Abstract
In recent years, researchers have found that epigenetics plays an important role in the occurrence and development of hepatocellular carcinoma (HCC). DNA methylation is involved in the proliferation and metastasis of HCC. However, the junctophilin 3 (JPH3) level and the potential regulatory mechanism of its DNA methylation in HCC remain uncertain. In the present study, 73 HCC samples were enrolled to analyze the expression of JPH3. Reverse-transcription quantitative PCR, western blotting and immunohistochemistry were used to detect the expression of JPH3 in HCC. Kaplan-Meier method and Cox regression analysis were applied to evaluate the prognostic impact of JPH3 on HCC patients. DNA methylation-specific PCR and bisulfite Sanger sequencing were used to detect the degree of DNA methylation of JPH3 in HCC. The demethylation drug 5-Aza-2′-deoxycytidine (5-Aza) was used to reduce the DNA methylation of JPH3. The role of JPH3 in the malignant biological behavior of HCC by promoting epithelial-mesenchymal transition (EMT) was confirmed by functional cell experiments. The results showed that JPH3 exhibited low levels in HCC tissues and cell lines. HCC patients with low expression of JPH3 had poor survival outcomes. JPH3 had higher DNA methylation levels in HCC tissues and cell lines. When the demethylation drug 5-Aza was used to reduce the degree of methylation of JPH3, its protein expression level was significantly increased and it significantly inhibited the malignant biological behavior of HCC cells. Additionally, effective increase in the expression of JPH3 through gene regulation technology also inhibited the proliferation, invasion and migration of HCC cells. After altering the DNA methylation level of JPH3, the EMT of HCC cells was also affected. Therefore, our study demonstrated the inactivation of JPH3 by promoter methylation and its function as a tumor suppressor in HCC. JPH3 may serve as a biomarker for early diagnosis and as a potential therapeutic target for HCC.
Keywords: hepatocellular carcinoma, methylation, proliferation, migration, junctophilin 3, JPH3, epithelial-mesenchymal transition
Introduction
Hepatocellular carcinoma (HCC) is the third most commonly diagnosed malignant tumor (1). The symptoms of patients are not obvious during the initial stage, and consequently, most patients with HCC are diagnosed at the middle and late stages of the disease, often accompanied by intrahepatic and extrahepatic metastasis (2). The prognosis is poor, which brings a heavy burden to these patients. Approximately 110,000 people die of liver cancer in China every year, accounting for 45% of the world's liver cancer deaths (3). The detection of serum α-fetoprotein (AFP) combined with ultrasound imaging can provide a reliable clinical basis for the early detection of liver cancer, which allows for its successful resection (3). Combined with active comprehensive treatment, resection can significantly improve the 5-year survival rate of liver cancer patients. However, recurrence and metastasis often occur in patients with liver cancer, resulting in low quality of life among patients with liver cancer. At present, there is a lack of effective prevention and treatment measures in the clinic. Therefore, it is of great significance to clarify the molecular mechanism of HCC and to find effective molecular therapeutic target genes.
The main function of junctophilin is to participate in the formation of coupled membrane complexes, maintain a very short distance between the cell membrane and endoplasmic reticulum, and provide a structural basis for signal transduction between the cell membrane and endoplasmic reticulum, such as calcium-induced aluminum release process of cardiomyocytes, voltage-dependent release in skeletal muscle, and library controlled calcium influx (4,5). As an important member of the junctophilin family, junctophilin 3 (JPH3 is widely involved in the pathophysiological process of cells, but its role in HCC is unknown (6).
In recent years, it has been found that epigenetics plays an important role in the malignant occurrence and development of HCC (7). Epigenetic changes refer to alterations in the functions and characteristics of genes in various ways without changing the target gene sequence, and they can be inherited through genomic modification during cell division and proliferation cycle including DNA methylation, genomic imprinting, maternal effects, and gene silencing (8,9). These epigenetic modifications are widely involved in the physiological and pathological processes, variability and adaptability of organisms and can be affected by gene and environmental factors (10).
JPH3 has been reported to play a tumor-suppressor role in a variety of gastrointestinal tumors (11). Previous literature has reported that DNA methylation regulates the expression of JPH3 and causes its low expression in tumors, which plays a biological role in promoting the malignant progression of tumors (6). However, it is still unclear whether it also displays the above phenomenon in HCC. Our results showed that JPH3 is expressed at a low level in HCC, thus DNA methylation is suspected to cause the low expression of JPH3. In the present study, the relationship between its DNA methylation level and protein expression was analyzed. Furthermore, the relationship between its expression level and the prognosis of HCC patients was also analyzed. In cell function experiments, we used demethylation drugs to observe the effect of JPH3 on the malignant biological behavior of HCC cells after altering the DNA methylation level of JPH3 and found that EMT regulates HCC.
Materials and methods
Tissue samples and related clinical data
Seventy-three patients with primary hepatocellular carcinoma were diagnosed at the Department of Hepatobiliary Pancreatic Surgery, Affiliated Hospital of Zunyi Medical University, and received surgical treatment (from May 2012 to November 2015; age range, 25–71 years; sex distribution, 34 male, 39 female). The collected data included HCC tissues and paired adjacent tissues. The present study was performed in accordance with the principles outlined in the Declaration of Helsinki. The Human Trial Ethics Committee of the Affiliated Hospital of Zunyi Medical University approved this study (ethical approval no. 2011035). Written informed consent was obtained from the patients who provided the specimens.
The detailed inclusion criteria were as follows: i) HCC was confirmed by postoperative pathology, and the histopathological diagnosis was clear; ii) patients were diagnosed with HCC for the first time without distant metastasis; iii) patients had not received any treatment prior to surgery; iv) no other serious malignant disease had been diagnosed; v) the clinical, pathological and surgical data were complete; vi) the follow-up information was complete and available.
The exclusion criteria were as follows: i) those who could not communicate normally; ii) patients with organic dysfunction of the heart, brain, lung and kidney; iii) patients who were diagnosed with HCC for the first time with distant metastasis; iv) patients with incomplete clinical and pathological data; v) patients with systemic diseases; vi) patients with autoimmune dysfunction; and vii) follow-up information was not complete and available.
Cell culture and 5-Aza
HCC cell lines Huh-7, MHCC-97H, MHCC-97L, Hep3B, and human normal hepatocytes THLE2 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM high glucose medium [containing 10% fetal bovine serum (FBS)] and cultured in an incubator at constant temperature of 37°C with 5% CO2. Huh-7 and Hep3B cells were treated with 5-aza-2′-deoxycytidine (5-Aza) (Sigma-Aldrich; Merck KGaA) at 1 µM for 4 days as previously reported (12). The culture medium and 5-Aza were replaced daily.
