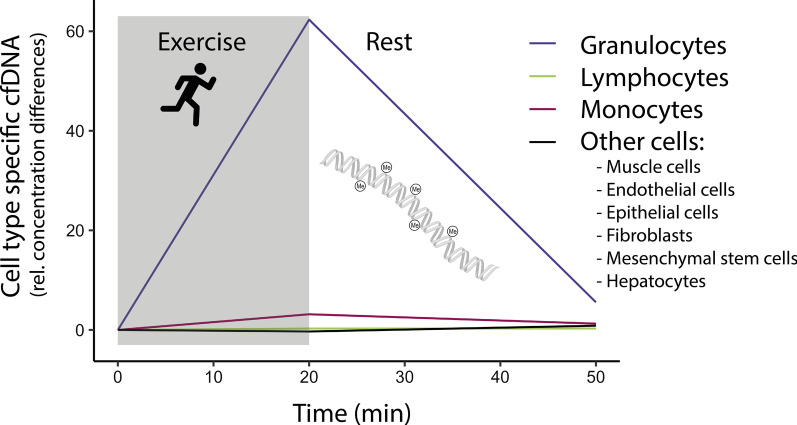
Abstract
Physical activity impacts immune homeostasis and leads to rapid and marked increase in cell-free DNA (cfDNA). However, the origin of cfDNA during exercise remains elusive and it is unknown if physical activity could improve or interfere with methylation based liquid biopsy. We analyzed the methylation levels of four validated CpGs representing cfDNA from granulocytes, lymphocytes, monocytes, and non-hematopoietic cells, in healthy individuals in response to exercise, and in patients with hematological malignancies under resting conditions. The analysis revealed that physical activity almost exclusively triggered DNA release from granulocytes, highlighting the relevance as a pre-analytical variable which could compromise diagnostic accuracy.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-022-01245-3.
Keywords: Cell-free DNA, cfDNA, Methylation, Tissue of origin, cfDNA release, Exercise, Physical activity, Hematological malignancies
Introduction
Circulating cell-free DNA (cfDNA) opened up a new horizon for genomic analyses, including absolute quantification, fragmentation profile analysis, detection of mutations and copy number variations, as well as epigenetic profiling [1]. Cancers, including hematological malignancies, are accompanied by increased levels of cfDNA [2]. More recently, research indicates that DNA methylation analyses are promising tools for diagnostic and prognostic purposes, and cfDNA can be utilized for studying global methylation profiles [3, 4]. Exercise impacts immune homeostasis and leads to rapid and markedly increased levels of cfDNA in blood. Depending on exercise modality, duration, and intensity cfDNA increases 2–20 fold [5–8]. Even submaximal exercise levels lead to 2–4 fold increases [5], showing a half-life of ~ 15 min [6]. However, the origin of cfDNA during exercise has not been studied in detail and the contributing source cells remain elusive. Hence, it remains unknown how acute physical activity affects the composition of cfDNA. Exercise-induced cfDNA releases could interfere with, or improve the diagnostic accuracy of methylation specific testing, depending on whether the DNA is derived from the clinically relevant cell type.
In a sex-mismatch transplantation model we already showed that the major part of cfDNA is released from cells of the hematopoietic lineage during exercise [9]. Since the occurrence of neutrophil extracellular traps (NETs) has been described following physical exhaustion [10], and cfDNA levels are correlated with markers of neutrophil activation including neutrophil elastase, and/or myeloperoxidase [10, 11], it can be considered that granulocytes contribute to the pool of cfDNA [12]. However, a more comprehensive picture is pending. Furthermore, we have shown that a targeted CpG methylation sequencing approach can be utilized to determine the cell types of origin reliably [13, 14].
