Abstract
Cultures of Helicobacter pylori obtained from the American Type Culture Collection (strain 43504) were grown as isolated colonies or lawns on blood agar plates and in broth culture with constant shaking. Examination of bacterial growth with Gram-stained fixed preparation and differential interference contrast microscopy on wet preparations revealed that bacteria grown on blood agar plates had a morphology consistent with that normally reported for H. pylori whereas bacteria from broth cultures had the morphologic appearance of Helicobacter heilmannii. Bacteria harvested from blood agar plates assumed an H. heilmannii-like morphology when transferred to broth cultures, and bacteria from broth cultures grew with morphology typical of H. pylori when grown on blood agar plates. Analysis by PCR of bacteria isolated from blood agar plates and broth cultures indicated that a single strain of bacteria (H. pylori) was responsible for both morphologies.
The presence of spiral bacteria in gastric mucosa was first reported over a century ago; however, it was not until the work of Warren and Marshall in the 1980s that the role of these bacteria in gastric disease became clear (4, 5, 12). It is now well established that Helicobacter pylori can be pathogenic in humans and is the cause of gastric disease in a significant percentage of the human population. A second bacterium, currently named Helicobacter heilmannii, has also been implicated as a potential cause of gastric disease in humans (6, 10). Infection with H. heilmannii is thought to be considerably less frequent than infection with H. pylori. Standard tests for gastric infection including detection of urease activity and antibody-based assays may not distinguish between these bacteria due to common enzymes and cross-reactivity (2, 13). Furthermore, there is no available routine method to culture H. heilmannii (1). Thus, differentiation between H. heilmannii and H. pylori as a cause of gastric infection is based primarily on morphological differences between the two organisms. Typically, H. pylori assumes a curved rod-like or short spiral morphology with up to three turns while H. heilmannii is reported to grow as a long spiral bacterium with four or more turns (6, 7).
Our laboratory has been working on the development of molecularly and serologically based assays for aid in diagnosing infection with H. pylori. As part of these studies, we adopted a broth culture technique to enhance our capabilities to produce and process H. pylori for use in our assays. Stock cultures of H. pylori were obtained from the American Type Culture Collection (ATCC) (Manassas, Va.) and grown on blood agar plates or in broth. Routine microscopic examination of cultured organisms revealed that growth in broth induced the H. pylori cells to grow as long spirals (5 to >20 turns) while morphology typical of H. pylori was observed for cultures grown on blood agar. Morphology appeared to be dependent on culture conditions, as it could be reversed by changing the culture method. These results suggest that reliance on morphology as the sole criterion for distinguishing between H. heilmannii and H. pylori may need to be reevaluated, as at least some reports of H. heilmannii may represent in vivo growth of H. pylori in its long spiral form.
MATERIALS AND METHODS
Broth culture.
Lyophilized H. pylori ATCC 43504 was reconstituted with 0.3 ml of brucella broth (Difco, Detroit, Mich.) with 1% cyclodextrin (BBCD) (8). This inoculum was added to 25 ml of BBCD in a 25-cm2 tissue culture flask and incubated at 37°C with rocking under microaerobic conditions with BBL Campy Pack Plus system (Becton Dickinson, Cockeysville, Md.). After 4 days, 8 ml of the culture was transferred to 100 ml of BBCD in a 150-ml filter unit receiver flask (Nalgene, Rochester, N.Y.) and incubated as described above. After 4 days, the bacteria were harvested by centrifugation at 10,000 × g for 10 min at 4°C. The pellet was resuspended in phosphate-buffered saline (PBS) with 5 mM MgCl2, centrifuged, and resuspended for a total of five washes. The protein concentration of the final suspension was determined with a Bradford microassay (Bio-Rad, Richmond, Calif.).
Plate culture.
The ATCC stock culture was reconstituted, transferred to the initial 25-ml broth culture, and incubated as described above. After 4 days, 20 plates of Trypticase soy agar with 5% sheep blood (Becton Dickinson) were each inoculated with 6 drops of broth culture. The plates were incubated inverted in a GasPak CO2 jar system (Becton Dickinson). After 7 days, the bacteria were harvested by scraping the colonies from the agar, suspending them in PBS with 5 mM MgCl2, washing them five times, and determining the protein concentration as described above.