Cell transfection
Plasmid JPH3 and the plasmid control were synthesized by Shanghai GenePharma Co., Ltd. Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) was used for transient transfection, following the manufacturer's instructions. The cells were harvested at 48 h following transfection.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted from 100 mg HCC tissues, paired adjacent tissue and HCC cell lines using TRIzol reagent (Takara Bio, Inc.). Reverse transcription of cDNA was performed using the PrimeScript RT Reagent Kit (Takara Bio, Inc.). The temperature protocol for RT was as follows: 35°C for 5 min, followed by 47°C for 30 min and 80°C for 5 min. In addition, qPCR was performed with SYBR Premix Ex Taq II (Takara Bio, Inc.) and a Light Cycler system (Roche Diagnostics GmbH). The thermocycling conditions for qPCR were as follows: initial activation step at 45°C for 30 min followed by 40 cycles of denaturation at 92°C for 15 sec, annealing at 65°C for 30 sec and extension at 72°C for 30 sec. The results were analyzed using the 2−ΔΔCq method (13). GAPDH was used as the internal standard. The following primer sequences were used for the qPCR: JPH3 forward, 5′-AATCCTTGCCTGTCGCTCTA-3′ and reverse, 5′-CCCAATCGTGTGGTTCTTCT-3′; and GAPDH forward, 5′-GGAGCGACATCCGTCCAAAAT −3′ and reverse, 5′-GGCTGTTGTCAATCTTCTCATGG-3′.
Western blot analysis
Total protein from 100 mg HCC tissues, paired adjacent tissue and HCC cell lines (1×106) was extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology Co., Ltd.). A Nanodrop spectrophotometer was used for quantification, and an equal amount 30 µg was added to each well of 12% gels and resolved by SDS-PAGE. Proteins were transferred to PVDF membranes and blocked by incubation for 1 h at 37°C with 5% nonfat powdered milk. The membranes were probed at 4°C overnight with antibodies against JPH3 (dilution 1:3,000; ab79063; Abcam), N-cadherin (dilution 1:5,000; ab76011; Abcam), vimentin (dilution 1:2,000; ab92547; Abcam), Ki-67 (dilution 1:5,000; ab92742; Abcam), E-cadherin (dilution 1:10,000; ab40772; Abcam), and GAPDH (dilution 1:5,000; ab9485; Abcam). The PVDF membranes were incubated with horseradish peroxidase-conjugated secondary antibody (dilution 1:5,000; ab6721; Abcam) at room temperature for 1 h. Protein expression levels were visualized using an enhanced chemiluminescence detection system (Bio-Rad Laboratories, Inc.). GAPDH was used as the internal loading control in the western blot analysis. Protein expression was quantified using Quantity One version 4.6.5 software (Bio-Rad Laboratories, Inc.).
Immunohistochemistry
The sample was dehydrated with xylene and alcohol. Antigen retrieval was performed in 10 mmol/l sodium citrate solution (pH 6.0) at 100°C for 15 min, and the samples were cooled for 30 min. All slides were incubated with 5% goat serum (OriGene Technologies, Inc.) for 15 min at room temperature to block non-specific binding at room temperature. The slides were incubated with antibodies against JPH3 (dilution 1:300; ab79063; Abcam) at 4°C overnight. The samples were incubated with biotinylated secondary antibody at 37°C for 30 min then stained with DAB (3,3-diaminobenzidine) and Mayer's hematoxylin.
JPH3 staining classifications were as follows: range 0–3: 0, negative; 1, weak; 2, moderate; and 3, strong; the percentage of positive cells ranged from 0–4: 0, negative or <5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. The percentage of positive cells and the intensity were used to determine the final staining scores. Grades <4 were defined as low JPH3 expression, while grades ≥4 were defined as high JPH3 expression.
Immunofluorescence
Huh-7 and Hep3B cells were fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100 solution for 30 min. Then, the cells were washed with PBS and blocked in 5% bovine serum albumin for 1 h at room temperature. The coverslips were incubated with antibodies against JPH3 (dilution 1:3,000; ab79063; Abcam) overnight at 4°C. Then, the cells were washed with PBS and incubated with appropriate secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI). Slides were imaged using an inverted fluorescence microscope. (magnification, ×200; Zeiss AG).
DNA extraction and methylation-specific PCR
DNA was isolated from HCC tissues (tumor and paired normal adjacent tissues) and HCC cell lines by using a DNA Isolation kit (Tiangen). Determination of bisulfite conversion was performed using the EpiTect Bisulfite Kit (Qiagen Inc.). Methylation-specific PCR (MSP) was performed with 2 µl of bisulfite-modified DNA (100 ng/50 µl) and 48 µl of PCR mixture consisting of 10X PCR Buffer (Mg2+ free), 25 mM MgCl2, dNTP mixture (each 2.5 mM), sense primer (20 µM), antisense primer (20 µM), and Takara EpiTaq HS (5 U/µl; Takara). PCR amplification was conducted using 40 cycles (95°C for 20 sec, 41°C for 30 sec, and 75°C for 30 sec). For parallel quality control, a plasmid containing a methylated JPH3 sequence and water without DNA template were used as positive and negative controls, respectively. The following primer sequences were used for the methylation-specific PCR: JPH3 M forward, 5′-AGACGTTGGTTAGGTTTCGC-3′ and JPH3 U forward, 5′-GAGATGTTGGTTAGGTTTTGT −3′.
Bisulfite Sanger sequencing (BSP)
Five hundred nanograms of genomic DNA extracted from HCC tissues (tumor tissues and paired normal adjacent tissues) was bisulfite converted using a MethylCode™ Bisulfite Conversion kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The JPH3 promoter was amplified by PCR with Taq DNA Polymerase (Invitrogen; Thermo Fisher Scientific, Inc.). The primer sequence was designed using Methyl Primer Express™ Software v1.0 (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR products were electrophoresed, purified using Spin-X tubes, and then cloned into the pUC-T vector (both from CWbiotech). Ten single products were sequenced for each sample.
Transwell invasion assays
Cells (1×105 cells/well) were resuspended in high-glucose DMEM containing 1% FBS and seeded into the upper Transwell chamber at a density of 1×105 cells/well. Tranwell inserts (Corning, Inc.) were precoated with Matrigel (1 mg/ml) at 37°C for 30 min. High-glucose DMEM (500 µl) of containing 10% FBS was added to the matched lower chamber. After 36 h incubation, the Transwell chambers were fixed with 4% paraformaldehyde at room temperature for 30 min, stained with 0.5% crystal violet at room temperature for 20 mins, and finally counted under an inverted optical microscope (×100 magnification; Olympus Corp.).
CCK-8
Hep3B and Huh-7 cells (1×103 cells/well) were seeded into 96-well plates and incubated at 37°C for 24, 48 or 72 h. Then, CCK-8 reagent (Dojindo Molecular Technologies, Inc.) (10 µl) was added to each well, and incubation at 37°C for 2 h. The absorbance was measured at 450 nm by a spectrophotometer (Bio-Rad Laboratories, Inc.).
Colony formation assay
Huh-7 and Hep3B cells were plated in 6-well plates (500 cells/well) containing 2.5 ml of medium and cultured for 20 days. Colonies formed by cell proliferation were stained with crystal violet, and colonies containing at least 50 cells were counted. Colonies were fixed with 20% methanol for 30 min and stained with 0.1% crystal violet for 5 min, and then counted. The numbers of colonies were then counted manually using a light microscope at ×20 magnification.
