Heparan sulfate proteoglycans carry long sugar chains that are heavily modified by different enzymes. Specific modifications may contribute to the emerging role of heparan sulfate in synapse organization. Here, Cizeron et al. show that distinct modifications are present at excitatory and inhibitory neuromuscular junctions in Caenorhabditis elegans, potentially contributing to this sugar code. Moreover, stabilization of the synaptic organizer MADD-4/Punctin requires specific modification patterns involving 3-O-sulfation, a modification of heparan sulfate chains particularly important for sugar-protein interactions.
Keywords: heparan sulfate modification enzymes, synapse, C. elegans, 3-O-sulfotransferase, MADD-4/Punctin, synaptomatrix, heparan sulfate proteoglycan, syndecan
Abstract
Heparan sulfate (HS) proteoglycans contribute to the structural organization of various neurochemical synapses. Depending on the system, their role involves either the core protein or the glycosaminoglycan chains. These linear sugar chains are extensively modified by HS modification enzymes, resulting in highly diverse molecules. Specific modifications of glycosaminoglycan chains may thus contribute to a sugar code involved in synapse specificity. Caenorhabditis elegans is particularly useful to address this question because of the low level of genomic redundancy of these enzymes, as opposed to mammals. Here, we systematically mutated the genes encoding HS modification enzymes in C. elegans and analyzed their impact on excitatory and inhibitory neuromuscular junctions (NMJs). Using single chain antibodies that recognize different HS modification patterns, we show in vivo that these two HS epitopes are carried by the SDN-1 core protein, the unique C. elegans syndecan ortholog, at NMJs. Intriguingly, these antibodies differentially bind to excitatory and inhibitory synapses, implying unique HS modification patterns at different NMJs. Moreover, while most enzymes are individually dispensable for proper organization of NMJs, we show that 3-O-sulfation of SDN-1 is required to maintain wild-type levels of the extracellular matrix protein MADD-4/Punctin, a central synaptic organizer that defines the identity of excitatory and inhibitory synaptic domains at the plasma membrane of muscle cells.
Introduction
The formation and maintenance of synapses involve a wide diversity of synaptic adhesion molecules as well as extracellular matrix (ECM) components (Yuzaki 2018). Among these diverse classes of molecules, heparan sulfate proteoglycans (HSPGs) have emerged as bona fide synapse organizers (Condomitti and de Wit 2018; Saied-Santiago and Bülow 2018). HSPGs are composed of a core protein and of one or several heparan sulfate (HS) glycan chains. Both protein and sugar moieties can interact with synaptic components, but in some cases, the glycosaminoglycan (GAG) chains were demonstrated to be required for the synaptic functions of HSPGs. For instance, in the mouse hippocampus, HS chains of the presynaptically bound HSPG glypican 4 are required to bind both the postsynaptic adhesion protein LRRTM4 (de Wit et al. 2013; Siddiqui et al. 2013) and the presynaptic co-receptor LARs (Ko et al. 2015). Thus, HS chains create a trans-synaptic bridge that is required for the synaptogenic activity resulting from this interaction. HS sugar chains are linear glycans of the repeating disaccharide hexuronic acid/N-acetyl-glucosamine, which are modified by a series of HS modification enzymes (HSMEs, Figure 1). Such modifications trigger an exquisite level of molecular diversity that contributes to the binding of specific partners. Based on the expression pattern of HSMEs, it is supposed that neurons express HS chains with different modifications, thus generating a “sugar code,” where precise HS modifications may contribute to specificity during nervous system development, including at the synapse (Holt and Dickson 2005; Poulain and Yost 2015).
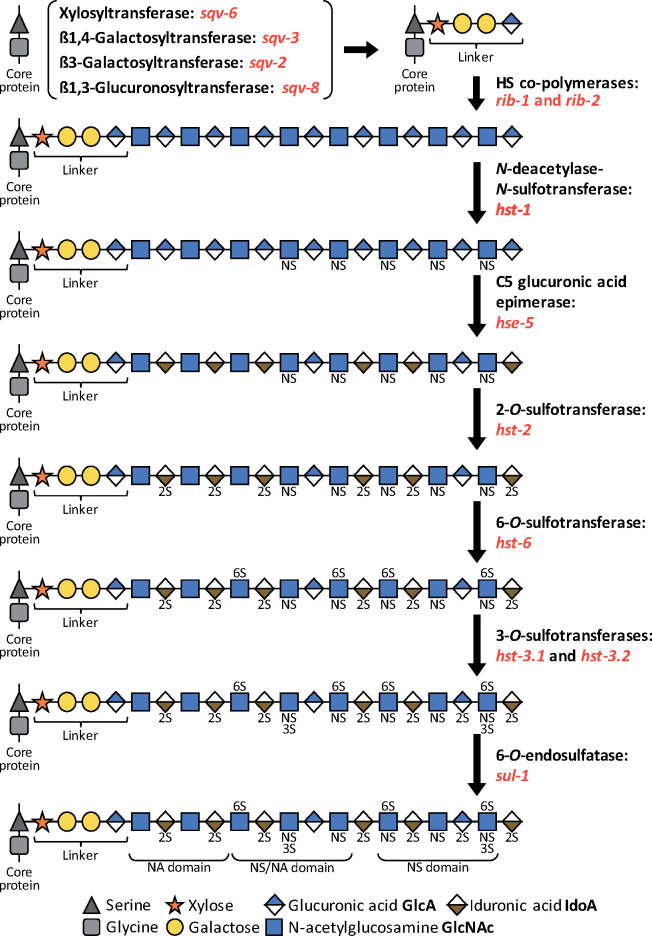
Figure 1.
Biosynthesis and modification of HS chains. HSPGs are composed of a core protein and linear GAG chain(s) attached at the level of serine residue(s). Prior to chain expansion, a tetrasaccharide linker is added on the core protein by four enzymes: a xylosyltransferase (sqv-6), a ß1,4-Galactosyltransferase (sqv-3), a ß3-Galactosyltransferase (sqv-2) and a ß1,3-Glucuronosyltransferase (sqv-8). Next, two co-polymerases (rib-1 and rib-2) synthetize the HS chain, which is composed of 50–150 repetitions of the disaccharides N-acetylglucosamine and glucuronic acid. Several modification enzymes can alter these chains. hst-1 deacetylates and adds a sulfate group to the nitrogen atom in N-acetylglucosamine. hse-5 catalyzes the C5 epimerization of the glucuronic acid into iduronic acid. hst-2 transfers a sulfate group in position 2 of the hexuronic acid. hst-6 transfers a sulfate group in position 6 of glucosamine residues. hst-3.1 and hst-3.2 transfer a sulfate group in position 3 of glucosamine residues. The 6-O-sulfatase sul-1 can then remove specific sulfate groups in position 6 of glucosamine residues. These modifications occur in a template free, yet nonrandom, manner and result in formation of different domains with specific properties and binding partners: N-acetylated (NA) domains, N-sulfated (NS) domains, and N-sulfated/N-acetylated (NA/NS) domains. This biosynthesis predominantly occurs in the Golgi apparatus, and some steps may occur simultaneously.
HS chains are enriched at synapses in both mammals and invertebrates (Ethell and Yamaguchi 1999; Ren et al. 2009; Attreed et al. 2012; Maïza et al. 2020). Enzymatic degradation of HS chains in cultured slices prevents long-term potentiation (LTP) in acute treatments (Lauri et al. 1999) and results in network impairments in chronic treatments (Korotchenko et al. 2014). Complete absence of HS chains is lethal in the mouse (Lin et al. 2000) and in C. elegans (Kitagawa et al. 2007). However, specific removal of the HS-copolymerase EXT1 in the nervous system results in abnormal brain morphogenesis (Inatani et al. 2003), while the absence of EXT1 from glutamatergic neurons in the forebrain impairs AMPA receptor-mediated transmission and induces behavioral deficits reminiscent of autism spectrum disorder (Irie et al. 2012). In humans, EXT1 mutations cause hereditary multiple exostoses (Liang et al. 2020) sometimes associated with autism (Li et al. 2002). At drosophila neuromuscular junctions (NMJ), the absence of HS chains or the complete lack of sulfation causes both morphological and physiological defects (Ren et al. 2009). In C. elegans, hypomorphic mutations of rib-1/EXT1 result in neuronal patterning defects (Blanchette et al. 2017). Moreover, removing HS chains from a single HSPG can have major consequences. For example, mutation of HS attachment sites in Neurexin compromises mouse survival and results in synaptic impairments (Zhang et al. 2018).
Specific modifications of HS chains likely contribute to their synaptic functions. For example, heparin inhibits LTP in hippocampal slices only if it contains 2-O- or 6-O-sulfated residues (Lauri et al. 1999). However, assessing the role of HS modifications at the synapse is difficult to address in mammals because many HSMEs are encoded by multiple paralogs. Lowering activities of HS 6-O-sulfotransferase (hs6st) or HS 6-O-endosulfatase (sulf1) at drosophila NMJ results in decreased or increased transmission, respectively (Dani et al. 2012). Here, we used the nematode C. elegans, which encodes single orthologs for most HSMEs (Figure 1), to systematically explore the synaptic roles of HS modifications individually and in combinations at the NMJ. This system provides a simple, genetically tractable model, where individual synapses from multiple innervations can be studied in live animals.
