Graphical abstract
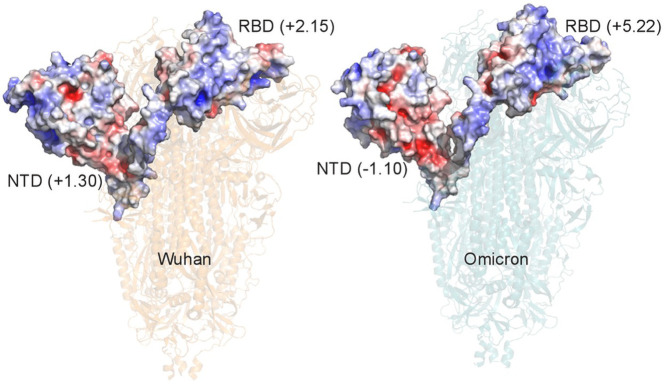
Comparison of the surface electrostatic potential of the N-terminal (NTD) and the Receptor Binding Domains (RBD) of the Wuhan and Omicron Spikes. The entire Spikes are represented with transparent ribbon models. Only the NTD and RBD surfaces of the chain A are represented. Domain net charges are reported within parenthesis beside the domain labels. Surface color ranges from -5.0 (red) to + 5.0 (blue) kT/e.
Abbreviations: EPrbd, Electrostatic potential of the Spike receptor binding domain; EPntd, Electrostatic potential of the Spike N-terminal domain
Dear Editor
Ibrahim and colleagues, in this Journal, published a predicted structure for the SARS-CoV-2 receptor binding site.1 We and others have focussed attention upon the electrostatic potential (EP) of the receptor binding (EPrbd) and N-terminal (EPntd) domains of SARS-CoV-2 Spike (S protein) that in all previously studied SARS-CoV-2 Variants of Concern (VOC) have a positive charge and strongly interact with negatively-charged patches of the ACE2 and other receptors.2, 3, 4, 5
SARS-CoV-2 Omicron variant is the last and most transmissible of all SARS-CoV-2 mutants. It has an unprecedented, high number of mutations in the receptor binding domain and a novel three amino acids (EPE) insertion in the N-terminal domain of the S Protein, which have determined a new SARS-CoV-2 pandemic trajectory.2 Initial observations of qualitative nature suggested increasingly positive EPrbd in the successive, worldwide-spreading VOC.4 We here particularly focus upon VOC which have been able to spread globally and overtake the previously dominant viral strain(s). We assign numerical values to the EPrbd and EPntd, and identify some peculiarities of the Omicron variant that could markedly impact virus transmissibility and pathogenesis.
VOC RBD and NTD, their modeling and energy minimization were as previously described.4 Net RBD and NTD charge at pH= 7.0 were predicted using the PROPKA3, a software allowing pK calculationa of protein ionizable residues, taking int account the influence of the local structural environment, inclusive of residue solvent exposure, presence of hydrogen bonds or interactions with other charged groups. The pKa values were used to predict the overall domain charge (EPrbd and EPntd). Surface electrostatic potential has been calculated for each variant RBD and NTD by means of the program APBS (Adaptive Poisson-Boltzmann Solver) within the PyMOL environment using default parameters. Electrostatic potential was mapped onto the domain accessibility surfaces and represented with a color gradient ranging from blue to red indicating positive and negative values, respectively.
The calculated pK were used to assign numerical values to the EPrbd and EPntd. Knowing the importance of the RBD-ACE2 binding strength for SARS-CoV-2 transmissibility, we also compared the EPrbd values to the estimated effective reproduction numbers (ERN) of each VOC.2 , 6 Confirming color gradient visualization4, the EPrbd was positive and linearly increased from the Wuhan strain (2.15) to successive variants, reaching its highest value (5.22) in Omicron VOC (Table 1 ). Other noticeable data in Table 1 are, first, that EPrbd values fall within the range of the estimated ERN of the Wuhan reference, Alpha and Delta but not Omicron variant which has a remarkably high ERN value range. The second is the equivalence of the EPrbd fold increase of Omicron relative the Wuhan strain (2.44) to the (around) 2.4 fold enhanced affinity of Omicron RBD binding to ACE2 relative to the Wuhan strain, as measured by surface plasmon resonance.7 The third is the negative EPntd of the Omicron variant, contrasting the EPntd positivity of all other SARS-CoV-2 strains. All this highlights the peculiarity of the Omicron VOC in such basic chemical and functionals parameters as the RBD and NTD electrostatic potentials.
Table 1.
