Abstract
Objectives
To use the nationwide Norwegian surveillance programme on resistant microbes in humans (NORM) to address longitudinal changes in the population structure of Klebsiella pneumoniae isolates from 2001–15, focusing on the emergence and dissemination of ESBL-producing K. pneumoniae in Norway.
Methods
Among blood (n = 6124) and urinary tract (n = 5496) surveillance isolates from 2001–15, we used Illumina technology to whole genome sequence 201 ESBL-producing isolates from blood (n = 130) and urine (n = 71), and 667 non-ESBL isolates from blood. Complete genomes for four isolates were resolved with Oxford Nanopore sequencing.
Results
In a highly diverse collection, Klebsiella variicola ssp. variicola caused 24.5% of Klebsiella pneumoniae species complex (KpSC) bacteraemias. ESBL production was limited to K. pneumoniae sensu stricto (98.5%). A diverse ESBL population of 57 clonal groups (CGs) were dominated by MDR CG307 (17%), CG15 (12%), CG70 (6%), CG258 (5%) and CG45 (5%) carrying blaCTX-M-15. Yersiniabactin was significantly more common in ESBL-positive (37.8%) compared with non-ESBL K. pneumoniae sensu stricto isolates (12.7%), indicating convergence of virulence and resistance determinants. Moreover, we found a significantly lower prevalence of yersiniabactin (3.0%, 37.8% and 17.3%), IncFIB (58.7%, 87.9% and 79.4%) and IncFII plasmid replicons (40.5%, 82.8% and 54.2%) in K. variicola ssp. variicola compared with ESBL- and non-ESBL K. pneumoniae sensu stricto isolates, respectively.
Conclusions
The increase in Norwegian ESBL-producing KpSC during 2010–15 was driven by CG307 and CG15 carrying blaCTX-M-15. K. variicola ssp. variicola was a frequent cause of invasive KpSC infection, but rarely carried ESBLs.
Introduction
Klebsiella pneumoniae is an important human pathogen1 and acknowledged as a key host for the spread of antimicrobial resistance (AMR).2,3 The global spread of MDR4K. pneumoniae is closely linked to the spread of extended-spectrum β-lactamases (ESBLs) and carbapenemases. This has been facilitated by successful clonal lineages or clonal groups (CGs) such as CG258, CG15 and CG307,5–7 and horizontal gene transfer (HGT),2,8 fuelled by antibiotic selection.9
While K. pneumoniae typically causes severe infections in vulnerable hospitalized patients,1 some hypervirulent (HV) clones cause community acquired invasive infections, often in healthy individuals.10 HV clones cluster in CG23, CG65 and CG86, and harbour capsular loci K1 or K2, siderophores and other virulence factors supporting colonization, tissue invasion and immune evasion.10,11 High-risk K. pneumoniae clones, categorized as either MDR or HV, rarely display both traits.12 However, in recent years, convergence of the two traits has been reported.13
K. pneumoniae is a highly diverse species, and the term K. pneumoniae species complex (KpSC) has been introduced to encompass seven closely related taxa,14 of which K. pneumoniae sensu stricto, Klebsiella variicola and Klebsiella quasipneumoniae are the most frequently reported in human clinical samples.15,16
Most molecular epidemiological studies of KpSC have focused on outbreaks or isolates with particular characteristics such as AMR or virulence, most often with a cross-sectional study design. Thus, there is a need for longitudinal studies, including both resistant and susceptible isolates, to improve our understanding of the population dynamics in clinical KpSC isolates.
In this study, we used the Norwegian surveillance programme on resistant microbes (NORM) from 2001–15. The aim of this study was to address the longitudinal dynamics of KpSC clinical isolates, dominant CGs and their associations with clinically important AMR and virulence determinants, with a particular focus on the emergence and dissemination of ESBLs.
Material and methods
Bacterial isolates
NORM monitors AMR in Klebsiella spp. isolated from blood and urine isolates. Antimicrobial susceptibility testing is performed and interpreted according to EUCAST guidelines and breakpoints.17 Isolates with reduced susceptibility to cefotaxime and/or ceftazidime are verified as ESBL carriers with phenotypic ESBL testing.18 Isolates are stored locally at −80°C at the participating laboratories.18
All putative ESBL-producing KpSC blood (n = 149) and urine isolates (n = 91) from 2001–15 registered in the NORM database were included in the study. For comparison, a subset of non-ESBL blood culture isolates (n = 815) were included. As the number of isolates per year increased during the study period, we took measures to achieve similar sized sample sets for each year. Consecutive isolates were selected from each laboratory for each year according to the following key: 2001, all isolates; 2005, every 2 out of 3; 2009, every 1 out of 2; and 2015, every 1 out of 3 (Figure S1, available as Supplementary data at JAC Online).
Isolates registered as either K. pneumoniae or Klebsiella spp. were included. Species identification was confirmed by MALDI-TOF MS (MBT Compass Library DB-6903, Bruker Daltonik), and subsequently by whole genome sequencing. Only KpSC isolates were included for further analyses. Antimicrobial susceptibility profiles, laboratory, year and source of isolation were retrieved from the NORM database. For isolates with discordant ESBL genotype and phenotype, the phenotype was confirmed using the combined disc method (Becton Dickinson, New Jersey, USA).19 Colistin MICs were determined by broth microdilution using Sensititre FRCOL plates (Thermo Fisher Scientific, East Grinstead, UK) according to manufacturer’s instructions.
