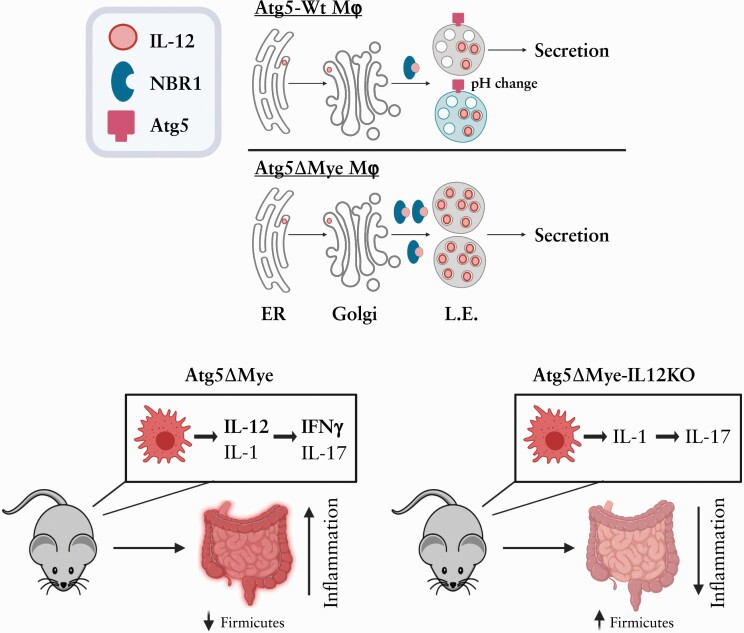
Abstract
Intestinal myeloid cells play a critical role in balancing intestinal homeostasis and inflammation. Here, we report that expression of the autophagy-related 5 [Atg5] protein in myeloid cells prevents dysbiosis and excessive intestinal inflammation by limiting IL-12 production. Mice with a selective genetic deletion of Atg5 in myeloid cells [Atg5ΔMye] showed signs of dysbiosis preceding colitis, and exhibited severe intestinal inflammation upon colitis induction that was characterised by increased IFNγ production. The exacerbated colitis was linked to excess IL-12 secretion from Atg5-deficient myeloid cells and gut dysbiosis. Restoration of the intestinal microbiota or genetic deletion of IL-12 in Atg5ΔMye mice attenuated the intestinal inflammation in Atg5ΔMye mice. Additionally, Atg5 functions to limit IL-12 secretion through modulation of late endosome [LE] acidity. Last, the autophagy cargo receptor NBR1, which accumulates in Atg5-deficient cells, played a role by delivering IL-12 to LE. In summary, Atg5 expression in intestinal myeloid cells acts as an anti-inflammatory brake to regulate IL-12, thus preventing dysbiosis and uncontrolled IFNγ-driven intestinal inflammation.
Keywords: Autophagy, macrophages, inflammation, cytokines, microbiota
Graphical Abstract
Graphical Abstract.
1. Introduction
Immune system dysregulation, intestinal barrier defects, and dysbiosis are believed to be driven in part by genetics, according to recent genome-wide association studies [GWAS].1 Many of the identified genes are involved in innate cell bacterial recognition and processing, and appear to contribute to the pathogenesis observed in inflammatory bowel disease [IBD], such as NOD2 and ATG16L1 which are linked to autophagy.1 Autophagy is one such process associated with IBD susceptibility.2–11 Autophagy is a conserved catabolic process that degrades protein aggregates, damaged organelles, and numerous pathogens.12 Autophagy has proven critical for intestinal homeostasis. Defects in the autophagic pathway, specifically in intestinal epithelial cell lineages, results in increased intestinal permeability and Paneth and goblet cell [GC] dysfunction.13–18 More recently, an IBD risk locus associated with autophagy was found to disrupt the microbiota, albeit it is unclear what cell type mediates this effect.19 Nevertheless, there is mounting evidence that autophagic genes are critical for intestinal homeostasis, and defects in the autophagic process can lead to increased susceptibility to intestinal pathogens and overall enhanced IBD susceptibility.6,20–22
Many of these IBD risk genes and pathways, including autophagy, are highly relevant to myeloid cell function in addition to that of epithelial cells.23–26 However, the specific role for autophagy and autophagy genes in myeloid cells in maintaining the balance between intestinal homeostasis and inflammation has yet to be fully explored. The prevailing hypothesis linking autophagy to IBD is through the IL-17 signalling pathway. IL-1, IL-17, and IL-23, all involved in IL-17-mediated inflammation, are upregulated in IBD.27–36 Our previous work, as well as that of others, has demonstrated that autophagy regulates the production of the proinflammatory cytokines IL-1α, IL-1β, and IL-23, primarily in infection models.37–42 This observation is the canonical pathway frequently described linking autophagy dysregulation to excess inflammation via IL-1 and IL-17. IL-1 and IL-23 are key regulators of IL-17-mediated inflammation.43,44 However, the role of IL-17 in IBD pathogenesis has recently been questioned, as therapeutic targeting of IL-17 exacerbates inflammation in both IBD patients and animal models of IBD,45–49 suggesting that IL-17 may not be the predominant driver of autophagy-linked IBD pathogenesis. Thus, there is a clear gap in knowledge regarding which inflammatory pathway might underlie IBD pathology with respect to autophagy dysregulation.
This study assessed the role of the autophagy gene Atg5 in myeloid cells in maintaining the balance between intestinal homeostasis and inflammation. Atg5’s most well-understood actions are via the Atg5-Atg12-Atg16L1 complex, which acts as the E3 enzyme conjugating PE to LC3, and along with the E1-like actions of Atg7 and the E2-like actions of Atg3, this pathway drives isolation membrane formation and eventual autophagosome maturation.50–53 Atg5 is also embedded in autophagosomal membranes, which allows it to interact with fusion proteins in lysosomal membranes such as Tectonic β-propeller repeat containing 1 [TECRP] that facilitate autophagosome-lysosome fusion.54,55 Thus, Atg5 plays a major role in selective and bulk autophagy which are critical for cell autonomous immunity. However, it is unclear how Atg5 expression, specifically in myeloid cells, functions outside of bacterial recognition and processing. Here, we show at steady state that mice with an Atg5-deficiency in myeloid cells [herein called Atg5ΔMye mice]37,56 show alterations in the gut microbiota as well as mucosal TH1 [IFNγ] skewing. Atg5ΔMye mice were susceptible to chemically-induced colitis that was characterised by an enhanced IFNγ response. Both TH1 skewing and microbiota changes were partly driven through IL-12 dysregulation in Atg5-deficient myeloid cells. Confirming our findings, Atg5ΔMye mice crossed with IL12p35-deficient mice resulted in restoration of the gut microbiota and protection from dextran sodium sulphate [DSS]-induced colitis through the reduction of the IL-12 pathway. In addition, faecal microbiota transplant of control microbiota into Atg5ΔMye mice reduced chemically-induced colitis, suggesting the Atg5ΔMye-associated microbiota also contributes to the exacerbated colitis phenotype in Atg5ΔMye mice. We further show IL-12 regulation was not mediated through autophagy but through interactions with sequestosome-1-like receptor NBR1 and Atg5 action on late endosomes [LE]. There has not been a single description of autophagic proteins regulating IL-12-driven immune responses. Thus, these data indicate a new autophagy-independent role for Atg5 and NBR1 in myeloid cells in influencing intestinal homeostasis through an IL-12 pathway.
2. Results
2.1. Altered colonic microbiota in mice with an Atg5-deficiency in myeloid cells
The gut microbiota is influenced by environmental factors, immune responses, and genetics which is highlighted in individuals with IBD. Numerous studies have reported a decrease in bacterial diversity in IBD patients, with major alterations in the phyla Firmicutes and Bacteroidetes.57–62 There is substantial evidence that intestinal myeloid cells are regulated by commensal microbiota.58,63–66 However, numerous studies have emerged demonstrating myeloid regulation of the microbiota.67–70 Autophagy genes appear to be involved in the latter as it was shown the autophagy protein, Atg16L1, expressed in myeloid cells from both humans and mice, altered IgA-coated intestinal bacteria at steady state and during inflammation,26 but it was not confirmed if this increase in IgA-coated bacteria affects the overall microbial community. More recently, another mouse model with a global knockin of the IBD risk allele ATG16L1 T300A had an altered gut microbiota preceding colitis induction.19
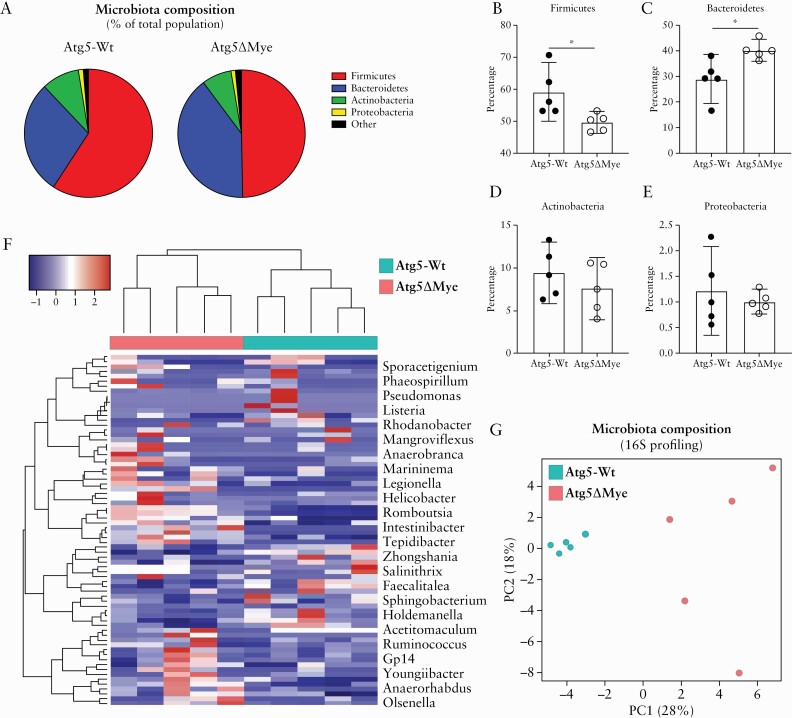
Using Atg5ΔMye mice [myeloid specific loss of autophagy-related 5, Atg5, gene], we investigated the importance of Atg5 expression in myeloid cells in maintaining the intestinal microbiota.37,56 In comparison with littermate controls [Atg5-Wild-type, Atg5-Wt], Atg5ΔMye mice showed significant differences in the intestinal microbial composition [Figure 1A]. Similar to what is observed in IBD patients,59–62,71 Atg5ΔMye mice had a decrease in the phylum Firmicutes [Figure 1B] and an increase in the phylum Bacteroidetes [Figure 1C] compared with Atg5-Wt mice. No significant changes were observed for phyla Actinobacteria and Proteobacteria [Figure 1D and E]. Additionally, the principal coordinate analysis [PCoA] plot and heatmap of bacterial communities revealed tight clustering of Atg5-Wt microbiota that was distinct from Atg5ΔMye microbiota at the genus level [Figure 1F and G], and this was also observed for female mice [Supplementary Figure 1A and B, available as Supplementary data at ECCO-JCC online]. Thus, Atg5 expression in myeloid cells is critical for the maintenance of the intestinal microbiota.