Wound healing assay
Huh-7 and Hep3B cells (5×105) were grown in 6-well plates, and grown until they formed a confluent monolayer. When the cells reached confluence, any nonadherent cells were washed away with phosphate-buffered saline (PBS). The cell monolayer was scratched with a pipette tip (10 µl) to generate 3 scratch wounds and then rinsed with PBS to remove nonadherent cells, and the medium was replaced with fresh DMEM high glucose medium without FBS. After 0 and 36 h, the distance between the wound sides was measured.
Flow cytometry analysis of cell apoptosis and cell cycle distribution
Huh-7 and Hep3B cells (1×105 cells) were seeded into 6-well plates. Once they reached confluence, the cells were collected and incubated with Annexin V-FITC (5 µl) and propidium iodide (PI) solution (5 µl; Biogot Technology Co., Ltd.) at room temperature for 15 min according to the manufacturer's instructions. Cells were subsequently suspended in 400 µl binding buffer. For cell cycle analysis, Huh-7 and Hep3B cells were collected and fixed in 75% ethanol at −20°C overnight. The fixed cells were washed with PBS and incubated with RNase A for 20 min at room temperature. These cells were stained with PI and incubated in the dark for 30 min at 4°C. Cell apoptosis and cell cycle progression were analyzed using flow cytometry (BD Biosciences). The percentages of cells within each phase of the cell cycle were analyzed with ModFit version 4.0 (Verity Software House, Inc.) and CellQuest version 5.1 (Thermo Fisher Scientific, Inc.).
Statistical analyses
Statistical data were analyzed using SPSS version 22.0 software (IBM Corp.) and GraphPad Prism version 5.0 software (GraphPad Software Inc.). Paired Student's t-tests were used to determine the expression of JPH3 in the HCC and the corresponding peritumor tissues. If not specified, the unpaired Student's t-test or analysis of variance was used, followed by Dunnett's multiple comparison test to compare differences between groups. Associations between clinicopathological parameters and JPH3 expression were analyzed using Pearson's Chi-squared test. Kaplan-Meier survival analysis and Cox regression assays were used to analyze the prognostic significance of JPH3. P<0.05 was considered to indicate a statistically significant difference. All data are presented as the means ± SD.
Results
JPH3 is downregulated in HCC
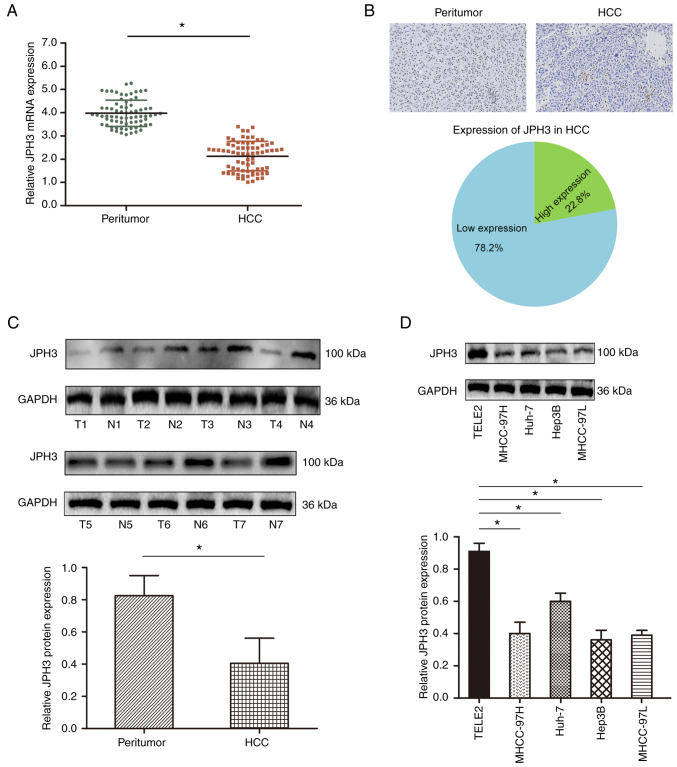
We first analyzed the expression levels of JPH3 in the HCC tissues. The mRNA expression of JPH3 was significantly lower in HCC samples than that in the paired normal adjacent tissues (Fig. 1A). Immunohistochemical staining showed that JPH3 expression was lower in the HCC samples than that in the paired normal adjacent tissues, and JPH3 expression was primarily localized in the nucleus (Fig. 1B). Western blot analysis also confirmed that JPH3 expression was significantly lower in the HCC samples than that in the paired normal adjacent tissues (Fig. 1C). In addition, the expression of JPH3 was also lower in HCC cell lines than in normal hepatocytes THLE2 (Fig. 1D).
Figure 1.
Expression of JPH3 is downregulated in HCC. (A) JPH3 mRNA expression in HCC samples and paired normal adjacent tissues assessed by RT-qPCR. (B) Immunohistochemical staining of JPH3 in HCC tissues and paired normal adjacent tissues (magnification, ×100). (C) Western blotting was used to analyze the protein expression of JPH3 in HCC tissues (T1-T4) and paired normal adjacent tissues (N1-N4). (D) Western blotting was used to analyze the protein expression of JPH3 in HCC cell lines and hepatocyte TELE2 cell line. *P<0.05. HCC, hepatocellular carcinoma; JPH3, junctophilin 3.
Hypermethylation of JPH3 promoter CpG island contributes to low JPH3 expression in HCC
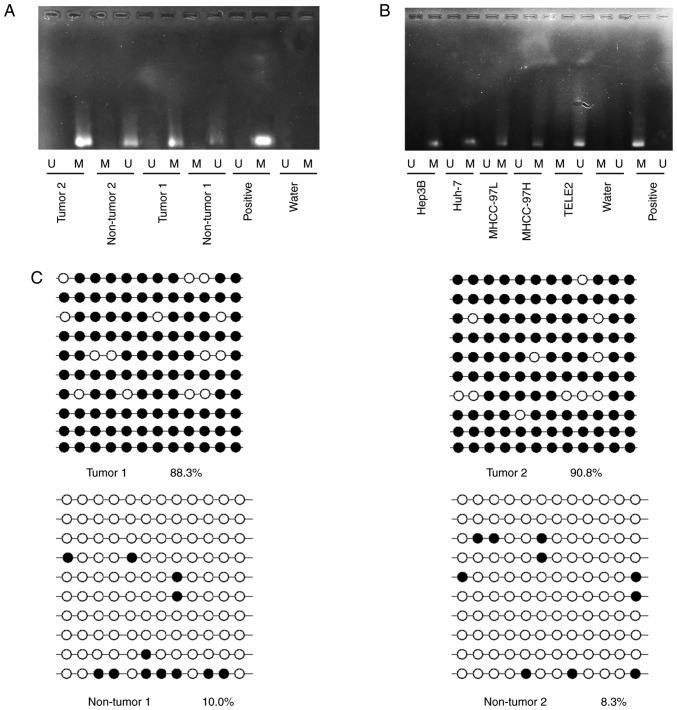
JPH3 has been reported to exhibit DNA methylation, causing low protein expression. The methylation status of JPH3 was assayed in randomly selected HCC tissues compared to paired normal adjacent tissues. JPH3 promoter methylation in HCC tissues was higher than that in the paired normal adjacent tissues (Fig. 2A). Analysis of TELE2, MHCC-97H, MHCC-97L, Hep3B, and Huh-7 cell lines revealed a significantly high degree of methylation in the HCC cell lines, but not in the TELE2 cells (Fig. 2B). Next, bisulfite sequence-PCR (BSP) was used to analyze the specific methylation of CpG island of JPH3, and the results showed that JPH3 promoter methylation in HCC tissues was higher than that in the paired normal adjacent tissues (Fig. 2C).