Whole genome sequencing
DNA was extracted using the MagNAPure 96 system (Roche Applied Science, Manheim, Germany) and sequencing libraries were prepared using Nextera XT DNA Library preparation protocol (Illumina, San Diego, CA, USA). Paired-end reads (2 × 150, 2 × 250 or 2 × 300 bp) were generated for all isolates using Illumina MiSeq with v3 chemistry. For isolates where only FASTQ files were received, sequencing was performed on an Illumina HiSeq 2500 at Eurofins Genomics (Eurofins Genomics Europe, Konstanz, Germany) generating 2 × 125 bp paired-end reads.
To achieve closed genomes, selected isolates were also long-read sequenced. DNA was extracted manually using the Beckman Coulter Life Science GenFind V3 kit (C34881) according to the supplemental protocol ‘DNA extraction from Bacteria using GenFind v3’ (Beckman Coulter, Brea, CA, USA). DNA libraries were prepared using the 1D Ligation sequencing kit (SQK-LSK108) and the Native barcoding kit (EXP-NBD103) [Oxford Nanopore Technologies (ONT), Oxford, UK] according to the ONT protocol ‘native barcoding genomic DNA’ or ‘genomic DNA by ligation’ without shearing to maximize the sequencing read length. Finally, libraries were loaded onto a R9.4.1 MinION flow cell (FLO-MIN106) or a R9.4.1 Flongle flow cell (FLO-FLG001) and sequenced on an ONT MinION Mk1B device (MIN-101B).
In silico analysis
Short-read sequences were trimmed based on quality and adapter content with TrimGalore v0.6.420 and de novo assembly was performed with Unicycler v0.4.8,21 which uses SPAdes v3.13.122 for assembly and Pilon v1.2323 for polishing. Kleborate v2.0.424 was used to identify species and determine multilocus sequence type (ST), virulence loci and AMR genes (CARD database v3.0.825) from assembled genomes. Kaptive26 was used to identify capsule (K) biosynthesis loci reporting calls with confidence level ‘Good’ or higher. Putative ESBL carriers with no definite ESBL gene had their read sets investigated using SRST2 v.0.2.027 with CARD database v3.0.8, as some reads may have been discarded in the assembly process. Only ESBL gene matches with 100% sequence coverage and identity were included in further analyses. Plasmid replicons were identified with SRST2 v0.2.0 using the Plasmidfinder database version 2021–01-13.28
A core chromosomal single-nucleotide polymorphism (SNP) alignment of the verified ESBL and non-ESBL genomes was generated to assess their relatedness. The short-reads were mapped to the chromosome of the ST23 reference genome NTUH-K2044 (NC_01273.1) with the RedDog V1beta.1129 pipeline, using Bowtie2 v2.3.4.230 for read mapping and SAMtools v1.931 for SNP calling. RedDog was used with default parameters as described previously,13 except for the read depth threshold which was set to ≥8 (default ≥10) to include all genomes. A maximum likelihood (ML) phylogeny was inferred from the resulting alignment (868 genomes, 867 815 SNPs) using FastTree v2.1.10 (gamma distribution of rate heterogeneity among sites).32
CGs were defined by patristic distances. This method was chosen as it has previously been used to cluster CGs in KpSC12 and a distance threshold of 0.04 was used as it grouped STs that have previously been identified as belonging to clinically distinct CGs. The CGs dominated by ST14 and ST340 were denoted CG15 and CG258, as these names are more commonly used.2,33
Long-read sequences were base called and de-multiplexed high-accuracy mode using Guppy Basecalling Software v3.2.434 followed by quality filtering with Filtlong v0.2.0.35 To resolve the complete genome sequence of these isolates, hybrid assembly with the corresponding short-read genomes using Unicycler v0.4.8 was performed. The completed genomes were subsequently annotated with the NCBI Prokaryotic Annotation Pipeline v5.136 using default parameters.
To assess the clonal relatedness of the ST307 genomes, an alignment was generated with RedDog, using the hybrid-assembled closed ST307 genome (Genbank accessionCP073627) as the mapping reference. The resulting alignment with 391 variant sites was screened for recombination events using Gubbins v2.3.437 with convergence method ‘weighted Robinson-Foulds’. This alignment was passed to RAxML v8.2.1038 to infer ML phylogeny. The best-scoring ML tree was chosen from five independent runs with the GTR+ nucleotide substitution model, followed by a rapid bootstrap analysis (100 replicates) to estimate branch support.
Data availability
The 868 KpSC short-read and four long-read sequence files have been deposited in the European Nucleotide Archive under BioProject PRJEB27256 (Table S1). The four hybrid-assembled completed genomes have been deposited in GenBank (Table S1) under accession numbers CP073791-CP073796, CP073627-CP073629, CP073783-CP073787 and CP073788-CP073790.
Definitions
MDR was defined as phenotypic resistance to agents in three antimicrobial classes.4 HV was defined as either (a) the presence of rmpA or rmpA2; and/or (b) the presence of aerobactin (iuc) and salmochelin (iro).39 ESBL isolates were defined as either having known ESBL genes (i.e. blaCTX-M, blaSHV-2, blaSHV-5, blaSHV-12, blaSHV-18 and blaSHV-24), or in absence of known ESBL genes, a confirmed ESBL phenotype. Isolates with plasmid-mediated AmpC genes only or carbapenemase-encoding genes (regardless of ESBL gene presence) were excluded from further analysis.