In C. elegans, each body-wall muscle cell is innervated by both excitatory cholinergic and inhibitory GABAergic motoneurons that form alternating en passant synapses along the ventral and dorsal cords (VNC and DNC, respectively). The synaptic organizer MADD-4/Punctin controls the cholinergic vs GABAergic identity of the postsynaptic domains (Pinan-Lucarré et al. 2014). MADD-4 is an evolutionarily conserved ECM protein that belongs to the ADAMTS-like (ADAMTSL) family and has 2 poorly characterized mammalian orthologs, ADAMTSL1/Punctin1 and ADAMTSL3/Punctin2. The madd-4 locus expresses long (MADD-4L) and short (MADD-4S) isoforms. MADD-4L is solely expressed by cholinergic neurons. Its secretion controls the synaptic localization of two types of ionotropic acetylcholine receptors (AChR), the heteromeric levamisole-sensitive L-AChRs and the homomeric nicotine-sensitive N-AChRs. MADD-4S is expressed by GABAergic and cholinergic motoneurons. It triggers the localization of type-A GABA receptors (GABARs) at GABA synapses and prevents their recruitment by MADD-4L at cholinergic synapses (Pinan-Lucarré et al. 2014; Maro et al. 2015; Tu et al. 2015).
We recently showed that MADD-4 is necessary for the localization of the HSPG syndecan SDN-1 at C. elegans NMJs (Zhou et al. 2020a). In contrast with mammals that express 4 syndecans, in C. elegans there is only one syndecan, a transmembrane core protein carrying at least 2 HS chains (Minniti et al. 2004; Rhiner et al. 2005). Our study revealed that SDN-1 is present at both cholinergic and GABAergic synapses. At cholinergic NMJs, SDN-1 triggers the formation of an intracellular scaffold that localizes N-AChRs at the synapse. SDN-1 is also required to maintain the synaptic content of L-AChRs and GABARs, likely through extracellular interactions with additional synaptic proteins. The synaptic localization of SDN-1 depends on MADD-4: in the absence of MADD-4, SDN-1 is almost completely depleted from NMJs. Reciprocally, MADD-4 levels are decreased by 30% in the absence of SDN-1. To date, SDN-1 is the only protein shown to regulate the amount of MADD-4 while not presenting gross presynaptic defects. Interestingly, point mutations of the GAG attachment sites in SDN-1 cause a 20% decrease of MADD-4 levels at the nerve cords. However, since these mutations also reduced SDN-1 levels at the synapse, it was difficult to conclude on the role of the HS chains in the NMJ organization.
Here, we investigated the synaptic functions of HS chains by systematically inactivating HS synthesis and modification enzymes and assessing their impact on MADD-4, L-AChR, and GABAR levels. We show that specific HS modifications are present at C. elegans nerve cords at both cholinergic and GABAergic synapses and are likely carried by SDN-1. Expression of antibodies recognizing specific HS chain modification patterns can increase synaptic protein levels, suggesting that these antibodies mask a negative regulation of HS chains. Moreover, epimerization, 2-O-sulfation and 6-O-sulfation modifications are individually largely dispensable for correct establishment of postsynaptic identity at C. elegans NMJ, while 3-O-sulfation is required to maintain normal levels of MADD-4 at the NMJ.
Materials and methods
Strains and genetics
All C. elegans strains were grown at 20°C on nematode growth medium (NGM) agar plates with Escherichia coli OP50 as a food source. All strains were originally derived from the wild-type Bristol N2 strain. A complete list of strains used in this study is provided in Supplementary Table S1 in Supplemental File S1.
Confocal microscopy
Animals were imaged at the young adult stage (24 hours post L4 larval stage), except for strains expressing scFv antibodies (and corresponding controls) that were acquired at the L4 stage. For in vivo imaging, live hermaphrodites were mounted on 2% agarose (w/v in water) dry pads immersed in 5% poly-lysine beads diluted in M9 buffer (3 g of KH2PO4, 6 g of Na2HPO4, 5 g of NaCl, and 0.25 g of MgSO4·7 H2O, distilled water up to 1 L). Confocal images were taken on an Andor spinning disk system (Oxford Instruments) installed on a Nikon-IX86 microscope (Olympus) equipped with a 60x/NA1.42 oil immersion objective and an Evolve EMCCD camera. For each animal, an image of the nerve cord was acquired as a stack of optical sections (0.2 μm apart). Images of the DNC and VNC were acquired around the mid-body, anterior to the vulva.
Whole fluorescence quantification
Quantification of the signal intensity at the cord was performed using a macro in the Fiji software (Schindelin et al. 2012; Rueden et al. 2017). Briefly, images of DNCs and VNCs were cropped as a rectangle containing solely the nerve cord and projected as a sum in Z direction. Then, the mean intensity was projected in x direction (orthogonal to the cord) and the intensity was measured as the area under the curve, after exclusion of the background.
Data representation and statistics
Confocal imaging of control and test groups were repeated at least 3 times for each experiment and the data were pooled together. Fluorescence intensity was normalized as a percentage of the mean value of the control group. Violin plots were generated using the Prism software (GraphPad) as follows: the median is shown by a solid dark line, quartiles are shown by two lighter lines, and individual worms are indicated by dots. N numbers are indicated above or below the genotype for each graph. Statistical testing was performed using the Prism software (GraphPad). For 2-group comparisons, the nonparametric Mann–Whitney test was applied and for >2 group comparisons, the nonparametric Kruskal Wallis test was performed with the Dunn’s multiple comparison test.
Data availability
All strains and reagents are available upon request. Supplementary Table S1 in Supplementary File S1 provides a complete list of strains used in this study. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.14533956.
Results
HS epitopes at NMJs are likely carried by SDN-1/syndecan
Chemical modifications of HS chains expand the versality of HSPG interactions. In order to visualize the pattern of specific HS modifications at C. elegans NMJs, we used two strains expressing the GFP-tagged single chain variable fragment (scFv) antibodies HS3A8 and HS4C3, which are expressed from mid-embryonic development to adulthood (Attreed et al. 2012). HS3A8 recognizes an HS epitope with 6-O-sulfated and 2-O-sulfated glucosamine, and HS4C3 binds an HS epitope with 3-O-sulfated and 6-O-sulfated glucosamine(s) (with minor contribution of 2-O-sulfated glucosamine) (Attreed et al. 2012). In these transgenic strains, scFv-GFP fusions are secreted in the pseudocoelomic cavity of the worm and bind specific HS epitopes in vivo while unbound antibodies are eliminated by scavenger cells that continuously filter the extracellular fluid. As previously reported, HS3A8 and HS4C3 strongly concentrate at the nerve cords and more specifically in synaptic regions [Figure 2, A and B and (Attreed et al. 2012)]. At high magnification, the HS3A8 signal appeared enriched at cholinergic synapses, as shown by its very strong colocalization with L-AChRs (Figure 2A). In contrast, HS4C3 was also detected in-between cholinergic synapses (Figure 2A, arrowheads), suggesting that it may be localized at both types of synapse (cholinergic and GABAergic). Indeed, HS4C3 appears colocalized with GABARs (Figure 2B). Thus, HSPGs with distinct HS modifications are present at different types of synapses.
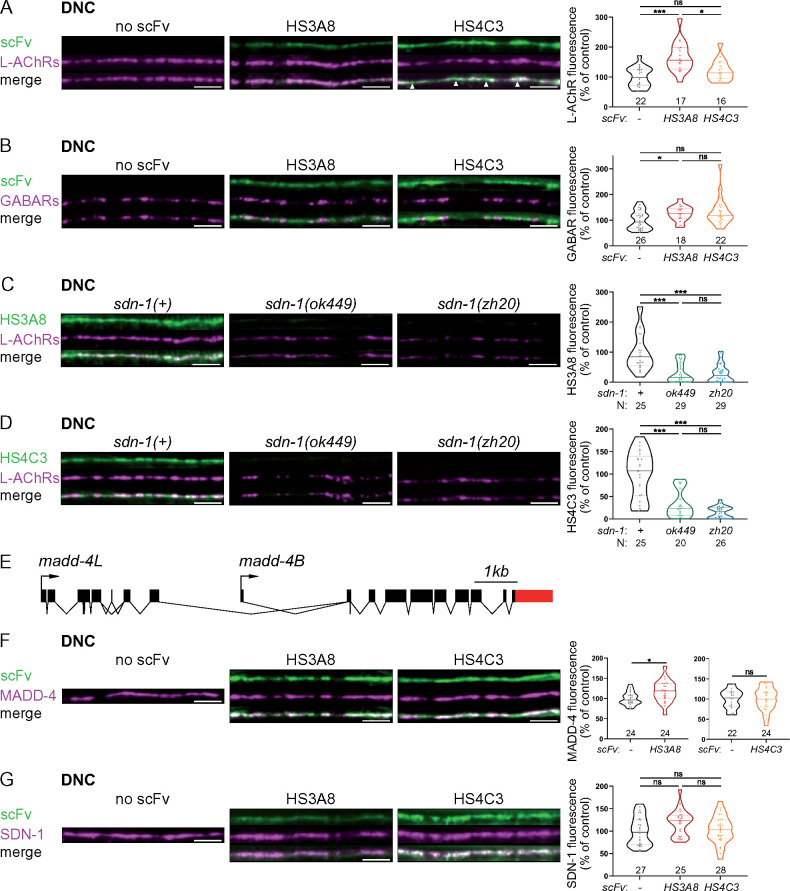
Figure 2.