Figure 2.
Aberrant DNA hypermethylation of JPH3 in HCC. (A) The methylation status of JPH3 was randomly detected in HCC tissues and paired normal adjacent tissues. (B) Methylation status of JPH3 was detected in HCC cell lines and hepatocyte TELE2 cell line. M, methylated; U, unmethylated. (C) Bisulfite sequencing analysis was performed on JPH3 promoter methylation in randomly detected HCC tissues and paired normal adjacent tissues. Black dots, methylation; white dots, unmethylated. HCC, hepatocellular carcinoma; JPH3, junctophilin 3.
Low expression of JPH3 is significantly associated with poor clinical prognosis in HCC
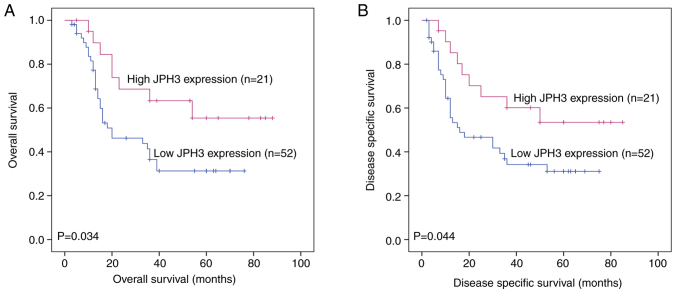
We further analyzed the association between JPH3 expression and the clinicopathological characteristics of the HCC patients. JPH3 expression was significantly associated with TNM stage (P=0.003) and tumor size (P=0.016) (Table I). The HCC patients with low JPH3 expression exhibited significantly shorter overall survival (OS) than the HCC patients with high JPH3 expression (P=0.034, Fig. 3A). Cox proportional hazards regression analysis indicated that TNM stage (P=0.014), tumor size (P=0.030) and JPH3 (P=0.010) were significantly associated with OS in HCC. In addition, multivariate analysis showed that TNM stage (P=0.017), tumor size (P=0.027) and JPH3 (P=0.015) were independent prognostic factors for OS in HCC (Table II). Moreover, HCC patients with low JPH3 expression exhibited significantly shorter DSS than HCC patients with high JPH3 expression (P=0.044, Fig. 3B). Cox proportional hazards regression analysis indicated that TNM stage (P=0.009), tumor size (P=0.024) and JPH3 (P=0.015) were significantly associated with DSS in HCC. In addition, multivariate analysis showed that TNM stage (P=0.007), tumor size (P=0.021) and JPH3 (P=0.019) were independent prognostic factors for DSS in HCC (Table III).
Table I.
Associations between JPH3 and clinicopathological features of the HCC patients.
| JPH3 expression | ||||
|---|---|---|---|---|
|
|
||||
| Variables | Cases | High (n=21) | Low (n=52) | P-value |
| Age, years | ||||
| <50 | 40 | 12 | 28 | 0.798 |
| ≥50 | 33 | 9 | 24 | |
| Sex | 0.527 | |||
| Male | 34 | 11 | 23 | |
| Female | 39 | 10 | 29 | |
| AFP (ng/ml) | 0.232 | |||
| ≤20 | 25 | 5 | 20 | |
| >20 | 48 | 16 | 32 | |
| HBsAg | 0.478 | |||
| Positive | 44 | 14 | 30 | |
| Negative | 29 | 7 | 22 | |
| Tumor size (cm) | 0.016 | |||
| ≤5 | 36 | 15 | 21 | |
| >5 | 57 | 6 | 31 | |
| TNM stage | 0.003 | |||
| I/II | 29 | 14 | 15 | |
| III/IV | 44 | 7 | 37 | |
| Multiplicity | 0.438 | |||
| Presence | 40 | 13 | 27 | |
| Absence | 33 | 8 | 25 | |
| Intrahepatic metastasis | 0.739 | |||
| Presence | 36 | 11 | 25 | |
| Absence | 37 | 10 | 27 | |
HCC, hepatocellular carcinoma; AFP, α-fetoprotein; HBsAg, hepatitis B surface antigen; JPH3, junctophilin 3.
Figure 3.
Association between JPH3 expression and poor prognosis as determined by Kaplan-Meier survival analysis in HCC. (A) Kaplan-Meier analysis of overall survival (OS) in HCC patients with high and low JPH3 expression. (B) Kaplan-Meier analysis of disease-specific survival (DSS) in HCC patients with high and low JPH3 expression. HCC, hepatocellular carcinoma; JPH3, junctophilin 3.
Table II.
Univariate and multivariate analysis of different prognostic variables influencing overall survival (OS) in the HCC patients.
| Univariate analysis model | Multivariate analysis model | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (years) | 0.336 (0.276-1.084) | 0.536 | |||
| <50 | 40 | ||||
| ≥50 | 33 | ||||
| Sex | 0.408 (0.339-1.127) | 0.506 | |||
| Male | 34 | ||||
| Female | 39 | ||||
| AFP (ng/ml) | 1.115 (0.869-2.064) | 0.726 | |||
| ≤20 | 25 | ||||
| >20 | 48 | ||||
| HBsAg | 1.046 (0.479-1.069) | 0.551 | |||
| Positive | 44 | ||||
| Negative | 29 | ||||
| TNM stage | 1.148 (0.486-2.394) | 0.014 | 1.241(0.556-2.047) | 0.017 | |
| I/II | 29 | ||||
| III/IV | 44 | ||||
| Tumor size, cm | 1.012 (0.784-1.840) | 0.030 | 1.047 (0.774-1.931) | 0.027 | |
| ≤5 | 36 | ||||
| >5 | 57 | ||||
| Intrahepatic metastasis | 0.756 (0.841-1.894) | 0.447 | |||
| Presence | 36 | ||||
| Absence | 37 | ||||
| JPH3 expression | 1.427 (0.841-3.004) | 0.010 | 1.439 (0.715-3.046) | 0.015 | |
| Low | 52 | ||||
| High | 21 | ||||
HR, hazard ratio; CI, confidence interval; HCC, hepatocellular carcinoma; AFP, α-fetoprotein; HBsAg, hepatitis B surface antigen; JPH3, junctophilin 3.
Table III.