Figure 1.
Alterations in the composition of the colonic microbial community in mice with a selective Atg5-deficiency in myeloid cells. Bacterial composition as assessed by 16S rRNA sequencing of DNA extracted from freshly collected stool of Atg5-Wt and Atg5ΔMye mice [n = 5 per group]. [A] Pie chart showing the average proportion of Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and all other phyla in 8-week old mice. [B‐E] Quantification of the phyla [B] Firmicutes, [C] Bacteroidetes, [D] Actinobacteria, and [E] Proteobacteria. [F] Heatmap of the relative abundance of colonic microbes [genus level]. [G] Principal coordinates analysis [PCoA] plot of microbiota composition [genus level]. Data are shown as mean (±95% confidence interval [CI]) and a t test was used to measure specific microbiome species abundance between groups. Adjusted p-value >0.05 was used as significant threshold.
2.2. Myeloid Atg5 expression regulates IFNγ response in the intestinal microenvironment
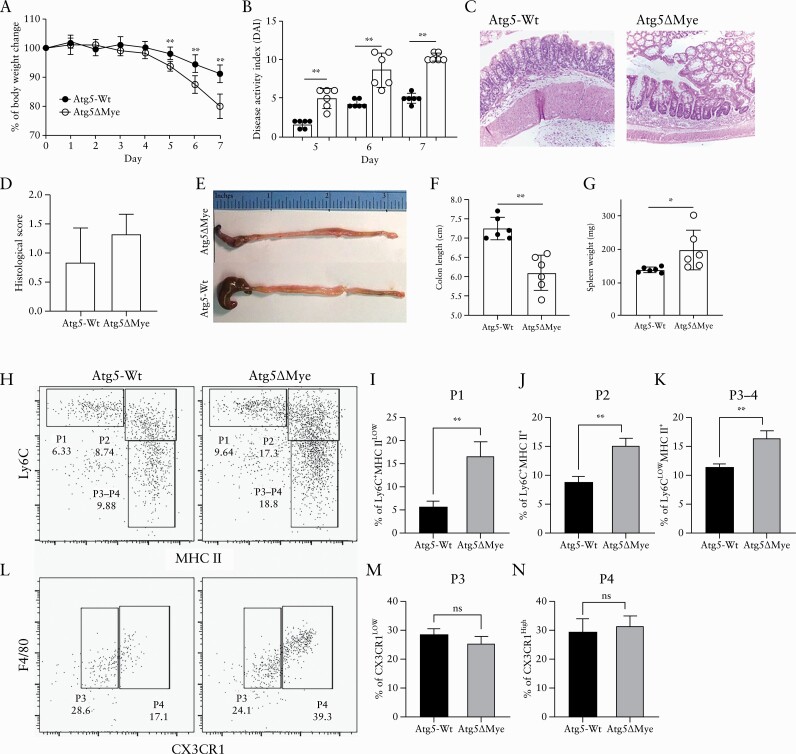
The acute DSS-induced colitis model is an innate/wound repair model that is sensitive to the gut microbiota.72,73 To examine the impact of the microbiota alteration in Atg5ΔMye mice during an inflammatory state, we subjected both Atg5ΔMye and Atg5-Wt mice to DSS-induced acute colitis. Similar to what was reported in mice with an Atg16l1- or Atg7-deficiency specifically in myeloid cells,26,74 colitis induction in Atg5ΔMye mice resulted in an exacerbated inflammatory response compared with Atg5-Wt mice. Intestinal epithelial disruption via DSS caused significant weight loss [Figure 2A] as well as acute diarrhoeal response and mucosal bleeding [disease activity index, DAI, Figure 2B] in Atg5ΔMye mice; however, histologically both groups showed inflammation [Figure 2C and D]. Severe inflammation from the caecum to the distal colon of Atg5ΔMye mice was observed [Figure 2E]. The length of the colon was also measured to assess colonic mucosal injury and revealed colonic shortening [Figure 2F]. We also found that the spleen weight of colitic Atg5ΔMye mice was increased [Figure 2G]. Examination of the cellular infiltration in the colonic lamina propria [LP] during DSS-induced colitis revealed a significant increase in polymorphonuclear [PMN] leukocytes [Ly6GhighCD11b+] in colitic Atg5ΔMye mice compared with colitic controls [Supplementary Figure 2A and B, available as Supplementary data at ECCO-JCC online]. We also found an increase in monocytes and macrophages that were distinguished by Ly6C, MHCII, and CX3CR1 expression [Figure 2H–N]. Colonic macrophages are replenished by monocytes through a differentiation process called the monocyte waterfall.75–78 Ly6ChighMHCII− [P1] monocytes acquire MHCII to become Ly6ChighMHCII+ monocytes [P2]. These monocytes then differentiate into macrophages Ly6C-MHCII+ [P3-4] after downregulating Ly6C and then become fully mature macrophages by upregulating CX3CR1 [P4]. We find P1 [Figure 2I], P2 [Figure 2J], and the P3-4 mixed [Figure 2K] subsets are significantly increased in the LP of colitic Atg5ΔMye mice compared with colitic control mice; however, P3 and P4 subsets were not significantly different [Figure 2L–N]. During colitis, pro-inflammatory Ly6C+MHCII+ monocytes [P2] accumulate in the colon,79 which we find are increased in Atg5ΔMye mice [Figure 2J]. P4 macrophages from Atg5ΔMye mice also showed a general increase in TLR4 [Supplementary Figure 2C and G], CD14 [Supplementary Figure 2E and I], and CD115 [Supplementary Figure 2F and J] expression while expressing lower levels of CD206 [Supplementary Figure 2D and H] compared with P4 macrophages isolated from colitic control mice; however, only the levels of CD115 were significantly increased in the LP of colitic Atg5ΔMye mice. Overall, these results indicate that Atg5 expression in myeloid cells, like Atg7 and Atg16L1, is critical to control acute intestinal inflammation.
Figure 2.
Myeloid Atg5 expression prevents excessive DSS-induced colitis and an accumulation of lamina propria myeloid cells. DSS-induced colitis and myeloid cell response in Atg5-Wt and Atg5ΔMye mice. [A] Percent weight loss between Atg5-Wt and Atg5ΔMye mice. [B] Disease activity index [DAI] as determined by weight loss, behaviour, acute diarrhoeal response, and mucosal bleeding. [C] H&E staining of colon sections. [D] Histological scores. [E] Representative gross anatomy of the caecum and colon of colitic Atg5-Wt and Atg5ΔMye mice. [F] Colon length after colitis. [G] Spleen weight after colitis. [H] Dot plots showing lamina propria [gated on CD45+CD11b+ cells] monocyte waterfall as determined by Ly6C and MHC II staining after colitis. [I] Graph showing percent of P1 [Ly6C+MHC IILow] cells. [J] Graph showing percent of P2 [Ly6C+MHC II+] cells. [K] Graph showing percent of P3-4 [Ly6CLowMHC II+] cells. [L] Dot plots showing F4/80 expression and CX3CR1 expression on P3-P4 gated cells in H. [M] Graph showing percent of P3 [Ly6CLowMHC II+CX3CR1Low] cells. [N] Graph showing percent of P4 [Ly6CLowMHC II+CX3CR1+] cells. Representative of three independent experiments, Graphs indicate mean [±SD]; *p <0.05, **p <0.01. Two-tailed unpaired Student’s t tests or by two-way ANOVA with Tukey’s post hoc test. DSS, dextran sodium sulphate; H&E, haematoxylin and eosin; SD, standard deviation; ANOVA, analysis of variance.
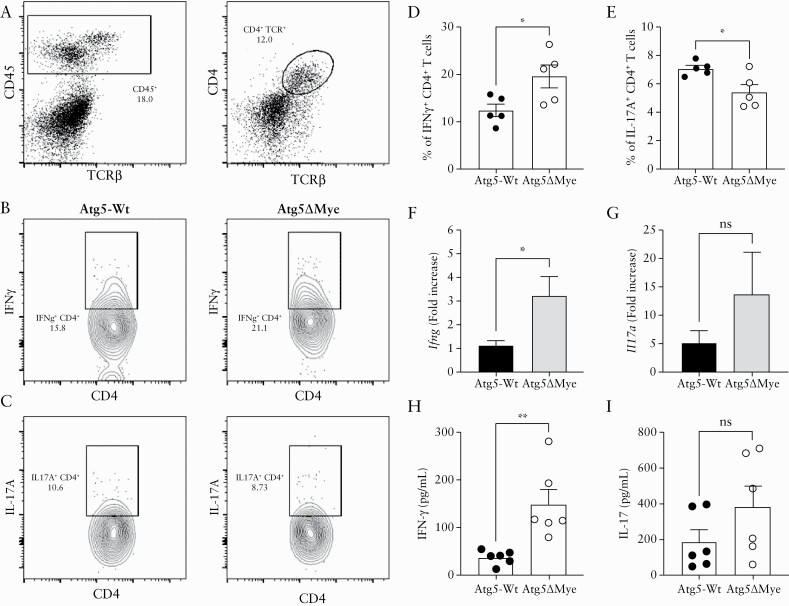
The proinflammatory cytokine IL-17 contributes to shaping and regulating the intestinal microbiota. We and others have shown that T cells from mice wherein myeloid cells lack Atg5 or other autophagy-related genes display skewing towards IL-17 polarisation.37,39–41 This T cell polarisation is mediated through the dysregulation of cytokines or cellular components that promote IL-17 responses.43,44 Microbiota differences can also alter T cell polarisation.80–84 Although the acute DSS model is not solely dependent on B and T cell responses, both participate in the exaggerated presentation of the disease. In fact, T cells have been shown to be the major driver of colonic inflammation in the acute DSS model by Day 4, with a peak at Day 8.85 Interestingly, we do not see major changes in DAI in Atg5ΔMye mice until Days 6 and 7 [Figure 2B] when we would expect T cells to be responding. To determine whether an enhanced level of IL-17-producing T cells populating the intestinal mucosa is influencing the microbiota of Atg5ΔMye mice and contributing to this excess inflammatory response, we isolated and stimulated CD4+ T cells from the colon lamina propria [LP] of mice during DSS treatment to determine TH polarisation. Interestingly, we observed an increase in IFNγ-producing CD4+ T cells isolated from the colonic LP of Atg5ΔMye mice [Figure 3A, B, and D]. Colitic colons from Atg5ΔMye mice also showed an increased IFNγ gene and protein expression compared with controls [Figure 3F and H]. Additionally, there was also an increase in IFNγ-producing CD4+ T cells isolated from the mesenteric lymph nodes (mLN) of colitic Atg5ΔMye mice [Supplementary Figure 3A, available as Supplementary data at ECCO-JCC online]. We also observed a significant reduction in IL-17-producing CD4+ T cells isolated from the colon of Atg5ΔMye mice compared with controls [Figure 3A, C, and E]. An although there was a difference in IL-17-producing CD4+ T cells in the colon, we found no significant difference in IL-17 gene and protein expression in the colon from both colitic Atg5ΔMye and Atg5-Wt mice [Figure 3G and I] or from mLN CD4+ T cells [Supplementary Figure 3B].
