cell lines. ***P<0.001 vs. MCF-10A. (B) Luciferase reporter gene assay was used to verify the relationship between DDX11-AS1 and miR-497. ***P<0.001 vs. Vector. RT-qPCR assay detected the expression levels of miR-497 in (C) MCF-7/PTX and (D) MDA-MB-231/PTX cells after transfection. ***P<0.001 vs. shRNA-NC. DDX11-AS1, DDX11 antisense 1; miR-497, microRNA-497; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; shRNA, short hairpin RNA.
Knockdown of DDX11-AS1 reduces the sensitivity of drug-resistant breast cancer cells to PTX via upregulation of miR-497 expression
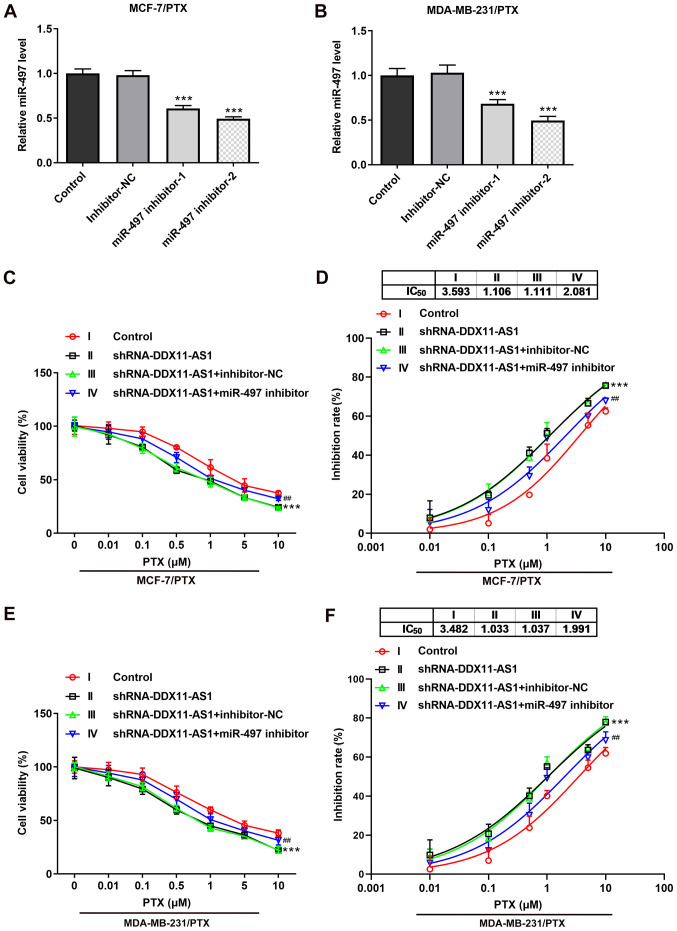
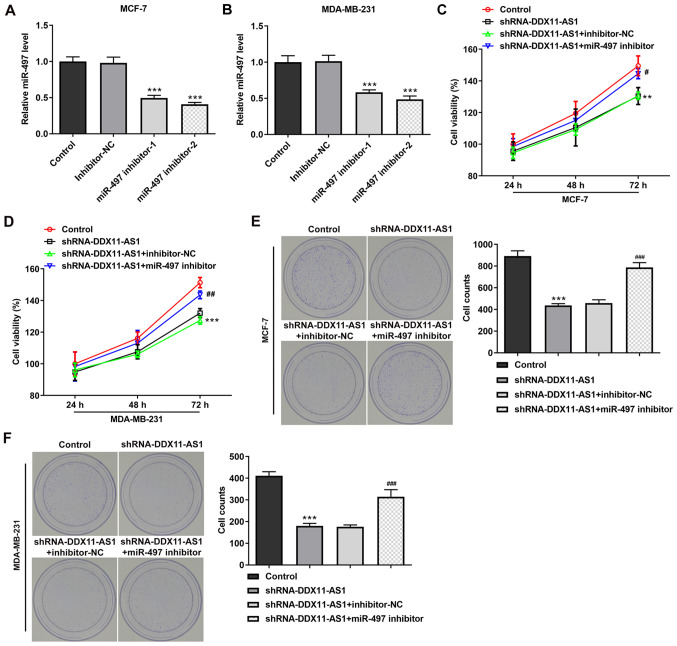
Transfection was used to knockdown miR-497 expression in PTX-resistant cell lines, and transfection efficiency was detected by RT-qPCR (Fig. 8A and B). miR-497 inhibitor-2 was selected for subsequent experiments. Cells were divided into control, shRNA-DDX11-AS1, shRNA-DDX11-AS1 + inhibitor-NC and shRNA-DDX11-AS1 + miR-497 inhibitor groups. The results of the CCK-8 assay demonstrated that the IC50 value of the shRNA-DDX11-AS1 + miR-497 inhibitor group was markedly increased compared with that of the shRNA-DDX11-AS1 + inhibitor-NC group in MCF-7/PTX (Fig. 8C and D) and MDA-MB-231/PTX (Fig. 8E and F) cells.