Data handling and statistical analysis
Data analysis and statistics were done using R version 4.0.2 (2020–06-22).40 Distribution differences were calculated with Fisher exact test, with Benjamini–Hochberg correction for multiple testing when necessary. P < 0.05 was considered statistically significant.
Ethics
The study was approved by the Regional Committee for Medical and Health Research Ethics (Reference: 2017/1185–3).
Results
We received 954/1055 (90.4%) of requested isolates, of which 223 putative ESBL-producing isolates (blood, n = 144; urine, n = 79) and 667 non-ESBL blood isolates were confirmed as KpSC by MALDI-TOF MS and WGS (Figure S1). Known ESBL genes were detected in 192/223 (86%) putative ESBL isolates. ESBL phenotype was confirmed in nine additional isolates, resulting in an ESBL group consisting of 201 isolates (blood, n = 130; urine, n = 71). Six isolates with carbapenemase genes and one isolate with a plasmid-mediated AmpC gene only were excluded (Figure S2). The dataset can be explored at https://microreact.org/project/4dBcaZsZmKoAzvatzPaGds.
Phylogenetic diversity in ESBL and non-ESBL KpSC populations
The species distributions in the ESBL and non-ESBL groups were different (P < 0.0001) (Table 1). The ESBL group consisted of 98.5% K. pneumoniae sensu stricto. In contrast, the non-ESBL group isolates comprised K. pneumoniae sensu stricto (69.1%), K. variicola ssp. variicola (24.5%), K. quasipneumoniae ssp. similipneumoniae (3.3%) and K. quasipneumoniae ssp. quasipneumoniae (3.1%). K. variicola ssp. tropica, K. africana or K. quasivariicola were not detected (Table 1, Figure 1).
Table 1.
ESBL and non-ESBL groups: species distribution, clonal groups (CGs), and sequence types (STs) numbers
| ESBL isolates |
Non-ESBL isolates |
||||
|---|---|---|---|---|---|
| Species identification | No. (%) | CGs/STs | No. (%) | CGs/STs | P valuea |
| K. pneumoniae sensu stricto | 198 (98.5) | 54/70 | 461 (69.1) | 136/222 | |
| K. variicola ssp. variicola | 1 (0.5) | 1/1 | 163 (24.5) | 80/115 | |
| K. quasipneumoniae ssp. similipneumoniae | 2 (1.0) | 2/2 | 22 (3.3) | 20/20 | |
| K. quasipneumoniae ssp. quasipneumoniae | – | – | 21 (3.1) | 10/20 | |
| K. pneumoniae species complex | 201 | 57/73 | 667 | 246/377 | <0.0001 |
Fisher exact test for difference in species distributions between groups.
Figure 1.
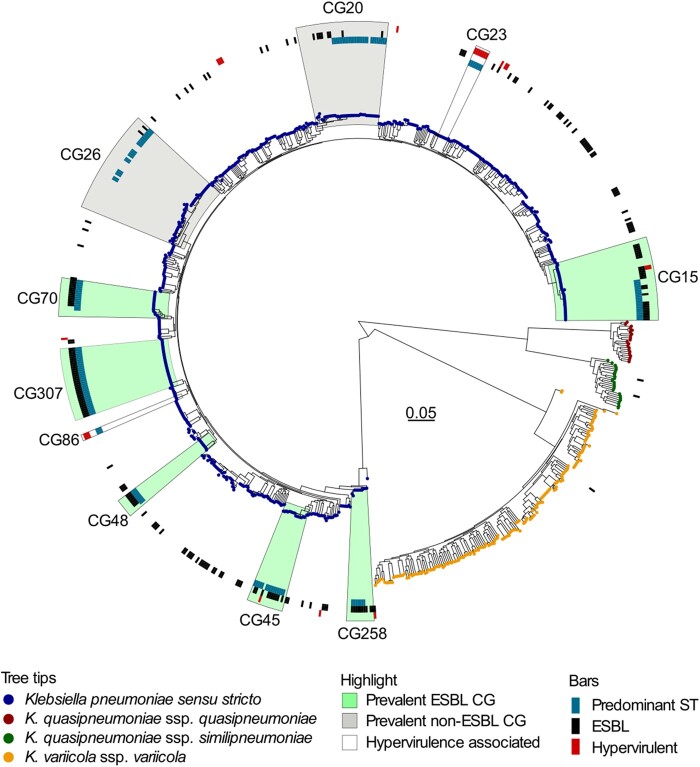
Maximum likelihood tree of the 868 Klebsiella pneumoniae species complex genomes. Tips are coloured according to the Klebsiella species. Prevalent CGs representing >5% of isolates, either in the ESBL or the non-ESBL group, are highlighted in green and grey, respectively. CGs commonly associated with hypervirulence are highlighted without colour. The highlighted areas include all genomes on the most recent common ancestor node of the CG indicated. The circles from inner to outer show genomes with the predominant sequence type (ST, blue bars) within the CG, presence of ESBL-encoding genes (black bars) and isolates meeting the study definition of hypervirulence (red bars).
The 868 KpSC isolates were phylogenetically highly diverse with a total of 413 different STs assigned to 261 CGs (Table S1). The Simpson’s diversity indices for STs were 0.95 for the ESBL group and 0.99 for the non-ESBL group, respectively.
The ESBL group (n = 201) consisted of 73 STs assigned to 57 CGs (mean number of isolates per CG 3.53, range 1–34). CG307 was the most prevalent (16.9%, n = 34; ST307) followed by CG15 (12.4%, n = 15; comprising ST14, ST15 and ST627), CG70 (6.5%, n = 13; ST70), CG258 (5.0%, n = 10; ST11, ST340 and ST437) and CG45 (5.0%, n = 10; ST45 and ST2954). The remaining CGs represented <5% of ESBL isolates each (Figure 1).