Figure 3.
Myeloid Atg5 expression prevents excessive IFNγ-mediated intestinal inflammation. Analysis of colonic IFNγ production after colitis induction. [A] Dot plots showing LP CD4+ T cells staining after colitis. [B] Intracellular IFNγ staining in LP CD4+ T cells. [C] Intracellular IL-17A staining in LP CD4+ T cells. [D] Graph showing the percent of IFNγ +CD4+ T cells isolated from the LP of colitic mice. [E] Graph showing the percent of IL-17A+CD4+ T cells isolated from the LP of colitic mice. [F] Colonic Ifng and [G] Il17a gene expression after colitis induction. [H] Colonic IFNγ and [I] IL-17 protein expression as determined by ELISA. Representative of three independent experiments. Graphs indicate mean [±SD]; *p <0.05, **p <0.01, Two-tailed unpaired Student’s t tests. LP, lamina propria; ELISA, enzyme-linked immunosorbent assay.
Our observation that CD4+ T cells are skewed towards a type 1 immune response in Atg5ΔMye mice after colitis induction prompted an examination of cytokine expression and TH skewing at steady state. Spontaneous colitis never occurred in Atg5ΔMye mice but, similar to what was observed during colitis, we observed an increase in IFNγ-producing CD4+ T cells isolated from the colonic LP of Atg5ΔMye mice [Supplementary Figure 3C and D] with no significant difference in the number of IL-17-producing CD4+ T cells [Supplementary Figure 3E and F] at steady state. This discrepancy in IL-17 expression could be tissue-specific, as models in which myeloid cells lack autophagic components and express high levels of IL-17 have been reported for only respiratory infections.37,40,41 However, others have reported TH1 skewing in ATG16L1T300A knockin mice as well as in conditional myeloid Atg7-deficient mice during intestinal inflammation.19,26 Furthermore, IFNγ has been shown to play an indispensable role in the initiation of acute DSS colitis, and IFNγ-deficient mice are protected from acute DSS-induced colitis.86 Thus, this model is sufficient to show IFNγ responses from T cells in Atg5ΔMye mice. Overall, at steady state and during inflammation, Atg5ΔMye mice show differences in CD4+ T cell polarisation in the colon particularly to a type 1 immune response.
2.3. Atg5 limits IL-12 production in myeloid cells independent of canonical autophagy
The current paradigm is that an autophagic defect [or loss of an autophagy-related gene] in myeloid cells leads to enhanced IL-1α/β expression.37–39 The excess production of IL-1 skews lymphocytes to produce IL-17, subsequently promoting IL-17-mediated inflammation. However, we found colitic Atg5ΔMye mice presented with increased IFNγ and no significant difference in IL-17 in the colon microenvironment. This suggests that colonic Atg5-deficient myeloid cells are likely to produce other factors that promote TH1 skewing and IFNγ production. IL-12p70 [hereafter called IL-12] is a major cytokine involved in TH1 polarisation and is a potent inducer of IFNγ.35,87–89 IL-12 is a heterodimeric protein comprising IL-12p35 and IL-12p40 [IL-12p40 also dimerises with IL-23p19 to form IL-23, a major cytokine maintaining TH17 cells] and is highly expressed by various myeloid cells including macrophages.35,90–93 We have previously shown that Atg5ΔMye mice also produce excess IL-12 during tuberculosis infection.37 Examination of the colon at steady state and during colitis revealed a general increase in IL-12 gene and protein expression [Supplementary Figure 3G–L] in Atg5ΔMye mice. This increase in colonic IL-12 suggests the cytokines required to promote type 1 immune responses are present in the colonic microenvironment of Atg5ΔMye mice. Nevertheless, it is unclear if this excess IL-12 is coming from Atg5-deficient myeloid cells.
We next set out to determine if Atg5-deficient myeloid cells produce excess IL-12. Using bone marrow-derived macrophages [BMM] from Atg5ΔMye and Atg5-Wt mice, we showed that Atg5-deficient macrophages secrete excess IL-12 upon LPS/IFN-γ stimulation compared with Atg5-Wt macrophages [Figure 4A]. For many cell types, low levels of IL-12p35 are constitutively expressed, whereas IL-12p40 expression occurs primarily in macrophages and dendritic cells, and both increase in response to microbial stimulation.90–93 We observed no difference in Il12a [IL-12p35] and Il12b [IL-12p40] gene expression [Supplementary Figure 4A and B, available as Supplementary data at ECCO-JCC online]. Furthermore, there was no difference in cell surface expression of IFNγ-receptor, TLR4/MD2, or CD14 between Atg5-Wt and Atg5-deficient BMM [Supplementary Figure 4C] that would enable an enhanced response. These data suggest that Atg5 has a post-transcriptional role in regulating IL-12 secretion.
Figure 4.
Atg5 regulates IL-12 secretion in macrophages. Analysis of IL-12 secretion in macrophages. [A] Detection of IL-12 by ELISA from Atg5-Wt or Atg5ΔMye BMM after LPS and IFNγ stimulation. [B and C] Detection of IL-12 by ELISA from Atg5-Wt [B] or Atg5ΔMye [C] BMM after LPS and IFNγ stimulation in the presence or absence of Baf. A1. Representative of two independent of experiments. [D‐F] 1–3 µm Z-stack images were performed on BMM stimulated with LPS, IFNγ, and Baf. A1 using immunofluorescence at 1.6 Zoom by a 63x oil immersion objective. [D] Atg5-Wt BMM were stained for LAMP1 [green] and IL-12p35 [red]. [E] Atg5-Wt BMM were stained for ATG5 [red] and IL-12p35 [green]. [F] Atg5ΔMye BMM were stained for LAMP1 [green] and IL-12p35 [red]. Representative from 65 images from 15 slides from two independent of experiments. Arrows in images indicate puncta co-localising with insets displaying enlargement of indicated region. Scale bars: 10 µm. Graphs indicate mean [±SD]; *p <0.05, **p <0.01,***p <0.001. Two-tailed unpaired Student’s t tests. ELISA, enzyme-linked immunosorbent assay; BMM, bone marrow-derived macrophages; LPS, lipopolysaccharide; SD,standard deviation.
As mentioned above, Atg5 assists in autophagosome formation and autophagosome-lysosome fusion. Thus, the genetic deletion of Atg5 would consequently affect several steps in autophagy. We next considered whether IL-12 was a direct target for autophagic removal. However, endogenous IL-12 and LC3 did not co-localise during stimulation and treatment with bafilomycin A1 [Baf. A1] [Supplementary Figure 4D] in Atg5-Wt or Atg5-deficient BMM, suggesting that IL-12 is unlikely to be a direct target for autophagic degradation. Baf. A1 is widely used to inhibit autophagic flux by targeting the V-ATPase ATP6V0C/V0 subunit c, but can also de-acidify endosome/lysosome vesicles through the same mechanism.94,95 Atg5 also regulates acidification and de-acidification of late endosomal compartments96 and, before stimulation, Atg5-deficient BMM display increased lysotracker staining compared with Atg5-Wt BMM [Supplementary Figure 4E], suggesting that the loss of Atg5 affects the regulation of vesicles’ pH levels.96,97 Interestingly, Baf. A1 treatment decreased IL-12 secretion in Atg5-Wt BMM [Figure 4B]. Furthermore, treatment of Atg5-deficient BMM with Baf. A1 also reduced IL-12 secretion [Figure 4C]. These data suggest that Atg5 regulates IL-12 secretion through vesicle acidification, and Baf. A1 can compensate for the loss of Atg5 in regulating IL-12 secretion.
We next examined the intracellular localisation of IL-12 after Baf. A1 treatment. As mentioned above, IL-12 consists of subunits IL-12p35 and IL-12p40. Whereas the IL-12p40 subunit has conventional secretory sequences and can be secreted in its homodimeric form, the IL-12p35 peptide is leaderless, cannot be secreted as a monomer, and conventional secretion cannot be induced by the addition of a secretory sequence.90,91,93 The purpose of IL-12p35’s constitutive expression remains poorly understood,90 but a recent report demonstrated that IL-12p35 is trafficked to late endosomes [LE] before secretion.98 As observed with total IL-12, Baf. A1 treatment resulted in IL-12p35 accumulation in LE as determined by IL-12p35 [red] and Lamp1 [green] co-localisation [Figure 4D]. White arrows in images indicate puncta co-localising with insets displaying enlargement of indicated region. Interestingly, IL-12p35 [green] co-localised with Atg5 [red] [Figure 4E]. IL-12 [red] co-localisation with Lamp1 [green] was more evident in Atg5-deficient BMM [Figure 4F]. Further analysis revealed that IL-12 [red] also co-localised with the LE marker Rab7 [green] [Supplementary Figure 4F], as previously reported.98 Taken together, our data suggest that Lamp1+ LEs99,100 contain IL-12, and the lack of Atg5 leads to increased levels of IL-12 in these vesicles.