Microscopy.
Wet preparations and Gram-stained (Becton Dickinson) smears were prepared for all cultures at times of transfer and harvest. These were examined with a Zeiss Axioplan photomicroscope with 63× and 100× objectives under bright-field, phase-contrast, and differential interference contrast settings.
Western blot.
Harvested bacteria from broth and plate cultures were diluted to a concentration of 4.0 mg/ml in sample buffer containing 0.1 M dithiothreitol as the reducing agent. The samples were electrophoresed on 11% polyacrylamide gels by a modification of the Laemmli method (3) with an LKB Multiphore II electrophoresis unit (Pharmacia Biotech, Piscataway, N.J.). Separated antigens were transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.) with a Nova blot semidry transfer system (Pharmacia Biotech). Membranes were blocked with 0.5% bovine serum albumin in PBS, cut into strips, dried, and stored desiccated until needed. Strips were incubated with polyclonal rabbit or human anti-H. pylori sera diluted 1:1,000 in PBS with 1% nonfat dry milk for 1 h at room temperature. Test strips were washed three times with PBS and then incubated with a 1:1,000 dilution of biotinylated goat anti-rabbit immunoglobulin G (heavy plus light chains) (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). Strips were washed as described above and then incubated with a 1:1,000 dilution of peroxidase-conjugated streptavidin for 1 h at room temperature. Strips were washed again and incubated with 4-chloro-1-naphthol substrate (7.8 mM 4-chloro-1-naphthol diluted 1:2 with 1:1,500 30% H2O2 in citrate phosphate buffer, pH 4.0). Strips were washed with distilled water, air dried, and evaluated for banding patterns.
PCR analysis.
PCR was carried out in a Perkin-Elmer 9600 thermocycler (Foster City, Calif.) with 10 ng of H. pylori genomic DNA per 100-μl reaction mixture unless otherwise noted. All oligonucleotides used were purchased from Integrated DNA Technologies (Coralville, Iowa).
Oligonucleotides HpF1 (5′ GATAAGTTGATGCTCCACTACGCTG 3′) and HpB25 (5′ CTCAATAGGGGTATGCACGGTTAC 3′) were used to amplify a 279-nucleotide (nt) product of the H. pylori urease A-encoding gene. Oligonucleotides HpF2 (5′ AAGCAGTAGCTTTGATTAGTGCCC 3′) and HpB5 (5′ CGCCATCCATCACATCATCTG 3′) are internal to HpF1 and HpB25 in the H. pylori urease A-encoding gene and were used in a secondary reaction to amplify a 120-nt product from the 279-nt primary amplification product.
A 212-nt product of the urease A-encoding gene (in a region not covered by the HpF1-HpB25 and HpF2-HpB5 primer pairs) was amplified with oligonucleotides HpF34 (5′ GTTCAAATCGGCTCACACTTCC 3′) and HpB36 (5′ TCGTTGTCTGCTTGTCTATCAACC 3′). Oligonucleotides Hpcag1 (5′ TTTCAAATACACCAACGCCTCC 3′) and HpcagB3 (5′ CCAACCAATTCTTTGTTGCTGC 3′) were used to amplify a 243-nt product of the H. pylori cytotoxin-associated gene (cagA). Following amplification, 20-μl samples of each reaction mixture were mixed with 5 μl of gel loading buffer (Sigma Biochemical, St. Louis, Mo.) and run in 3.75% Nusieve 3:1 agarose (FMC Bioproducts, Rockland, Maine) gels.
Urease gene fingerprinting.
A 2.4-kb fragment encompassing the H. pylori urease A- and urease B-encoding genes was amplified with oligonucleotides designated HpR1 (5′ AGGAGAATGAGATGA 3′) and HpR2 (5′ ACTTTATTGGCTGGT 3′). Each reaction mixture contained 20 μg of H. pylori genomic DNA, and each was run in the thermocycler as described above.