Figure 8.
Knockdown of DDX11-AS1 reduces the sensitivity of drug-resistant breast cancer cells to PTX by targeting the upregulation of miR-497. RT-qPCR assay detected the expression of miR-497 in (A) MCF-7/PTX and (B) MDA-MB-231/PTX cells after transfection. ***P<0.001 vs. Inhibitor-NC. CCK-8 assay detected (C) cell viability and (D) cell inhibition rate in the MCF-7/PTX cell line. CCK-8 assay detected (E) cell viability and (F) cell inhibition rate in the MDA-MB-231/PTX cell line. ***P<0.001 vs. Control; ##P<0.01 vs. shRNA-DDX11-AS1 + inhibitor-NC. DDX11-AS1, DDX11 antisense 1; miR-497, microRNA-497; NC, negative control; PTX, paclitaxel; shRNA, short hairpin RNA.
Knockdown of DDX11-AS1 inhibits the proliferation, invasion and migration of breast cancer cells via upregulation of miR-497
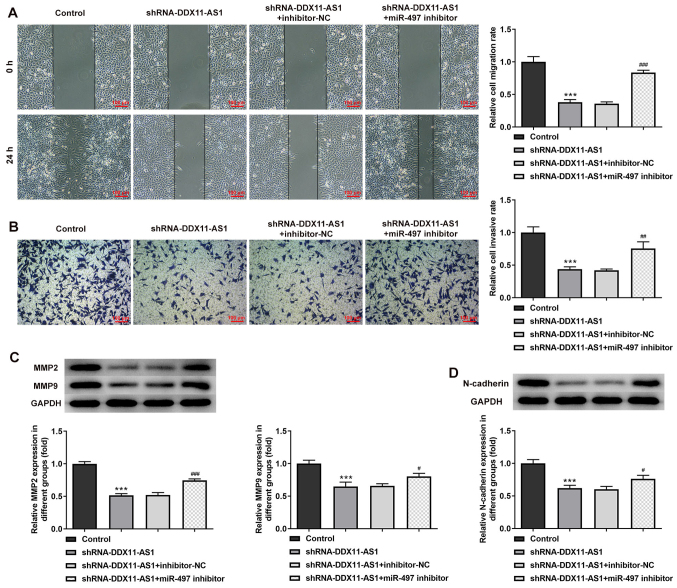
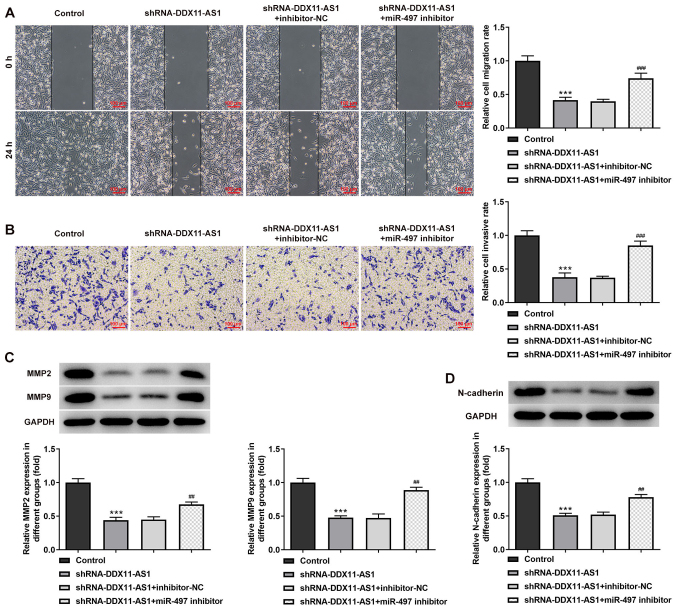
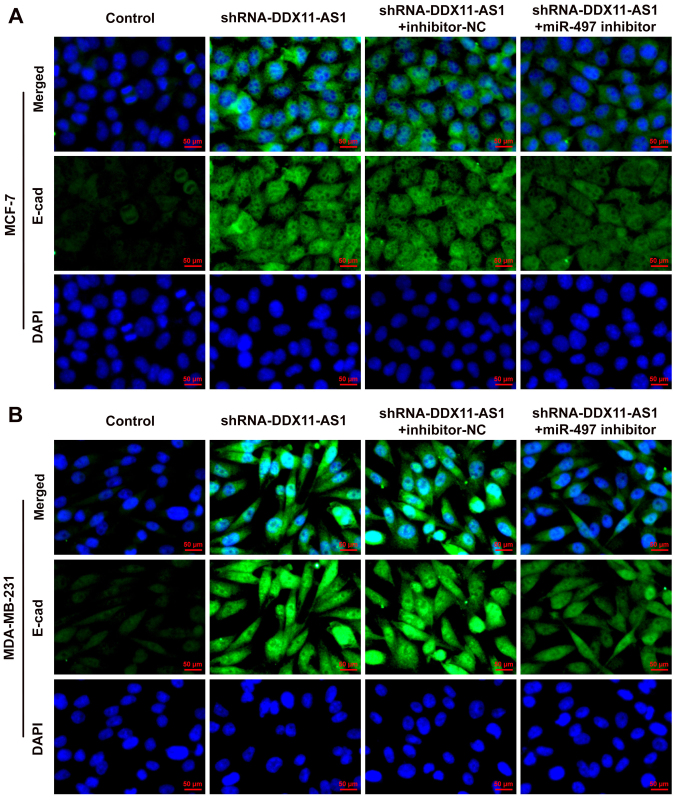
Cell transfection was used to interfere with miR-497 expression in breast cancer parental cells, and the transfection efficiency was detected by RT-qPCR (Fig. 9A and B). miR-497 inhibitor-2 was selected for subsequent experiments. The cells were divided into control, shRNA-DDX11-AS1, shRNA-DDX11-AS1 + inhibitor-NC and shRNA-DDX11-AS1 + miR-497 inhibitor groups. The results of MTT (Fig. 9C and D) and colony formation (Fig. 9E and F) assays demonstrated that the proliferative ability of cells in the shRNA-DDX11-AS1 + miR-497 inhibitor group was increased compared with that of cells in the shRNA-DDX11-AS1 + inhibitor-NC group. In addition, in MCF-7 cells, the invasion (Fig. 10A) and migration (Fig. 10B) of cells in the shRNA-DDX11-AS1 + miR-497 inhibitor group were markedly increased