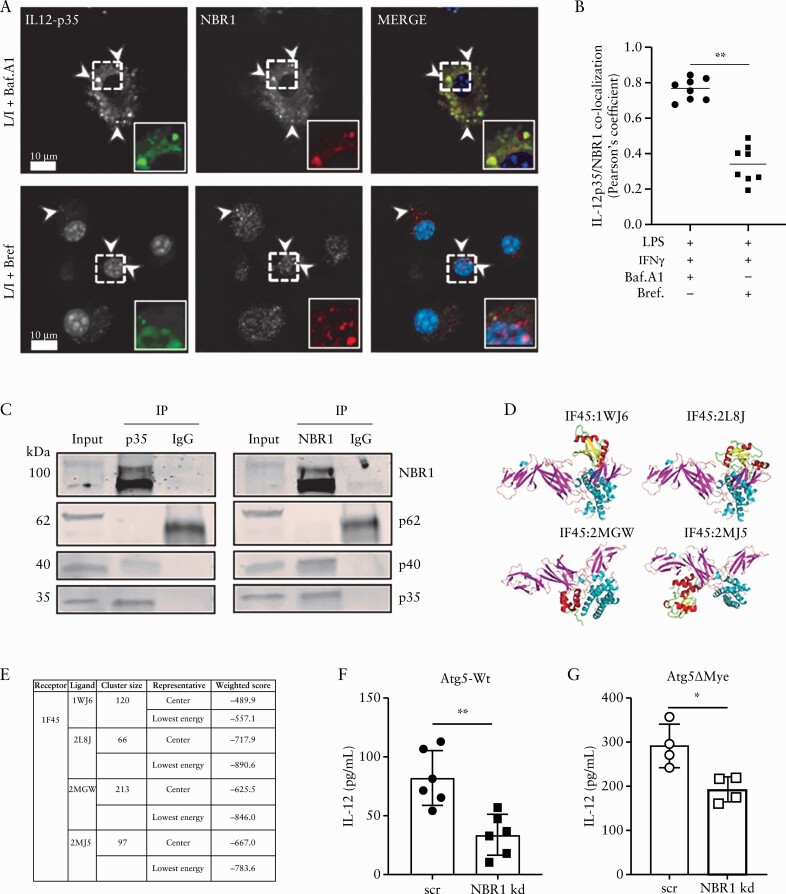
2.4. The sequestosome-1-like receptor NBR1 is involved in IL-12 secretion
Most cytokines are directed by their signal sequence through endoplasmic reticulum [ER]-golgi complex pathway for processing and trafficking, but some inflammatory cytokines, including IL-1β and IL-18, are known to be excreted via alternative strategies.38,101–105 For IL-1β, the selective autophagy cargo receptor TRIM16 directs IL-1β to LC3-II+ sequestration membranes for secretion.103 This suggests that other selective cargo receptors, such as the sequestosome-1-like receptors [SLRs: p62/SQSTM1 and NBR1], could act as possible cargo receptors for alternative secretion.102,105,106 We considered that NBR1 might be responsible for delivering IL-12 to LE for secretion given a homologue of the mammalian NBR1; Nbr1 from Schizosaccharomyces pombe was shown to deliver proteins to LE,107 and NBR1 accumulates upon autophagy inhibition.108 Indeed, confocal imaging revealed NBR1 [red] co-localised with IL-12p35 [green] in WT BMM after LPS/IFN-γ and Baf. A1 treatment [Figure 5A, top row, and B]; white arrows indicate puncta co-localising. However, this co-localisation was reduced upon brefeldin A [BrefA] treatment, suggesting that NBR1 [red] and IL-12 [green] interaction occurs after IL-12 leaves the ER [Figure 5A, bottom row, and B]. This reduction in co-localisation was not due to enhanced secretion, as IL-12 was undetectable by enzyme-linked immunosorbent assay [ELISA] after LPS/IFNγ/brefA treatment [data not shown]. To verify NBR1/IL-12 interaction in macrophages, cell lysates were immunoprecipitated with anti-IL12p35, anti-NBR1, or isotype control antibodies, and subjected to western blot with anti-IL-12p35, anti-IL-12p40, anti-NBR1, and anti-p62 antibodies. Both IL-12p40 and NBR1 co-precipitated with anti-IL-12p35 antibodies [Figure 5C]. Reciprocally, IL-12p35 and IL-12p40 co-precipitated with anti-NBR1 antibodies [Figure 5C]; however, neither probe co-precipitated p62/SQSTM1 [Figure 5C]. This interaction was further verified in silico through ClusPro docking,109 as IL-12 [1F45] interacted with multiple peptide fragments [1WJ6, 2L8J, 2MGW, and 2MJ5] of NBR1 [Figure 5D and E]. Lastly, a knockdown of NBR1 via small interfering RNA [siRNA] in Atg5-Wt BMM reduced IL-12 secretion compared with scrambled siRNA control [Figure 5F; Supplementary Figure 4G]. A reduction in IL-12 secretion was also observed in Atg5-deficient BMM after NBR1 knockdown, suggesting this interaction is independent of Atg5 [Figure 5G; Supplementary Figure 4G]. Taken together, our data suggest NBR1 is partially responsible for IL-12 secretion whereby it functions to direct IL-12p35 to LE for secretion.
Figure 5.
NBR1 and IL-12 interaction. Analysis of NBR1 and IL-12 interaction in macrophages. [A] Atg5-Wt BMM were stained for NBR1 [red] and IL-12p35 [green] after LPS and IFNγ stimulation and the addition of Bafilomycin A1 [top row] or Brefeldin A [bottom row]. [B] Quantification was performed using Pearson’s correlation coefficient [co-localisation] using image analysis. Representative of two independent of experiments and of 10 images from five slides. Arrows in images indicate puncta co-localising with insets displaying enlargement of indicated region. Scale bars: 10 µm. [C] Anti-IL-12p35 antibody co-immunoprecipitated NBR1 and IL-12p40, and anti-NBR1 antibody co-immunoprecipitated IL-12p35 and IL-12p40. [D and E] Protein docking between IL12A [PDB ID: 1F45] and NBR1. [D] The docking models and [E] ClusPro docking results for four candidate interaction models between 1F45 and four peptide fragments of NBR1 [PDB IDs: 1WJ6, 2L8J, 2MGW, and 2MJ5]. [F and G] Effects of NBR1 knockdown on IL-12 secretion in Atg5-Wt or Atg5ΔMye BMM. Graphs indicate mean [±SD]. * p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t tests. BMM, bone marrow-derived macrophages; LPS, lipopolysaccharide; SD,standard deviation.
2.5. IL-12 and the Atg5ΔMye gut microbiota contribute to the exacerbated intestinal inflammation
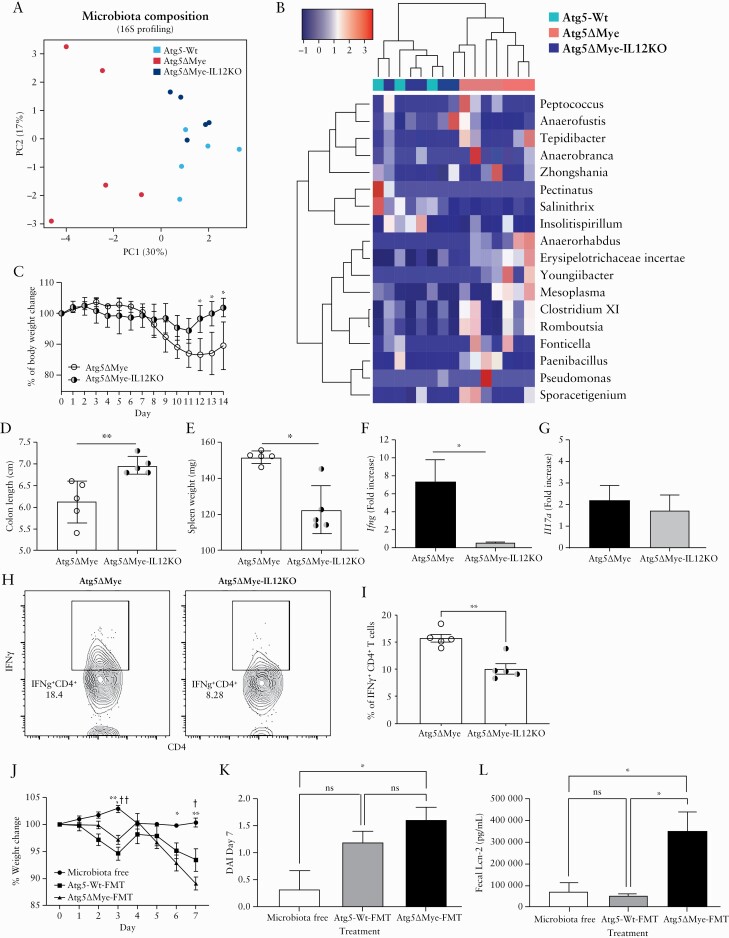
We found the exacerbated intestinal inflammation in Atg5ΔMye mice coincides with dysregulated IL-12 levels. Although this is the first finding linking Atg5 to IL-12 secretion, IL-12 has been associated with intestinal inflammation87,110 and it has been targeted therapeutically to attenuate intestinal inflammation. In both chemically induced colitis and genetic-deficient mouse models of colitis, antibodies against IL-12 prevented intestinal inflammation.110–114 Clinical trials have also been performed in individuals with IBD, using antibodies against the IL-12p40 subunit.48,115–119 These antibodies target both IL-12 and IL-23 pathways and has proven to be a safe and effective treatment approach in patients with IBD. Antibodies against IL-12/IL-23 also show efficacy in patients who failed treatment with anti–TNF-α agents, and the efficacy is more pronounced among secondary non-responders. Nevertheless, it unclear if IL-12 is leading to the changes in the gut microbiota and susceptibility to DSS-induced colitis we observe in Atg5ΔMye mice. Given the antibodies target both IL-12 and IL-23, we decided to target only IL-12 by crossing Atg5ΔMye mice with IL-12p35-deficient mice120 to generate Atg5ΔMye-IL12KO mice. We first examined the colonic microbiota at steady state in all three groups of mice [i.e., Atg5-Wt, Atg5ΔMye and Atg5ΔMye-IL12KO]. Atg5ΔMye-IL12KO mice showed a global change in the intestinal microbial composition in comparison with Atg5ΔMye mice, particularly an increase in the phylum Firmicutes [Supplementary Figure 5A, available as Supplementary data at ECCO-JCC online]. Furthermore, the PCoA plots and heatmap show clustering of the bacterial communities [at genus level] of the Atg5ΔMye-IL12KO and Atg5-Wt colonic mice which was separated from the bacterial community in the Atg5ΔMye mice [Figure 6A and B].
Figure 6.