The aim of our exploratory study was to identify the source cells and kinetics of cfDNA in healthy subjects during physical activity. To this end, we used targeted methylation analysis with the pre-validated CpG sites to assess the origin of cfDNA in healthy participants with a longitudinal design—before, after, and 30 min after exercise—to consider the inter-individual variation. For comparison we also analyzed a small sub-cohort of patients with hematological malignancies under resting conditions.
Methods
Study collective
The study comprised of 10 healthy participants with a mean age of 26.0 ± 5.6 years and 6 patients aged 57.4 ± 11.6 with hematologic malignancies, including low risk myelodysplastic syndrome (LR MDS), chronic myelomonocytic leukemia (CMML), myelodysplastic/myeloproliferative neoplasm (MDS/MPN), myelofibrosis, and two patients with polycythemia vera (PV). All diagnoses were confirmed by a hematopathologist according to the 2016 WHO criteria [15]. The participants gave their informed consent to participate. All experimental procedures were approved by the Human Ethics Committee Rhineland-Palatinate and conformed to the standards of the Declaration of Helsinki of the World Medical Association.
Exercise testing of healthy participants
The 10 healthy participants conducted an incremental running test on a treadmill, starting at 4 km/h. The speed was increased every three min by 1.5 km/h until volitional exhaustion as described by Ochmann et al. [16]. Venous blood samples were collected before (Pre), immediately after (Post), and 30 min after the test (+ 30′). Heart rate (electrocardiogram), as well as oxygen uptake and carbon dioxide release (spiroergometry), were recorded continuously (Geratherm Respiratory). Gas exchange and heart rate were measured continuously. After each step of the test the participants were asked for their rating of perceived exhaustion (RPE) [16].
Sample preparation, DNA extraction and bisulfite conversion
Whole blood samples were collected in tripotassium-EDTA covered Monovettes® (Sarstedt) and were processed within < 3 h after sampling. Whole blood was centrifuged at 2500×g for 15 min. Separated plasma was centrifuged a second round at 2500×g for 15 min and stored at − 80 °C before further processing. DNA was extracted from 4 ml of plasma using QIAamp Circulating Nucleic Acid Kit (Qiagen) and eluted in 55 µl of UltraPure™ DNase/RNase-Free Distilled Water (Invitrogen). 50 µl of the eluate was bisulfite converted using the EZ DNA Methylation Kit (Zymo Research).
Selection of cell type specific CpGs and deconvolution
Deconvolution of cell types is based on CG dinucleotides (CpGs) that are specifically hypomethylated in different cell types. Pre-validated CpG methylation sites were selected to estimate the origin of cfDNA from lymphocytes (cg17587997, FYN protooncogene (FYN)) [14], monocytes (cg10480329, centromere protein A (CENPA)) [14], and granulocytes (cg05398700, WD repeat domain 20 (WDR20)) [14], and to differentiate leukocytes from other cells (cg10673833, myosin IG (MYO1G)) [13], including endothelial cells, epithelial cells, fibroblasts, mesenchymal stem cells, hepatocytes, and muscle cells, described in Schmidt et al. [13]. We then generated a reference-based non-negative least-squares (NNLS) algorithm for the four CpGs of these cellular categories [13, 14]. An Excel calculation tool for cell type deconvolution based on the pyrosequencing measurements is provided in Additional file 1.
Quantification of cfDNA and pyrosequencing
The cfDNA concentration was measured applying a pre-validated qPCR assay described in Neuberger et al. [17]. For pyrosequencing 4 µl of the bisulfite converted DNA were amplified with region-specific biotinylated/unmodified primer pairs (see Additional file 2) using the PyroMark PCR kit (Qiagen) according to the manufacturer’s instructions using the following protocol: Initial activation at 95 °C for 15 min, 45 cycles of 30 s at 94 °C, 30 s at 56 °C, and 30 s at 72 °C followed by a final extension at 72 °C for 10 min. Pyrosequencing was performed on the PyroMark Q96 ID with the respective reagents (Qiagen).
