Abstract
Background
Echinocandin resistance represents a great concern, as these drugs are recommended as first-line therapy for invasive candidiasis. Echinocandin resistance is conferred by mutations in FKS genes. Nevertheless, pathways are crucial for enabling tolerance, evolution, and maintenance of resistance. Therefore, understanding the biological processes and proteins involved in the response to caspofungin may provide clues indicating new therapeutic targets.
Objectives
We determined the resistance mechanism and assessed the proteome response to caspofungin exposure. We then evaluated the phenotypic impact of calcineurin inhibition by FK506 and cephalosporine A (CsA) on caspofungin-resistant Candida glabrata isolates.
Methods
Twenty-five genes associated with caspofungin resistance were analysed by NGS, followed by studies of the quantitative proteomic response to caspofungin exposure. Then, susceptibility testing of caspofungin in presence of FK506 and CsA was performed. The effects of calcineurin inhibitor/caspofungin combinations on heat stress (40°C), oxidative stress (0.2 and 0.4 mM menadione) and on biofilm formation (polyurethane catheter) were analysed. Finally, a Galleria mellonella model using blastospores (1 × 109 cfu/mL) was developed to evaluate the impact of the combinations on larval survival.
Results
F659-del was found in the FKS2 gene of resistant strains. Proteomics data showed some up-regulated proteins are involved in cell-wall biosynthesis, response to stress and pathogenesis, some of them being members of calmodulin–calcineurin pathway. Therefore, the impact of calmodulin inhibition was explored. Calmodulin inhibition restored caspofungin susceptibility, decreased capacity to respond to stress conditions, and reduced biofilm formation and in vivo pathogenicity.
Conclusions
Our findings confirm that calmodulin-calcineurin-Crz1 could provide a relevant target in life-threatening invasive candidiasis.
Introduction
The echinocandins are recommended as first-line therapy for invasive candidiasis because of their low toxicity and high efficacy, especially against azole-resistant Candida isolates.1,2 Echinocandin resistance in Candida spp is associated with treatment failures, and is conferred by mutations in ‘hot spot’ regions of the target FKS genes that lead to amino acid substitutions in the 1,3-β-d-glucan synthase enzyme.3,4 In Candida albicans, substitutions in Ser641 and Ser645 are the most frequent FKS mutations and cause the most pronounced resistance phenotypes.5 In Candida glabrata, the principal reported substitutions were Ser629 in Fks1, and Ser663 and Phe659 in Fks2,3,6,7 while natural polymorphisms have been described in Candida parapsilosis and Candida guilliermondii FKS1.8 The ultimate consequence of these mutations is a significant decrease in the echinocandin affinity for the enzyme target and high MIC values. It is important to highlight that in some resistant isolates, no FKS mutations were identified and isolates with the same FKS mutations exhibit different resistance profiles indicating that other resistance mechanisms and putative target genes may be implicated.9
In addition to these described resistance mechanisms, there is also a new hypothesis in which regulators of cellular stress responses could be crucial for enabling the evolution and maintenance of drug resistance.4,10,11 Indeed, some cellular stress responses are governed by signalling pathways, the most-studied pathways being cAMP, calmodulin-calcineurin (CaM/CaL), TOR (target of rapamycin), and mitogen-activated protein kinase (MAPK).11–14 The CaM/CaL pathway, formed by a complex of the proteins Cnb1, Cna1, Hsp90 and the transcription factor Crz1 in yeast, is involved in calcium homeostasis, cell-wall biosynthesis, protein trafficking, adaptation to environmental changes and even more importantly, in the response to antifungal drug pressure.11,15,16 Crz1 is found downstream in the CaM/CaL pathway and is one of the main antifungal targets since Crz1 is not present in human cells. Once the transcription factor is activated by dephosphorylation, mediated by the CaM/CaL-Hsp90 complex, it moves to the cell nucleus. Crz1 contains a C2H2 zinc finger motif that binds to a specific calcineurin-dependent response element (CDRE) in the gene promoters; in C. glabrata Crz1 initiates activation of ∼87 genes, among which is the FKS2 gene, which is involved in resistance to caspofungin.9,15
The spread of antifungal resistance and the limited number of available antifungal drugs amplifies the need to identify new fungal targets for development of novel therapeutic alternatives. Calcineurin inhibitors such as tacrolimus (FK506) and cyclosporine A (CsA), which bind to the immunophilins FKBP12 and cyclophilin A, respectively, are well recognized as immunosuppressive drugs with potential antifungal properties.17,18
In attempts to identify protein targets, proteomic approaches should be employed. This approach has been previously applied in C. albicans to study many aspects, including adaptive responses to osmotic stress, macrophage interaction, and antifungal exposure. These studies evidenced that pathways such as the MAPK signalling pathway played a significant role in several biological responses.19–24
To our knowledge, few proteomic studies have been done in C. glabrata. Among them, studies explored the implication of hyperadhesive proteins in host–pathogen interaction and biofilm formation, mechanisms of drug resistance (mainly in biofilms) and response to antifungal drugs such as clotrimazole and 5-flucytosine.25–28 However, similar approaches to study the antifungal response to caspofungin have not been previously described.
Based on this, the first objective of the present study was to identify resistance mechanisms in echinocandin-resistant C. glabrata clinical isolates. We then investigated the proteomic response to caspofungin exposure and determined the impact of calcineurin inhibition on susceptibility, stress tolerance, biofilm formation, and assessed pathogenicity in the Galleria mellonella model.