Reaction mixtures were analyzed on 0.7% SeaKem GTG agarose (FMC Bioproducts) gels to verify successful amplification of the expected 2.4-kb product. Amplified products were ethanol precipitated and resuspended in 20 μl of Tris-EDTA buffer. The DNA was digested with 10 to 20 U of restriction enzyme HaeIII (Promega, Madison, Wis.) in the appropriate buffer for 4 h at 37°C. Samples were electrophoresed on 3.75% Nusieve 3:1 agarose gels to compare restriction patterns.
Genomic DNA fingerprinting.
Three micrograms of purified genomic DNA from broth- or plate-grown bacteria was digested separately with 10 to 20 U of restriction endonucleases HaeIII, HindIII, and DraI (Promega). Reaction mixtures were incubated at 37°C for 4 h. Five microliters of gel loading buffer was added to each 20-μl reaction mixture. Samples were run on a 0.8% SeaKem GTG agarose gel in order to compare restriction patterns.
RESULTS
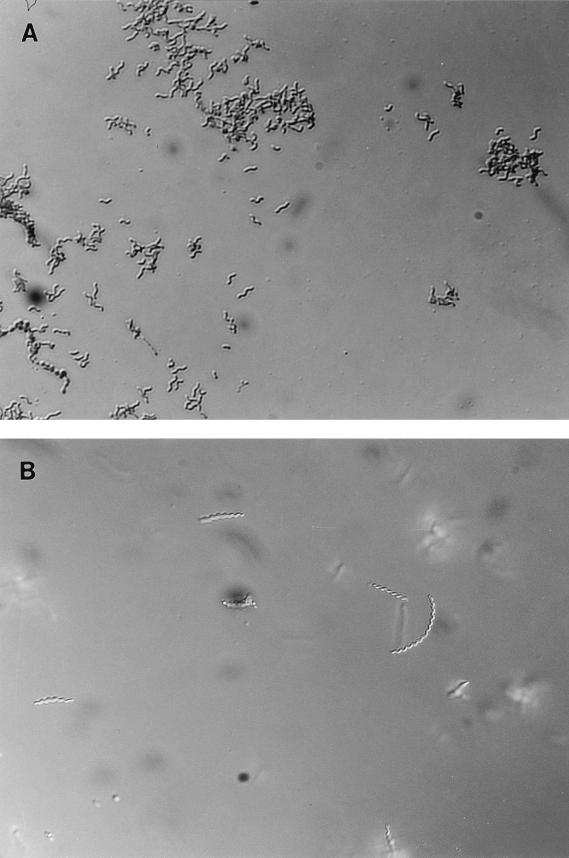
Examination of Gram-stained smears and wet preparations from blood agar- and broth-grown cultures revealed that growth on agar resulted in a bacterial morphology consistent with H. pylori (Fig. 1A) while bacteria grown in broth assumed a morphology consistent with that reported for H. heilmannii (Fig. 1B). Individual colonies from blood agar plates used to inoculate broth resulted in cultures containing long spiral forms resembling H. heilmannii. By the second or third passage in broth cultures, microscopic examination revealed only long spiral and degenerate coccoid forms. When broth cultures (third or higher passage) were used to streak blood agar plates, all resultant colonies examined showed bacteria with a morphology typical of H. pylori. The use of individual colonies from blood agar plates to start new agar plates and broth cultures was repeated four times. In each instance, resultant growth was typical of H. pylori when blood agar plate colonies were tested and typical of H. heilmannii when broth cultures were tested.
FIG. 1.
Photomicrographs show the morphologic differences which occur as a result of growing H. pylori (ATCC 43504) on blood agar plates (A) or in broth (B). Bacteria harvested from the appropriate culture source were prepared as wet preparations and photographed with a 63× objective with differential interference contrast optics.