Data analysis and statistics
Statistical analyses were conducted with R version 4.0.2, using tidyverse version 1.3.0, and rstatix version 0.6.0. Graphical illustrations were prepared with ggplot2 version 3.3.2 and corrplot package version 0.85. Continuous data were log 10 transformed and tested for normal distribution with Shapiro–Wilk test. Non-normal distributed data were expressed as median (25th; 75th percentiles). Global significant differences were tested with Friedman rank sum test. Paired or unpaired Wilcoxon rank sum tests with Bonferroni corrections, were used to compare within and between group differences.
Results and discussion
Analysis of cfDNA methylation is a promising approach with potential clinical applications in the field of oncology and hemato-oncology [1]. Since physical activity increases the levels of cfDNA at low intensities [6], exercise is a relevant pre-analytical variable for liquid biopsy. It could be applied to elevate the low levels of cfDNA, but exercise could also interfere with the clinical accuracy of methylation diagnostic. Similar to other studies, we identified significantly increased absolute cfDNA concentrations in patients with myeloid neoplasms or acute leukemia [18, 19].
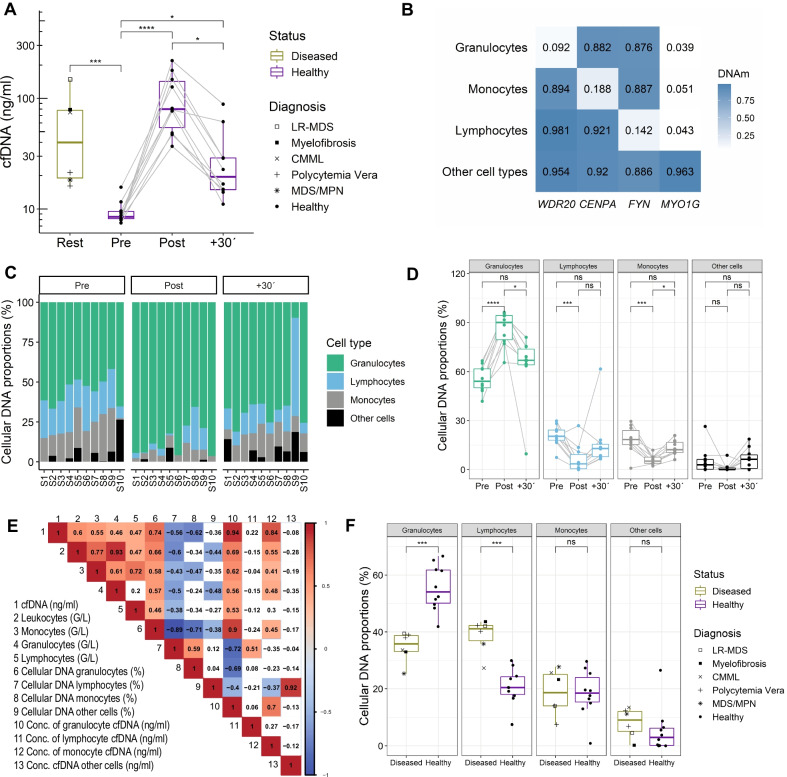
Patients with hematological malignancies showed elevated cfDNA concentrations of 48.1 (19.1; 78.0) ng/ml compared to 8.5 (8.1; 9.5) ng/ml in healthy untrained individuals. PV and MDS/MPN showed lower cfDNA levels than patients with LR MDS, or myelofibrosis (Fig. 1A).
Fig. 1.