Materials and methods
Microorganisms
One susceptible and two caspofungin-resistant C. glabrata isolates were studied. The first one, C. glabrata PUJ/HUSI 0916 was recovered from a blood culture of a haematopoietic stem cell transplant recipient admitted to the Hospital Universitario San Ignacio Bogota, Colombia. The caspofungin-resistant isolates CAGL1875 and CAGL1256 were obtained from blood and urine cultures of hospitalized patients in the ICU of the Centre Hospitalier Universitaire de Nantes, France and identified by ITS sequencing in a previous study by our research team.11,29 Isolates were categorized as susceptible or resistant to caspofungin according to the interpretative breakpoints of CLSI M60 (MIC >0.5 mg/L indicates a resistant strain). In addition, the reference C. glabrata ATCC 2001, C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 were used in specific experiments, as described in the results.
Sequencing and identification of molecular resistance mechanism
Twenty-five genes associated with antifungal drug resistance (Table 1) of C. glabrata isolates were sequenced using an Illumina paired-end sequencing platform with a read length of 300 bp and an average read depth coverage of 300×. The obtained raw read sequences were cleaned using fastp30 and assembled with SPAdes v3.12.0.31 Gene prediction was conducted with Prodigal V2.6.332 and the coding sequences obtained were annotated using blastn,33 against the genome of C. glabrata ATCC 2001 reference strain. For each of these genes, multiple sequence alignment (nucleotide and amino acid) and mutation identification was performed using T-Coffee34 and JalView32 for visualization.
Table 1.
Evaluation of genes associated with caspofungin resistance
| Gene symbol | Systematic name | Mutations found | Mutation in resistant isolates (1875–1256) | Resistance-associated mutation (+/−) |
|---|---|---|---|---|
| CEK1 | CAGL0K04169g | – | – | − |
| CDC55 | CAGL0L06182g | – | – | − |
| CDC6 | CAGL0K00605g | R117K, V163A, K268R, R80K | – | − |
| DOT6 | CAGL0I05060g | P104S | – | − |
| FKS1 | CAGL0G01034g | G14S | – | − |
| FKS2 | CAGL0K04037g | F659-Del, T926P | F659-Del | + |
| FKS3 | CAGL0M13827g | A42V, T1676S | – | − |
| MKT1 | CAGL0J05566g | N512K, A643T | – | − |
| MOH1 | CAGL0F04631g | S15N | – | − |
| MPH1 | CAGL0F04895g | – | – | − |
| MRPL11 | CAGL0J09724g | – | – | − |
| MSH2 | CAGL0I07733g | – | – | − |
| PDR1 | CAGL0A00451g | V91I, L98S, D243N | – | − |
| PHO4 | CAGL0D05170g | S327N | – | − |
| SNQ2 | CAGL0I04862g | P1104H | – | − |
| SUI2 | CAGL0B03795g | – | – | − |
| TCB1 | CAGL0J08591g | Q437E, K585R, N622K | – | − |
| TCB3 | CAGL0L11440g | – | – | − |
| TOD6 | CAGL0A04257g | P64S, D81N, N85D | – | − |
| TPK2 | CAGL0M08404g | T132A, T158A | – | − |
| CRZ1 a | CAGL0M06831g | – | – | − |
| SLT2 a | CAGL0J00539g | – | – | − |
| SRP1 a | CAGL0J11440g | – | – | − |
| DBP5 a | CAGL0L110021g | – | – | − |
| SWI1 a | CAGL0C01683g | – | – | − |
Genes encoding proteins related to the caspofungin exposure response found in this study.
Computational transmembrane region predictions were carried out using RaptorX,35,36 Sable,37–40 TMHMM v.2.0,41 TOPCONS,42 TMpred,43 CCTOP,44,45 HMMTOP46 and Phobius.47 InterProScan548 was used for primary protein structure analysis.
Proteomic analysis
CAGL1875 (one of the two resistant isolates, both have the same mutation) was resuspended into both fresh yeast extract peptone dextrose (YPD) broth (10 g/L yeast extract, 20 g/L bacto peptone, 20 g/L dextrose) and YPD plus caspofungin at 5 mg/L (OD600nm 0.3) and incubated at 30°C for 7 h under shaking until achieving a density of 107cells/mL (OD 0.8–1.0). Fifty mL of cell culture was collected and processed for sample preparation; four biological replicates were performed under each condition. Cell viability was assessed through propidium iodide staining. Cell extracts were obtained by suspending cells in lysis buffer and disrupting them by centrifugation with glass beads (0.4–0.6 mm diameter) in a Fast-Prep system (Bio101, Savant) applying five 20 s rounds at 5.5 speed with intermediate ice cooling. Cell extracts were separated from glass beads by centrifugation and the supernatant was collected and cleared by centrifugation.49 Protein concentration was measured by Bradford protein assay. Digestion and desalting of peptides were carried out in gel with trypsin, according to Sechi and Chait.50
The desalted protein digest was analysed by RP-LC-ESI-MS/MS in an EASYnLC1000 System coupled to the Q-Exactive-HF mass spectrometer through the Nano-Easy spray source (Thermo Scientific, Canada). Peptide identification was carried out using the Mascotv.2 search engine through the Protein Discoverer Software. Database search was performed against SwissProt and Mascot scores were adjusted using a percolator algorithm. Acceptance criteria for protein identification were a false discovery rate (FDR) <1% and at least one peptide identified with high confidence (CI > 95%). To determine the abundances of the identified peptides and proteins, a label-free processing workflow was initiated. As an estimation of the relative protein abundances the normalized spectral abundance factor (NSAF) was used, and the average of the normalized values was calculated.51 The recalibration of the masses was performed through a rapid search in Sequest HT against the database. Subsequently, alignment of the retention times between the different samples analysed for the quantification of the precursor ions was performed, taking into account unique peptides and razor peptides (i.e. peptides that can be assigned to more than one protein). Finally, the results were normalized to the total amount of the peptides, equalling the total abundance among the different samples. After the analyses were finalized, a final report presented the list of peptide groups and proteins with scaled abundances and selected ratios. The Proteome Discoverer application includes a feature for assessing the significance of differential expression by providing P values for those ratios (P value <0.05). The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE,52,53 with dataset identifier PXD021578.