Among the 667 non-ESBL blood culture isolates there were 377 STs assigned to 246 CGs (mean number of isolates per CG 2.71, range 1–42). CG26 (6.3%, n = 42; comprising 11 STs) and CG20 (5.2%, n = 35; comprising 6 STs) were the most prevalent. In the remaining 244 CGs (88.5%, n = 590 isolates) each CG represented <5% of the isolates. Ten (1.5%) non-ESBL HV-associated isolates, CG23 (n = 7) and CG86 (n = 3), were observed. CG307 was absent in the non-ESBL group, while the other major ESBL CGs were present in low numbers; CG15 (2.1%, n = 14), CG70 (0.7%, n = 7), CG45 (1%, n = 7) and CG258 (0.4%, n = 3). In total, 33 of 57 CGs (57.9%) in the ESBL group were present in the non-ESBL group.
Temporal trends
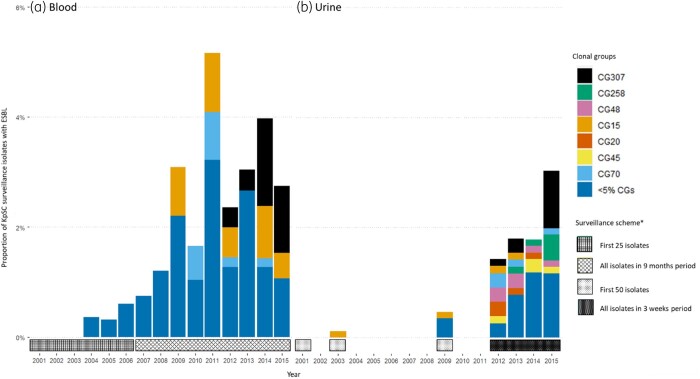
The first occurrence in this study of an ESBL-producing isolate was the globally successful ESBL CG15 in 2003 in a urine specimen. Between 2009 and 2012 it was the most prevalent CG among our blood culture samples. From 2012, the increase in ESBLs was associated with the emergence of CG307. Urine isolates exhibited greater diversity of CGs compared with blood, but all prevalent urine CGs were also represented in blood cultures isolates, albeit several in low numbers (Figure 2). There were no apparent CG trends in the non-ESBL group.
Figure 2.
Temporal distribution of Klebsiella pneumoniae species complex (KpSC) ESBL clonal groups (CGs) in blood (a) and urine (b) as proportion of the surveillance for each year. Distribution of CGs in blood and urine separated by most prevalent CGs and other CGs. *The surveillance starts in January each year.
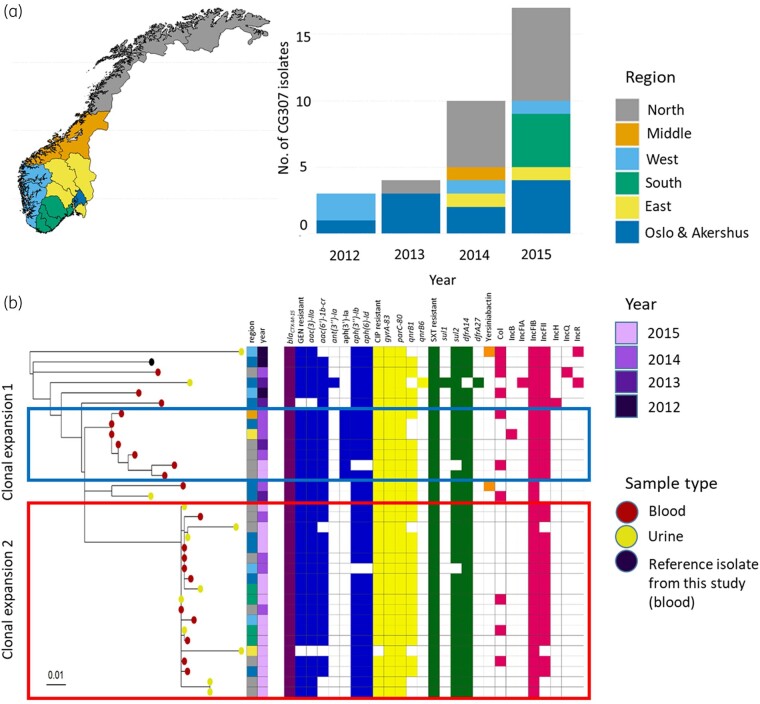
CG307 was first observed in 2012 in two of the six NORM surveillance regions, and was present in five regions by 2014 (Figure 3a), representing 44.4% of blood ESBL isolates and 34.6% of urine ESBL isolates in 2015. CG70, CG258 and CG45 emerged in the same period, but did not expand to the same degree. Chromosomal SNPs, temporal and geographical distribution of the most prevalent CGs are shown in Table S2. A core genome phylogenetic analysis of CG307 indicated unrelated occurrences of isolates in 2012–13, while isolates from 2014–15 seem to represent two clonal expansions (red and blue boxes in Figure 3b). This is supported by the Bayesian phylodynamic analysis of global CG307 (including 30 of the genomes reported here), adapted from Wyres et al.,6 showing that the most recent common ancestor for the two proposed clonal expansions dates back to 2009 (Figure S3).
Figure 3.