Concentration and origin of cfDNA in healthy persons during exercise and patients with hematological malignancies. A cfDNA concentration in blood plasma. B Heatmap of the percentage of methylation of the given CpGs in the reference matrix. C, D Deconvolution results indicating the origin of cfDNA samples before and after exercise in healthy subjects. E Spearman correlation matrix between cfDNA, blood counts, and deconvolution results. F Differences in cell type specific cfDNA proportions between healthy and diseased persons at rest. In all analysis P < .05 was considered significant. *P < .05, **P < .01, ***P < .001, ****P < .0001, ns: not significant
During the all-out exercise test, which lasted 20.1 ± 5.4 min (mean ± SD), cfDNA levels increased ~ tenfold from 8.5 (8.2; 9.5) ng/ml to 80 (55.6; 142.7) ng/ml, decreasing to 19.8 (14.9; 28.9) ng/ml after 30 min rest, with a typical half-life of ~ 15 min [6] (Fig. 1A). Typically, healthy individuals show higher cfDNA elevations in response to exercise compared to patients, which is likely associated with exercise duration and intensity [9, 17]. However, Tug et al. studied cfDNA kinetics in hematopoietic stem cell transplantation patients showing that short-term treadmill exercise increased cfDNA levels up to sixfold, with a mean of ~ threefold [9]. 90 min after exercise the cfDNA values were reduced to resting levels again.
To estimate the cellular composition of cfDNA during exercise, we used a non-negative least squares (NNLS) deconvolution algorithm, referring to the mean DNA methylation levels of the selected CpGs from different cell types (Fig. 1B) [13]. As shown by Schmidt et al., the approach allows reliable estimation for the cellular composition by targeted analysis of the individual CpGs [13], showing similar results to previously described deconvolution results from another working group [3].
Our exploratory study demonstrates that exercise particularly triggered release of cfDNA from granulocytes. We show that granulocytes are the major source of cfDNA during exercise. Cell type specific proportions increase significantly from 54.1% (50.1; 61.8) to 90.2% (79.7; 94.4) after exercise (Fig. 1C, D). The correlation between the cell counts and cfDNA concentrations does not reflect this result (Fig. 1E). Monocyte and lymphocyte specific DNA proportions decreased significantly, but the relative DNA concentration, which was calculated by multiplying the cell type proportion with the cfDNA concentration, as measured by qPCR, indicated that a minor but significant proportion seems to be released from monocytes (see Table 1). During acute exercise and 30 min after exhaustion, no DNA was released from other cell types including muscle cells, endothelial cells, epithelial cells, fibroblasts, mesenchymal stem cells, and hepatocytes. In line with previous findings in non-exercising cohorts, granulocyte specific DNA represents the largest fraction in cfDNA, and the proportions of monocyte and lymphocyte specific cfDNA are similar, whereas the proportion from non-hematopoietic cells is lower [3]. While we provide evidence that granulocytes are the major source of cfDNA during exercise, the restricted number of represented cell types is a limitation and the inclusion of further cell types and cellular sub-characterization could be considered. The activation of granulocytes in response to exercise, as well as the fast release mechanisms of cfDNA are not clarified in detail, however, the association with oxidative stress, metabolic demands, neurohormonal stressors, acute phase proteins, and platelet activation, can be suggested [10].
Table 1.