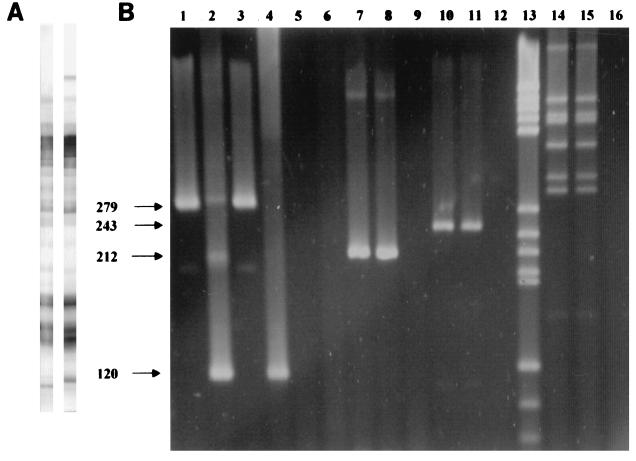
Protein profiles from polyacrylamide gel electrophoresis gels for blood agar- and broth-grown bacteria did not reveal any readily discernible differences. Western blot results (Fig. 2A) indicate that there are no or only minor differences in production or expression of immunodominant proteins associated with shifts in morphology.
FIG. 2.
(A) Western blots prepared from broth-cultured bacteria (left) and blood agar-grown bacteria (right) were reacted with polyclonal rabbit anti-H. pylori sera. No major differences in antigen profile were observed. The high-molecular-weight band present on the blot strip on the right is detected in approximately 50% of blots made from blood agar-grown bacteria. The reason for the variability in expression and/or detection is not known. (B) PCR analysis of H. pylori genomic DNA. Lanes 1 to 12 are PCRs with DNA from H. pylori grown on agar plates (lanes 1, 2, 7, and 10), DNA from H. pylori grown in broth (lanes 3, 4, 8, and 11), and water negative controls (lanes 5, 6, 9, and 12). Lane 13 shows DNA molecular mass marker set V (Boehringer Mannheim, Indianapolis, Ind.), size range, 587 to 89 bp. Lanes 14 (plate-grown H. pylori), 15 (broth-grown H. pylori), and 16 (negative control) show restriction patterns resulting from HaeIII digestion of the 2.4-kb PCR product obtained from amplification of genomic DNA with primers HpR1 and HpR2. Primer pairs used for amplification are HpF1-HpB25 (lanes 1, 3, and 5), HpF2-HpB5 (lanes 2, 4, and 6), HpF34-HpB36 (lanes 7 to 9), and HpcagF1-HpcagB3 (lanes 10 to 12).
Molecular analysis for the urease and cytotoxin-associated genes by PCR and nested PCR and genomic and urease gene fingerprinting revealed no differences between blood agar-grown and broth-grown cultures. Results of PCR and urease gene fingerprinting are shown in Fig. 2B.
DISCUSSION
Bacteria belonging to the Helicobacter genus have been detected in the gastrointestinal tracts of humans and a wide variety of other mammalian species. The presence of these bacteria in the gastric mucosa is associated with gastritis and other, more serious, gastric diseases. Morphologically, members of this genus appear quite diverse, occurring in apparently nonviable coccoid forms or as rods, curved rods, and short and long spiral forms. Many species are identified primarily by their morphology. This is particularly true for H. heilmannii, which cannot be routinely cultured. During efforts to scale up production of H. pylori antigen for use in immunoassays, we noted that culturing our ATCC-derived strain in BBCD with shaking yielded bacterial growth which resembled that reported for H. heilmannii. The present study was performed to confirm our observations that culture conditions were responsible for the change in bacterial morphology. Results obtained demonstrate that morphologic changes are associated with culture conditions and are completely reversible. We further demonstrated that no overt changes in production of immunologically relevant proteins are associated with changes in morphology, and finally, we provide molecular evidence that a single bacterial strain is present in the cultures.
The significance of our findings with relation to the genus Helicobacter and gastrointestinal disease remains to be determined. While growth of H. pylori as a long spiral bacterium may be limited to the specific growth conditions used in this study, it seems more likely that this morphology is also assumed in vivo under some as-yet-unknown conditions. If the latter is the case, some reported H. heilmannii infections may actually be due to H. pylori, since, by microscopic analysis, the two organisms appear to be indistinguishable. This could also explain some of the conflicts between reports on H. heilmannii. Perusal of the literature reveals that some investigators feel that H. heilmannii infection in humans represents zoonotic transmission from cats, dogs, or possibly swine (9, 11, 13). These conclusions are based in part on morphologic and in part on molecular analysis. There is also a considerable variation in reported levels of urease activity, immunologic cross-reactivity with H. pylori, and the localization of H. pylori versus H. heilmannii (stomach epithelial surface or lumen and gastric pits, respectively) (4, 10, 11). Based on the findings reported in this study, it seems reasonable to speculate that at least some reports of H. heilmannii infection are in fact in vivo detection of H. pylori in its long spiral form, which the organism may assume under different in vivo microenvironments.