Gene ontology (GO) FungiDB (http://fungidb.org) was used to search for enriched GO terms in the input list of the identified C. glabrata gene products compared with the genes from the C. glabrata CBS138 genome. Terms with a P value <0.04 from a calculated and curated evidence list were included.
Antifungal susceptibility testing
Antifungal susceptibility testing was carried out using broth microdilution method (BMD), following the CLSI M27-A3 guidelines with slight modifications for the combination of caspofungin with the calcineurin inhibitors.54 Briefly, isolates were subcultured on yeast YPD and grown for 24 h at 35°C. The yeast suspensions were prepared in Roswell Park Memorial Institute medium (RPMI) 1640 (Sigma–Aldrich) to a final concentration of 103 cells/mL. Yeast inoculum (100 μL) was added to a 96-well plate containing serial two-fold dilutions of caspofungin with or without inhibitors FK506 or CsA (15 mg/L, which was the concentration selected after screening that did not generate significant growth changes and did not show toxicity). MICs were visualized, and densitometry (530 nm, microplate reader, Thermo Scientific) was used to determine the lowest concentration of drug that caused a significant decrease (MIC/2 or ≥50%) compared with that of the drug-free growth control after 48 h of incubation. Quality control was ensured by testing the CLSI-recommended strains.55
Stress-related phenotypic assays
For heat-shock stress, droplet tests were performed by spotting serial dilutions of C. glabrata cells (106 to 103 cells/mL) onto YPD agar plates with FK506 or CsA (15 mg/L), caspofungin (1 mg/L) or both compounds. The plates were incubated at 37°C or 40°C for 24 h. For oxidative stress, YPD plates were prepared as previously, except that the medium was supplemented with the naphthoquinone menadione (0.2 and 0.4 mM). The plates were incubated at 37°C for 24 h.11,24
Biofilm formation
The C. glabrata isolates were grown on Sabouraud dextrose agar (SDA) and incubated at 30°C for 24 h. Two hundred μL of Candida cell suspensions (106 cells/mL) in RPMI-1640 with MOPS were dispensed in 96-well microdilution wells with or without GDHK-1325 250 mm Gam-polyurethane catheter pieces (Hechingen, Germany) and allowed to adhere for 24 h at 37°C. The non-adherent cells were removed by washing with 300 μL PBS. Caspofungin was added at 1 mg/L with or without 15 mg/L of the calcineurin inhibitors for 24 h incubation at 37°C for the biofilm adhesion phase. Then wells or catheter pieces were washed twice with PBS and finally 100 μL of RPMI-1640 plus 10 μL of 700 μM resazurin (Sigma–Aldrich) was added to each well and incubated at 37°C for 4 h. The biofilm was quantified indirectly by measuring the fluorescent water-soluble resorufin product that results when resazurin is reduced by reactions associated with respiration. Fluorescence was then measured at 560 nm with emission at 590 nm. The results were expressed in arbitrary fluorescence units (AU).11,56
Invertebrate Galleria mellonella model
Killing assays were performed in G. mellonella as described by Fallon et al.57 Briefly, final (sixth) instar larvae weighing approximately 300 mg were used. Suspensions of individual Candida isolates that had been grown on SDA for 48 h at 35°C were harvested by gently scraping colony surfaces with sterile plastic loops, washed twice in sterile phosphate-buffered saline (PBS), counted in haemocytometers and adjusted to 1 × 109 cfu/mL. Larvae received a 10 μL inoculum and 10 μL of caspofungin (1 μg/larva), FK506 and CsA (15 mg/L), or their combination by injection into the last left and right proleg using a 0.5 mL BD syringe. After inoculation, larvae were placed in Petri dishes and incubated in darkness at 37°C. To compare mortality, three biological replicates were performed with 10 larvae for each isolate evaluated. A group of 10 larvae was used for each of the controls: absolute (uncleaned, uninoculated), disinfection (cleaned with ethanol 70%), and inoculation (received 10 μL sterile PBS). The larvae were monitored for 10 days, and survival outcome was determined; larvae were considered dead when no response was observed following touch stimulation.
Statistical analysis
All experiments were performed on three independent biological replicates; survival curves were constructed using the method of Kaplan and Meier, then the curves were compared using the Log-Rank (Mantel-Cox) test. Statistical models were constructed and analysed using PRISM software version 7.
Data availability
All experimental data are provided in the manuscript and in supplementary files, or are available via ProteomeXchange with identifier PXD021578 (10.6019/PXD021578) and in the NCBI BioProject database with the accession number PRJNA692260.