Hematological responses following exercise
| Pre | Post | + 30′ | Chi-square | P | |
|---|---|---|---|---|---|
| cfDNA (ng/ml) | 8.5 (8.2; 9.5) | 80 (55.6; 142.7) | 19.8 (14.9; 28.9) | 20 | 4.54E−05 |
| Leukocytes (G/L) | 6 (5.6; 6.1) | 9.5 (8.7; 12) | 7.1 (6; 7.5) | 15.8 | 0.000371 |
| Monocytes (G/L) | 0.4 (0.4; 0.6) | 0.7 (0.6; 0.8) | 0.5 (0.4; 0.6) | 15.2 | 0.0005 |
| Granulocytes (G/L) | 3.7 (3.1; 4.3) | 5.4 (4.4; 8.1) | 5.1 (3.5; 5.4) | 16.2 | 0.000304 |
| Lymphocytes (G/L) | 1.7 (1.4; 1.9) | 3.1 (2.4; 3.7) | 1.5 (1.3; 1.5) | 18.2 | 0.000112 |
| Cellular DNA monocytes (%) | 18.5 (15.3; 23.9) | 5.2 (3.8; 7.6) | 12.2 (10.4; 16.7) | 14.6 | 0.000676 |
| Cellular DNA granulocytes (%) | 54.1 (50.1; 61.8) | 90.2 (79.7; 94.4) | 66.9 (64.2; 73.8) | 18.2 | 0.000112 |
| Cellular DNA lymphocytes (%) | 20.4 (18; 24.2) | 3.5 (0.6; 9.2) | 12.9 (7.9; 15.5) | 11.4 | 0.003346 |
| Cellular DNA other cell types (%) | 3 (0.2; 6.2) | 0 (0; 1) | 6.3 (0.7; 9) | 2.6 | 0.272532 |
| Conc. of monocyte cfDNA (ng/ml) | 1.7 (1.4; 2.2) | 4.9 (3.6; 6.9) | 3 (2.3; 4.7) | 9.8 | 0.007447 |
| Conc. of granulocyte cfDNA (ng/ml) | 5.3 (4.4; 5.4) | 67.6 (46.7; 136.1) | 10.8 (9.3; 19.8) | 20 | 4.54E−05 |
| Conc. of lymphocyte cfDNA (ng/ml) | 1.9 (1.5; 2.3) | 2.1 (0.5; 8) | 2.1 (1.9; 3.7) | 0.6 | 0.740818 |
| Conc. cfDNA other cells (ng/ml) | 0.3 (0; 0.7) | 0 (0; 0.8) | 1.2 (0.1; 1.6) | 1.4 | 0.496585 |
Data are expressed as median (25th, 75th percentiles). The relative concentration of cfDNA from different cell types was calculated by multiplying the cfDNA concentration with the cellular DNA amount (%). Global statistical differences were calculated with nonparametric Friedman rank sum test
Under resting conditions, healthy and diseased persons show similar cfDNA proportions from monocytes and other cell types, whereas the levels of granulocyte and lymphocyte specific DNA differ significantly (Fig. 1F, Table 1). The cfDNA levels of lymphocytes specific, hypomethylated cg17587997, which occur in the 5’UTR of FYN, are markedly increased and may be linked to pathogenesis [20]. Recently, Sontag et al. demonstrated that epigenetic lymphocyte counts in AML patients might be affected by aberrant DNA methylation in the FYN proto-oncogene [21]. Since epigenetic changes crucially contribute to hematological malignancies [22], further analyses of relevant methylation sites have a great potential for monitoring disease burden and treatment response.
Our exploratory study has several limitations. (1) The number of samples is relatively low, but the longitudinal analysis of the healthy donors excludes inter-individual variation. To gain further insights for which type of hemato-oncological diseases exercise might evoke additional alterations of the cfDNA profile a much larger number of patient samples would be required. Notably, our study did not aim to validate relevant methylation-based disease classifiers. (2) The age groups were not adequately matched. Since biological processes such as age can affect the methylation profile, the validity of the results can be compromised. However, in another recent study we determined the impact of age on the methylation status of cell-type specific CpGs used in this study. The stratification into younger ≤ 30 years and older ≥ 60 did not indicate significant differences [21]. However, it is conceivable that in elderly donors the exercise-induced cell-type specific release of cfDNA is slightly different and this should be further analyzed in the future. (3) We only used one exercise setting in a controlled design. To determine to what extend exercise affects cfDNA-based liquid biopsy, different clinically relevant scenarios, e.g., cycling for 30 min at moderate intensity or walking stairs, should be tested in further studies. Even low intensity physical activity with low workload, comparable to walking stairs, can increase cfDNA levels more than twofold, thereby affecting the diagnostic accuracy of cfDNA methylation based testing if resting periods are too short [9]. (4) Genome wide DNA methylation analysis might provide additional insight into the composition of other leukocyte subsets or non-hematopoietic cells [3]. The targeted analysis at cell-type specific CpGs provides a trade-off with reduced possibility for bioinformatics deconvolution and a more standardized and cost-effective approach [21].