HS modifications carried by SDN-1 contribute to NMJ organization. (A, B) Confocal images of scFv antibodies coupled with GFP (green), together with L-AChRs (UNC-29-tagRFPt knock-in, magenta, panel A) or GABARs (UNC-49-tagRFPt knock-in, magenta, panel B) at the DNC of worms expressing none (left panels), HS3A8-GFP (middle panels) or HS4C3-GFP (right panels). Arrowheads indicate HS4C3 signal in-between L-AChR clusters (A). Quantifications show the L-AChR (A) or GABAR (B) fluorescence intensity at DNC in each genotype and results of Dunn’s multiple comparison tests. (C, D) Confocal images of scFv antibodies coupled with GFP (green; C, HS3A8-GFP; D, HS4C3-GFP), together with L-AChRs (UNC-29-tagRFPt knock-in, magenta) at the DNC of control (left), sdn-1(ok449) (middle) and sdn-1(zh20) (right) worms. Quantifications show the scFv fluorescence intensity (C, HS3A8-GFP; D, HS4C3-GFP) at DNC in each genotype and results of Dunn’s multiple comparison test. (E) Schematics of the madd-4 locus modified in the kr373 knock-in allele, resulting in C-terminal tagging of both MADD-4S and MADD-4L isoforms with TagRFP-T. (F) Confocal images of scFv antibodies coupled with GFP (green), together with MADD-4-tagRFPt knock-in (magenta) at the DNC of worms expressing none (left panels), HS3A8-GFP (middle panels) or HS4C3-GFP (right panels). Quantifications show the MADD-4 fluorescence intensity at DNC in each genotype and results of Mann–Whitney tests. (G) Confocal images of scFv antibodies coupled with GFP (green) and SDN-1-BFP knock-in (magenta ) at the DNC of worms expressing none (left panels), HS3A8-GFP (middle panels) or HS4C3-GFP (right panels). Quantifications show the SDN-1-BFP fluorescence intensity at DNC in each genotype and results of Dunn’s multiple comparison test. In all figures, data distribution in each group is represented as violin plots where the median is shown by a solid dark line, quartiles by two lighter lines and individual worms are represented by dots. In all figures, quantifications are represented as a percentage of the control group, N numbers are indicated above or below each genotype and significance of statistical tests is indicated as follows: ns, not significant, *P<0.05, **P<0.01, ***P<0.001. In all figures, scale bars are 5 µm.
We previously showed that the HSPG syndecan SDN-1 concentrates at NMJs (Zhou et al. 2020a). To test if HS epitopes detected by the HS-specific scFv-GFP fusions are carried by SDN-1, we looked at HS3A8 and HS4C3 expression in two different sdn-1 mutant strains, containing either a null allele (zh20) (Rhiner et al. 2005) or a predicted truncated protein missing the GAG chain attachment sites (ok449) (Minniti et al. 2004). In both mutants, the fluorescence of these two scFv antibodies was abolished at the nerve cords (Figure 2, C and D). Thus, distinct HS modification patterns recognized by these two scFv antibodies are likely carried by SDN-1 at C. elegans NMJs. This may reflect a sugar code where SDN-1 carries distinct HS modifications between cholinergic and GABAergic NMJs, which may be important for synapse specificity.
HS3A8 antibodies increase L-AChR, GABAR, and MADD-4 synaptic content
Since HS3A8 and HS4C3 differentially localize at cholinergic and GABAergic NMJs, we wondered if the binding of scFv antibodies themselves might differentially impact the localization of synaptic proteins. To test this possibility, we quantified the levels of L-AChRs and GABARs, respectively, as well as the level of the synapse organizer MADD-4, using knock-in lines expressing RFP-tagged endogenous proteins. The madd-4 knock-in allele involves a C-terminal tag that labels both MADD-4S and MADD-4L isoforms (Figure 2E). Remarkably, we observed that HS3A8 induced a 70% increase of L-AChR levels at NMJs while HS4C3 had no effect (Figure 2A). HS3A8 expression also increased the levels of GABARs and MADD-4, by 30 and 20%, respectively, while HS4C3 did not impact GABAR nor MADD-4 levels (Figure 2, B and F). Two main hypotheses could explain the impact of HS3A8 on synaptic protein content. First, HS3A8 binding could neutralize negative regulatory functions provided by the GAG modification patterns that it recognizes. Alternatively, the scFv antibody could trap SDN-1 at the synapse. To distinguish between these hypotheses, we measured SDN-1 levels, using a knock-in allele with a N-terminal mNeonGreen (mNG) tag, in strains expressing HS3A8 or HS4C3. We found that SDN-1 levels were not significantly changed by the presence of HS3A8 nor HS4C3 (Figure 2G). These findings suggest that the HS3A8 antibody displays neutralizing activity, rather than artificially retaining SDN-1 at the synapse.
Synaptic proteins are decreased in sqv-6 mutants
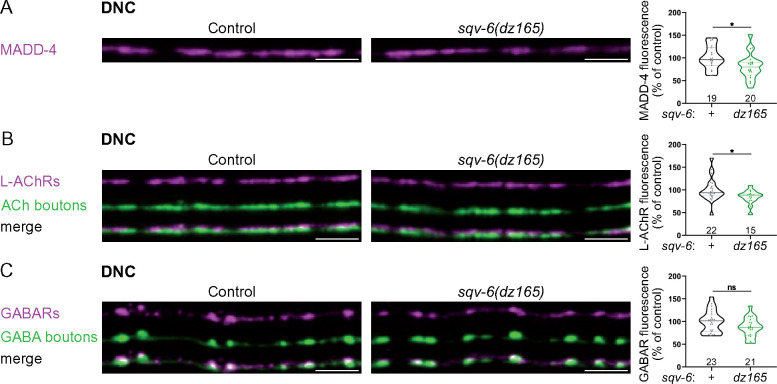
Our results indicate that HS chains participate in the organization of NMJs. To assess the synaptic impact of removal of GAG chains, we first analyzed sqv-6 mutants, in which the tetrasaccharide linker is not added to core proteins (Figure 1). As null alleles of sqv-6 are maternal lethal (Hwang et al. 2003), we used the hypomorphic allele dz165, a splice acceptor mutation in the sqv-6 gene (Díaz-Balzac et al. 2014). In these mutants, MADD-4-RFP and L-AChR-RFP levels were decreased by 19% and 16%, respectively, at the DNC (Figure 3, A and B). GABAR-RFP levels were significantly decreased by 22% at the VNC of sqv-6 mutants as compared to control animals (controls, N = 17; sqv-6, N = 24, Mann–Whitney test P = 0.01), but failed to reach statistical significance at the DNC (Mann–Whitney test P = 0.12; Figure 3C). Despite these decreases, both L-AChRs and GABARs appeared correctly localized opposite the corresponding presynaptic boutons in sqv-6 mutants (Figure 3, B and C). These results suggest that GAG chains are necessary to maintain correct levels of several synaptic proteins at the NMJ, but did not distinguish between HS and chondroitin sulfate (CS) chains, since sqv-6 is required for the synthesis of the tetrasaccharide linker (Figure 1) that is common to both HS and CS biosynthesis.
Figure 3.
sqv-6 is necessary for correct NMJ organization. (A) Confocal images of MADD-4-tagRFPt knock-in (magenta) at the DNC of control (left) and sqv-6(dz165) (right) worms. Quantifications show the MADD-4 fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (B) Confocal images of L-AChRs (UNC-29-tagRFPt knock-in, magenta) and cholinergic (ACh) boutons (SNB-1-BFP, green) at the DNC of control (left) and sqv-6(dz165) (right) worms. Quantifications show the L-AChR fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (C) Confocal images of GABARs (UNC-49-tagRFPt knock-in, magenta) and GABA boutons (SNB-1-GFP, green) at the DNC of control (left) and sqv-6(dz165) (right) worms. Quantifications show the GABAR fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. Scale bars, 5 µm.