Results
Next-generation sequencing (NGS) revealed FKS2 nucleotide deletion
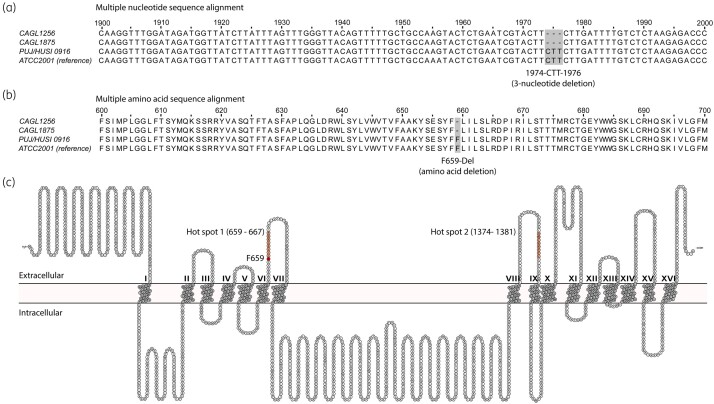
To identify the caspofungin resistance mechanisms of C. glabrata isolates, 25 genes associated with antifungal drug resistance were screened for mutations. Although some mutations were identified in our resistant isolates (CAGL1256 and CAGL1875), only the FKS2 gene exhibited a 3 nucleotide deletion (1974-CTT-1976), which has been previously reported to be associated with echinocandin-resistant phenotypes. All other observed mutations were found in both resistant and susceptible isolates (Table 1). The 3 nucleotide deletion detected (Figure 1a), which preserves the same open reading frame, explained the single amino acid deletion at Fks2 (F659-del; Figure 1b) observed in the two resistant isolates. This amino acid deletion, which confers resistance to the echinocandins, resides within the Fks2 hot spot 1 (Figure 1c).
Figure 1.
Multiple alignments of Fks2 (β-1,3-glucan synthase catalytic subunit 2) nucleotide and amino acid sequences from C. glabrata resistant and susceptible strains. Multiple nucleotide (a) and amino acid sequence alignments (b) that show a 3 nucleotide deletion (1974-CTT-1976) and a single amino acid deletion (F659-Del), respectively, occurring only in resistant strains of C. glabrata. (c) A consensus C. glabrata Fks2 membrane protein structure and topology model predicted by seven different membrane protein secondary structure prediction servers and visualized with Protter.88 Hot spots 1 and 2 are showed in red, as well the position F659 at which the single amino acid deletion associated with resistance to echinocandins occurs. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Proteomic analysis of resistant C. glabrata treated with caspofungin revealed an increase of proteins related to stress adaptation and cell-wall organization
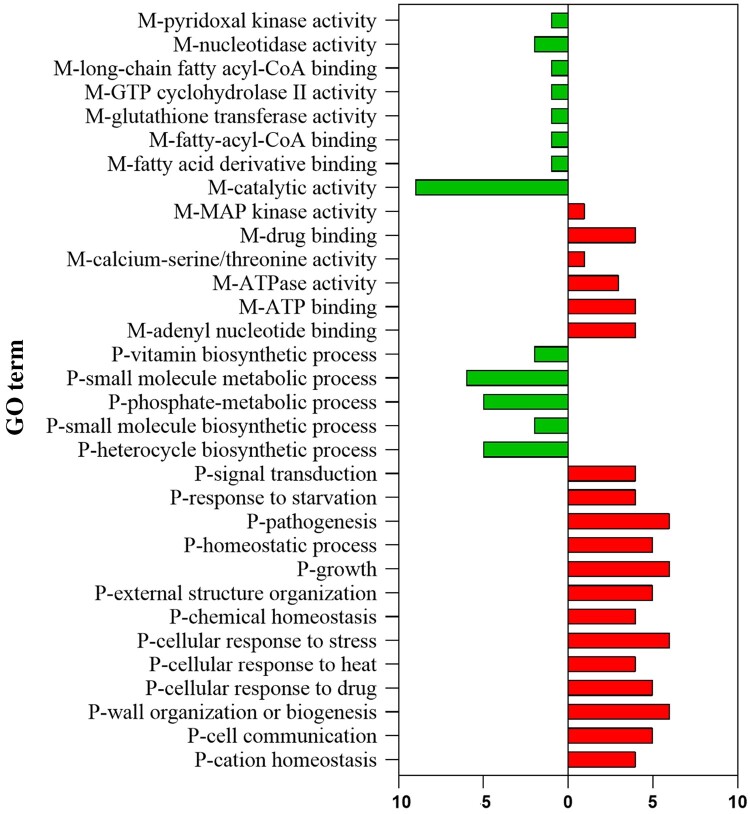
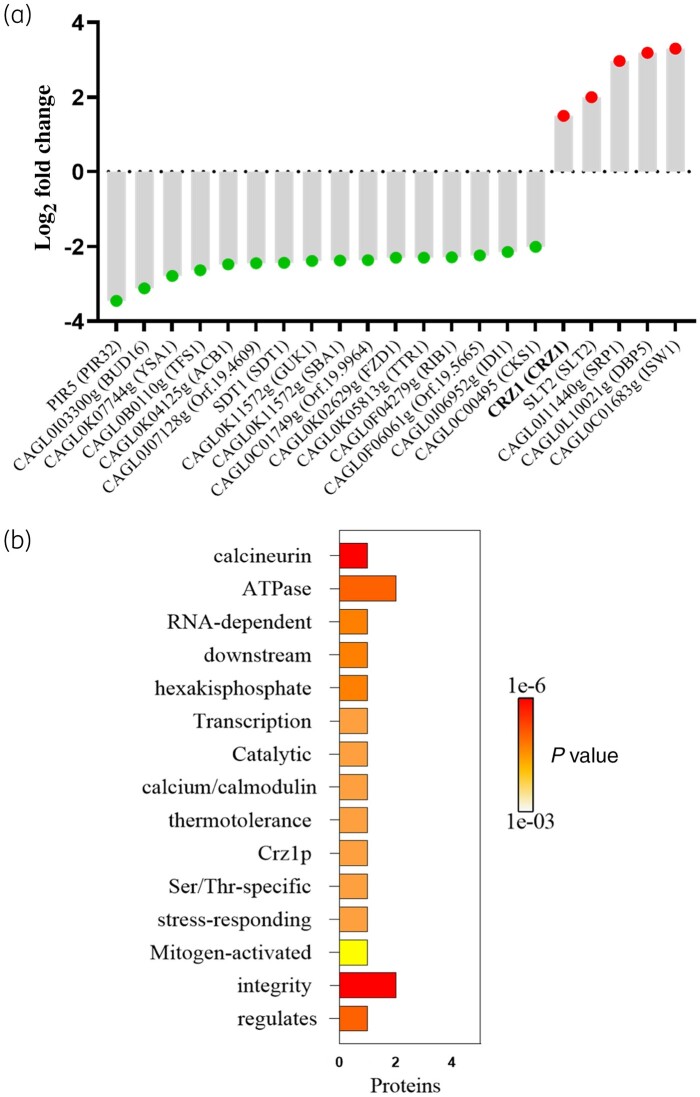
The CAGL1875 isolate was cultured at a concentration of caspofungin indicating resistance, then a label-free quantification proteomic method was performed to determine the number of proteins and level expression in response to caspofungin. A total of 1796 proteins were identified (Table S1, available as Supplementary data at JAC Online). While 1509 of them were encoded by uncharacterized ORFs, 287 were encoded by well-characterized genes. Among these proteins, 16 were identified as less abundant (i.e. downregulated) and 5 proteins were identified as more abundant (i.e. upregulated) after caspofungin exposure (>1.5-fold change and P value < 0.05). Using GO enrichment tools, several GO terms were found to be enriched from these 21 proteins. In the molecular functions, 14 terms were enriched. Among them, MAP-kinase activity, drug binding, calcium-serine/threonine, ATPase activity, and ATP-binding were over-represented. Regarding biological process categories, 18 terms were enriched, including signal transduction, pathogenesis, response to stress/drug and wall-biogenesis were over-represented (Figure 2). Following caspofungin exposure, the