Univariate and multivariate analysis of different prognostic variables influencing disease-specific survival (DSS) of the HCC patients.
| Univariate analysis | Multivariate analysis model | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Variable | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (years) | 0.304 (0.365-1.203) | 0.630 | |||
| <50 | 40 | ||||
| ≥50 | 33 | ||||
| Sex | 0.348 (0.340-1.178) | 0.405 | |||
| Male | 34 | ||||
| Female | 39 | ||||
| AFP (ng/ml) | 0.842 (0.634-1.778) | 0.836 | |||
| ≤20 | 25 | ||||
| >20 | 48 | ||||
| HBsAg | 1.135 (0.782-1.842) | 0.641 | |||
| Positive | 44 | ||||
| Negative | 29 | ||||
| TNM stage | 1.004 (0.447-1.236) | 0.009 | 1.047 (0.486-1.445) | 0.007 | |
| I/II | 29 | ||||
| III/IV | 44 | ||||
| Tumor size (cm) | 1.148 (0.556-2.014) | 0.024 | 1.214 (0.336-2.174) | 0.021 | |
| ≤5 | 36 | ||||
| >5 | 57 | ||||
| Intrahepatic metastasis | 1.047 (0.660-1.930) | 0.475 | |||
| Presence | 36 | ||||
| Absence | 37 | ||||
| JPH3 expression | 1.410 (0.559-2.014) | 0.015 | 1.507 (0.663-2.474) | 0.019 | |
| Low | 52 | ||||
| High | 21 | ||||
HR, hazard ratio; CI, confidence interval; HCC, hepatocellular carcinoma; AFP, α-fetoprotein; HBsAg, hepatitis B surface antigen; JPH3, junctophilin 3.
Restoration of JPH3 expression by treatment with 5-Aza demethylation inhibits the proliferation, migration and invasion abilities in HCC
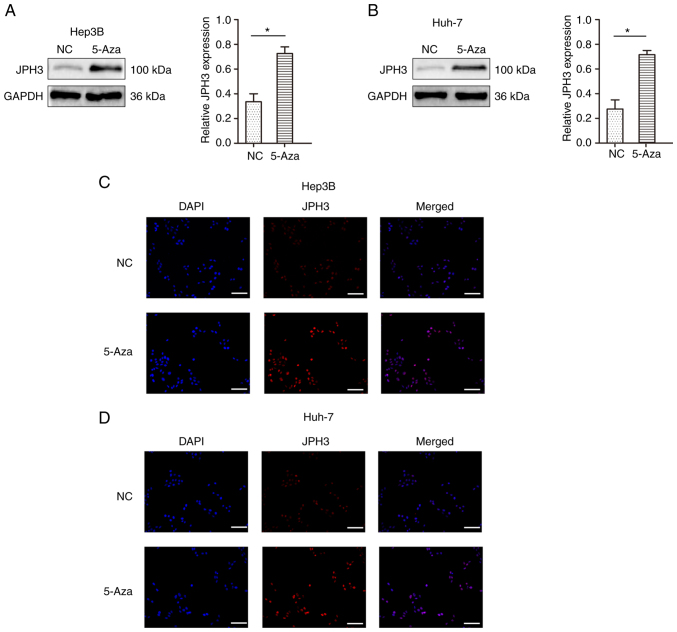
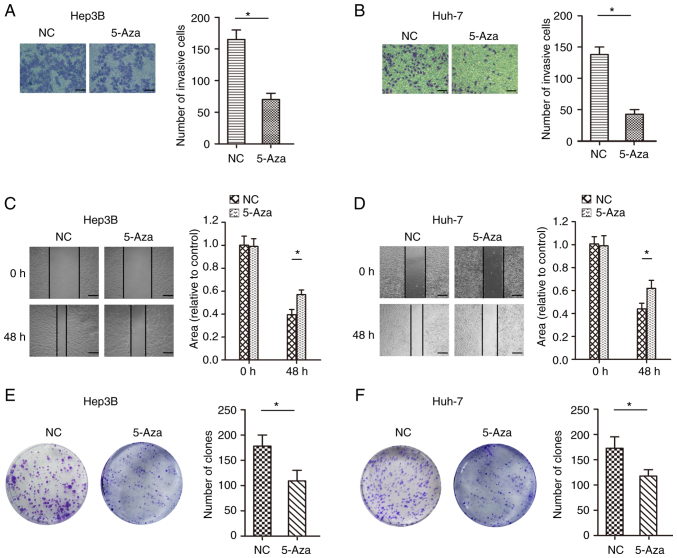
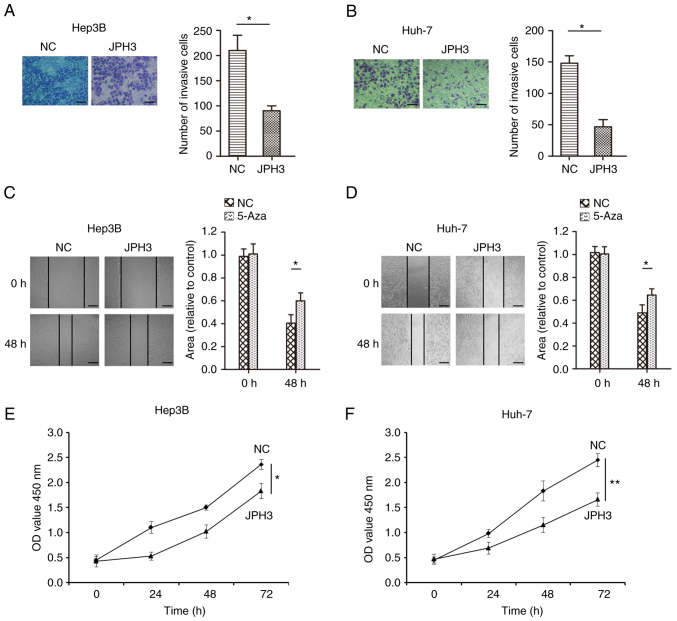
To test whether methylation directly induces JPH3 silencing, the JPH3 gene was demethylated in the Huh-7 and Hep3B cell lines with 5-Aza, an inhibitor of DNA methyltransferases. As shown in Fig. 4A and B, after treatment with 1 µM 5-Aza for 96 h, the expression of the JPH3 protein was restored. The immunofluorescence results confirmed the above results (Fig. 4C and D). We analyzed the effect of JPH3 on proliferation, migration and invasion in HCC. The invasive ability of cells treated with 5-Aza was less than that of the control Huh-7 and Hep3B cell lines (Fig. 5A and B). In addition, the migration ability of the cells treated with 5-Aza was less than that of the control Huh-7 and Hep3B cell lines (Fig. 5C and D). The colony formation experiment also showed that treatment with 5-Aza significantly inhibited cell proliferation in the Huh-7 and Hep3B cell lines (Fig. 5E and F). Moreover, increasing the expression of JPH3 using JPH3 plasmid also inhibited the proliferation, invasion and migration of HCC cells (Fig. 6A-F). The above results indicate that demethylation of JPH3 can restore the protein expression of JPH3 and inhibit proliferation, migration and invasion in HCC.
Figure 4.
Restoration of JPH3 expression by treatment with 5-Aza demethylation in HCC. (A) Protein expression of JPH3 after treatment with 5-Aza-2′-deoxycytidine (5-Aza) demethylation was detected by western blot analysis in the Hep3B cell line. (B) Protein expression of JPH3 after treatment with 5-Aza demethylation was detected by western blot analysis in the Huh-7 cell line. *P<0.05. (C) Protein expression of JPH3 after treatment with 5-Aza demethylation was detected by immunofluorescence in the Hep3B cell line (magnification, ×100). (D) Protein expression of JPH3 after treatment with 5-Aza demethylation was detected by immunofluorescence in the Huh-7 cell line (magnification, ×100). HCC, hepatocellular carcinoma; JPH3, junctophilin 3.