Epidemiology and phylogeny of CG307. (a) Map of Norway with surveillance regions as defined by the Norwegian surveillance programme on resistant microbes (NORM), with bar plot showing number of CG307 isolates per region per year. (b) CG307 core genome phylogeny with metadata, distribution of AMR determinants, virulence determinants and replicon families. Red and blue boxes mark proposed clonal expansions. A study isolate from blood was used as reference isolate (Genbank accessionCP073627).
Antimicrobial resistance: phenotype and genotype
All ESBL group isolates had an ESBL phenotype, whereas five isolates had no detectable ESBL genotype.
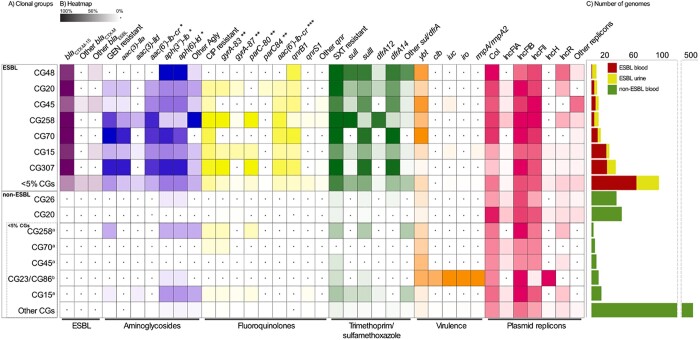
Overall, blaCTX-M-15 was the most prevalent genotype (n = 146; 72.6%), and was more dominant in high prevalence CGs (93.5%) compared with other CGs (55%, P < 0.001). The remaining ESBL genotypes comprised blaTEM-3 (n = 2; 2.0%) and variants of blaCTX-M (n = 22; 10.9%) and blaSHV (n = 22; 10.9%) (Figure 4 and Table S3). Reduced susceptibility to meropenem was found in two ESBL isolates (CG258) and one non-ESBL isolate (CG515), all without carbapenemase genes.
Figure 4.
Distribution of resistance determinants, virulence factors and plasmid replicons among clonal groups (CGs). For data see Table S3. (a) Distribution of CGs in the ESBL and non-ESBL groups, separated by most prevalent CGs (each representing ≥5% of isolates in group) and other CGs. In the non-ESBL group, <5% CGs which were the (i) most prevalent CGs from the ESBL group and the (ii) hypervirulence-associated CG23 and CG86 are shown separately. (b) The intensity of the box shading indicates the percentage of genomes harbouring the respective determinant. White shading with a black dot indicates that there are no determinants present. For each antibiotic class, the presence of resistant phenotype and resistance determinants are indicated. GEN, gentamicin, CIP, ciprofloxacin, SXT, trimethoprim/sulfamethoxazole, Agly, acquired aminoglycoside resistance genes; *, does not confer resistance to gentamicin; **, chromosomal mutation position; ***, may reduce susceptibility to both aminoglycosides and fluoroquinolones. (c) Total number of genomes in the ESBL and non-ESBL groups.
Prevalent AMR determinants are shown in Figure 4 and Table S3. Of note, armA (n = 1) and rmtG (n = 1) encoding 16S-rRNA methylases were rare, while aac(6′)-Ib-cr, which may reduce susceptibility to both aminoglycosides and quinolones was found in 48.3% (n = 97) of the ESBL isolates. Interestingly, aac(6')-Ib-cr was frequently found along with other determinants, in particular in 82.5% (n = 66) and 84.0% (n = 68) of isolates carrying aac(3)-IIa and qnrB1, respectively. Colistin resistance determinants were rare. Three ESBL isolates carried mcr-9.1 (n = 2) or a truncated chromosomal mgrB gene (n = 1) of these only the isolate with truncated mgrB showed an elevated MIC of 16 mg/L for colistin.
Phenotypic MDR was frequent in the ESBL group, (70.1%, n = 141) compared with the non-ESBL group (0.3%, n = 2), (P < 0.001). Notably, 83.0% (n = 117) of MDR ESBL isolates were carrying blaCTX-M-15. Only 11.2% (n = 18) of ESBL isolates were susceptible to all three of gentamicin, ciprofloxacin and trimethoprim/sulfamethoxazole.
Diversity in capsule loci and virulence determinants
Capsule loci (KLs) were identified in 73.6% (n = 148, 38 KLs) of the ESBL isolates and 54.3% (n = 362, 87 KLs) of the non-ESBL isolates. KL102 (13.4%, n = 27) was the most prevalent KL among the ESBL isolates, mainly associated with CG307 (n = 25). The HV-associated KL1 and KL2 were rare, being detected in 5.5% of ESBL (KL2, n = 11) and 3.7% of non-ESBL (KL1, n = 11; KL2, n = 14) isolates, respectively (Table S4).
Eighteen isolates (2.1%), all K. pneumoniae sensu stricto, met the HV definition, of which 7 of 15 non-ESBL HV isolates belonged to CG23 (Table S5). However, hypervirulence was found in three ESBL isolates. Long-read sequencing of one CG133 (ST420) (Genbank accessionCP073783-CP073787) ESBL isolate showed blaCTX-M-15 to be situated on an IncFII plasmid without any of the virulence loci, while the remaining two ST15 isolates (Genbank accessionCP034045-CP034052 and CP034053-CP034057) have previously been reported by Lam et al.41 to carry virulence genes and blaCTX-M-15 situated on the same plasmid.