compared with those of cells in the shRNA-DDX11-AS1 + inhibitor-NC group, and MMP2 and MMP9 expression was increased (Fig. 10C). Finally, the expression levels of EMT-related proteins (E-cadherin and N-cadherin) were detected. The expression levels of N-cadherin (Fig. 10D) in MCF-7 cells in the shRNA-DDX11-AS1 + miR-497 inhibitor group were also increased compared with those in cells in the shRNA-DDX11-AS1 + inhibitor-NC group. In MDA-MB-231 cells, the trend of invasion and migration was consistent with that in MCF-7 cells (Fig. 11A-D). Subsequently, the expression levels of the EMT-related protein E-cadherin were detected by immunofluorescence, and the results revealed that the expression levels of E-cadherin in cells in the shRNA-DDX11-AS1 + miR-497 inhibitor group were decreased compared with those in cells in the shRNA-DDX11-AS1 + inhibitor-NC group (Fig. 12A and B). These results suggested that knockdown of DDX11-AS1 inhibited the proliferation, invasion and migration of breast cancer cells via upregulation of miR-497 expression.
Figure 9.
Knockdown of DDX11-AS1 inhibits the proliferation of breast cancer cells by targeting miR-497. Reverse transcription-quantitative PCR assay detected the expression of miR-497 in (A) MCF-7 and (B) MDA-MB-231 cells after transfection. ***P<0.001 vs. Inhibitor-NC. MTT detected cell viability in (C) MCF-7 and (D) MDA-MB-231 cells. Colony formation assay was used to detect the reproductive capacity of (E) MCF-7 and (F) MDA-MB-231 cells. Clones were counted. **P<0.05, ***P<0.001 vs. Control; #P<0.05, ##P<0.01, ###P<0.001 vs. shRNA-DDX11-AS1 + inhibitor-NC. DDX11-AS1, DDX11 antisense 1; miR-497, microRNA-497; NC, negative control; shRNA, short hairpin RNA.