In summary, our study provides evidence that the additional release of cfDNA during exercise is particularly attributed to granulocytes. Consequently, exercise is a relevant pre-analytical variable, which affects the composition of cfDNA. For liquid biopsy it may be advantageous to take samples at a resting state without previous exercise.
Supplementary Information
Additional file 1. NNLS deconvolution table.
Acknowledgements
We thank all participants for their contributions to this study.
Abbreviations
- CENPA
Centromere protein A
- cfDNA
Cell-free DNA
- CMML
Chronic myelomonocytic leukemia
- EDTA
Ethylenediamine tetraacetic acid
- FYN
FYN proto-oncogene
- LR MDS
Low risk myelodysplastic syndrome
- MDS/MPN
Myelodysplastic/myeloproliferative neoplasm
- MYO1G
Myosin IG
- NETs
Neutrophil extra cellular traps
- NNLS
Non-negative least-squares
- PV
Polycythemia vera
- UTR
Untranslated region
- WDR20
WD repeat domain 20
Authors' contributions
PS, WW, MPR, EN designed the research. KFAP and AB performed exercise testing and sample preparation. SS, EN measured the samples and analyzed the data. EN drafted the manuscript with input from all authors. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. The study was supported by intramural funding (Stufe 1) of the Johannes Gutenberg University Mainz (PS), by the German Ministry of Education and Research (WW: VIP+, 03VP06120), and the German Research Foundation (MPR: CRC156/2 KS01, CRC1066/3 TP B18, CRC1292/2 TP21N).
Availability of data and materials
The datasets analyses of the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All experimental procedures were approved by the Human Ethics Committee Rhineland-Palatinate and conformed to the standards of the Declaration of Helsinki of the World Medical Association. The participants gave their informed consent to participate.
Consent for publication
Not applicable.
Competing interests
W.W. is cofounder of Cygenia GmbH that can provide service for analysis of epigenetic signatures (www.cygenia.com).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372. [DOI] [PubMed]
- 2.Lim JK, Kuss B, Talaulikar D. Role of cell-free DNA in haematological malignancies. Pathology. 2021;53:416–426. doi: 10.1016/j.pathol.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9:5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caggiano C, Celona B, Garton F, Mefford J, Black BL, Henderson R, et al. Comprehensive cell type decomposition of circulating cell-free DNA with CelFiE. Nat Commun. 2021;12:1–13. doi: 10.1038/s41467-021-22901-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haller N, Tug S, Breitbach S, Jörgensen A, Simon P. Increases in circulating cell-free DNA during aerobic running depend on intensity and duration. Int J Sports Physiol Perform. 2017;12:455–462. doi: 10.1123/ijspp.2015-0540. [DOI] [PubMed] [Google Scholar]
- 6.Breitbach S, Tug S, Simon P. Circulating cell-free DNA: an up-coming molecular marker in exercise physiology. Sports Med. 2012;42:565–586. doi: 10.2165/11631380-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 7.Neuberger EWI, Hillen B, Mayr K, Simon P, Krämer-Albers E-M, Brahmer A. Kinetics and topology of DNA associated with circulating extracellular vesicles released during exercise. Genes. 2021;12. [DOI] [PMC free article] [PubMed]
- 8.Stawski R, Walczak K, Kosielski P, Meissner P, Budlewski T, Padula G, et al. Repeated bouts of exhaustive exercise increase circulating cell free nuclear and mitochondrial DNA without development of tolerance in healthy men. PLoS ONE. 2017;12:e0178216. [DOI] [PMC free article] [PubMed]