Limiting IL-12 in mice with a selective Atg5-deficiency in myeloid cells restores the intestinal microbiota and alleviates inflammation. Bacterial composition as assessed by 16S rRNA sequencing from freshly collected stool of Atg5ΔMye and Atg5ΔMye-IL12KO mice before colitis induction [n = 5 per group]. [A] Principal coordinates analysis [PCoA] plot of microbiota composition [genus level]. [B] Heatmap of the relative abundance of colonic microbes [genus level]. A t test was used to measure specific microbiome species abundance between groups. Adjusted p-value >0.05 was used as significant threshold. [C] Percent weight loss between Atg5ΔMye and Atg5ΔMye-IL12KO mice. [D] Colon length after colitis. [E] Spleen weight after colitis. [F] Colonic Ifng and [G] Il17a gene expression after colitis induction. [H] Intracellular IFNγ staining in LP CD4+ T cells after colitis induction. [I] Graph showing the percent of IFNγ +CD4+ T cells isolated from the LP of colitic mice. [J] Percent weight loss between Atg5ΔMye mice receiving [●] no microbiota, FMT of [■] Atg5-Wt microbiota, or [▲] Atg5ΔMye microbiota [ƗAtg5-WT FMT vs no FMT, *Atg5ΔMye FMT vs no FMT]. [K] Disease activity index [DAI] as determined by weight loss, behaviour, acute diarrhoeal response and mucosal bleeding at Day 7. [L] Faecal Lcn-2 levels as detected by ELISA from FMT Atg5ΔMye groups. Representative of two‐three independent experiments. Graphs indicate mean [±SD]. * or Ɨp <0.05, ** or ƗƗp <0.01. Two-tailed unpaired Student’s t tests or by one- or two-way ANOVA with Tukey’s post hoc test. LPS, lipopolysaccharide; SD,standard deviation; LP, lamina propria; ELISA, enzyme-linked immunosorbent assay; FMT, faecal microbiota transplant; ANOVA, analysis of variance.
We next examined Atg5ΔMye-L12KO mice response to DSS-induced colitis. Atg5ΔMye-IL12KO mice were protected from DSS-induced colitis compared with Atg5ΔMye mice. Atg5ΔMye-IL12KO mice suffered minimal weight loss, showed no shortening of the colon after colitis induction, and had reduced spleen weights [Figure 6C–E]; however, no major changes were observed by histology [Supplementary Figure 5B and C]. There was also a decrease in Ifng gene expression in the colon after colitis induction in Atg5ΔMye-IL12KO mice, but no change in Il17a gene expression between both groups of mice [Figure 3F and G]. Isolation and stimulation of colonic LP CD4+ T cells showed a decrease in IFNγ-producing CD4+ T cells from Atg5ΔMye-IL12KO mice compared with Atg5ΔMye mice [Figure 6H and I]. Additionally, we observed a decrease in the percent of IFNγ-producing CD4+ T cells from the mLN of Atg5ΔMye-IL12KO mice compared with Atg5ΔMye mice [Supplementary Figure 5D]. However, Atg5ΔMye-IL12KO mLN CD4+ T cells did produce more IL-17 [Supplementary Figure 5E]. This change in IL-17 expression is likely due to Atg5-deficient myeloid cells producing excess IL-1.37 Nevertheless, genetic deletion of IL-12p35 from mice in which myeloid cells also lack Atg5 restores the microbiota and protects against DSS-induced colitis.
The DSS-induced colitis model is sensitive to the gut microbiota.72,73 The restoration of the microbiota in Atg5ΔMye-IL12KO mice [Figure 6A and B] which coincides with attenuated intestinal inflammation suggest that the Atg5ΔMye-associated gut microbiota may play a role in the exacerbated inflammation observed in Atg5ΔMye mice. Therefore, we performed a faecal microbiota transplant [FMT] in Atg5ΔMye mice to examine the pathogenic potential of the Atg5ΔMye-associated gut microbiota. First, the microbiota was ablated in a several groups of Atg5ΔMye mice via antibiotic treatment and microbiota was collected from fresh stool samples from either Atg5ΔMye or Atg5-Wt mice. Antibiotic-treated Atg5ΔMye mice were then orally gavaged with Atg5-Wt-associated microbiota, Atg5ΔMye-associated microbiota, or carrier solution. One week after FMT, mice were subjected to DSS-supplemented water for 7 days. Daily assessment revealed significant weight loss in Atg5ΔMye mice receiving Atg5ΔMye microbiota compared with microbiota-free Atg5ΔMye mice [Figure 6J]. This was also observed in Atg5ΔMye mice receiving Atg5-Wt microbiota when compared with microbiota-free Atg5ΔMye mice [Figure 6J]. Interestingly, microbiota-free Atg5ΔMye mice subjected to DSS had minimal weight loss. Furthermore, the DAI revealed a significant difference between Atg5ΔMye mice receiving FMT of Atg5ΔMye microbiota compared with microbiota-free Atg5ΔMye mice at Day 7 [Figure 6J; Supplementary Figure 5F]. Assessment of the stool for lipocalin-2 [Lcn-2], a marker of intestinal inflammation,121–123 at Day 7 revealed FMT of Atg5ΔMye microbiota had significantly increased faecal Lcn-2 levels compared with Atg5ΔMye mice with no FMT as well as mice receiving a FMT of Atg5-Wt microbiota [Figure 6K]. Additionally, proximal colons were immediately removed, cleaned, and then incubated for 4 h in RPMI.124 After the incubation period, the colon and supernatant [concentrated through centrifugation] were prepared for IL-12p35 immunoblot [Supplementary Figure 5G–I]. No significant change was observed for the colon between FMT-treated mice, but the concentrated supernatant from the colon of Atg5ΔMye mice receiving FMT of Atg5ΔMye microbiota showed an increase in IL-12 [Supplementary Figure 5G and I]. Overall, this suggest that the gut microbiota of Atg5ΔMye mice also contributes to the increased IL-12 and exacerbated colitis we have observed [Figure 2].
3. Discussion
This work identifies a role for Atg5 in myeloid cells in unconventional cytokine secretion that consequently affects intestinal homeostasis. Additionally, it adds to the plethora of functions described for Atg5.96,97,125–129 Along with these studies, this work establishes a new non-autophagy, immunological role for Atg5 in promoting TH1 responses. In the context of IBD and autophagy-related proteins [Atg], GWAS have only identified ATG16L1 variants.3 Nevertheless, animal models with an Atg5-deficiency display a similar phenotype to Atg16L1-deficient mice.14 Interestingly, the levels of ATG5 and the function of autophagy [and possibly other ATG5 functions] are decreased in IBD patient samples. This decrease in ATG5 expression and autophagy activity in IBD patients is linked to an increased expression of the microRNA miR30C that acts to downregulate ATG5 expression.55,130 Therefore, inhibiting Atg5 expression and function, described here in only myeloid cells, ultimately alters the intestinal microenvironment.
As previously reported, Atg5-deficient myeloid cells produce excess pro-inflammatory cytokines including IL-1α and IL-1β.37,38 Thus, we cannot dismiss that either cytokine contributes to the excess IFNγ observed in Atg5cKO mice, as IL-1 can synergise with IL-12 to enhance IFNγ production and TH1 skewing.131,132 Although we and others did not find any changes in colonic IL-17-producing cells in mice with a genetic deletion of Atg5 or Atg7 in myeloid cells,37,74 both colonic TH17 and TH1 cells were found to be increased in ATG16L1 T300A knockin mice.19 Given that Atg5ΔMye-IL12KO mice were protected against colitis induction, these results together suggest that IL-12 is a key cytokine driving intestinal pathology and TH1 skewing in our model.
This similarilty in intestinal TH1 responses between these mouse models is likely linked to the functional role of Atg5, Atg7, and Atg16L1. All three proteins are involved in some aspect of the autophagy process as well as endosome and lysosome regulation.12,107,133 The role of the TH17 response in driving intestinal pathology is up for debate as there is strong evidence suggesting that IL-17 may have a protective role in the intestine. Exacerbated intestinal inflammation was reported in Il17a-deficient mice or after in vivo blockade of IL-17 during DSS-induced colitis.47,49 Furthermore, in clinical trials for IBD patients, targeting IL-17 or IL-17R worsened symptoms, leading to early clinical trial termination.46,134 Nevertheless, this does not rule out the effects of increased levels of IL-17 during intestinal inflammation.30,35,45
Alterations in the gut microbiota have also been observed in ATG16L1 T300A knockin mice. Similar to our findings, ATG16L1 T300A knockin mice also had a decrease in the phylum Firmicutes and an increase in the phylum Bacteroidetes.19 It is unclear if these microbiota changes in ATG16L1 T300A knockin mice are due specifically to myeloid cells, since all cells including the intestinal epithelium express the ATG16L1 T300A gene in this mouse model. Furthermore, it is unclear if a certain genus that is lost/decreased [Firmicutes] or gained/increased [Bacteroidetes] is the driver of intestinal inflammation in Atg5Mye mice. A caveat of our FMT study is the potential loss of strictly anaerobic microorganisms after stool collection and processing. Nevertheless, the identification of the microorganism[s] driving intestinal inflammation could prove beneficial in individuals who have IBD associated with an autophagy risk loci134; however, this is out of the scope of this study. Additionally, IL-17 also plays a major role in regulating mucosal IgA production.136,137 Interestingly, increased IgA-coated bacteria were found in mice that lack Atg16L1 in myeloid cells, as well as in the stool of Crohn’s disease patients who were homozygous for the ATG16L1 T300A risk allele.26 Additionally, deletions or polymorphisms of autophagy genes in myeloid cells alter bacterial clearance, suggesting that there are multiple mechanisms by which autophagy genes regulate host-microbiota interactions. Our data suggest that myeloid cells likely have a strong contribution in maintaining the microbiota through Atg5’s action on IL-12 secretion.
The autophagic process and the genes that regulate autophagy are crucial for intestinal homeostasis. Autophagy is required for the maintenance of tight junction integrity, gut-commensal homeostasis, and control of invasive bacteria at the intestinal epithelium.13–18,20–22,74,138 Multiple lines of evidence also suggest that autophagy regulates inflammatory cytokines.37,38,139–141 These attributes make modulation of autophagy or single autophagic proteins an excellent therapeutic target. Nevertheless, it is critical to understand the function of these individual proteins, given they can modulate different TH responses. In fact, reagents that are known to target autophagy activation, such as rapamycin, increase IL-12 secretion.142,143 Additionally chloroquine, a known inhibitor of autophagosome-lysosome fusion, reduces IL-12 secretion144 likely through a similar mechanism as does Baf. A1. These reagents have been used with some clinical efficacy in IBD patients and animal models of IBD.145–148 It is unclear if these effects were due to IL-12 regulation, but targeting IL-12, specifically the IL-12p40 subunit that is shared with IL-23, has also proven beneficial for some IBD patients.35,48,149 Therefore, understanding how autophagy or single autophagic proteins regulate pro-inflammatory cytokines will allow precision in modulating the immune system in chronic inflammatory conditions like IBD. Last, targeting these autophagy proteins or their functions could be an alternative in mitigating problems associated with biologics, including loss of function, immunogenicity, and cost.150
In conclusion, our data support a novel role for Atg5 expression in myeloid cells in regulating intestinal homeostasis. Atg5 expression in myeloid cells controls the IL-12-IFNγ pathway that influences the microbiota and limits this pathway during colitis [Supplementary Figure 5J]. Mechanistically, we show that both Atg5 and the cargo receptor NBR1 regulate IL-12 secretion in myeloid cells. Specifically, we propose that NBR1 shuttles IL-12 to LE, whereby Atg5 functions to control the pH of these IL-12-containing vesicles for secretion. A genetic deletion of Atg5 results in dysregulation of IL-12 secretion as well as the accumulation of SLRs such as NBR1,108 which could potentially allow for increased accumulation of IL-12 in LE. Consequently, these attributes increase IL-12 secretion. A reduction in IL-12 secretion could be achieved by removing NBR1 or reducing LE/lysosomal pH with Baf. A1,- even in the absence of Atg5 expression. Nevertheless, the dysregulation of IL-12 in Atg5ΔMye mice leads to alterations of the microbiota and severe colitis when the intestinal barrier is disrupted.