Predicted net charge of the Spike RBD and NTD variants in the folded state calculated with the PROPKA program, and compared with the ranges of the estimated effective reproduction number (ERN)a.
| SARS-CoV-2 | Pango Lineage | EPrbd | EPntd | Estimated ERN (range)a |
|---|---|---|---|---|
| Wuhan | B.1 | 2.15 | 1.30 | 1.4–2.5 |
| Alpha | B.1.1.7 | 3.18 | 1.69 | 1.8–4.7 |
| Delta | B.1.617.2 | 4.15 | 1.28 | 3.4–8.6 |
| Omicron | B.A.1 | 5.22 | -1.10 | 9.7--13.0 |
Our data suggest that EPrbd could be a proxy of the binding affinity RBD/ACE2, then impacting on virus transmissibility. Hypothetically, the higher the EPrbd, the higher the binding affinity to ACE2, the more transmissible the VOC. This hypothesis appears to hold with the Wuhan reference strain, Alpha and Delta VOC as their EPrbd values do linearly increase and parallel the value range of their effective reproduction numbers. as estimated by epidemiological and clinical studies.2 , 6 However, it does not hold for the Omicron variant since its EPrbd value is substantially lower than the estimated ERN, This VOC is characterized by 37 mutations in the sequence of the Spike (S protein), of which fifteen are just located in its RBD.7, 8, 9 While they appear to have preserved if not increased (as our EPrbd data and a previous report suggests; 9) the binding strength to ACE2 receptor, there is both laboratory and clinical evidence that this mutational array allows SARS-CoV-2 escaping from neutralizing antibodies of vaccinated and COVID-19 convalescent subjects, as well as the great majority of the monoclonal antibodies in clinical use.9 , 10 Thus, enhanced transmissibility and immuno-escape are the two features of Omicron worldwide spread.
Mutations in other molecular constituents, both within and outside the S protein could also contribute to virus transmissibility. One is the electrostatic potential of S-protein N-terminal domain . The negative value of the Omicron EPntd is rather unusual, in clear contrast with the positivity of all other variants, as already reported.3 Rather than affecting RBD-ACE2 interaction, the S protein-NTD could play a role in virus binding to other host cell surface constituent(s). SARS-CoV-2 has been shown to use the NTD domain to recognize and bind to several host receptors or candidate-co-receptors 3 , 6 , 11 If EP positivity is important for SARS-CoV-2 binding to the above receptors, as it is for RBD-ACE2 interaction, then the negative EPntd value would imply a less efficient receptor binding of the Omicron variant relative to that of Wuhan, Alpha and Delta variants. Given that at least one receptor is markedly expressed in lung and bronchial cells 11, the EPntd negative value could be one of the factors contributing to explain the supposed lower pathogenicity of the Omicron variant for the low respiratory tract.2
We recognize the limitations of our in silico data, though they are tempered by the similarity between our ratio EPrbd Omicron variant /Wuhan strain to that measured by Cameroni et al.8 using a surface plasmon resonance technique. We also recognize the rather speculative nature of some of our suggestions. Experimental confirmations are needed. All above considered, our data highlight some special features of S protein RBD and NTD moieties possessed by the most transmissble and contagious Omicron variant. We invite to consider accurate EP determinations as a help in disentangling the complex mechanisms of SARS-CoV-2 transmissibility and infectivity.
Declaration of Competing Interest
We report no competing interests.
References
- 1.Ibrahim I.M., Abdelmalek I.H., Elshalat M.E., Elfiky A.A. COVID-19 spike- host cell receptor GRP78 binding site prediction. J Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization W.H. Worls Health Organization; 2022. Enhances Responses to Omicron SARS-CoV-2 variant. Technical brief and Priority For Member States. Updated #6:21. [Google Scholar]
- 3.Fantini J., Noura Y., Fodil A., Chahiniana H. Sctrutural dynamics of SARS-CoV-2 variants: a health monitoring strategyfor anticipating Covid-19 outbreaks. J.Infect. 2021;83:197–206. doi: 10.1016/j.jinf.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascarella S., Ciccozzi M., Bianchi M., Benvenuto D., Cauda R., Cassone A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: a hint to higher transmissibility? J Med Virol. 2021 doi: 10.1002/jmv.27528. [DOI] [PubMed] [Google Scholar]
- 5.Nassar A., Ibrahim I.M., Amin F.G., Magdy M., Elgharib A.M., Azzam E.B., et al. A review of human coronaviruses’ receptors: the host-cell targets for the crown-bearing viruses. Molecules. 2021;26:6455–6462. doi: 10.3390/molecules26216455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26(24) doi: 10.2807/1560-7917.ES.2021.26.24.2100509. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameroni E., Bowen J.E., Rosen I.E., Saliba S.K., Zepeda K., Culab K., et al. Broadly neutralizin antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021 doi: 10.1038/1041/s.586-021-043862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mannar D., Saville J.W., Zhou X., Srnivastava S.S., Berezuk A.M., Tuttle K.S., et al. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022 doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S.V., Pollard A.J., et al. Reduced neutralization of SARS-CoV-2 omicron B.1.1.529 variant by post immunization serum. Lancet. 2022 doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S., Qiu Z., Hou Y., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]