Considerable work needs to be done before the significance of our finding can be determined. It is necessary to evaluate multiple well-defined strains of H. pylori to learn if the ability to assume a long spiral morphology is a common trait, and efforts must be made to isolate and culture long spiral bacteria from patient material to determine if these can be cultured by the broth procedure reported here and, if successful, to determine if growth on semisolid media will result in a morphology consistent with H. pylori.
The growing literature on H. heilmannii concerning infection rates, pathology, treatments, and diagnosis which is based almost exclusively on morphologic identification should be viewed with some caution based on the findings reported herein. We provide clear evidence, based on growth in vitro and molecular analysis, that H. pylori, under appropriate conditions, can be indistinguishable from H. heilmannii if morphology is used as a sole criterion. We are currently performing further studies to determine the role, if any, that long spiral forms of H. pylori play in human gastric disease.
REFERENCES
- 1.Andersen L P, Norgaard A, Holck S, Blom J, Elsborg L. Isolation of a “Helicobacter heilmanii”-like organism from the human stomach. Eur J Clin Microbiol Infect Dis. 1996;15:95–96. doi: 10.1007/BF01586196. [DOI] [PubMed] [Google Scholar]
- 2.Debongnie J C, Donnay M, Mairesse J. Gastrospirillum hominis (“Helicobacter heilmanii”): a cause of gastritis, sometimes transient, better diagnosed by touch cytology? Am J Gastroenterol. 1995;90:411–416. [PubMed] [Google Scholar]
- 3.Fawcett P T, Gibney K M, Rose C D, Dubbs S B, Doughty R A. Frequency and specificity of antibodies that crossreact with Borrelia burgdorferi antigens. J Rheumatol. 1992;19:582–587. [PubMed] [Google Scholar]
- 4.Lee A, O’Rourke J. Gastric bacteria other than Helicobacter pylori. Gastroenterol Clin N Am. 1993;22:21–42. [PubMed] [Google Scholar]
- 5.Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 6.McNulty C A M, Dent J C, Curry A, Uff J S, Ford G A, Gear M W L, Wilkinson S P. New spiral bacterium in gastric mucosa. J Clin Pathol. 1989;42:585–591. doi: 10.1136/jcp.42.6.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris A, Ali M R, Thomsen L, Hollis B. Tightly spiral shaped bacteria in the human stomach: another cause of active chronic gastritis? Gut. 1990;31:139–143. doi: 10.1136/gut.31.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivieri R, Bugnoli M, Armellini D, Bianciardi S, Rappuoli R, Bayeli P F, Abate L, Esposito E, De Gregorio L, Aziz J, Basagni C, Figura N. Growth of Helicobacter pylori in media containing cyclodextrins. J Clin Microbiol. 1993;31:160–162. doi: 10.1128/jcm.31.1.160-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Queiroz D M M, Rocha G A, Mendes E N, Moura S B, Oliveira A M R, Miranda D. Association between Helicobacter and gastric ulcer disease of the pars esophagea in swine. Gastroenterology. 1996;111:19–27. doi: 10.1053/gast.1996.v111.pm8698198. [DOI] [PubMed] [Google Scholar]
- 10.Solnick J, O’Rourke J, Lee A, Paster B, Dewhirst F, Tompkins L. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 11.Solnick J V, O’Rourke J, Lee A, Tompkins L S. Molecular analysis of urease genes from a newly identified uncultured species of Helicobacter. Infect Immun. 1994;62:1631–1638. doi: 10.1128/iai.62.5.1631-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren J R. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273. [PubMed] [Google Scholar]
- 13.Yeomans N D, Kolt S D. Helicobacter heilmannii (formerly Gastrospirillum): association with pig and human gastric pathology. Gastroenterology. 1996;111:244–247. doi: 10.1053/gast.1996.v111.agast961110244. [DOI] [PubMed] [Google Scholar]