Epimerization, 2-O-sulfation and 6-O sulfation are dispensable for NMJ organization
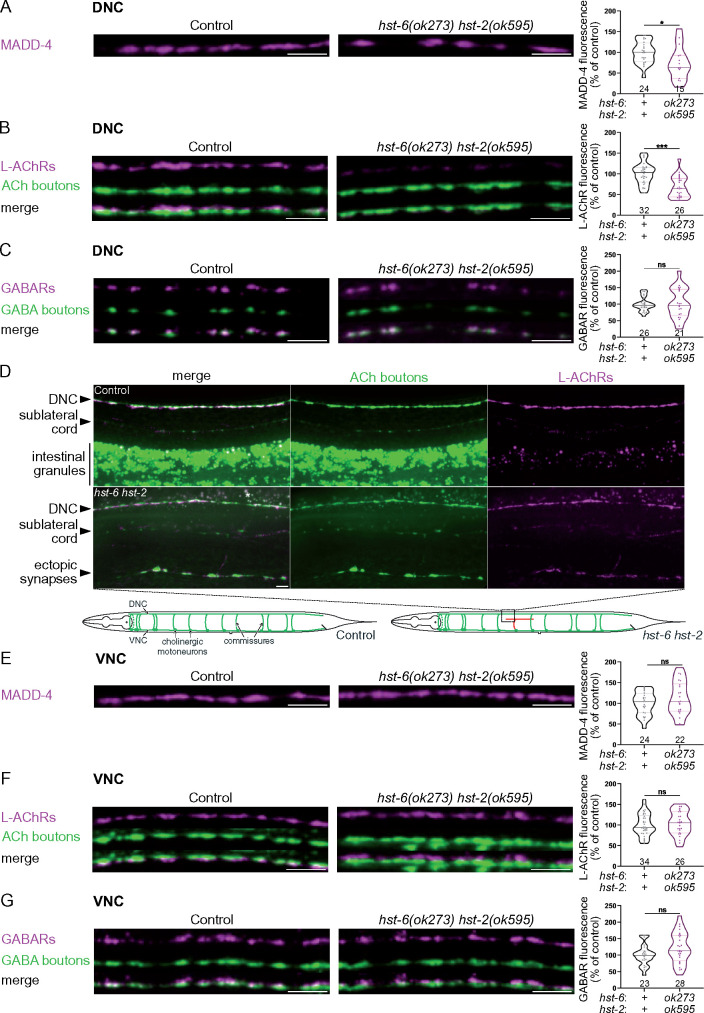
To more specifically test the function of HS at the NMJ, we analyzed MADD-4, L-AchR, and GABAR levels in mutants of HSMEs, including the hse-5 epimerase, the hst-2 2-O-sulfotransferase and the hst-6 6-O-sulfotransferase. The mutant alleles (tm472, ok595, and ok273, respectively) are large deletions (Bülow and Hobert 2004) that correspond to functionally null alleles (Townley and Bülow 2011). No single mutant had an impact on NMJ organization (Table 1), suggesting that these modifications are individually dispensable for correct expression and localization of MADD-4 and neurotransmitter receptors at NMJs. However, it has been demonstrated that impairing the activity of one HSME can increase other HSME activities (Townley and Bülow 2011). Specifically, loss of hst-2 results in increased 6-O-sulfation and, conversely, loss of hst-6 in increased 2-O-sulfation. Since HS3A8 recognizes 2-O-sulfated and 6-O-sulfated HS chains and induces a phenotype at NMJs, we hypothesized that either 6-O- or 2-O-sulfation might modulate synaptic protein content. We thus analyzed synaptic organization in hst-2 hst-6 double mutants. We observed a significant decrease of MADD-4-RFP and L-AChR-RFP, but not GABAR-RFP levels at the DNC of hst-2 hst-6 double mutants (Figure 4, A–C). However, we often observed axon guidance defects and a disorganization of cholinergic boutons in these mutants. Thus, presynaptic defects at the DNC may explain the decrease in L-AChR-RFP and MADD-4-RFP levels, as well as the reduced number of MADD-4 clusters at the DNC. Both cholinergic and GABAergic motoneurons are located in the VNC and extend commissural neurites toward the DNC (Figure 4D). In some hst-2 hst-6 worms, cholinergic commissures were not properly formed and we could sometimes observe ectopic cholinergic boutons forming outside the DNC (Figure 4D). We, therefore, quantified the levels of MADD-4-RFP, L-AChR-RFP, and GABAR-RFP at the VNC, which should be exempt from this commissural defect, and we observed no postsynaptic defects, even though we cannot exclude the presence of presynaptic defects (Figure 4, E and F). Thus, hst-2 hst-6 double mutants may have fewer or disorganized cholinergic synapses at the DNC accounting for the overall decreased receptor content. Therefore, epimerization, 2-O-sulfation and 6-O-sulfation are likely not critical for the postsynaptic organization of NMJs.
Table 1.
hse-5, hst-2, hst-6, and sul-1 are individually dispensable for NMJ organization
| MADD-4 |
L-AChRs (UNC-29) |
GABARs (UNC-49) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls |
Mutants |
P-value | Controls |
Mutants |
P-value | Controls |
Mutants |
P-value | |||||||
| Mean ± SEM (%) | N | Mean ± SEM (%) | N | Mean ± SEM (%) | N | Mean ± SEM (%) | N | Mean ± SEM (%) | N | Mean ± SEM (%) | N | ||||
| hse-5(tm472) | 100 ± 7 | 25 | 100 ± 7 | 25 | 0.99 | 100 ± 4 | 23 | 108 ± 4 | 25 | 0.08 | 100 ± 7 | 32 | 81 ± 3 | 31 | 0.06 |
| hst-2(ok595) | 100 ± 11 | 18 | 124 ± 10 | 18 | 0.08 | 100 ± 6 | 24 | 99 ± 6 | 16 | 0.94 | 100 ± 19ª | 24 | 110 ± 19ª | 26 | 0.46 |
| hst-6(ok273) | 100 ± 5 | 26 | 107 ± 4 | 36 | 0.26 | 100 ± 5 | 30 | 90 ± 6 | 28 | 0.11 | 100 ± 4 | 34 | 93 ± 4 | 29 | 0.32 |
| sul-1(gk151) | 100 ± 6 | 24 | 110 ± 5 | 21 | 0.28 | 100 ± 5 | 26 | 101 ± 4 | 21 | 0.81 | 100 ± 4 | 20 | 114 ± 5 | 22 | 0.01 |
Quantification of the fluorescence intensity of MADD-4 (MADD-4-tagRFPt knock-in), L-AChRs (UNC-29-tagRFPt knock-in), and GABARs (UNC-49-tagRFPt knock-in), at the DNC, in hse-5(tm472), hst-2(ok595), hst-6(ok273), and sul-1(gk151) mutants, except for hst-2(ok595), where GABARs were quantified in UNC-49-pHluorin knock-in (a). Quantifications are expressed as a percentage of mean control intensity ± standard error of the mean (SEM) and P-values are calculated using the Mann–Whitney test.
Figure 4.
Postsynaptic NMJ organization is unchanged in hst-2 hst-6 double mutants. (A) Confocal images of MADD-4-tagRFPt knock-in (magenta) at the DNC of control and hst-6(ok273) hst-2(ok595) worms. Quantifications show the MADD-4 fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (B) Confocal images of L-AChRs (UNC-29-tagRFPt knock-in, magenta) and cholinergic (ACh) boutons (SNB-1-BFP, green) at the DNC of control and hst-6(ok273) hst-2(ok595) worms. Quantifications show the L-AChR fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (C) Confocal images of GABARs (UNC-49-tagRFPt knock-in, magenta) and GABA boutons (SNB-1-GFP, green) at the DNC of control and hst-6(ok273) hst-2(ok595) worms. Quantifications show the GABAR fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (D) Confocal images of L-AChRs (UNC-29-tagRFPt knock-in, magenta) and cholinergic (ACh) boutons (SNB-1-BFP, green) in controls (top panels) and hst-6(ok273) hst-2(ok595) worms (bottom panels), showing an example of commissures that did not reach the DNC and formed ectopic synapses outside the main and sublateral cords. * Indicates autofluorescent signal coming from intestinal granules as the intestine is located below the DNC in this image. The schematics show an example of defect of cholinergic motoneurons projections: in the control, cholinergic motoneurons are located ventrally and project axons in the VNC, as well as commissural axons toward the DNC. In hst-6(ok273) hst-2(ok595) mutants, commissural axons sometimes fail to reach the DNC and can form ectopic synapses, as illustrated here in red. The black square indicates the approximate location of the image shown above. (E) Confocal images of MADD-4-tagRFPt knock-in (magenta) at the VNC of control and hst-6(ok273) hst-2(ok595) worms. Quantifications show the MADD-4 fluorescence intensity at VNC in each genotype and results of Mann–Whitney test. (F) Confocal images of L-AChRs (UNC-29-tagRFPt knock-in, magenta) and cholinergic (ACh) boutons (SNB-1-BFP, green) at the VNC of control and hst-6(ok273) hst-2(ok595) worms. Quantifications show the L-AChR fluorescence intensity at VNC in each genotype and results of Mann–Whitney test. (G) Confocal images of GABARs (UNC-49-tagRFPt knock-in, magenta) and GABA boutons (SNB-1-GFP, green) at the VNC of control and hst-6(ok273) hst-2(ok595) worms. Quantifications show the GABAR fluorescence intensity at VNC in each genotype and results of Mann–Whitney test. Scale bars, 5 µm.