Figure 5.
Restoration of JPH3 expression by treatment with 5-Aza demethylation inhibits proliferation, migration and invasion in HCC cells. (A) Transwell assays were used to analyze the effect of restoration of JPH3 expression by treatment with 5-Aza-2′-deoxycytidine (5-Aza) on the invasion of Hep3B cells (magnification, ×200). (B) Transwell assays were used to analyze the effect of restoration of JPH3 expression by treatment with 5-Aza on the invasion of Huh-7 cells (magnification, ×200). (C) A wound healing assay was used to analyze the effect of restoration of JPH3 expression by treatment with 5-Aza on the migration of Hep3B cells (magnification, ×100). (D) A wound healing assay was used to analyze the effect of restoration of JPH3 expression by treatment with 5-Aza on the migration of Huh-7 cells (magnification, ×100). (E) Colony formation experiments were also used to analyze the restoration of JPH3 expression by treatment with 5-Aza on the proliferation of Hep3B cells. (F) Colony formation experiments were also used to analyze the restoration of JPH3 expression by treatment with 5-Aza on the proliferation of Huh-7 cells. *P<0.05. HCC, hepatocellular carcinoma; JPH3, junctophilin 3.
Figure 6.
Increased JPH3 expression inhibits the proliferation, migration and invasion in HCC. (A) Transwell assays were used to analyze the effect of increasing JPH3 expression on the invasion of Hep3B cells (magnification, ×200). (B) Transwell assays were used to analyze the effect of increasing JPH3 expression on the invasion of Huh-7 cells (magnification, ×200). (C) A wound healing assay was used to analyze the effect of increasing JPH3 expression via plasmid on the migration of Hep3B cells (magnification, ×100). (D) A wound healing assay was used to analyze the effect of increasing JPH3 expression on the migration of Huh-7 cells (magnification, ×100). (E) CCK-8 experiments were also used to analyze the effect of increasing JPH3 expression on the proliferation of Hep3B cells. (F) CCK-8 experiments were also used to analyze effect of increasing JPH3 expression on the proliferation of Huh-7 cells. *P<0.05, **P<0.01. HCC, hepatocellular carcinoma; JPH3, junctophilin 3.
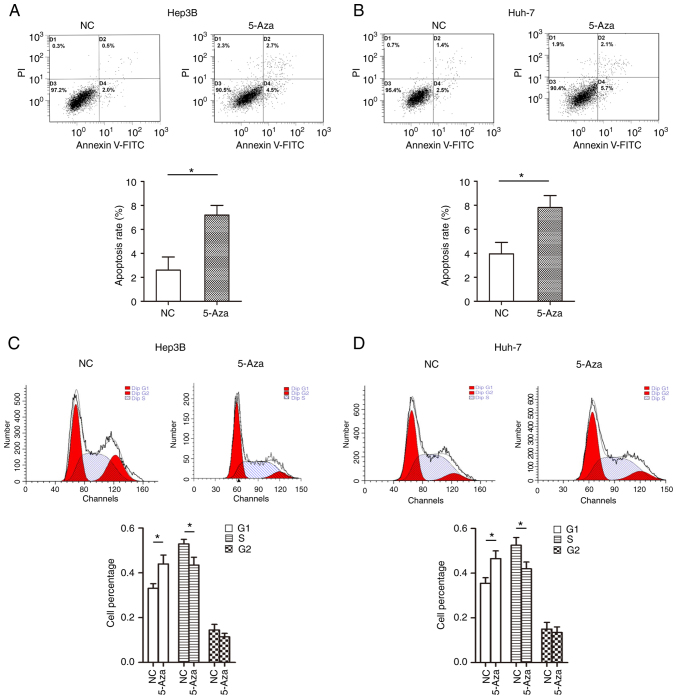
Restoration of JPH3 expression by treatment with 5-Aza demethylation regulates the cell cycle and apoptosis in HCC cells
We further analyzed the effect of JPH3 on cell cycle distribution and apoptosis in the HCC cells. The results showed that treatment with 5-Aza significantly promoted cell apoptosis in the Huh-7 and Hep3B cell lines (Fig. 7A and B). In addition, the G1 stage of cells in the 5-Aza-treated group was higher than that noted in the control group of Huh-7 and Hep3B cell lines (Fig. 7C and D). Moreover, increasing the expression of JPH3 promoted cell apoptosis and prevented cell growth in the Huh-7 and Hep3B cell lines (Fig. S1). These results indicate that demethylation of JPH3 induced Huh-7 and Hep3B cell cycle arrest in G1 stage and promoted apoptosis.
Figure 7.
Restoration of JPH3 expression by treatment with 5-Aza demethylation regulates cell cycle distribution and promotes apoptosis in HCC. (A) Flow cytometric apoptosis detection was used to analyze the effect of restoration of JPH3 expression by treatment with 5-Aza-2′-deoxycytidine (5-Aza) on the apoptosis of Hep3B cells. (B) Flow cytometric apoptosis detection was used to analyze the effect of restoration of JPH3 expression by treatment with 5-Aza on apoptosis of Huh-7 cells. (C) Flow cytometry detection was used to analyze the effect of the restoration of JPH3 expression by treatment with 5-Aza on cell cycle distribution of Hep3B cells. (D) Flow cytometry detection was used to analyze the effect of restoration of JPH3 expression by treatment with 5-Aza on cell cycle distribution of Huh-7 cells. *P<0.05. HCC, hepatocellular carcinoma; JPH3, junctophilin 3.
JPH3 affects the biological behavior of HCC by regulating EMT
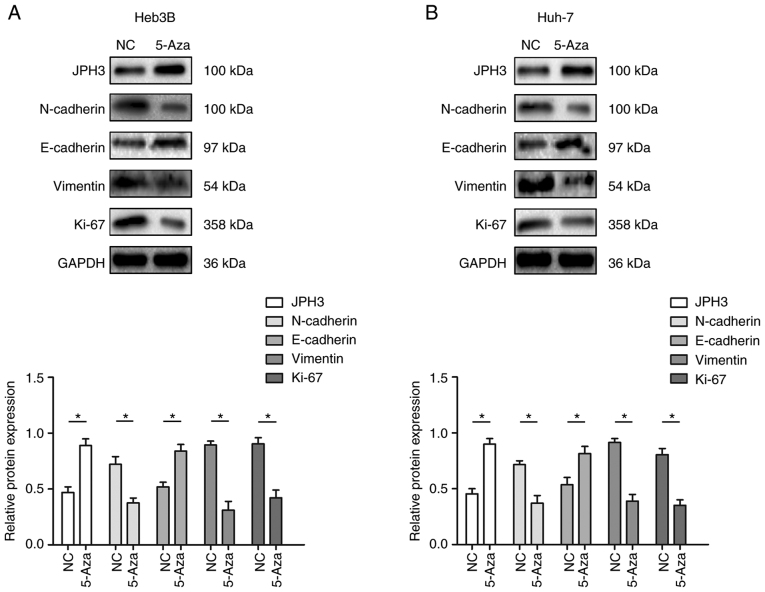
Based on the above research results, we further analyzed whether JPH3 affects the malignant biological behavior of HCC by regulating EMT. Western blot results revealed that restoration of JPH3 expression by 5-Aza significantly suppressed the expression of N-cadherin, vimentin and KI-67, which were increased by downregulated expression of JPH3, and 5-Aza treatment significantly increased the expression of E-cadherin, which was inhibited by downregulated expression of JPH3 (Fig. 8A and B). Moreover, increasing the expression of JPH3 significantly decreased the expression of N-cadherin, vimentin and KI-67, which were increased by downregulated expression of JPH3, and the expression of E-cadherin, which was inhibited by downregulated expression of JPH3 was increased (Fig. S2).