- 9.Tug S, Helmig S, Deichmann ER, Schmeier-Jürchott A, Wagner E, Zimmermann T, et al. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc Immunol Rev. 2015;21:164–173. [PubMed] [Google Scholar]
- 10.Beiter T, Fragasso A, Hudemann J, Schild M, Steinacker J, Mooren FC, et al. Neutrophils release extracellular DNA traps in response to exercise. J Appl Physiol. 2014;117:325–333. doi: 10.1152/japplphysiol.00173.2014. [DOI] [PubMed] [Google Scholar]
- 11.Tug S, Mehdorn M, Helmig S, Breitbach S, Ehlert T, Simon P. Exploring the potential of cell-free-DNA measurements after an exhaustive cycle-ergometer test as a marker for performance-related parameters. Int J Sports Physiol Perform. 2017;12:597–604. doi: 10.1123/ijspp.2016-0157. [DOI] [PubMed] [Google Scholar]
- 12.Beiter T, Fragasso A, Hartl D, Nieß AM. Neutrophil extracellular traps: a walk on the wild side of exercise immunology. Sport Med. 2015;45:625–640. doi: 10.1007/s40279-014-0296-1. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt M, Maié T, Dahl E, Costa IG, Wagner W. Deconvolution of cellular subsets in human tissue based on targeted DNA methylation analysis at individual CpG sites. BMC Biol. 2020;18:1–13. doi: 10.1186/s12915-020-00910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frobel J, Božić T, Lenz M, Uciechowski P, Han Y, Herwartz R, et al. Leukocyte counts based on DNA methylation at individual cytosines. Clin Chem. 2018;64:566–575. doi: 10.1373/clinchem.2017.279935. [DOI] [PubMed] [Google Scholar]
- 15.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 16.Ochmann DT, Philippi KFA, Zeier P, Sandner M, Hillen B, Neuberger EWI, et al. Association of innate and acquired aerobic capacity with resilience in healthy adults: protocol for a randomized controlled trial of an 8-week web-based physical exercise intervention. JMIR Res Protoc. 2021;10:e29712. [DOI] [PMC free article] [PubMed]
- 17.Neuberger EWI, Brahmer A, Ehlert T, Kluge K, Philippi KFA, Boedecker SC, et al. Validating quantitative PCR assays for cfDNA detection without DNA extraction in exercising SLE patients. Sci Rep. 2021;11:13581. doi: 10.1038/s41598-021-92826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz AK, Stanulla M, Cario G, Flohr T, Sutton R, Möricke A, et al. Quantification of free total plasma DNA and minimal residual disease detection in the plasma of children with acute lymphoblastic leukemia. Ann Hematol. 2009;88:897–905. doi: 10.1007/s00277-009-0698-6. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Gisbert N, Fernández-Ibarrondo L, Fernández-Rodríguez C, Gibert J, Andrade-Campos M, Arenillas L, et al. Circulating cell-free DNA improves the molecular characterisation of Ph-negative myeloproliferative neoplasms. Br J Haematol. 2021;192:300–309. doi: 10.1111/bjh.17087. [DOI] [PubMed] [Google Scholar]
- 20.Chougule RA, Kazi JU, Rönnstrand L. FYN expression potentiates FLT3-ITD induced STAT5 signaling in acute myeloid leukemia. Oncotarget. 2016;7:9964–9974. doi: 10.18632/oncotarget.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sontag S, Bocova L, Hubens WHG, Nüchtern S, Schnitker M, Look T, Schröder KM, et al. Toward clinical application of leukocyte counts based on targeted DNA methylation analysis. Clin Chem. 2022 doi: 10.1093/clinchem/hvac006. [DOI] [PubMed] [Google Scholar]
- 22.Blecua P, Martinez-Verbo L, Esteller M. The DNA methylation landscape of hematological malignancies: an update. Mol Oncol. 2020;14:1616–1639. doi: 10.1002/1878-0261.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. NNLS deconvolution table.
Data Availability Statement
The datasets analyses of the current study are available from the corresponding author on reasonable request.