Furthermore, we checked if the 6-O-sulfatase sul-1 could have a role at synapses. We find that sul-1 mutants have normal levels of MADD-4 and L-AChRs at the synapse (Table 1). We measured a slight increase of GABARs at the DNC (+14%, Table 1), which was not seen at the VNC (4% increase, controls N = 25, sul-1 mutants N = 24, Mann–Whitney test P = 0.98). Altogether, this suggests that selectively removing or adding 6-O-sulfated groups may not be critical for the regulation of receptor content at the NMJ.
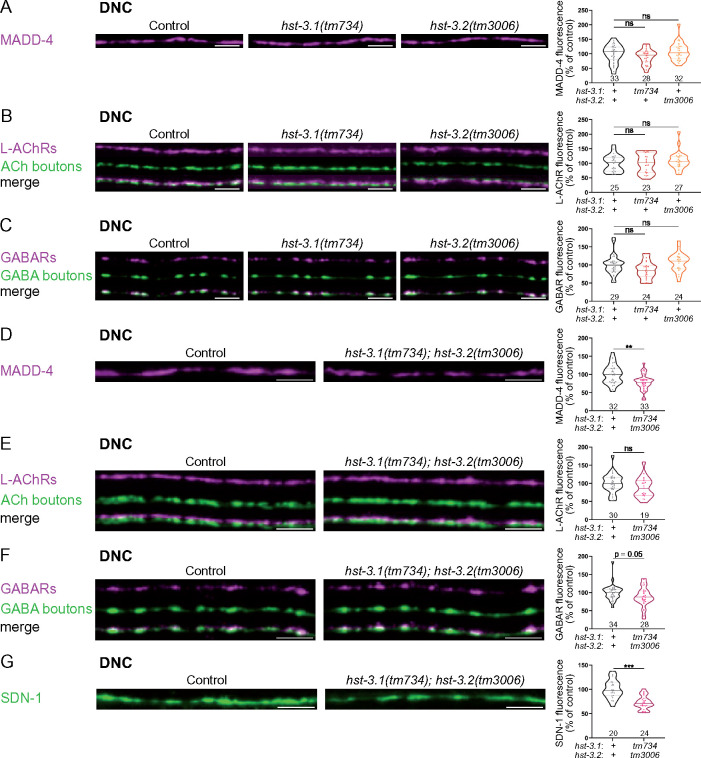
3-O-sulfation is necessary for MADD-4 stabilization at NMJs
We then tested the requirement of 3-O-sulfation for NMJ organization. Individual removal of 3-O-sulfotransferase enzymes hst-3.1 or hst-3.2 did not impact the levels of MADD-4-RFP, L-AChR-RFP or GABAR-RFP (Figure 5, A–C). As the two enzymes may serve partially redundant functions, we also tested the hst-3.1; hst-3.2 double mutant. These mutants showed a 22% decrease in MADD-4-RFP levels at the DNC (Figure 5D) while there was no difference for L-AChR-RFP (Figure 5E) and a 12% decrease of GABAR-RFP (Figure 5F). We also quantified the levels of synaptic proteins at the VNC. MADD-4-RFP levels were decreased by 15% (controls, N = 39; hst-3.1; hst-3.2, N = 29; Mann–Whitney test P = 0.04), confirming the effect seen at the DNC, whereas L-AChR-RFP and GABAR-RFP levels were not significantly changed at the VNC (UNC-29: 9% increase, controls N = 27 and hst-3.1; hst-3.2 mutants N = 18, Mann–Whitney test P = 0.38; UNC-49: 11% decrease, controls N = 37 and hst-3.1; hst-3.2 mutants N = 27, Mann–Whitney test P = 0.19). So far, syndecan SDN-1 is the only protein known to regulate MADD-4 synaptic content without having major presynaptic defects (Zhou et al. 2020a). We, therefore, wondered if the absence of 3-O-sulfation also impacted the levels of SDN-1 at NMJs. Indeed, the level of mNG-SDN-1 was decreased by 27% at the DNC in hst-3.1; hst-3.2 double mutants (Figure 5G).
Figure 5.
3-O-sulfation is required for the stabilization of MADD-4 and SDN-1 at the NMJs. (A) Confocal images of MADD-4-tagRFPt knock-in (magenta) at the DNC of control (left), hst-3.1(tm734) (middle) and hst-3.2(tm3006) (right) worms. Quantifications show the MADD-4 fluorescence intensity at DNC in each genotype and results of Dunn’s multiple comparison test. (B) Confocal images of L-AChRs (UNC-29-tagRFPt knock-in, magenta) and cholinergic (ACh) boutons (SNB-1-BFP, green) at the DNC of control (left), hst-3.1(tm734) (middle) and hst-3.2(tm3006) (right) worms. Quantifications show the L-AChR fluorescence intensity at DNC in each genotype and results of Dunn’s multiple comparison test. (C) Confocal images of GABARs (UNC-49-tagRFPt knock-in, magenta) and GABA boutons (SNB-1-GFP, green) at the DNC of control (left), hst-3.1(tm734) (middle) and hst-3.2(tm3006) (right) worms. Quantifications show the GABAR fluorescence intensity at DNC in each genotype and results of Dunn’s multiple comparison test. (D) Confocal images of MADD-4-tagRFPt knock-in (magenta) at the DNC of control and hst-3.1(tm734); hst-3.2(tm3006) worms. Quantifications show the MADD-4 fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (E) Confocal images of L-AChRs (UNC-29-tagRFPt knock-in, magenta) and cholinergic (ACh) boutons (SNB-1-BFP, green) at the DNC of control and hst-3.1(tm734); hst-3.2(tm3006) worms. Quantifications show the L-AChR fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (F) Confocal images of GABARs (UNC-49-tagRFPt knock-in, magenta) and GABA boutons (SNB-1-GFP, green) at the DNC of control and hst-3.1(tm734); hst-3.2(tm3006) worms. Quantifications show the GABAR fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. (G) Confocal images of mNG-SDN-1 knock-in (green) at the DNC of control and hst-3.1(tm734); hst-3.2(tm3006) worms. Quantifications show the SDN-1 fluorescence intensity at DNC in each genotype and results of Mann–Whitney test. Scale bar, 5 µm.
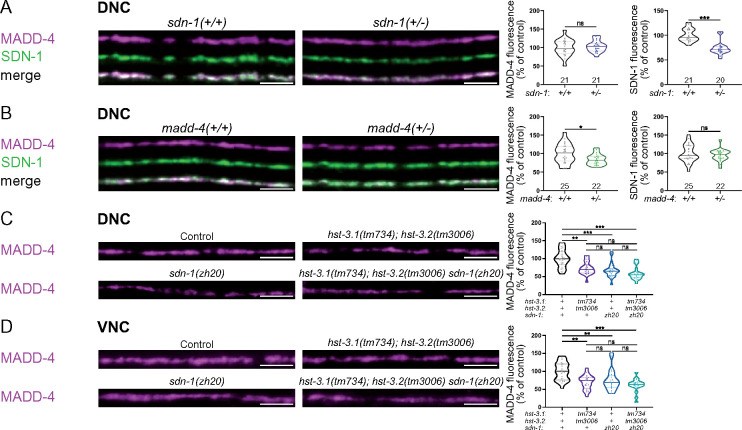
Since MADD-4 levels are decreased by about 30% in sdn-1(0) (Zhou et al. 2020a), we wondered if the small decrease of SDN-1 in hst-3.1; hst-3.2 double mutants could explain the MADD-4 phenotype in these mutants. To test this hypothesis, we generated heterozygous sdn-1(+/-) animals, expressing only one copy of mNG-SDN-1, and checked whether these animals presented a change in MADD-4-RFP levels. In these animals, mNG-SDN-1 levels are decreased by 27% at the DNC, while MADD-4-RFP levels remain unchanged (Figure 6A). Thus, the comparable reduction of SDN-1 levels in hst-3.1; hst-3.2 double mutants does not explain the decrease of MADD-4. Since MADD-4 is absolutely required for SDN-1 localization at C. elegans NMJs (Zhou et al. 2020a), we tested the opposite hypothesis, i.e., whether the MADD-4 decrease is responsible for the drop in SDN-1 levels in hst-3.1; hst-3.2 double mutants. We followed the same strategy and observed a 16% decrease of MADD-4-RFP levels in madd-4(+/-) heterozygotes while SDN-1 levels remain unchanged (Figure 6B). These results suggest that the phenotypes observed for SDN-1 and MADD-4 reporters do not result from their mutual changes. Thus, 3-O-sulfation is required to stabilize MADD-4 at the synapse. In addition, 3-O-sulfation is required for SDN-1 stability or trafficking to the synapse. Consistent with this hypothesis, sdn-1(ΔGAG) worms, in which SDN-1 does not carry HS chains, also have decreased levels of SDN-1 at the NMJ (Zhou et al. 2020a). To test the genetic interaction between the 3-O-sulfotransferases and sdn-1, we generated hst-3.1; hst-3.2 sdn-1 triple mutants. At both DNC and VNC, we observed around 40% decrease in MADD-4 levels in triple mutants, which was not significantly different from the 30% decrease seen in hst-3.1; hst-3.2 and sdn-1 mutants (Figure 6, C and D). This suggests that the 3-O-sulfation modifications required to stabilize MADD-4 are specifically located on SDN-1 chains. Indeed, if modification of other HSPGs were involved, we would expect a substantially larger reduction of MADD-4 levels in triple mutants compared to sdn-1(0) animals.