Figure 10.
Knockdown of DDX11-AS1 inhibits the invasion and migration of MCF-7 cells by targeting miR-497. (A) Wound-healing assay was used to detect cell migration. (B) Transwell assay was used to detect cell invasion. Western blotting was used to detect the protein expression levels of (C) MMP2 and MMP9, and (D) N-cadherin. ***P<0.001 vs. Control; #P<0.05, ##P<0.01, ###P<0.001 vs. shRNA-DDX11-AS1 + inhibitor-NC. DDX11-AS1, DDX11 antisense 1; miR-497, microRNA-497; MMP, matrix metalloproteinase; NC, negative control; shRNA, short hairpin RNA.
Figure 11.
Knockdown of DDX11-AS1 inhibits the invasion and migration of MDA-MB-231 cells by targeting miR-497. (A) Wound-healing assay was used to detect cell migration. (B) Transwell assay was used to detect cell invasion. Western blotting was used to detect the protein expression levels of (C) MMP2 and MMP9, and (D) N-cadherin. ***P<0.001 vs. Control; ##P<0.01, ###P<0.001 vs. shRNA-DDX11-AS1 + inhibitor-NC. DDX11-AS1, DDX11 antisense 1; miR-497, microRNA-497; MMP, matrix metalloproteinase; NC, negative control; shRNA, short hairpin RNA.
Figure 12.
Expression of E-cadherin in breast cancer cell lines. Immunofluorescence was used to detect the expression of E-cadherin in (A) MCF-7 and (B) MDA-MB-231 cells. DDX11-AS1, DDX11 antisense 1; miR-497, microRNA-497; NC, negative control; shRNA, short hairpin RNA.
Discussion
Chemotherapy has gradually become the main method of breast cancer treatment. Cisplatin, carboplatin, docetaxel, gemcitabine, PTX, tamoxifen and other chemotherapy drugs are able to improve the quality of life and overall survival of patients to a certain extent (23). However, chemotherapy resistance greatly reduces the efficacy of targeted therapy for breast cancer, and causes the phenomenon of drug resistance in breast cancer cells (24). Research has demonstrated that due to drug resistance, the 5-year survival rate of patients with breast cancer is still <20% worldwide (25). Therefore, it is necessary to study the drug resistance markers and related mechanisms of breast cancer in order to reverse chemotherapy drug resistance.
The newly discovered lncRNA DDX11-AS1 has been reported to be abnormally highly expressed in a number of malignant tumors, such as liver cancer (26), colorectal cancer (13) and bladder cancer (27). These findings suggested that DDX11-AS1 may be a tumor marker or therapeutic target. In addition, it has been reported that the elimination of DDX11-AS1 reduced the resistance of esophageal cancer cells to PTX (14). This finding indicated that the drug resistance of tumor cells may be overcome by regulating DDX11-AS1.
The present study revealed that lncRNA DDX11-AS1 expression was not only abnormally increased in breast cancer cells, but also markedly increased in PTX-resistant breast cancer cells. The present experimental results were consistent with those of Zhang et al (14). Interfering with DDX11-AS1 inhibited the PTX resistance of breast cancer cells, and suppressed the proliferation, invasion and migration of breast cancer cells.
In terms of the underlying mechanism, it was revealed that DDX11-AS1 could regulate miR-497 expression. The present study revealed that lncRNA DDX11-AS1 could target miR-497 through the LncBase website. Subsequently, the binding relationship between the two was verified using the luciferase reporter gene assay. To the best of our knowledge, there has been no previous study on the relationship between lncRNA DDX11-AS1 and miR-497. To the best of our knowledge, the present study is the first to verify the relationship between lncRNA DDX11-AS1 and miR-497, and study the regulatory effects of the two on the proliferation, invasion and drug resistance of breast cancer cells. miR-497 has been shown to be abnormally expressed in osteosarcoma (28), melanoma (29), squamous cell carcinoma (30) and may be involved in regulation of the proliferation, invasion and apoptosis of cancer cells. miR-497 expression was previously revealed to be markedly downregulated in breast cancer tissue samples and cell lines (31,32). Furthermore, a previous study demonstrated that miR-497 reduced cisplatin resistance of ovarian cancer cells by targeting mTOR/P70S6K1 (19). In addition, upregulation of miR-497 induced resistance of human glioma cells to temozolomide by targeting mTOR/Bcl-2 (33), which showed opposite results compared with the current study. These findings suggested that miR-497 may serve an important role in tumor resistance. Therefore, it was hypothesized that lncRNA DDX11-AS1 may affect breast cancer cells and drug resistance by regulating miR-497. The present study revealed that knockdown of DDX11-AS1 enhanced the sensitivity of drug-resistant breast cancer cells to PTX, and inhibited the proliferation, invasion and migration of breast cancer cells via upregulation of miR-497 expression.