The distribution of virulence determinants is shown in Figure 4, Table S3 and Table S5. Yersiniabactin was the most prevalent acquired siderophore in K. pneumoniae sensu stricto isolates, and more dominant in ESBL isolates (37.8%, n = 75) compared with non-ESBL isolates (17.3%, n = 80) (P < 0.001). Notably, among K. pneumoniae sensu stricto non-ESBL isolates, there was higher prevalence of yersiniabactin in CGs that were also found in the ESBL group compared with other CGs (29.4% and 11.0% respectively, P < 0.0009). Only 5/163 (3.1%) non-ESBL K. variicola spp. variicola had yersiniabactin. While no ESBL isolates had the genotoxin colibactin, it was present in 11 non-ESBL isolates (CG23, n = 7; CG133, n = 2; CG417; CG643).
Plasmid replicon patterns in ESBL and non-ESBL KpSC populations
Fifteen plasmid replicon families were identified in the ESBL group. IncFIB (87.9%, n = 176) and IncFII (82.8%, n = 164) were the most common. In the non-ESBL group, fourteen plasmid replicon families were identified dominated by IncFIB (72.0%, n = 480), IncFII (48.7%, n = 325), IncFIA (15.0%, n = 100), and IncR (14.5%, n = 97). Twelve plasmid replicon families were found in both groups (Table S6).
Interestingly, IncFIB and IncFII were more abundant in K. pneumoniae sensu stricto in the ESBL group compared with the non-ESBL group (P < 0.001 and P < 0.001, respectively). Additionally, in the non-ESBL group IncFIB and IncFII were significantly more common in K. pneumoniae sensu stricto compared with the other species (P < 0.001 and P < 0.03, respectively, Table 2).
Table 2.
Distribution of prevalent replicon types in Klebsiella pneumoniae sensu stricto and K. variicola ssp. variicola
| ESBL |
Non-ESBL |
P valuea |
|||
|---|---|---|---|---|---|
| Replicon type | (A) K. pneumoniae sensu stricto (n = 198) | (B) K. pneumoniae sensu stricto (n = 461) | (C) K. variicola ssp. variicola (n = 163) | A vs B | B vs C |
| Col | 63.1% (125) | 63.3% (292) | 47.9% (78) | NS | 0.006 |
| IncFIA | 6.1% (12) | 14.3% (66) | 17.8% (29) | 0.004 | NS |
| IncFIB | 87.9% (174) | 79.4% (366) | 58.3% (95) | 0.001 | 0.001 |
| IncFII | 82.8% (164) | 54.2% (250) | 40.5% (66) | 0.001 | 0.03 |
| IncR | 18.2% (36) | 14.1% (65) | 12.3% (20) | NS | NS |
Fisher exact test with Benjamini & Hochberg correction for multiple testing. NS, not significant.
Discussion
We have used the nationwide Norwegian AMR surveillance framework to perform a population structure analysis of all ESBL-producing KpSC blood and urine isolates as well as a representative collection of non-EBSL blood isolates from 2001–15 for comparison. The combined use of WGS and national registry data allowed the analysis of temporal and geographical trends in the species distribution, phylogeny, AMR and virulence determinant content in KpSC clinical isolates during a period when ESBL-producing Enterobacterales gained a foothold in Norway.
Firstly, we noted a significant difference in species distribution between the ESBL and non-ESBL group. While the ESBL group was essentially dominated by K. pneumoniae sensu stricto, K. variicola ssp. variicola accounted for 24.5% of the non-ESBL group. This is in line with findings in ESBL-producing or MDR KpSC strain collections from the USA42 and the British Isles,42,43 dominated by K. pneumoniae sensu stricto. Two studies (Sweden 2007–09, single centre16 and Japan 2014–17, two centre44) of consecutive blood KpSC isolates, both with low prevalence of ESBL, showed similar species distributions compared with our results.
As gastrointestinal colonization is considered the primary source of the majority of KpSC bloodstream infections,45,46 we expect that the observed proportion of K. variicola ssp. variicola among blood isolates reflects the ratio of gut colonization in the patient population. This is supported by a recent Norwegian study where 16.3% of 2975 healthy adults had KpSC in faecal screening samples, of which 28% were K. variicola spp. variicola.47 While K. pneumoniae sensu stricto is the predominant KpSC species reported in other gut carriage studies, carriage rates in the range of 10%–20% of K. variicola have been shown for intensive care patients45 and pregnant women in low-income countries.39 As a frequent gut resident, one could expect K. variicola spp. variicola to acquire ESBL-encoding plasmids and genes in vivo. To our knowledge there are no experimental studies supporting any mechanisms explaining the low abundance of ESBL in K. variicola spp. variicola compared with K. pneumoniae sensu stricto.
ESBL rates increased in clinical KpSC isolates during the study period, from 0% in 2001 to 3.1% in 2015.18 Our temporal data show an increasing predominance of blaCTX-M-15 accompanied by an increasing co-resistance to other clinical important antibiotics. An overall increase in ESBL and MDR rates in clinical isolates of KpSC was observed in other European countries in the same period.48 AMR determinants, as well as phenotypic resistance against gentamicin, ciprofloxacin and trimethoprim-sulfamethoxazole, were rare among non-ESBL isolates in our study. This observation strongly suggests that the overall increase in Norwegian MDR KpSC is driven by the expansion of ESBL-producing K. pneumoniae sensu stricto, in particular blaCTX-M-15 carrying CGs, such as CG15 and CG307.