downregulated proteins were mainly of enzymatic groups: CAGL0I03300g (homologue to Candida albicans Bud16, here named as Ca.Bud16), CAGL0K07744g (Ca.Ysa1), CAGL0K05813g (Ca.Ttr1), CAGL0J06952g (Ca.Idi1), CAGL0H09218g (Cg.Sdt1). The protein with the most negative differential ratio was CAGL0M08514g (Cg.Pir5), a protein associated with β-1,3-glucan strengthening. The proteins that were more abundant after caspofungin exposure were involved in DNA binding, i.e. CAGL0M06831g (Cg.Crz1) CAGL0J11440g (Ca.Srp1), CAGL0L10021g (Ca.Dbp5) and CAGL0C01683g (Ca.Isw1). Of interest are the proteins involved in antifungal responses, CAGL0M06831g (Cg.Crz1) (CaM/CaL-pathway), CAGL0J00539g (Cg.Slt2) (PKC-pathway) and CAGL0J11440g (Ca.Srp1). The corresponding protein abundance is presented in Figure 3, and description of these proteins is provided in Table S2. An additional analysis was carried out with proteins identified as more abundant if the change in abundance ratio caspofungin: control was >1 after caspofungin exposure (Figures S1 and S2, and Table S3).
Figure 2.
Gene Ontology (GO) analysis of the proteins considered differentially abundant after caspofungin treatment (>1.5-fold change and p-value <0.05). M indicates a molecular function; P indicates a biological process; the x-axis indicates the number of associated proteins. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Figure 3.
Label-free quantitative proteomics results. (a) Protein abundance profile. Down-regulated proteins are marked with a green spot and up-regulated are marked with a red spot (Candida albicans orthologue names are given inside the parentheses). (b) Word enrichment that was created using the P values (Fisher's exact test) and the full terms from the enrichment analysis via a program called GO summaries available at the FungiDB website (https://fungidb.org/fungidb/app/). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Considering the important role of the CaM/CaL in antifungal response, and the significant change of Crz1 expression (to date there are no Crz1 inhibitors) after caspofungin exposure, we focused this study on targeting upstream CaM/CaL proteins using commercial inhibitors. Additionally, the genes corresponding to the five most-abundant proteins following caspofungin exposure were evaluated for mutations, however, none was observed (Table 1).
Calcineurin inhibition restored susceptibility of caspofungin-resistant C. glabrata
Pharmacological inhibition of calcineurin by FK506 and CsA did not show any statistically significant decrease of susceptible (PUJ/HUSI0916 and ATCC 2001) or resistant (CAGL1256 and CAGL1875) C. glabrata growth. Otherwise, in the presence of caspofungin, the inhibitors allowed susceptibility restoration in resistant clinical isolates, with a significant reduction in MIC values from >16 mg/L to 0.25 and 0.5 mg/L, respectively (Figure 4).
Figure 4.
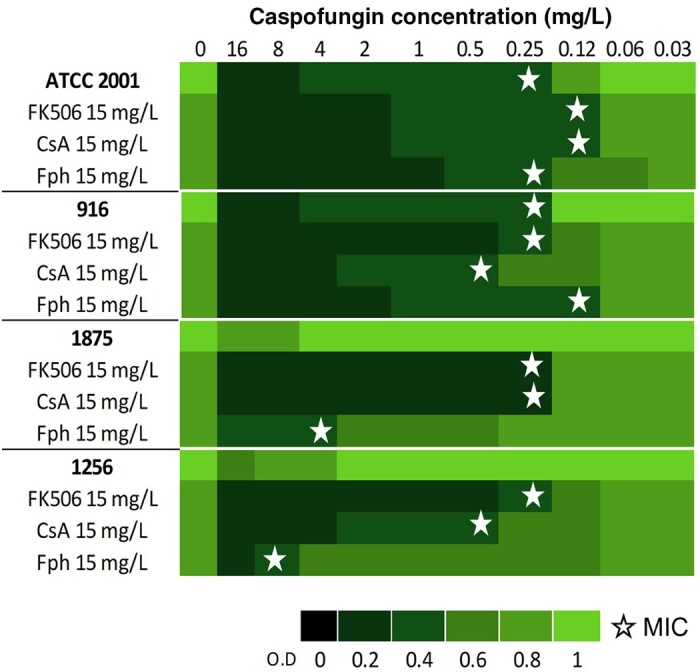
Caspofungin MICs (indicated by stars) under basal and calcineurin inhibition (FK506, CsA, Fph). Resistant strains have MICs >0.5 mg/L (1875 and 1256). The green bars indicate relative fold growth.