Figure 6.
3-O-sulfation of SDN-1 is required for stabilization of MADD-4 at NMJs. (A) Confocal images of MADD-4-tagRFPt knock-in (magenta) and mNG-SDN-1 knock-in (green) at the DNC of sdn-1(+/-) heterozygous worms. Quantifications show the MADD-4 (left) and SDN-1 (right) fluorescence intensity at DNC in each genotype and corresponding results of Mann–Whitney test. (B) Confocal images of MADD-4-tagRFPt knock-in (magenta) and mNG-SDN-1 knock-in (green) at the DNC of madd-4(+/-) heterozygous worms. Quantifications show the MADD-4 (left) and SDN-1 (right) fluorescence intensity at DNC in each genotype and corresponding results of Mann–Whitney test. (C, D) Confocal images of MADD-4-tagRFPt knock-in at the DNC (C) and VNC (D) of control, hst-3.1(tm734); hst-3.2(tm3006), sdn-1(zh20), and hst-3.1(tm734); hst-3.2(tm3006) sdn-1(zh20) worms. Quantifications show the MADD-4 fluorescence intensity at DNC (C) and VNC (D) in each genotype and corresponding results of Dunn’s multiple comparison test. Scale bar, 5 µm.
Removing 6-O-sulfation can compensate for the absence of 3-O-sulfation
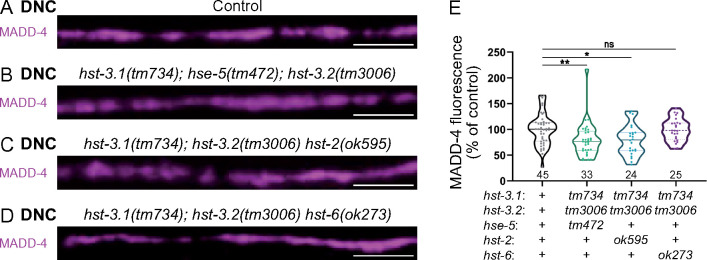
In the absence of a given HSME, other types of modifications can be changed and potentially induce compensatory effects. The use of C. elegans provides a means to test if inactivating other HSME enzymes could potentiate the effect of the absence of 3-O-sulfation on MADD-4 levels. We, therefore, measured MADD-4-RFP levels in triple mutants. We find that in hst-3.1; hse-5; hst-3.2 and in hst-3.1; hst-3.2 hst-2 triple mutants, MADD-4RFP levels are decreased by about 20% (Figure 7, A–C and E), which is comparable to the hst-3.1; hst-3.2 double mutant alone (Figure 5D). However, a MADD-4 decrease is no longer observed in hst-3.1; hst-3.2 hst-6 triple mutants (Figure 7, D and E), indicating that removing 6-O-sulfation could compensate the absence of 3-O-sulfation. One potential explanation for this effect is that the lack of 3-O-sulfation in hst-3.1; hst-3.2 double mutants may result in a compensatory increase in 6-O-sulfation. Alternatively, the increase in N-sulfation or in 2-O-sulfation observed in the absence of hst-6 (Townley and Bülow 2011) may compensate for the decrease in 3-O-sulfation.
Figure 7.
Removing 6-O-sulfation rescues MADD-4 levels in the absence of 3-O-sulfation. (A–D) Confocal images of MADD-4-tagRFPt knock-in (magenta) at the DNC of control (A) and triple mutants: hst-3.1(tm734); hse-5(tm472); hst-3.2(tm3006) (B), hst-3.1(tm734); hst-3.2(tm3006) hst-2 (ok595) (C), hst-3.1(tm734); hst-3.2(tm3006) hst-6(ok273) (D). (E) Quantifications show the MADD-4 fluorescence intensity at DNC in each genotype and results of Dunn’s multiple comparison test. Scale bar, 5 µm.
Discussion
Here, we have systematically assessed a potential role for HSMEs in the synaptic organization of C. elegans NMJs. We show that HS chains recognized by HS3A8 and HS4C3 antibodies are enriched at the synapse and most likely carried by SDN-1/syndecan. HS3A8 binding causes a strong increase of L-AChR synaptic content, as opposed to HS4C3, suggesting that specific HS modifications might negatively regulate L-AChR clustering at synapses. Partial removal of HS and CS chains in sqv-6 hypomorphic mutants results in decreased levels of MADD-4, L-AchRs, and GABARs. Epimerization, 2-O-sulfation and 6-O-sulfation of HS seem individually dispensable for proper recruitment of these synaptic proteins. However, disruption of 3-O-sulfation in hst-3.1; hst-3.2 double mutants resulted in decreased levels of the synaptic organizer MADD-4, which can be compensated by further removal of the 6-O-sulfotransferase hst-6. Additional removal of sdn-1 in hst-3.1; hst-3.2 double mutants did not further reduce the levels of MADD-4 at NMJs. Altogether, our results point to a specific role of 3-O-sulfotransferases in synapse organization and support a role for the sugar code in synapse specificity.
Regulation of MADD-4 levels by HS chains
Here, we identify a role for HS chains and, more specifically, for 3-O-sulfation in the maintenance of synaptic levels of the synapse organizer MADD-4/Punctin. MADD-4 is secreted by motoneurons and concentrates in the synaptomatrix of C. elegans NMJ. Interestingly, both MADD-4 isoforms contain the same Ig-like domain whose structural model shows a highly electropositive surface pocket of a suitable size for lodging heparin with high affinity. Experimentally, a truncated version of MADD-4, retaining only 4 thrombospondin repeats (TSP-1) and the Ig-like domain, binds heparin with an estimated Kd value of 10 nM (Platsaki et al. 2020), showing that MADD-4 is indeed able to bind HS chains in vitro. Together with the co-localization of MADD-4 with HS chains at cholinergic and GABAergic NMJ, MADD-4 fulfills the criteria of a bona fide HS binding protein (Xu and Esko 2014).
MADD-4 was shown to position several components at the post-synapse based on demonstrated physical interactions (UNC-40/DCC, NLG-1/neuroligin) (Maro et al. 2015; Tu et al. 2015; Zhou et al. 2020b) or by phenotypic analysis of madd-4 mutants (LEV-9, LEV-10, OIG-4, AchRs, and GABARs) (Gally et al. 2004; Gendrel et al. 2009; Rapti et al. 2011; Pinan-Lucarré et al. 2014). Yet, how MADD-4 itself localizes at cholinergic and GABAergic synapses is not known. MADD-4 might interact with presynaptic partner(s) that remain to be identified. Alternatively, interactions with ECM components might limit the diffusion of MADD-4 after secretion by presynaptic boutons and subsequently organize the post-synaptic membrane compartment in register with neurotransmitter release sites. 3-O-sulfated HS chains of syndecan seem to provide such binding sites to stabilize MADD-4 in the synaptic cleft. First, MADD-4 synaptic levels are reduced by 30% in sdn-1(zh20) mutants (Zhou et al. 2020a), which is comparable to the phenotype observed in hst-3.1; hst-3.2 double mutants. Second, MADD-4 is also decreased in sdn-1(ΔGAG) mutants, in which HS chain attachment sites were mutated (Zhou et al. 2020a). The interpretation of these data was initially complicated by the decrease of SDN-1 itself in these mutant contexts. However, sdn-1(+/-) worms, which express only one copy of sdn-1, have a similar decrease of SDN-1 as in hst-3.1; hst-3.2 double mutants but retain wild-type amounts of MADD-4 at the synapse. Moreover, the hst-3.1; hst-3.2 sdn-1 triple mutants do not show a further reduction of MADD-4 levels compared to hst-3.1; hst-3.2 double mutants and sdn-1 mutants. This suggests that MADD-4 interacts with syndecan 3-O-sulfated HS chains rather than with the core protein. The requirement of syndecan HS chains for MADD-4 stabilization is reminiscent of the requirement of HS chains on glypican 4 for the binding of the synaptic organizers LRRTM4 and GPR158 in vitro (de Wit et al. 2013; Condomitti et al. 2018).