Our KpSC strain collection is characterized by a large clonal diversity, both in the ESBL and non-ESBL groups, throughout the study period. The most striking shift in the temporal data, in addition to increasing ESBL rates, is the introduction and subsequent spread of CG307 since 2012. The CG307 phylogenetic analyses based on our dataset reveal nationwide expansion of this clone, which seems to be closely related to international isolates as previously shown by Wyres et al.6 Notably, we have not observed non-ESBL CG307. In contrast, clinical isolates of CG15, a frequent carrier of ESBLs, were also present without ESBLs.
The observed clonal diversity within the ESBL group and the increasing abundance of CG307 and CG15, is in line with results reported by Moradigaravand et al.43 in the British isles (2001–11) and Long et al.42 in the USA (2011–15). In contrast to their findings, CG258 was less frequently detected in our study. CG307 is still playing an important role in the dissemination of ESBL KpSC in Norway, as confirmed by WGS of blood culture ESBL isolates reported to NORM in 2019, where CG307 (29.2%) remains the dominant CG.18 While the prevalence of carbapenemase-producing KpSC in Norway is still low,18 the establishment of ESBL CG307 is a cause for concern, as this clone has proved to be a well prepared host for carbapenemase genes in other settings.7
Our results suggest that HV KpSC, including CG23 and CG86, are rare in clinical isolates in Norway. This is also in line with the recent population structure analysis of Norwegian KpSC faecal carrier isolates.47 Yersiniabactin, usually transferred by integrative conjugative elements (ICEs),49 is the most prevalent virulence-associated gene, mainly found in K. pneumoniae sensu stricto isolates. Notably, we observed a significantly higher prevalence of yersiniabactin in ESBL (37.8%) compared with non-ESBL (17.3%) K. pneumoniae sensu stricto isolates, in contrast to previously published results.49 Other virulence determinants were uncommon and played no dominant role in the examined KpSC population. Convergence of ESBL and hypervirulence were found in only three isolates.
Our data demonstrate that the replicon families IncFIB and IncFII, frequently associated with ESBL genes,50 were common both in ESBL-carrying (87.9% and 82.8%) and non-ESBL-carrying (79.7% and 54.2%) K. pneumoniae sensu stricto isolates. These replicons were also present in K. variicola ssp. variicola (58.3% and 40.5%), but were significantly less prevalent compared with non-ESBL K. pneumoniae sensu stricto.
Importantly, our data support the notion that there seems to be a higher propensity for certain K. pneumoniae sensu stricto clonal groups to acquire mobile genetic elements, represented by ESBL-encoding plasmids and yersinabactin-linked ICEs, compared with K. variicola ssp. variicola. This is concordant with Wyres et al.13 showing some KpSC clones to be generally better at acquiring genetic material via horizontal gene transfer than others. These observations need further investigation, including experimental studies on the underlying mechanisms.
The strengths of this study lie in the use of the comprehensive unselected national surveillance data collected over a 15 year period, encompassing the introduction of ESBLs, and the use of WGS to gain detailed insight into genomic epidemiological features. As we opted for temporal and geographical diversity, we have not done a randomized selection of non-ESBL isolates, which may have introduced a bias in estimating the prevalence of significant CGs or genetic determinants in recent years. The lack of urine ESBL isolates in the periods 2004–08 and 2010–11 may also conceal the early appearance of significant CGs.
In conclusion, the increase of ESBLs and clinically relevant co-resistance in K. pneumoniae sensu stricto in Norway during the study period is closely linked to blaCTX-M-15-carrying CGs, where CG307 and CG15 have played key roles. Yersinabactin and ESBL-encoding mobile genetic elements are uncommon in clinical isolates of K. variicola ssp. variicola compared with K. pneumoniae sensu stricto. Susceptible K. variicola ssp. variicola, however, is a significant pathogen causing one out of every four cases of KpSC bacteraemia in Norway.
Supplementary Material
Acknowledgements
We thank The Norwegian Surveillance Program on Resistant Microbes (NORM) for making data available for this study and the participants in The Norwegian Study Group on Klebsiella pneumoniae for providing isolates.
Members of the Norwegian Study Group on Klebsiella pneumoniae
Ståle Tofteland, Paul Christoffer Lindemann, Nina Handal, Åshild Marvik Rødland, Aleksandra Jakovljev, Sandra Åsheim, Karianne Wiger Gammelsrud, Rolf Arne Sandnes, Einar Tollaksen Weme, Angela Kümmel, Einar Nilsen, Belinda Langnes Lindstad, Anne C. Hollekim, Reidar Hjetland, Anne R. Oseid, Liv Jorunn Hafne.
Funding
This work was supported the Western Norway Regional Health Authority (fellowship numbers 912037 to I.H.L. and 912119 to A.F., and grant number 912050 to I.H.L.).
Transparency declarations
None to declare.
Author contributions
Conceived the study: A.F., Ø.S., A.S., G.S.S., I.H.L.; whole genome sequencing: R.B. and E.B.; data analysis: A.F. and M.A.K.H.; manuscript: A.F. and M.A.K.H. All authors contributed to data interpretation, read and commented on the manuscript. Study group members provided isolates and commented on the final manuscript.
Supplementary data
Figures S1 to S3 and Tables S1 to S6 are available as Supplementary data at JAC Online.