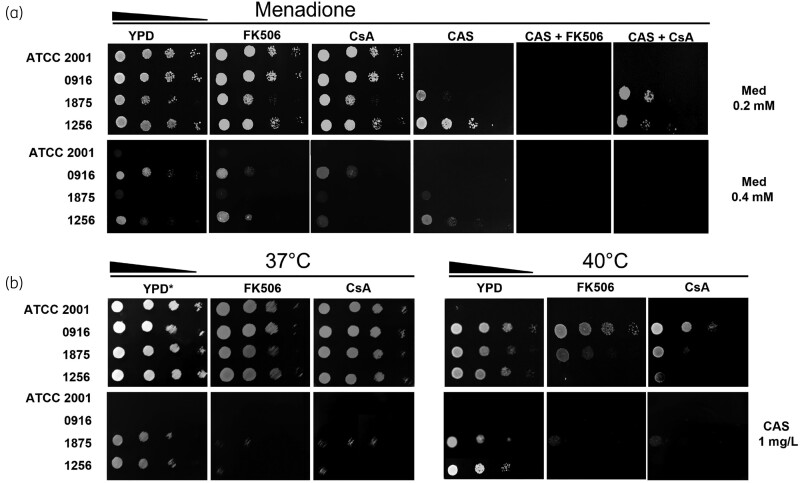
Response to oxidative stress is independent of caspofungin resistance phenotype and not correlated with calcineurin signalling
To understand the oxidative stress response in resistant C. glabrata, growth was assessed in the presence of menadione, a cytotoxic quinone that generates superoxide. The four isolates grew in up to 0.2 mM menadione and addition of FK506 and CsA did not show any significant modification. The significant growth reduction of the resistant isolate CAGL1875 suggested higher susceptibility to caspofungin in presence of 0.2 mM menadione. Since combinations of caspofungin and calcineurin inhibitors lead to complete growth inhibition without any additional stress, the impact of 0.2 mM menadione addition could not be interpreted. In the presence of 0.4 mM menadione, PUJ/HUSI0916 and CAGL1256 strains maintained a similar growth rate, independent of the caspofungin resistance phenotype (Figure 5a).
Figure 5.
Stress responses under CaM/CaL inhibition. (a) Strains grown in the presence of 0.2 and 0.4 mM menadione (Med). (b) Strains were streaked onto YPD and incubated at 37°C and 40°C, with or without 1 mg/L caspofungin (CAS), with or without 15 mg/L Fk506 and CsA. YPD at 37°C was used as no drug growth control (see the asterisk).
Calcineurin inhibitors compromised growth of caspofungin-resistant C. glabrata in heat-shock conditions
The spot test at 37°C confirmed previous BMD CLSI results concerning the effect of calcineurin inhibitors on caspofungin-resistant isolates (Figure 4). Heat shock at 40°C did not have an impact on isolate growth for all but ATCC 2001. Interestingly, at this temperature, the growth of caspofungin-resistant isolates was noticeably compromised by calcineurin inhibitors. At both 37°C and 40°C the inhibitor/caspofungin combination strongly affected growth (Figure 5b).
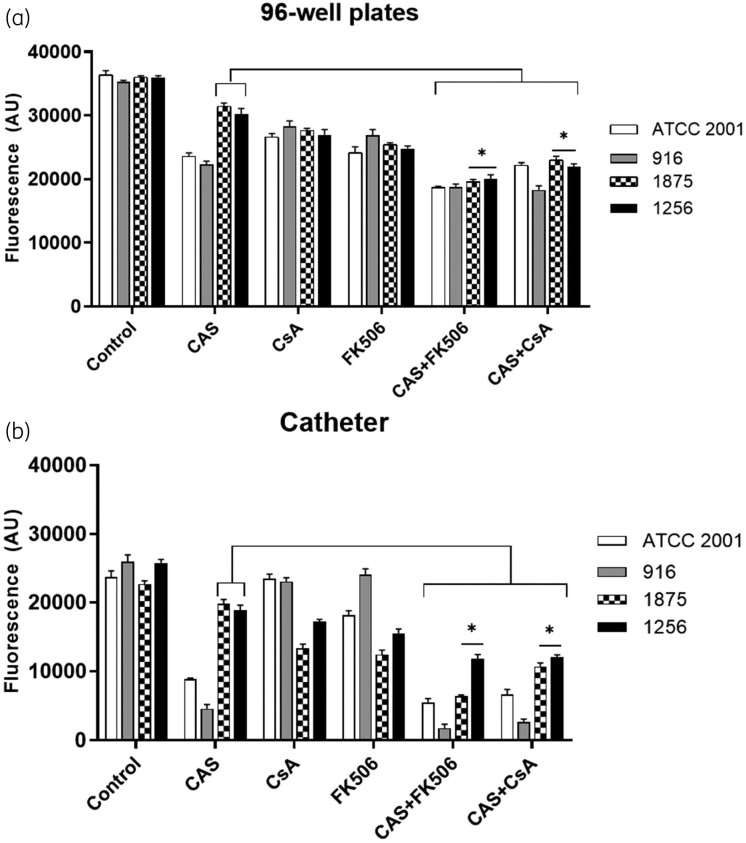
Calcineurin inhibitors significantly reduced biofilm-forming capacity
The four isolates had the capacity to form biofilm, but biofilm formation was lower in the catheter model. Caspofungin treatment reduced biofilm formation in susceptible isolates especially on catheter, whereas no significant activity was detected for resistant isolates (Figure 6a and b). In contrast, addition of calcineurin inhibitors to caspofungin significantly reduced the biofilm-formation capacity of resistant isolates, regardless of the model used (P < 0.05).