Figure 8.
Effect of restoration of JPH3 expression by treatment with 5-Aza demethylation on EMT in HCC. (A) Western blotting was used to analyze the protein expression of JPH3, E-cadherin, N-cadherin, vimentin and Ki-67 in Hep3B cells after treatment with 5-Aza-2′-deoxycytidine (5-Aza). (B) Western blotting was used to analyze the protein expression of JPH3, E-cadherin, N-cadherin, vimentin and Ki-67 in Huh-7 cells after treatment with 5-Aza. *P<0.05. HCC, hepatocellular carcinoma; JPH3, junctophilin 3; EMT, epithelial-mesenchymal transition.
Discussion
Hepatocellular carcinoma (HCC) is a malignant tumor with a high recurrence and mortality rate particularly in China (14). Most patients are in the middle and late stages of the disease at the time of diagnosis, accompanied by extensive intrahepatic and extrahepatic metastasis, and the patient prognosis is poor (15). Fundamentally speaking, tumors are a disease of tissue growth regulation, and the malignant biological behavior of a tumor is an important manifestation of abnormal growth regulation (16). These regulatory changes often involve gene mutations, deletion and chromosome abnormalities, and epigenetic changes are among the most common causes of gene function changes (16). The malignant growth of liver cancer is a complex process involving many regulatory factors, including gene mutations and epigenetic changes, changes in cell surface signal transduction molecules and adhesion ability, abnormal cell metabolism, and changes in tumor cells and the surrounding microenvironment (17–20).
DNA methylation leads to the formation of 5-methylcytosine (5 mC) in DNA through the methyl donor S-adenosine methionine (SAM) to the methyl of carbon 5 in the cytosine pyridine ring. DNA methyltransferase family can catalyze this reaction (21). In mammalian somatic cells, this stable synthetic epigenetic marker exists only in the dinucleotide of the CpG cytosine residue, while in embryonic stem cells, DNA methylation occurs in both CpG and non-CpG sequences (22). A large number of studies have shown that DNA methylation plays an important role in the proliferation and metastasis of HCC (7,23). DNA methylation is a chemical modification process that transfers S-adenosylmethionine as active methyl to a specific base in the DNA chain under the catalysis of DNA methyltransferase (24). It causes changes in DNA conformation, DNA stability and the mode of interaction between DNA and protein, and controls gene expression (25). DNA methylation during the occurrence and development of HCC is mainly manifested in the hypomethylation activation of proto-oncogenes and the methylation inactivation of tumor-suppressor genes (26). Studies have found that the methylation level of proto-oncogenes Ras and myc in the promoter region of HCC are significantly low, which enhances the expression of c-myc and c-N-Rras and promotes the growth, survival and metastasis of HCC cells (27). In HCC, hypermethylation of tumor-suppressor gene promoter CpG island interferes with the transcription of tumor-suppressor genes, resulting in abnormal cell proliferation, which is closely related to tumorigenesis (28). It has been found that inactivation of DLL3 gene methylation in HCC can promote the proliferation and metastasis of HCC cells through the Notch signaling pathway (29). In addition, Smad nuclear interacting protein-1 (snip1), whose promoter is subjected to hypermethylation in HCC cells, can mediate epithelial-mesenchymal transformation (EMT) and promote the metastasis of HCC (30). In the present study, we demonstrated that junctophilin 3 (JPH3) was also modified by DNA methylation in HCC, resulting in its low expression in HCC tissues. Its low expression in HCC was found to be significantly related to the poor prognosis of patients.
When analyzing the role of DNA methylation of JPH3 in the malignant progression of HCC, we found that after treating HCC cells with 5-Aza, the expression of JPH3 was significantly increased, the proliferation, invasion and migration of HCC were significantly inhibited, and the apoptosis level of HCC cells was increased. These results suggest that DNA methylation and protein expression of JPH3 may affect the signaling pathways regulating the invasion and metastasis of HCC. Thus, we focused on EMT. EMT refers to the process of epithelial cell phenotype transition to mesenchymal cells under specific physiological or pathological conditions (31). The main morphological characteristics of EMT are when epithelial cells lose their typical intercellular junction structure, reorganize the cytoskeleton, and change from a polygonal to a spindle-shaped fibroblast-like morphology (32). After EMT, the cells become isolated, their movement capacity is enhanced, and they are resistant to apoptosis (33). In HCC, TDP-43 inhibits GSK3β protein translation and activates the Wnt/β-catenin pathway, regulating EMT to induce proliferation and metastasis (34). In addition, miR-186 was found to regulate EMT changes in HCC cells by targeting CDK6 and this inhibits their proliferation, migration and invasion (35). MST4 inactivation was found to induce EMT in HCC cells, promote their migration and invasion potential in vitro, and promote intrahepatic metastasis in vivo(36). MST4 inactivation was also found to increase the expression and nuclear translocation of the key EMT transcription factor Snail1 through the PI3K/Akt signaling pathway to induce the EMT phenotype of HCC cells and enhance their invasion and metastasis potential (36). Our research confirmed that DNA methylation regulates the protein expression of JPH3 and this regulates the proliferation, migration, invasion and apoptosis of HCC; the regulatory mechanism may be related to the changes in EMT in tumor cells.
It has been reported that JPH3 is highly methylated in gastrointestinal tumors, resulting in decreased expression of its mRNA and protein levels. JPH3 can also upregulate the cytoplasmic Ca2+ level and unfolded protein response due to endoplasmic reticulum stress, induce calpain activation and subsequent mitochondrial membrane depolarization and apoptosis, and promote the malignant progression of gastrointestinal tumors (6). Our results are consistent with these findings. In the present study, the expression of JPH3 was analyzed in HCC for the first time and it was demonstrated that DNA methylation may be associated with the low expression of JPH3 in HCC. Hypermethylated JPH3 in HCC, which caused a decrease in the expression of JPH3, affected related signaling pathways and regulated the malignant progression of tumors. Of course, our research still has certain limitations. First, 5-Aza is a broad-spectrum and comprehensive DNA methyltransferase inhibitor, which can cause DNA demethylation (hypomethylation) and then promote gene expression. However, the specific targeting sites of 5-Aza acting on JPH3 are largely unknown. In addition, evidence verifying that the biological effects of JPH3 are related to 5-Aza, is not solid enough. The limitations will be further investigated. Understanding the above questions will better explain the important role of JPH3 and its DNA methylation in HCC.