Contributor Information
The Norwegian Study Group on Klebsiella pneumoniae:
Ståle Tofteland, Paul Christoffer Lindemann, Nina Handal, Åshild Marvik Rødland, Aleksandra Jakovljev, Sandra Åsheim, Karianne Wiger Gammelsrud, Rolf Arne Sandnes, Einar Tollaksen Weme, Angela Kümmel, Einar Nilsen, Belinda Langnes Lindstad, Anne C Hollekim, Reidar Hjetland, Anne R Oseid, and Liv Jorunn Hafne
References
- 1. Podschun R, Ullmann U.. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998; 11: 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Navon-Venezia S, Kondratyeva K, Carattoli A.. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 2017; 41: 252–75. [DOI] [PubMed] [Google Scholar]
- 3. Tacconelli E, Carrara E, Savoldi A. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018; 18: 318–27. [DOI] [PubMed] [Google Scholar]
- 4. Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 5. Wyres KL, Holt KE.. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 2016; 24: 944–56. [DOI] [PubMed] [Google Scholar]
- 6. Wyres KL, Hawkey J, Hetland MAK. et al. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother 2019; 74: 577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Villa L, Feudi C, Fortini D. et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom 2017; 3: e000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. David S, Cohen V, Reuter S. et al. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci USA 2020; 117: 25043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laxminarayan R, Duse A, Wattal C. et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013; 13: 1057–98. [DOI] [PubMed] [Google Scholar]
- 10. Russo TA, Marr CM.. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 2019; 32: e00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lam MMC, Wyres KL, Duchêne S. et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 2018; 9: 2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bialek-Davenet S, Criscuolo A, Ailloud F. et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 2014; 20: 1812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wyres KL, Wick RR, Judd LM. et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet 2019; 15: e1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodrigues C, Passet V, Rakotondrasoa A. et al. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res Microbiol 2019; 170: 165–70. [DOI] [PubMed] [Google Scholar]
- 15. Rodrigues C, Passet V, Rakotondrasoa A. et al. Identification of Klebsiella pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and Related Phylogroups by MALDI-TOF Mass Spectrometry. Front Microbiol 2018; 9: 3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maatallah M, Vading M, Kabir MH. et al. Klebsiella variicola is a frequent cause of bloodstream infection in the stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS One 2014; 9: e113539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. EUCAST. Clinical breakpoints - breakpoints and guidance. https://www.eucast.org/clinical_breakpoints/.
- 18. NORM/NORM-VET 2019. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. 2020. https://www.vetinst.no.
- 19. Tofteland S, Haldorsen B, Dahl KH. et al. Effects of phenotype and genotype on methods for detection of extended-spectrum-β-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J Clin Microbiol 2007; 45: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krueger F. TrimGalore. https://github.com/FelixKrueger/TrimGalore.
- 21. Wick RR, Judd LM, Gorrie CL. et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walker BJ, Abeel T, Shea T. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014; 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lam MMC, Wick RR, Watts SC. et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 2021; 12: 4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alcock BP, Raphenya AR, Lau TTY. et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 2020; 48: D517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wyres KL, Wick RR, Gorrie C. et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2016; 2: e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inouye M, Conway TC, Zobel J. et al. Short read sequence typing (SRST): multi-locus sequence types from short reads. BMC Genomics 2012; 13: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carattoli A, Zankari E, García-Fernández A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holt K, RedDog. https://github.com/katholt/reddog.
- 30. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Handsaker B, Wysoker A. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Price MN, Dehal PS, Arkin AP.. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valencia-Bacca J, Silva MM, Cerdeira L. et al. Detection and Whole-Genome Analysis of a High-Risk Clone of Klebsiella pneumoniae ST340/CG258 Producing CTX-M-15 in a Companion Animal. Microb Drug Resist 2020; 26: 611–5. [DOI] [PubMed] [Google Scholar]
- 34. Guppy. https://community.nanoporetech.com.
- 35. Wick RR. Filtlong. https://github.com/rrwick/Filtlong.
- 36. Tatusova T, DiCuccio M, Badretdin A. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 2016; 44: 6614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Croucher NJ, Page AJ, Connor TR. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huynh BT, Passet V, Rakotondrasoa A. et al. Klebsiella pneumoniae carriage in low-income countries: antimicrobial resistance, genomic diversity and risk factors. Gut Microbes 2020; 11: 1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. R Foundation for Statistical Computing. R: A language and environment for statistical computing. 2020. https://www.r-project.org/
- 41. Lam MMC, Wyres KL, Wick RR. et al. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J Antimicrob Chemother 2019; 74: 1218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Long SW, Olsen RJ, Eagar TN. et al. Population genomic analysis of 1,777 extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: unexpected abundance of Clonal Group 307. MBio 2017; 8: e00489-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moradigaravand D, Martin V, Peacock SJ. et al. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio 2017; 8: e01976-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Imai K, Ishibashi N, Kodana M. et al. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014-2017. BMC Infect Dis 2019; 19: 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gorrie CL, Mirceta M, Wick RR. et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 2017; 65: 208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin RM, Cao J, Brisse S. et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 2016; 1: e00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raffelsberger N, Hetland MAK, Svendsen K. et al. Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microbes 2021; 13: 1939599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. ECDC. Antimicrobial resistance surveillance in Europe 2015. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). 2017. https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2015.
- 49. Lam MMC, Wick RR, Wyres KL. et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 2018; 4: e000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol 2013; 303: 298–304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 868 KpSC short-read and four long-read sequence files have been deposited in the European Nucleotide Archive under BioProject PRJEB27256 (Table S1). The four hybrid-assembled completed genomes have been deposited in GenBank (Table S1) under accession numbers CP073791-CP073796, CP073627-CP073629, CP073783-CP073787 and CP073788-CP073790.