In conclusion, knockdown of lncRNA DDX11-AS1 may inhibit the proliferation, migration and PTX resistance of breast cancer cells by upregulating miR-497 expression. The current study provides a foundation for clinical breast cancer treatment and targeted therapy of breast cancer resistance.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ML and BZ wrote the manuscript and analyzed the data. MW and JJ performed the experiments and supervised the study. ML searched the literature and revised the manuscript for important intellectual content. ML and BZ confirmed the authenticity of all the raw data. All authors read and approved the final manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bevers TB, Helvie M, Bonaccio E, Calhoun KE, Daly MB, Farrar WB, Garber JE, Gray R, Greenberg CC, Greenup R, et al. Breast cancer screening and diagnosis, version 3.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2018;16:1362–1389. doi: 10.6004/jnccn.2018.0083. [DOI] [PubMed] [Google Scholar]
- 2.Falzone L, Salomone S, Libra M. Evolution of cancer pharmacological treatments at the turn of the third millennium. Front Pharmacol. 2018;9:1300. doi: 10.3389/fphar.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christofi T, Baritaki S, Falzone L, Libra M, Zaravinos A. Current perspectives in cancer immunotherapy. Cancers (Basel) 2019;11:1472. doi: 10.3390/cancers11101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong CWS, Wu M, Cho WCS, To KKW. Recent advances in the treatment of breast cancer. Front Oncol. 2018;8:227. doi: 10.3389/fonc.2018.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mustacchi G, De Laurentiis M. The role of taxanes in triple-negative breast cancer: Literature review. Drug Des Devel Ther. 2015;9:4303–4318. doi: 10.2147/DDDT.S86105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telli M. Evolving treatment strategies for triple-negative breast cancer. J Natl Compr Canc Netw. 2015;13((Suppl 5)):S652–S654. doi: 10.6004/jnccn.2015.0194. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Wu KJ, Jia QJ, Ding XF. Roles of miRNA and lncRNA in triple-negative breast cancer. J Zhejiang Univ Sci B. 2020;21:673–689. doi: 10.1631/jzus.B1900709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. Spec No: 1. [DOI] [PubMed] [Google Scholar]
- 9.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Polo A, Crispo A, Cerino P, Falzone L, Candido S, Giudice A, Petro GD, Ciliberto G, Montella M, Budillon A, Costantini S. Environment and bladder cancer: Molecular analysis by interaction networks. Oncotarget. 2017;8:65240–65252. doi: 10.18632/oncotarget.18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falzone L, Grimaldi M, Celentano E, Augustin LSA, Libra M. Identification of modulated MicroRNAs associated with breast cancer, diet, and physical activity. Cancers (Basel) 2020;12:2555. doi: 10.3390/cancers12092555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19:508–517. [PubMed] [Google Scholar]
- 13.Tian JB, Cao L, Dong GL. Long noncoding RNA DDX11-AS1 induced by YY1 accelerates colorectal cancer progression through targeting miR-873/CLDN7 axis. Eur Rev Med Pharmacol Sci. 2019;23:5714–5729. doi: 10.26355/eurrev_201907_18309. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Jiang H, Xu Z, Jiang Y, She Y, Huang X, Feng S, Chen W, Chen S, Chen Y, et al. The resistance of esophageal cancer cells to paclitaxel can be reduced by the knockdown of long noncoding RNA DDX11-AS1 through TAF1/TOP2A inhibition. Am J Cancer Res. 2019;9:2233–2248. [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z, Cai X, Huang C, Xu J, Liu A. miR-497 suppresses angiogenesis in breast carcinoma by targeting HIF-1α. Oncol Rep. 2016;35:1696–1702. doi: 10.3892/or.2015.4529. [DOI] [PubMed] [Google Scholar]