4. Materials and Methods
4.1. Animals
The transgenic Atg5ΔMye mice [myeloid specific Atg5 deletion] and Atg5-Wt mice have previously been characterised.56 B6.129S1-Il12atm1jm/J [IL-12p35-deficient mice] were purchased from JAX [002692]119 and crossed to Atg5ΔMye mice to generate Atg5ΔMye-IL12KO mice. All mice were genotyped for the presence of Atg5, Il12a or Cre expression by Transnetyx Inc. All experiments were approved by the Institutional Animal Care and Use Committee of the University of New Mexico Health Sciences Center, in accordance with the National Institutes of Health guidelines for use of live animals. The University of New Mexico Health Sciences Center is accredited by the American Association for Accreditation of Laboratory Animal Care.
4.2. Intestinal inflammation model
For dextran sodium sulphate [DSS]-induced colitis, mice were provided 2.5% DSS [colitis grade, ~30 000–50 000 MW; MP Biomedicals] in drinking water ad libitum. Reagents used for in vivo treatment as well as sample collection and stimulation are included in Table S1, available as Supplementary data at ECCO-JCC online.
4.3. Microbiota analysis
Fresh faecal samples from Atg5-Wt, Atg5ΔMye, and Atg5ΔMye-IL-12KO mice were collected fresh in sterile tubes and flash-frozen. Microbial communities were determined by sequencing of the 16S rRNA as previously reported,151 with minor modifications described below. Microbial DNA was isolated from faeces using the ZymoBIOMICS DNA Miniprep Kit [Zymo Research] following manufacturer’s recommendations. Variable regions V-3 through V-4 of the 16S rRNA gene were amplified by polymerase chain reaction [PCR] using 100 ng input of DNA for each sample in duplicate, using primers 319F- [5’-ACTCCTRCGGGAGGCAGCAG-3’] and 806R- [5’-GACAGGACTACHVGGGTATCTAATCC-3’] containing Nextera adapter overhangs. A second PCR was performed with Nextera® XT Index Kit v2 Set A [Illumina] to complete the adapter and add unique sample-specific barcodes. After each PCR, a clean-up with AxyPrep Fragment Select-I magnetic beads [Axygen Biosciences] was completed, and all PCR reactions were run on an Applied Biosystems 2720 Thermal Cycler. Indexed samples were combined to yield duplicate 300 ng pools, followed by the creation and denaturation of a 4-nM library, and paired 250-bp sequencing runs were completed on the Illumina MiSeq using v3 sequencing chemistry [Illumina]. All reagents and kits are listed in Table S2, available as Supplementary data at ECCO-JCC online.
4.4. Microscopy and image analysis
For confocal microscopy, macrophages were plated at 100 k cells per well on 18-mm glass coverslips and stimulated with LPS [500 ng/mL] and IFNɣ [10 ng/mL] for 8 h and treated with BrefA [10 nM] or Baf. A1 [10 nM] for 2 h. Cells were then fixed with 4% PFA followed by a wash with 1x PBS. Blocking buffer contained PBS with 50% FBS, 2% BSA, and 0.1% saponin. After a 1-h stain in primary antibody, cells were washed with PBS then followed by 1-h stain in blocking buffer containing secondary antibody. Cells were mounted using ProLong Gold Antifade with DAPI and imaged using the Zeiss LSM 800 Airyscan Confocal microscope with a 63x oil objective lens. Images were processed using Zen Software and Adobe Photoshop [version CC 2019]. All primary and secondary antibodies used for confocal staining are listed in Table S3, available as Supplementary data at ECCO-JCC online.
4.5. Statistical analysis
Statistical analysis was performed as described in figure legends, and graphs generated display mean (± standard deviation [SD]) and were obtained using Prism software. Microbiome data were sequenced and processed by Illumina’s service laboratory using their in-house analysis pipeline. Cluster analysis was performed using heatmap3152 package in R. T testing was used to measure specific microbiome species abundance between conditions. Adjusted p-value >0.05 was used as significant threshold. Principal coordinate analysis was conducted in R. Confocal images statistical analysis and additional software Pearson’s correlation coefficient were acquired from BMM images using Huygens’s Deconvolution Scientific Volume Image Software [UNM Fluorescence Microscopy and Cell Imaging shared resource]. Quantification figures were also made using Prism, and confocal image figures were constructed using Adobe Illustrator [version CC 2019]. All other data were analysed using one-way analysis of variance [ANOVA] or two-tailed unpaired Student’s t test [Prism].
Supplementary Material
Funding
Supported in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health [NIH] through grant no. UL1TR001449 [EFC], KL2TR001448 [EFC] and in part by NIH grant P20GM121176 [EFC], and the Bioinformatics Shared Resource at University of New Mexico Comprehensive Cancer Center with grant P30CA118100 [YG]. SMG was supported in part by the Infectious Disease and Inflammation Program pre-doctoral T32 training grant, National Institutes of Health/National Institute of Allergy and Infectious Diseases grant T32AI007538.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
SDM and SMG performed all analysis with the help from YG, ZERW, JRB, RRG, AJH, JGI, and SBB. SDM, ZERW, YG, KCS, and DLD contributed to microbiota sequencing and analysis. SMG, JRB, and SBB participated in imaging and analysis. JGI, SBB, and VD provided reagents and animals. SDM and SMG participated in writing the manuscript. EFC designed the study, analysed data and wrote the paper. All authors approved the final version of the manuscript.
References
- 1. Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 2019;17:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brest P, Lapaquette P, Souidi M, et al. . A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet 2011;43:242–5. [DOI] [PubMed] [Google Scholar]
- 3. Hampe J, Franke A, Rosenstiel P, et al. . A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 2007;39:207–11. [DOI] [PubMed] [Google Scholar]
- 4. Henckaerts L, Cleynen I, Brinar M, et al. . Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm Bowel Dis 2011;17:1392–7. [DOI] [PubMed] [Google Scholar]
- 5. Lahiri A, Hedl M, Abraham C. MTMR3 risk allele enhances innate receptor-induced signaling and cytokines by decreasing autophagy and increasing caspase-1 activation. Proc Natl Acad Sci U S A 2015;112:10461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lassen KG, Xavier RJ. Mechanisms and function of autophagy in intestinal disease. Autophagy 2018;14:216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell 2019;176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCarroll SA, Huett A, Kuballa P, et al. . Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet 2008;40:1107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rioux JD, Xavier RJ, Taylor KD, et al. . Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 2007;39:596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roberts RL, Gearry RB, Hollis-Moffatt JE, et al. . IL23R R381Q and ATG16L1 T300A are strongly associated with Crohn’s disease in a study of New Zealand Caucasians with inflammatory bowel disease. Am J Gastroenterol 2007;102:2754–61. [DOI] [PubMed] [Google Scholar]
- 11. Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, Hata A. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet 2007;52:575–83. [DOI] [PubMed] [Google Scholar]
- 12. Galluzzi L, Baehrecke EH, Ballabio A, et al. . Molecular definitions of autophagy and related processes. EMBO J 2017;36:1811–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cadwell K, Liu JY, Brown SL, et al. . A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008;456:259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cadwell K, Patel KK, Komatsu M, Virgin HW 4th, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 2009;5:250–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, et al. . Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 2017;214:3687–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nighot PK, Hu CA, Ma TY. Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J Biol Chem 2015;290:7234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel KK, Miyoshi H, Beatty WL, et al. . Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J 2013;32:3130–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong M, Ganapathy AS, Suchanec E, Laidler L, Ma T, Nighot P. Intestinal epithelial tight junction barrier regulation by autophagy-related protein ATG6/beclin 1. Am J Physiol Cell Physiol 2019;316:C753–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lavoie S, Conway KL, Lassen KG, et al. . The Crohn’s disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. Elife 2019;8:e39982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bel S, Pendse M, Wang Y, et al. . Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 2017;357:1047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamin JL, Sumpter R Jr, Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe 2013;13:723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burger E, Araujo A, López-Yglesias A, et al. . Loss of paneth cell autophagy causes acute susceptibility to Toxoplasma gondii-mediated inflammation. Cell Host Microbe 2018;23:177–90.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell 2015;58:507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology 2010;139:1630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006;313:1438–41. [DOI] [PubMed] [Google Scholar]
- 26. Zhang H, Zheng L, McGovern DP, et al. . Myeloid ATG16L1 facilitates host-bacteria interactions in maintaining intestinal homeostasis. J Immunol 2017;198:2133–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Angelidou I, Chrysanthopoulou A, Mitsios A, et al. . REDD1/autophagy pathway is associated with neutrophil-driven IL-1β inflammatory response in active ulcerative colitis. J Immunol 2018;200:3950–61. [DOI] [PubMed] [Google Scholar]
- 28. Coccia M, Harrison OJ, Schiering C, et al. . IL-1β mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4[+] Th17 cells. J Exp Med 2012;209:1595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dragasevic S, Stankovic B, Sokic-Milutinovic A, et al. . Importance of TLR9-IL23-IL17 axis in inflammatory bowel disease development: gene expression profiling study. Clin Immunol 2018;197:86–95. [DOI] [PubMed] [Google Scholar]
- 30. Jiang W, Su J, Zhang X, et al. . Elevated levels of Th17 cells and Th17-related cytokines are associated with disease activity in patients with inflammatory bowel disease. Inflamm Res 2014;63:943–50. [DOI] [PubMed] [Google Scholar]
- 31. Kanai T, Mikami Y, Sujino T, Hisamatsu T, Hibi T. RORγt-dependent IL-17A-producing cells in the pathogenesis of intestinal inflammation. Mucosal Immunol 2012;5:240–7. [DOI] [PubMed] [Google Scholar]
- 32. Liu Z, Yadav PK, Xu X, et al. . The increased expression of IL-23 in inflammatory bowel disease promotes intraepithelial and lamina propria lymphocyte inflammatory responses and cytotoxicity. J Leukoc Biol 2011;89:597–606. [DOI] [PubMed] [Google Scholar]