In conclusion, our research indicates that JPH3 is expressed at low levels in HCC and its low expression is caused by DNA methylation. The low expression of JPH3 in HCC was significantly correlated with a poor prognosis of patients. Mechanistically, JPH3 can inhibit proliferation, invasion and migration and promote the apoptosis of HCC, and its mechanism of action may be through the regulation of EMT. The present study provides a scientific basis for further exploring the role of JPH3 in HCC and also provides novel ideas for the precise treatment of HCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This study was supported by the National Natural Science Foundation of China (grant no. 81960125).
Availability of data and materials
The datasets used and or/analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YH and LZ conceived and designed the experiments. ZY, MZ, XY and HH performed the experiments and analyzed the data. YH and LZ drafted and revised the manuscript. All authors have read and approved the final manuscript for publication. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work including the data are appropriately investigated and resolved.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Affiliated Hospital of Zunyi Medical University. Written informed consent was obtained from all patients (ethical approval no. 2011035).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Couri T, Pillai A. Goals and targets for personalized therapy for HCC. Hepatol Int. 2019;13:125–137. doi: 10.1007/s12072-018-9919-1. [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Jiang J, Tang M, Huang Z, Chen L. Junctophilins emerge as novel therapeutic targets. J Cell Physiol. 2019;234:16933–16943. doi: 10.1002/jcp.28405. [DOI] [PubMed] [Google Scholar]
- 5.Margolis RL, Rudnicki DD. Pathogenic insights from Huntington's disease-like 2 and other Huntington's disease genocopies. Curr Opin Neurol. 2016;29:743–748. doi: 10.1097/WCO.0000000000000386. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Kuang Y, Li L, Tang H, Shi Q, Shu X, Zhang Y, Chan FK, Tao Q, He C. Epigenomic and functional characterization of junctophilin 3 (JPH3) as a novel tumor suppressor being frequently inactivated by promoter CpG methylation in digestive cancers. Theranostics. 2017;7:2150–2163. doi: 10.7150/thno.18185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han TS, Ban HS, Hur K, Cho HS. The epigenetic regulation of HCC metastasis. Int J Mol Sci. 2018;19:3978. doi: 10.3390/ijms19123978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Nebbioso A, Tambaro FP, Dell'Aversana C, Altucci L. Cancer epigenetics: Movin forward. PLoS Genet. 2018;14:e1007362. doi: 10.1371/journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55. doi: 10.1007/978-981-15-3449-2_1. [DOI] [PubMed] [Google Scholar]
- 11.Zhang D, Zhao J, Han C, Liu X, Liu J, Yang H. Identification of hub genes related to prognosis in glioma. Biosci Rep. 2020;40:BSR20193377. doi: 10.1042/BSR20193377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KH, He J, Wang DL, Cao JJ, Li MC, Zhao XM, Sheng X, Li WB, Liu WJ. Methylation-associated inactivation of LATS1 and its effect on demethylation or overexpression on YAP and cell biological function in human renal cell carcinoma. Int J Oncol. 2014;45:2511–2521. doi: 10.3892/ijo.2014.2687. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Huang A, Yang XR, Chung WY, Dennison AR, Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:146. doi: 10.1038/s41392-020-00264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, Han QJ, Zhang J. Hepatocellular carcinoma: Mechanisms of progression and immunotherapy. World J Gastroenterol. 2019;25:3151–3167. doi: 10.3748/wjg.v25.i25.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol. 2018;15:353–365. doi: 10.1038/s41571-018-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahn LM. Effects of the tumor microenvironment. Science. 2017;355:1386–1388. doi: 10.1126/science.355.6332.1386-l. [DOI] [PubMed] [Google Scholar]
- 18.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 19.Xie DY, Ren ZG, Zhou J, Fan J, Gao Q. 2019 Chinese clinical guidelines for the management of hepatocellular carcinoma: Updates and insights. Hepatobiliary Surg Nutr. 2020;9:452–463. doi: 10.21037/hbsn-20-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganne-Carrié N, Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol. 2019;70:284–293. doi: 10.1016/j.jhep.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Skvortsova K, Stirzaker C, Taberlay P. The DNA methylation landscape in cancer. Essays Biochem. 2019;63:797–811. doi: 10.1042/EBC20190037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. doi: 10.1186/s13072-017-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, Kim Y, Friso S, Choi SW. Epigenetics in non-alcoholic fatty liver disease. Mol Aspects Med. 2017;54:78–88. doi: 10.1016/j.mam.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18:1–14. doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 25.Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777–786. doi: 10.1016/S0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 26.Toh TB, Lim JJ, Chow EK. Epigenetics of hepatocellular carcinoma. Clin Transl Med. 2019;8:13. doi: 10.1186/s40169-019-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raggi C, Invernizzi P. Methylation and liver cancer. Clin Res Hepatol Gastroenterol. 2013;37:564–571. doi: 10.1016/j.clinre.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura M, Chiba T, Kanayama K, Kanzaki H, Saito T, Kusakabe Y, Kato N. Epigenetic dysregulation in hepatocellular carcinoma: An up-to-date review. Hepatol Res. 2019;49:3–13. doi: 10.1111/hepr.13250. [DOI] [PubMed] [Google Scholar]
- 29.Maemura K, Yoshikawa H, Yokoyama K, Ueno T, Kurose H, Uchiyama K, Otsuki Y. Delta-like 3 is silenced by methylation and induces apoptosis in human hepatocellular carcinoma. Int J Oncol. 2013;42:817–822. doi: 10.3892/ijo.2013.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acun T, Oztas E, Yagci T, Yakicier MC. SIP1 is downregulated in hepatocellular carcinoma by promoter hypermethylation. BMC Cancer. 2011;11:223. doi: 10.1186/1471-2407-11-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Saitoh M. Involvement of partial EMT in cancer progression. J Biochem. 2018;164:257–264. doi: 10.1093/jb/mvy047. [DOI] [PubMed] [Google Scholar]
- 33.Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med. 2019;216:1016–1026. doi: 10.1084/jem.20181827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo F, Wang H, Jiang M, Yang Q, Xiang Q, Zhou H, Hu X, Hao K, Yang J, Cao H, Shen Z. TDP-43 induces EMT and promotes hepatocellular carcinoma metastasis via activating Wnt/β-catenin signaling pathway. Am J Cancer Res. 2020;10:3285–3301. [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, Zhao Z, Ma Y. miR-186 represses proliferation, migration, invasion, and EMT of hepatocellular carcinoma via directly targeting CDK6. Oncol Res. 2020;28:509–518. doi: 10.3727/096504020X15954139263808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dian MJ, Li J, Zhang XL, Li ZJ, Zhou Y, Zhou W, Zhong QL, Pang WQ, Lin XL, Liu T, et al. MST4 negatively regulates the EMT, invasion and metastasis of HCC cells by inactivating PI3K/AKT/Snail1 axis. J Cancer. 2021;12:4463–4477. doi: 10.7150/jca.60008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and or/analyzed during the current study are available from the corresponding author on reasonable request.