- 16.Tian F, Zhan Y, Zhu W, Li J, Tang M, Chen X, Jiang J. MicroRNA-497 inhibits multiple myeloma growth and increases susceptibility to bortezomib by targeting Bcl-2. Int J Mol Med. 2019;43:1058–1066. doi: 10.3892/ijmm.2018.4019. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Wu S, Wang L, Kang S, Zhao B, He F, Liu X, Zeng Y, Liu J. Prognostic value of microRNA-497 in various cancers: A systematic review and meta-analysis. Dis Markers. 2019;2019:2491291. doi: 10.1155/2019/2491291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo G, He K, Xia Z, Liu S, Liu H, Xiang G. Regulation of microRNA-497 expression in human cancer. Oncol Lett. 2021;21:23. doi: 10.3892/ol.2020.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu S, Fu GB, Tao Z, OuYang J, Kong F, Jiang BH, Wan X, Chen K. miR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget. 2015;6:26457–26471. doi: 10.18632/oncotarget.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panayotopoulou EG, Müller AK, Borries M, Busch H, Hu G, Lev S. Targeting of apoptotic pathways by SMAC or BH3 mimetics distinctly sensitizes paclitaxel-resistant triple negative breast cancer cells. Oncotarget. 2017;8:45088–45104. doi: 10.18632/oncotarget.15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Lin T, Liao X, Li Z, Lin R, Qi X, Chen G, Sun L, Lin L. Network pharmacology based research into the effect and mechanism of Yinchenhao Decoction against Cholangiocarcinoma. Chin Med. 2021;16:13. doi: 10.1186/s13020-021-00423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan MS, Ansari J, Spooner D, Hussain SA. Chemotherapy for breast cancer (Review) Oncol Rep. 2010;24:1121–1131. doi: 10.3892/or_00000963. [DOI] [PubMed] [Google Scholar]
- 24.Chun KH, Park JH, Fan S. Predicting and overcoming chemotherapeutic resistance in breast cancer. Adv Exp Med Biol. 2017;1026:59–104. doi: 10.1007/978-981-10-6020-5_4. [DOI] [PubMed] [Google Scholar]
- 25.Malhotra A, Jain M, Prakash H, Vasquez KM, Jain A. The regulatory roles of long non-coding RNAs in the development of chemoresistance in breast cancer. Oncotarget. 2017;8:110671–110684. doi: 10.18632/oncotarget.22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Zhuang W, Huang M, Li X. Long noncoding RNA DDX11-AS1 epigenetically represses LATS2 by interacting with EZH2 and DNMT1 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2019;514:1051–1057. doi: 10.1016/j.bbrc.2019.05.042. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Chen J, Gao J, Zhang Y, Ma Y, Wei W, Wei Y. LncRNA DDX11-AS1 promotes bladder cancer occurrence via protecting LAMB3 from downregulation by sponging miR-2355-5p. Cancer Biother Radiopharm. 2020;35:319–328. doi: 10.1089/cbr.2019.3021. [DOI] [PubMed] [Google Scholar]
- 28.Pang PC, Shi XY, Huang WL, Sun K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20:3765–3769. [PubMed] [Google Scholar]
- 29.Chai L, Kang XJ, Sun ZZ, Zeng MF, Yu SR, Ding Y, Liang JQ, Li TT, Zhao J. miR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res. 2018;10:989–1003. doi: 10.2147/CMAR.S163335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei XH, Gu XL, Zhou XT, Ma M, Lou CX. miR-497 promotes the progression of cutaneous squamous cell carcinoma through FAM114A2. Eur Rev Med Pharmacol Sci. 2018;22:7348–7355. doi: 10.26355/eurrev_201811_16272. [DOI] [PubMed] [Google Scholar]
- 31.Zhong H, Yang J, Zhang B, Wang X, Pei L, Zhang L, Lin Z, Wang Y, Wang C. LncRNA GACAT3 predicts poor prognosis and promotes cell proliferation in breast cancer through regulation of miR-497/CCND2. Cancer Biomark. 2018;22:787–797. doi: 10.3233/CBM-181354. [DOI] [PubMed] [Google Scholar]
- 32.Bai J, Zhao WY, Li WJ, Ying ZW, Jiang DQ. Long noncoding RNA LINC00473 indicates a poor prognosis of breast cancer and accelerates tumor carcinogenesis by competing endogenous sponging miR-497. Eur Rev Med Pharmacol Sci. 2019;23:3410–3420. doi: 10.26355/eurrev_201904_17705. [DOI] [PubMed] [Google Scholar]
- 33.Zhu D, Tu M, Zeng B, Cai L, Zheng W, Su Z, Yu Z. Up-regulation of miR-497 confers resistance to temozolomide in human glioma cells by targeting mTOR/Bcl-2. Cancer Med. 2017;6:452–462. doi: 10.1002/cam4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.