Figure 6.
Biofilm formation by C. glabrata isolates grown (a) in microplate wells and (b) on catheter pieces, exposed to CsA, FK506, caspofungin and their combinations. Fluorescence was measured at 560 nm with emission at 590 nm. Data are expressed in arbitrary fluorescence unit (AU). An asterisk indicates a P value <0.05.
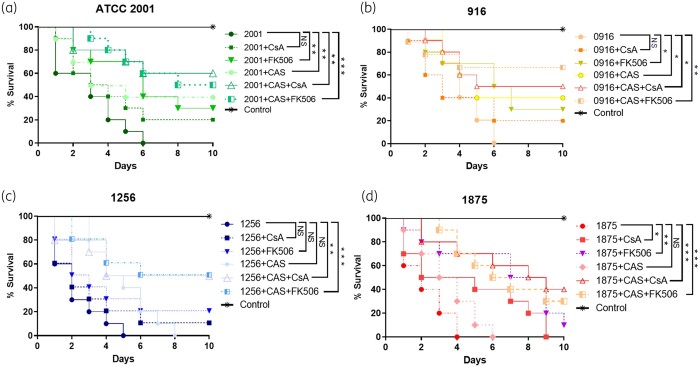
Calcineurin inhibition reduced C. glabrata pathogenicity in the invertebrate G. mellonella
C. glabrata isolates typically lead to complete mortality of G. mellonella by 4–6 days post-infection. Treatment with caspofungin (1 μg/larva) increased the larval survival when infected with susceptible isolates but did not exhibit, as expected, any statistically significant change for caspofungin-resistant isolates. However, addition of calcineurin inhibitors to caspofungin proved to be effective in prolonging survival (P < 0.05). No larval mortality was observed in control larvae injected with an equivalent volume of PBS (Figure 7).
Figure 7.
Galleria time–kill curves of caspofungin-susceptible (a, b) and resistant (c, d) C. glabrata isolates exposed to, caspofungin (1 μg/larva), calcineurin inhibitors (15 mg/L) and their combinations. The data are expressed as the percentages of survival. Log-rank (Mantel–Cox) test with P values of <0.05 was used to indicate statistical significance as follows: *P < 0.05, **P < 0.02 and ***P < 0.001.
Discussion
C. glabrata is one of the most prominent Candida species detected in bloodstream isolates worldwide, typically exhibiting intrinsic resistance to azoles.58–61 Moreover, echinocandin resistance in C. glabrata has increased, causing a serious clinical challenge.62 Different mechanisms of resistance to echinocandins have been described, mainly associated with FKS gene alterations.6,9 In this work, we employed NGS to provide a view of mutations involved in clinical caspofungin-resistant isolates targeting genes previously associated with echinocandin resistance.9,63,64 To date, only a single FKS2 gene deletion associated with caspofungin resistance has been found; however, a larger comprehensive comparative analysis is ongoing.
Herein, we describe the first proteome description of resistant C. glabrata after caspofungin exposure. Considering that caspofungin causes osmotic disruption of the fungal cell, enrichment of molecular functions and biological processes, as expected, were associated with antifungal response, cell wall biogenesis, and modulation of PKC and CaM/CaL pathways. These results, similar to those previously reported in C. albicans, confirm the association of these processes in the growth of C. glabrata with echinocandin exposure.13,65 Furthermore, we observed that caspofungin exposure resulted in increased GO annotations related to stress adaptation, such as chemical and cation homeostasis, cell wall organization or biogenesis, and response to heat. These features have been described by Hoehamer et al.19 as important changes in the C. albicans proteome in response to ketoconazole, amphotericin B, and caspofungin treatments.