Interestingly, in the absence of HS chains, SDN-1 still localizes at NMJs (Zhou et al. 2020a). Since MADD-4 is required for SDN-1 localization at NMJs (Zhou et al. 2020a), this implies that MADD-4 can still localize syndecan at NMJs in the absence of HS chains. This probably reflects the dual function of MADD-4 that binds, localizes, and activates the netrin receptor UNC-40/DCC, which in turn is able to nucleate the formation of an intracellular scaffold that recruits syndecan at synaptic sites (Zhou et al. 2020a, b). These intracellular interactions were demonstrated to be critical for the synaptic localization of N-AChRs (Zhou et al. 2020a), which unlikely depends on extracellular modifications of the SDN-1 HS chains, as opposed to L-AChRs, which clustering depends solely on an extracellular scaffold (Gally et al. 2004; Gendrel et al. 2009; Rapti et al. 2011). The crosstalk between extra- and intracellular scaffolds adds degrees of freedom to increase the functional diversity of synapses in the brain using a limited number of structural components. It will be of major interest in the future to test a potentially conserved interaction between the MADD-4 ortholog ADAMTSL3/Punctin2 and GAG chains in vertebrates, either at synapses or in perineuronal nets, which control major features of network plasticity during development.
A conserved role for 3-O-sulfation at synapses?
We performed here a comprehensive analysis of HSMEs and it is interesting to see that 3-O-sulfation is the only HS modification that is individually required in our system, as it represents a very rare modification (Thacker et al. 2014). Despite this rarity, mammals express 7 different 3-O-sulfotransferases, with distinct properties and spatiotemporal regulations (Yabe et al. 2005; Thacker et al. 2014). During evolution, the diversification of 3-O-sulfotransferases correlates with the development of a centralized nervous system (Tecle et al. 2013). Moreover, this modification is the last one to be added in the HS biosynthesis pathway, suggesting that it constitutes a final refinement of the HS code (Tecle et al. 2013). 3-O-sulfation is linked to high-affinity binding of proteins (Lindahl and Li 2009) and can organize a network of ionic and nonionic interactions as documented for the binding of heparin to antithrombin (Xu and Esko 2014). In drosophila, knock-down of Hs3st-B results in neurogenic phenotypes linked to aberrant Notch signaling (Kamimura et al. 2004). Moreover, hst-3.1 was also shown to be required for correct establishment of synapses between B-type ray neurons and EFs in C. elegans males (Lázaro-Peña et al. 2018). In addition, in a recent publication, expression of 2 different 3-O-sulfotransferases, Hs3st4 and Hs3st2, was detected at pre- and postsynaptic sites, respectively, in neuronal cell cultures (Maïza et al. 2020). This study also showed that a peptide, which is able to bind HS chains containing 3-O-sulfated residues, induces a decrease in glutamatergic transmission in acute treatments and has an impact on synapse assembly when used chronically to treat hippocampal neuron cultures. Altogether, these results highlight a prominent and conserved function of 3-O-sulfation at synapses.
Surprisingly, removing 6-O-sulfation by itself has no impact on MADD-4 levels, while it suppresses MADD-4 decrease in the hst-3.1; hst-3.2 double mutant. We speculate that the increase in N-sulfation or in 2-O-sulfation observed in the absence of hst-6 (Townley and Bülow 2011) may compensate for the decrease in 3-O-sulfation. Similar rescue has been observed previously between HSMEs regarding the migration of hermaphrodite specific neurons (HSN) defects in hst-2 mutants. While additional removal of hst-6 worsens the phenotype, further removal of hst-3.1 partially rescues the phenotypes in hst-3.1; hst-6 hst-2 compared to hst-6 hst-2 double mutants (Kinnunen 2014).
Is there a sugar code at C. elegans NMJs?
We have observed a differential patterning of two scFv antibodies (HS3A8 and HS4C3) at the NMJ. Indeed, HS3A8 appeared preferentially enriched at cholinergic synapses, while HS4C3 is also present at GABAergic synapses. The functional impact of the two scFvs was unambiguous, with a strong increase of L-AChR content in animals expressing HS3A8, and no impact of HS4C3. These two antibodies were characterized in vitro and HS3A8 seems to recognize 2-O- and 6-O-sulfation containing HS modification patterns while HS4C3 mostly binds a 3-O- and 6-O-sulfated glucosamine-containing epitope (Dennissen et al. 2002). We cannot exclude that the difference of effect may be due to distinct affinities of HS3A8 and HS4C3 for HS chains in vivo, but it is reasonable to propose that scFvs selectively disrupt relevant HS interactions and differentially impact the content of synaptic proteins.
Both scFv antibodies recognize epitopes carried by the core protein syndecan, which implies that syndecan HS chains may be differentially modified between excitatory and inhibitory synapses. What could lead to the differential segregation of distinct HS modification patterns between cholinergic and GABAergic synapses in muscle? One possibility is that additional modifications of the HS chains occur in the extracellular space. For example, the 6-O-endosulfatase QSulf1 has been shown to function extracellularly (Ai et al. 2006). Similarly, both 6-O-and 3-O-sulfotransferases are secreted in cell culture (Habuchi et al. 1998; Nagai et al. 2007), raising the possibility that these enzymes could also act extracellularly at specific synapses, thereby creating SDN-1 with different HS modification patterns in spatially distinct domains. Alternatively, differentially modified SDN-1 in muscle could be segregated by specific extracellular interactions into different domains, i.e., synapses. Using tissue-specific degradation of syndecan, we previously demonstrated that about 70% of the syndecan present at NMJs is synthesized in muscle cells (Zhou et al. 2020a). Therefore, distinct HS modifications could also be present on neuronal syndecan, which represents about 20% of its synaptic content (Zhou et al. 2020a). Indeed, it remains unknown whether HSMEs are expressed differentially in cholinergic versus GABAergic motoneurons in C. elegans. Whether HS chain modifications are segregated among synapses in the mammalian brain and which mechanisms might underlie these processes represent very challenging questions to address in vivo. The simplicity of the C. elegans NMJ might provide a valuable system to meet this challenge.
Acknowledgments
The authors thank Camille Vachon and Camilla Luccardini for performing exploratory experiments, the CIQLE Imaging facility for support and access to equipment.
Conceptualization: J.L.B., M.C., and H.E.B.; Methodology, M.C., H.E.B, and J.L.B.; Investigation, M.C. and L.G.; Writing, M.C., J.L.B., and H.E.B.; Funding Acquisition, J.L.B. and H.E.B.
Funding
This work was supported by the European Research Council (ERC_Adg C.NAPSE #695295), within the framework of the LABEX CORTEX (ANR-11-LABX-0042) of Université de Lyon operated by the French National Research Agency (ANR). Work in the Bülow lab is supported by grants from the National Institute of Health (NIH) (RC1 GM-090825 and R01 GM-101313 to H.E.B.).
Conflicts of interest
None declared.