- 33. Mao L, Kitani A, Strober W, Fuss IJ. The role of NLRP3 and IL-1β in the pathogenesis of inflammatory bowel disease. Front Immunol 2018;9:2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menghini P, Corridoni D, Butto LF, et al. . Neutralization of IL-1alpha ameliorates Crohn’s disease-like ileitis by functional alterations of the gut microbiome. Proc Natl Acad Sci U S A 2019;116:26717‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moschen AR, Tilg H, Raine T. IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol 2019;16:185–96. [DOI] [PubMed] [Google Scholar]
- 36. Shouval DS, Biswas A, Kang YH, et al. . Interleukin 1β mediates intestinal inflammation in mice and patients with interleukin 10 receptor deficiency. Gastroenterology 2016;151:1100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Castillo EF, Dekonenko A, Arko-Mensah J, et al. . Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A 2012;109:E3168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 2011;30:4701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peral de Castro C, Jones SA, Ni Cheallaigh C, et al. . Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol 2012;189:4144–53. [DOI] [PubMed] [Google Scholar]
- 40. Reed M, Morris SH, Owczarczyk AB, Lukacs NW. Deficiency of autophagy protein Map1-LC3b mediates IL-17-dependent lung pathology during respiratory viral infection via ER stress-associated IL-1. Mucosal Immunol 2015;8:1118–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012;150:803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang M, Kenny SJ, Ge L, Xu K, Schekman R. Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife 2015;4:e11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chung Y, Chang SH, Martinez GJ, et al. . Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 2009;30:576–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McGeachy MJ, Chen Y, Tato CM, et al. . The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol 2009;10:314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujino S, Andoh A, Bamba S, et al. . Increased expression of interleukin 17 in inflammatory bowel disease. Gut 2003;52:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hueber W, Sands BE, Lewitzky S, et al. ; Secukinumab in Crohn’s Disease Study Group. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 2012;61:1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogawa A, Andoh A, Araki Y, Bamba T, Fujiyama Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin Immunol 2004;110:55–62. [DOI] [PubMed] [Google Scholar]
- 48. Feagan BG, Sandborn WJ, Gasink C, et al. ; UNITI–IM-UNITI Study Group. Ustekinumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med 2016;375:1946–60. [DOI] [PubMed] [Google Scholar]
- 49. Yang XO, Chang SH, Park H, et al. . Regulation of inflammatory responses by IL-17F. J Exp Med 2008;205:1063–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanada T, Noda NN, Satomi Y, et al. . The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007;282:37298–302. [DOI] [PubMed] [Google Scholar]
- 51. Mizushima N, Noda T, Yoshimori T, et al. . A protein conjugation system essential for autophagy. Nature 1998;395:395–8. [DOI] [PubMed] [Google Scholar]
- 52. Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep 2013;14:206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sakoh-Nakatogawa M, Matoba K, Asai E, et al. . Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol 2013;20:433–9. [DOI] [PubMed] [Google Scholar]
- 54. Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell 2012;45:629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ye X, Zhou XJ, Zhang H. Exploring the role of autophagy-related gene 5 [ATG5] yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front Immunol 2018;9:2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao Z, Fux B, Goodwin M, et al. . Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 2008;4:458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Imhann F, Vich Vila A, Bonder MJ, et al. . Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018;67:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kang B, Alvarado LJ, Kim T, et al. . Commensal microbiota drive the functional diversification of colon macrophages. Mucosal Immunol 2020;13:216–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 2012;9:599–608. [DOI] [PubMed] [Google Scholar]
- 60. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. . Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006;55:205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol 2017;14:573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Willing BP, Dicksved J, Halfvarson J, et al. . A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010;139:1844–54.e1. [DOI] [PubMed] [Google Scholar]
- 63. Bain CC, Bravo-Blas A, Scott CL, et al. . Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 2014;15:929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 2014;111:2247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mortha A, Chudnovskiy A, Hashimoto D, et al. . Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 2014;343:1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Singh N, Gurav A, Sivaprakasam S, et al. . Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014;40:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bader JE, Enos RT, Velázquez KT, et al. . Macrophage depletion using clodronate liposomes decreases tumorigenesis and alters gut microbiota in the AOM/DSS mouse model of colon cancer. Am J Physiol Gastrointest Liver Physiol 2018;314:G22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Niess JH, Brand S, Gu X, et al. . CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 2005;307:254–8. [DOI] [PubMed] [Google Scholar]
- 69. Rescigno M, Urbano M, Valzasina B, et al. . Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2001;2:361–7. [DOI] [PubMed] [Google Scholar]
- 70. Vallon-Eberhard A, Landsman L, Yogev N, Verrier B, Jung S. Transepithelial pathogen uptake into the small intestinal lamina propria. J Immunol 2006;176:2465–9. [DOI] [PubMed] [Google Scholar]
- 71. Seksik P. [Gut microbiota and IBD]. Gastroenterol Clin Biol 2010;34[Suppl 1]:S44–51. [DOI] [PubMed] [Google Scholar]
- 72. Brinkman BM, Becker A, Ayiseh RB, et al. . Gut microbiota affects sensitivity to acute DSS-induced colitis independently of host genotype. Inflamm Bowel Dis 2013;19:2560–7. [DOI] [PubMed] [Google Scholar]
- 73. Roy U, Gálvez EJC, Iljazovic A, et al. . Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep 2017;21:994–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee HY, Kim J, Quan W, et al. . Autophagy deficiency in myeloid cells increases susceptibility to obesity-induced diabetes and experimental colitis. Autophagy 2016a;12:1390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bain CC, Schridde A. Origin, differentiation, and function of intestinal macrophages. Front Immunol 2018;9:2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bain CC, Scott CL, Uronen-Hansson H, et al. . Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol 2013;6:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schridde A, Bain CC, Mayer JU, et al. . Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol 2017;10:1387–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tamoutounour S, Henri S, Lelouard H, et al. . CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol 2012;42:3150–66. [DOI] [PubMed] [Google Scholar]
- 79. Nakanishi Y, Sato T, Takahashi K, Ohteki T. IFN-γ-dependent epigenetic regulation instructs colitogenic monocyte/macrophage lineage differentiation in vivo. Mucosal Immunol 2018;11:871–80. [DOI] [PubMed] [Google Scholar]
- 80. Atarashi K, Tanoue T, Shima T, et al. . Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011;331:337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen L, Sun M, Wu W, et al. . Microbiota metabolite butyrate differentially regulates Th1 and Th17 Cells’ differentiation and function in induction of colitis. Inflamm Bowel Dis 2019;25:1450–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ivanov II, Atarashi K, Manel N, et al. . Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ivanov II, Frutos Rde L, Manel N, et al. . Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008;4:337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sun M, Wu W, Chen L, et al. . Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun 2018;9:3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nunes NS, Kim S, Sundby M, et al. . Temporal clinical, proteomic, histological and cellular immune responses of dextran sulfate sodium-induced acute colitis. World J Gastroenterol 2018;24:4341–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ito R, Shin-Ya M, Kishida T, et al. . Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin Exp Immunol 2006;146:330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Eftychi C, Schwarzer R, Vlantis K, et al. . Temporally distinct functions of the cytokines IL-12 and IL-23 drive chronic colon inflammation in response to intestinal barrier impairment. Immunity 2019;51:367–80.e4. [DOI] [PubMed] [Google Scholar]
- 88. Perussia B, Chan SH, D’Andrea A, et al. . Natural killer [NK] cell stimulatory factor or IL-12 has differential effects on the proliferation of TCR-alpha beta+, TCR-gamma delta+ T lymphocytes, and NK cells. J Immunol 1992;149:3495–502. [PubMed] [Google Scholar]
- 89. Trinchieri G, Wysocka M, D’Andrea A, et al. . Natural killer cell stimulatory factor [NKSF] or interleukin-12 is a key regulator of immune response and inflammation. Prog Growth Factor Res 1992;4:355–68. [DOI] [PubMed] [Google Scholar]
- 90. Abdi K, Singh NJ. Making many from few: IL-12p40 as a model for the combinatorial assembly of heterodimeric cytokines. Cytokine 2015;76:53–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Carra G, Gerosa F, Trinchieri G. Biosynthesis and posttranslational regulation of human IL-12. J Immunol 2000;164:4752–61. [DOI] [PubMed] [Google Scholar]
- 92. D’Andrea A, Rengaraju M, Valiante NM, et al. . Production of natural killer cell stimulatory factor [interleukin 12] by peripheral blood mononuclear cells. J Exp Med 1992;176:1387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Reitberger S, Haimerl P, Aschenbrenner I, Esser-von Bieren J, Feige MJ. Assembly-induced folding regulates interleukin 12 biogenesis and secretion. J Biol Chem 2017;292:8073–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Harada M, Shakado S, Sakisaka S, et al. . Bafilomycin A1, a specific inhibitor of V-type H+-ATPases, inhibits the acidification of endocytic structures and inhibits horseradish peroxidase uptake in isolated rat sinusoidal endothelial cells. Liver 1997;17:244–50. [DOI] [PubMed] [Google Scholar]
- 95. Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H[+]-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 1991;266:17707–12. [PubMed] [Google Scholar]
- 96. Guo H, Chitiprolu M, Roncevic L, et al. . Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev Cell 2017;43:716–30.e7. [DOI] [PubMed] [Google Scholar]
- 97. Peng J, Zhang R, Cui Y, et al. . Atg5 regulates late endosome and lysosome biogenesis. Sci China Life Sci 2014;57:59–68. [DOI] [PubMed] [Google Scholar]