Literature cited
- Ai X, Do A-T, Kusche-Gullberg M, Lindahl U, Lu K, et al. 2006. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 281:4969–4976. [DOI] [PubMed] [Google Scholar]
- Attreed M, Desbois M, van Kuppevelt TH, Bülow HE.. 2012. Direct visualization of specifically modified extracellular glycans in living animals. Nat Methods. 9:477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette CR, Thackeray A, Perrat PN, Hekimi S, Bénard CY.. 2017. Functional requirements for heparan sulfate biosynthesis in morphogenesis and nervous system development in C. elegans, (A. D. Chisholm, Ed.). PLoS Genet. 13:e1006525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow HE, Hobert O.. 2004. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 41:723–736. [DOI] [PubMed] [Google Scholar]
- Condomitti G, de Wit J.. 2018. Heparan sulfate proteoglycans as emerging players in synaptic specificity. Front Mol Neurosci. 11:14. 10.3389/fnmol.2018.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condomitti G, Wierda KD, Schroeder A, Rubio SE, Vennekens KM, et al. 2018. An input-specific orphan receptor GPR158-HSPG interaction organizes hippocampal mossy fiber-CA3 synapses. Neuron 100:201–215.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N, Nahm M, Lee S, Broadie K.. 2012. A targeted glycan-related gene screen reveals heparan sulfate proteoglycan sulfation regulates WNT and BMP trans-synaptic signaling. PLoS Genet. 8:e1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennissen MABA, Jenniskens GJ, Pieffers M, Versteeg EMM, Petitou M, et al. 2002. Large, Tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J Biol Chem. 277:10982–10986. [DOI] [PubMed] [Google Scholar]
- de Wit J, O’Sullivan ML, Savas JN, Condomitti G, Caccese MC, et al. 2013. Unbiased discovery of glypican as a receptor for LRRTM4 in regulating excitatory synapse development. Neuron. 79:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Balzac CA, Lázaro-Peña MI, Tecle E, Gomez N, Bülow HE.. 2014. Complex cooperative functions of heparan sulfate proteoglycans shape nervous system development in Caenorhabditis elegans. G3 (Bethesda). 4:1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell IM, Yamaguchi Y.. 1999. Cell surface heparan sulfate proteoglycan syndecan-2 induces the maturation of dendritic spines in rat hippocampal neurons. J Cell Biol. 144:575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally C, Eimer S, Richmond JE, Bessereau J-L.. 2004. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature. 431:578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Rapti G, Richmond JE, Bessereau J-L.. 2009. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 461:992–996. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Kobayashi M, Kimata K.. 1998. Molecular characterization and expression of Heparan-sulfate 6-Sulfotransferase. J Biol Chem. 273:9208–9213. [DOI] [PubMed] [Google Scholar]
- Holt CE, Dickson BJ.. 2005. Sugar codes for Axons? Neuron. 46:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H-Y, Olson SK, Brown JR, Esko JD, Horvitz HR.. 2003. The Caenorhabditis elegans genes sqv-2 and sqv-6, which are required for vulval morphogenesis, encode glycosaminoglycan galactosyltransferase II and xylosyltransferase. J Biol Chem. 278:11735–11738. [DOI] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump A, Tessier-Lavigne M, Yamaguchi Y.. 2003. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 302:1044–1046. [DOI] [PubMed] [Google Scholar]
- Irie F, Badie-Mahdavi H, Yamaguchi Y.. 2012. Autism-like socio-communicative deficits and stereotypies in mice lacking heparan sulfate. Proc Natl Acad Sci USA. 109:5052–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K, Rhodes JM, Ueda R, McNeely M, Shukla D, et al. 2004. Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J Cell Biol. 166:1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen TK. 2014. Combinatorial roles of heparan sulfate proteoglycans and heparan sulfates in Caenorhabditis elegans neural development. PLoS One. 9:e102919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Izumikawa T, Mizuguchi S, Dejima K, Nomura KH, et al. 2007. Expression of rib-1, a Caenorhabditis elegans homolog of the human tumor suppressor EXT Genes, is indispensable for heparan sulfate synthesis and embryonic morphogenesis. J Biol Chem. 282:8533–8544. [DOI] [PubMed] [Google Scholar]
- Ko JS, Pramanik G, Um JW, Shim JS, Lee D, et al. 2015. PTPσ functions as a presynaptic receptor for the glypican-4/LRRTM4 complex and is essential for excitatory synaptic transmission. Proc Natl Acad Sci USA. 112:1874–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotchenko S, Cingolani LA, Kuznetsova T, Bologna LL, Chiappalone M, et al. 2014. Modulation of network activity and induction of homeostatic synaptic plasticity by enzymatic removal of heparan sulfates. Philos Trans R Soc Lond B Biol Sci. 369:20140134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauri SE, Kaukinen S, Kinnunen T, Ylinen A, Imai S, et al. 1999. Regulatory role and molecular interactions of a cell-surface heparan sulfate proteoglycan (N-syndecan) in hippocampal long-term potentiation. J Neurosci. 19:1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Peña MI, Díaz-Balzac CA, Bülow HE, Emmons SW.. 2018. Synaptogenesis is modulated by heparan sulfate in Caenorhabditis elegans. Genetics. 209:195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yamagata T, Mori M, Momoi MY.. 2002. Association of autism in two patients with hereditary multiple exostoses caused by novel deletion mutations of EXT1. J Hum Genet. 47:262–265. [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Wei Y, Dong Y, Zhang Z.. 2020. Identification of novel EXT mutations in patients with hereditary multiple exostoses using whole‐exome sequencing. Orthop Surg. 12:990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Wei G, Shi Z, Dryer L, Esko JD, et al. 2000. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 224:299–311. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Li J.. 2009. Chapter 3 Interactions between heparan sulfate and proteins—design and functional implications. In: International Review of Cell and Molecular Biology, Vol. 276. Elsevier Inc, p. 105–159. [DOI] [PubMed] [Google Scholar]
- Maïza A, Sidahmed-Adrar N, Michel PP, Carpentier G, Habert D, et al. 2020. 3-O-sulfated heparan sulfate interactors target synaptic adhesion molecules from neonatal mouse brain and inhibit neural activity and synaptogenesis in vitro. Sci Rep. 10:19114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro GS, Gao S, Olechwier AM, Hung WL, Liu M, et al. 2015. MADD-4/Punctin and Neurexin organize C. elegans GABAergic postsynapses through Neuroligin. Neuron. 86:1420–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minniti AN, Labarca M, Hurtado C, Brandan E.. 2004. Caenorhabditis elegans syndecan (SDN-1) is required for normal egg laying and associates with the nervous system and the vulva. J Cell Sci. 117:5179–5190. [DOI] [PubMed] [Google Scholar]
- Nagai N, Habuchi H, Kitazume S, Toyoda H, Hashimoto Y, et al. 2007. Regulation of heparan sulfate 6-O-Sulfation by β-secretase activity. J Biol Chem. 282:14942–14951. [DOI] [PubMed] [Google Scholar]
- Pinan-Lucarré B, Tu H, Pierron M, Cruceyra PI, Zhan H, et al. 2014. C. elegans Punctin specifies cholinergic versus GABAergic identity of postsynaptic domains. Nature. 511:466–470. [DOI] [PubMed] [Google Scholar]
- Platsaki S, Zhou X, Pinan-Lucarré B, Delauzun V, Tu H, et al. 2020. The Ig-like domain of Punctin/MADD-4 is the primary determinant for interaction with the ectodomain of neuroligin NLG-1. J Biol Chem. 295:16267–16279. 10.1074/jbc.RA120.014591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain FE, Yost HJ.. 2015. Heparan sulfate proteoglycans: a sugar code for vertebrate development? Development. 142:3456–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti G, Richmond J, Bessereau J-L.. 2011. A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans: role of C. elegans OIG-4 in AChR clustering. EMBO J. 30:706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Kirkpatrick CA, Rawson JM, Sun M, Selleck SB.. 2009. Cell type-specific requirements for heparan sulfate biosynthesis at the Drosophila neuromuscular junction: effects on synapse function, membrane trafficking, and mitochondrial localization. J Neurosci. 29:8539–8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhiner C, Gysi S, Fröhli E, Hengartner MO, Hajnal A.. 2005. Syndecan regulates cell migration and axon guidance in C. elegans. Development. 132:4621–4633. [DOI] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE.. 2017. ImageJ2: imageJ for the next generation of scientific image data. BMC Bioinformatics. 18:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saied-Santiago K, Bülow HE.. 2018. Diverse roles for glycosaminoglycans in neural patterning: Glycosaminoglycans in Neural development. Dev Dyn. 247:54–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Tari PK, Connor SA, Zhang P, Dobie FA, et al. 2013. An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron. 79:680–695. [DOI] [PubMed] [Google Scholar]
- Tecle E, Diaz-Balzac CA, Bülow HE.. 2013. Distinct 3- O -sulfated heparan sulfate modification patterns are required for kal-1 −dependent neurite branching in a context-dependent manner in Caenorhabditis elegans. G3 (Bethesda). 3:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker BE, Xu D, Lawrence R, Esko JD.. 2014. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 35:60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley RA, Bülow HE.. 2011. Genetic analysis of the heparan modification network in Caenorhabditis elegans. J Biol Chem. 286:16824–16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H, Pinan-Lucarré B, Ji T, Jospin M, Bessereau J-L.. 2015. C. elegans punctin clusters GABAA receptors via neuroligin binding and UNC-40/DCC recruitment. Neuron. 86:1407–1419. [DOI] [PubMed] [Google Scholar]
- Xu D, Esko JD.. 2014. Demystifying heparan sulfate–protein interactions. Annu Rev Biochem. 83:129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Hata T, He J, Maeda N.. 2005. Developmental and regional expression of heparan sulfate sulfotransferase genes in the mouse brain. Glycobiology. 15:982–993. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. 2018. Two classes of secreted synaptic organizers in the central nervous system. Annu Rev Physiol. 80:243–262. [DOI] [PubMed] [Google Scholar]
- Zhang P, Lu H, Peixoto RT, Pines MK, Ge Y, et al. 2018. Heparan sulfate organizes neuronal synapses through neurexin partnerships. Cell 174:1450–1464.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Vachon C, Cizeron M, Romatif O, Bülow HE, et al. 2020a. The HSPG Syndecan is a core organizer of cholinergic synapses in C. elegans. bioRxiv. 10.1101/2020.11.25.395806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Gueydan M, Jospin M, Ji T, Valfort A, et al. 2020b. The netrin receptor UNC-40/DCC assembles a postsynaptic scaffold and sets the synaptic content of GABAA receptors. Nat Commun. 11:2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and reagents are available upon request. Supplementary Table S1 in Supplementary File S1 provides a complete list of strains used in this study. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.14533956.