- 98. Chiaruttini G, Piperno GM, Jouve M, et al. . The SNARE VAMP7 regulates exocytic trafficking of interleukin-12 in dendritic cells. Cell Rep 2016;14:2624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cheng XT, Xie YX, Zhou B, Huang N, Farfel-Becker T, Sheng ZH. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J Cell Biol 2018;217:3127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dunster K, Toh BH, Sentry JW. Early endosomes, late endosomes, and lysosomes display distinct partitioning strategies of inheritance with similarities to Golgi-derived membranes. Eur J Cell Biol 2002;81:117–24. [DOI] [PubMed] [Google Scholar]
- 101. Abdel Fattah E, Bhattacharya A, Herron A, Safdar Z, Eissa NT. Critical role for IL-18 in spontaneous lung inflammation caused by autophagy deficiency. J Immunol 2015;194:5407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Claude-Taupin A, Bissa B, Jia J, Gu Y, Deretic V. Role of autophagy in IL-1β export and release from cells. Semin Cell Dev Biol 2018;83:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kimura T, Jia J, Kumar S, et al. . Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J 2017;36:42–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Murai H, Okazaki S, Hayashi H, et al. . Alternaria extract activates autophagy that induces IL-18 release from airway epithelial cells. Biochem Biophys Res Commun 2015;464:969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Claude-Taupin A, Jia J, Mudd M, Deretic V. Autophagy’s secret life: secretion instead of degradation. Essays Biochem 2017;61:637–47. [DOI] [PubMed] [Google Scholar]
- 106. Jiang S, Dupont N, Castillo EF, Deretic V. Secretory versus degradative autophagy: unconventional secretion of inflammatory mediators. J Innate Immun 2013;5:471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mardakheh FK, Yekezare M, Machesky LM, Heath JK. Spred2 interaction with the late endosomal protein NBR1 down-regulates fibroblast growth factor receptor signaling. J Cell Biol 2009;187:265–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kirkin V, Lamark T, Sou YS, et al. . A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009;33:505–16. [DOI] [PubMed] [Google Scholar]
- 109. Kozakov D, Hall DR, Xia B, et al. . The ClusPro web server for protein-protein docking. Nat Protoc 2017;12:255–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Uhlig HH, McKenzie BS, Hue S, et al. . Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity 2006;25:309–18. [DOI] [PubMed] [Google Scholar]
- 111. Berg DJ, Davidson N, Kühn R, et al. . Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4[+] TH1-like responses. J Clin Invest 1996;98:1010–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Davidson NJ, Hudak SA, Lesley RE, Menon S, Leach MW, Rennick DM. IL-12, but not IFN-gamma, plays a major role in sustaining the chronic phase of colitis in IL-10-deficient mice. J Immunol 1998;161:3143–9. [PubMed] [Google Scholar]
- 113. Fuss IJ, Marth T, Neurath MF, Pearlstein GR, Jain A, Strober W. Anti-interleukin 12 treatment regulates apoptosis of Th1 T cells in experimental colitis in mice. Gastroenterology 1999;117:1078–88. [DOI] [PubMed] [Google Scholar]
- 114. Neurath MF, Fuss I, Kelsall BL, Stüber E, Strober W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J Exp Med 1995;182:1281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Khorrami S, Ginard D, Marín-Jiménez I, et al. . Ustekinumab for the treatment of refractory Crohn’s disease: the Spanish experience in a large multicentre open-label cohort. Inflamm Bowel Dis 2016;22:1662–9. [DOI] [PubMed] [Google Scholar]
- 116. Mannon PJ, Fuss IJ, Mayer L, et al. ; Anti-IL-12 Crohn’s Disease Study Group. Anti-interleukin-12 antibody for active Crohn’s disease. N Engl J Med 2004;351:2069–79. [DOI] [PubMed] [Google Scholar]
- 117. Sandborn WJ, Feagan BG, Fedorak RN, et al. ; Ustekinumab Crohn’s Disease Study Group. A randomized trial of Ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with moderate-to-severe Crohn’s disease. Gastroenterology 2008;135:1130–41. [DOI] [PubMed] [Google Scholar]
- 118. Sandborn WJ, Gasink C, Gao LL, et al. ; CERTIFI Study Group. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med 2012;367:1519–28. [DOI] [PubMed] [Google Scholar]
- 119. Wils P, Bouhnik Y, Michetti P, et al. ; Groupe d’Etude Thérapeutique des Affections Inflammatoires du Tube Digestif. Subcutaneous ustekinumab provides clinical benefit for two-thirds of patients with Crohn’s disease refractory to anti-tumor necrosis factor agents. Clin Gastroenterol Hepatol 2016;14:242–50.e1–2. [DOI] [PubMed] [Google Scholar]
- 120. Mattner F, Magram J, Ferrante J, et al. . Genetically resistant mice lacking interleukin-12 are susceptible to infection with Leishmania major and mount a polarized Th2 cell response. Eur J Immunol 1996;26:1553–9. [DOI] [PubMed] [Google Scholar]
- 121. Alpízar-Alpízar W, Laerum OD, Illemann M, et al. . Neutrophil gelatinase-associated lipocalin [NGAL/Lcn2] is upregulated in gastric mucosa infected with Helicobacter pylori. Virchows Arch 2009;455:225–33. [DOI] [PubMed] [Google Scholar]
- 122. Playford RJ, Belo A, Poulsom R, et al. . Effects of mouse and human lipocalin homologues 24p3/lcn2 and neutrophil gelatinase-associated lipocalin on gastrointestinal mucosal integrity and repair. Gastroenterology 2006;131:809–17. [DOI] [PubMed] [Google Scholar]
- 123. Raffatellu M, George MD, Akiyama Y, et al. . Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 2009;5:476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ray AL, Castillo EF, Morris KT, et al. . Blockade of MK2 is protective in inflammation-associated colorectal cancer development. Int J Cancer 2016;138:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Inomata M, Into T, Niida S, Murakami Y. Atg5 regulates formation of MyD88 condensed structures and MyD88-dependent signal transduction. Biochem Biophys Res Commun 2013;437:509–14. [DOI] [PubMed] [Google Scholar]
- 126. Lee HK, Mattei LM, Steinberg BE, et al. . In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 2010;32:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ndoye A, Budina-Kolomets A, Kugel CH 3rd, et al. . ATG5 mediates a positive feedback loop between Wnt signaling and autophagy in melanoma. Cancer Res 2017;77:5873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Simon HU, Yousefi S, Schmid I, Friis R. ATG5 can regulate p53 expression and activation. Cell Death Dis 2014;5:e1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Yousefi S, Perozzo R, Schmid I, et al. . Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006;8:1124–32. [DOI] [PubMed] [Google Scholar]
- 130. Nguyen HT, Dalmasso G, Müller S, Carrière J, Seibold F, Darfeuille-Michaud A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology 2014;146:508–19. [DOI] [PubMed] [Google Scholar]
- 131. Cooper MA, Fehniger TA, Ponnappan A, Mehta V, Wewers MD, Caligiuri MA. Interleukin-1beta costimulates interferon-gamma production by human natural killer cells. Eur J Immunol 2001;31:792–801. [DOI] [PubMed] [Google Scholar]
- 132. Tominaga K, Yoshimoto T, Torigoe K, et al. . IL-12 synergizes with IL-18 or IL-1beta for IFN-gamma production from human T cells. Int Immunol 2000;12:151–60. [DOI] [PubMed] [Google Scholar]
- 133. Fraser J, Simpson J, Fontana R, Kishi-Itakura C, Ktistakis NT, Gammoh N. Targeting of early endosomes by autophagy facilitates EGFR recycling and signalling. EMBO Rep 2019;20:e47734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Targan SR, Feagan B, Vermeire S, et al. . A randomized, double-blind, placebo-controlled phase 2 study of brodalumab in patients with moderate-to-severe Crohn’s disease. Am J Gastroenterol 2016;111:1599–607. [DOI] [PubMed] [Google Scholar]
- 135. Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020;16:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Dann SM, Manthey CF, Le C, et al. . IL-17A promotes protective IgA responses and expression of other potential effectors against the lumen-dwelling enteric parasite Giardia. Exp Parasitol 2015;156:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kumar P, Monin L, Castillo P, et al. . Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity 2016;44:659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wu Y, Wang Y, Zou H, et al. . Probiotic Bacillus amyloliquefaciens SC06 induces autophagy to protect against pathogens in macrophages. Front Microbiol 2017;8:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lee JP, Foote A, Fan H, et al. . Loss of autophagy enhances MIF/macrophage migration inhibitory factor release by macrophages. Autophagy 2016b;12:907–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Merkley SD, Chock CJ, Yang XO, Harris J, Castillo EF. Modulating T cell responses via autophagy: the intrinsic influence controlling the function of both antigen-presenting cells and T cells. Front Immunol 2018;9:2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Saitoh T, Fujita N, Jang MH, et al. . Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008;456:264–8. [DOI] [PubMed] [Google Scholar]
- 142. Macedo C, Turnquist HR, Castillo-Rama M, et al. . Rapamycin augments human DC IL-12p70 and IL-27 secretion to promote allogeneic Type 1 polarization modulated by NK cells. Am J Transplant 2013;13:2322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ohtani M, Nagai S, Kondo S, et al. . Mammalian target of rapamycin and glycogen synthase kinase 3 differentially regulate lipopolysaccharide-induced interleukin-12 production in dendritic cells. Blood 2008;112:635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Said A, Bock S, Lajqi T, Müller G, Weindl G. Chloroquine promotes IL-17 production by CD4+ T cells via p38-dependent IL-23 release by monocyte-derived Langerhans-like cells. J Immunol 2014;193:6135–43. [DOI] [PubMed] [Google Scholar]
- 145. Goenka MK, Kochhar R, Tandia B, Mehta SK. Chloroquine for mild to moderately active ulcerative colitis: comparison with sulfasalazine. Am J Gastroenterol 1996;91:917–21. [PubMed] [Google Scholar]
- 146. Kanvinde S, Chhonker YS, Ahmad R, et al. . Pharmacokinetics and efficacy of orally administered polymeric chloroquine as macromolecular drug in the treatment of inflammatory bowel disease. Acta Biomater 2018;82:158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Nagar J, Ranade S, Kamath V, et al. . Therapeutic potential of chloroquine in a murine model of inflammatory bowel disease. Int Immunopharmacol 2014;21:328–35. [DOI] [PubMed] [Google Scholar]
- 148. Park TY, Jang Y, Kim W, et al. . Chloroquine modulates inflammatory autoimmune responses through Nurr1 in autoimmune diseases. Sci Rep 2019;9:15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sands BE, Sandborn WJ, Panaccione R, et al. ; UNIFI Study Group. Ustekinumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med 2019;381:1201–14. [DOI] [PubMed] [Google Scholar]
- 150. D’Haens G. Risks and benefits of biologic therapy for inflammatory bowel diseases. Gut 2007;56:725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Komesu YM, Richter HE, Dinwiddie DL, et al. . Methodology for a vaginal and urinary microbiome study in women with mixed urinary incontinence. Int Urogynecol J 2017;28:711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhao S, Guo Y, Sheng Q, Shyr Y. Advanced heat map and clustering analysis using heatmap3. Biomed Res Int 2014;2014:986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.