C. glabrata exposure to caspofungin resulted in an increased abundance of MAP kinase Slt2 and Crz1 proteins which, being part of PKC and CaM/CaL pathways, respectively, have been implicated in cell-wall biogenesis and integrity. This compensation phenomenon, also observed in Saccharomyces cerevisiae, C. albicans (Mkc1), and recently published in C. glabrata, constitutes a mechanism of tolerance to caspofungin.13,14,66–68 Mutants lacking SLT2/MKC1 and CRZ1 are both susceptible to echinocandins in in vitro assays.14,69 Nevertheless, Slt2 overexpression leads to hypervirulence.70 Another three proteins (CAGL0C01683g, CAGL0L10021g, CAGL0J11440g) were found with higher abundance after exposure to caspofungin, however, these proteins have not been characterized in C. glabrata to date. The protein CAGL0C01683g (Isw1) homologue in C. albicans and S. cerevisiae has been described as a chromatin remodelling factor involved in the repression of the initiation of transcription. ISW1 also works in parallel with the NuA4 and Swr1 complexes in the repression of stress-induced genes.71 Inhibitors of DNA methyltransferases are attractive compounds for epigenetic drug discovery. Therefore, the Isw1 function and its role in the antifungal drug response need further studies. The protein CAGL0L10021g (C.a, S.c Dpb5) is an ATP-dependent cytoplasmic RNA helicase involved in translation termination along with Sup45p (eRF1); it also has a role in the cellular response to heat stress.72 Finally, the protein CAGL0J11440g (C.a, Sc Srp1) (importin-α) has nuclear import signal receptor activity and is involved in the degradation of proteins. Loss of Srp1 is lethal, although several temperature-sensitive mutants have been described.73,74 To date, there are no drugs against these proteins, however, as some host and yeast enzymes are not identical, traditional medicinal chemistry and structure-based drug design can exploit these differences to synthesize drugs with high specificity for the yeast. Conversely, Pir5 protein was decreased in abundance in response to caspofungin exposure. Pir proteins are a structural constituent of the cell wall and are associated with cell-wall organization owing to linkage to multiple β-1,3-glucan chains. The changes in the Pir proteins are a consequence of the activation of the cell-wall integrity pathway.75–77 This decrease of Pir5 abundance appears to be part of a general compensatory mechanism in response to cell-wall weakening caused by caspofungin; consequently, the cell increases chitin and/or mannan production, a phenomenon reported in S. cerevisiae and Candida spp.75,78
Given the importance of the CaM/CaL-Crz1 pathway in several biological processes, the impact of FK506 and CsA calcineurin inhibitors was studied in temperature and oxidative stress conditions. Similar to previous studies, we confirm that the CaM/CaL pathway is involved in thermotolerance, mainly at higher temperatures.79 By contrast, according to our results, the inhibition of calcineurin does not appear to affect the growth of C. glabrata in oxidative stress. The antioxidant capacity of C. glabrata, mainly associated with the catalase Cta1, is higher than that of S. cerevisiae and C. albicans. Cta1 is controlled by the transcription factors Yap1, Msn2, and Msn4 and modulated by pathways other than CaM/CaL.80
Biofilm formation is another important factor in the understanding of cellular disruption. Biofilms are thought to provide ecologic advantages such as protection from the environment, nutrient availability, metabolic cooperation, and acquisition of new traits. In general, C. glabrata biofilms possessed a higher density of cells comparatively to C. tropicalis and C. parapsilosis biofilms. This may be implicated in the typical high degree of resistance of C. glabrata biofilms to azole antifungals and amphotericin B.81 Biofilm eradication as a therapeutic approach is generally effective using echinocandins, as long as the isolate is drug susceptible.82 In our study, planktonic cells of caspofungin-resistant isolates maintain this characteristic in biofilm community state, even in the presence of high doses of caspofungin. Nevertheless, this situation can be reversed by addition of CaM/CaL inhibitors, as we demonstrated in the clinical-relevant model using polyurethane catheter pieces.
On the other hand, we believe that the effect of FK506 and CsA on heat-shock tolerance or susceptibility restoration to caspofungin could contribute to their in vivo activity. Indeed, in Galleria the use of CaM/CaL inhibitors reduces the mortality caused by all isolates, as well the addition of inhibitors to caspofungin enhances its efficacy, allowing a significant increase in larval survival. We believe that treatment with the inhibitors plus the immune response of G. mellonella (antimicrobial peptides, lytic enzymes, and melanin) enhances the defence of the larvae to the C. glabrata infection. Meaning that the antifungal activity of the inhibitors and the immune system work together, resulting in greater larvae survival. This is in concordance with previous data showing the role of CaM/CaL pathway in virulence of fungal species.79,83
Despite these promising findings, non-immunosuppressive analogs of both FK506 and CsA with no cross-activity with calcineurin in human cells must be developed.17,84,85 With regards to the CaM/CaL pathway, the challenge also will lie in focusing on the transcription factor Crz1, as recently explored for Rhizoctonia solani.86 Transcription factors are now attractive as antifungal drug targets since they are evolutionarily divergent between fungi and humans and therefore can be exploited as selective targets.87
In conclusion, our study provides proteomic evidence that proteins of CaM/CaL pathway, such as Crz1, are more abundant after caspofungin exposure. In addition, inhibition of this pathway in the clinical isolates with an FKS2 gene mutation changed their planktonic and biofilm susceptibility, thermotolerance, and finally pathogenicity. Synthesis of more specific antifungal compounds targeting this stress response pathway could be a successful therapeutic strategy for fighting life-threatening fungal diseases.
Supplementary Material
Acknowledgements
We thank the Colombian Science Technology and Innovation Department (Minciencias) Call 757, Campus France (Eiffel scholarship) and the Complutense University of Madrid (member of ProteoRed-ISCIII network). Finally, we thank Diego Vesga and Eduardo Romeu for their support in obtaining proteomic data.
Funding
This study was financially supported by ECOS Nord C17S01, the research vice rectory of the Pontificia Universidad Javeriana in Bogotá, Colombia ID20291, and MINCIENCIAS 807–2018, Code 120380763646, contract RC No. 715–2018.
Transparency declarations
P.L.P. received grants from Astellas, Basilea, MSD and Pfizer and speaker’s fees from Gilead, Basilea, Pfizer and MSD. The other authors none to declare.
Author contributions
A.C.-G. performed experiments and wrote the main manuscript, A.C.-G., L.M. and C.G. designed proteomic experiments and analysed proteomic data, C.A.-M. analysed clinical implications, N.E.V.-V. conducted bioinformatics analysis of genomic data, and prepared Figure 1. D.M.E. and J.B. sequenced isolates. A.C., P.L.P. and C.M.P.-G. designed the experiments and wrote the final manuscript, and P.L.P., C.M.P.-G. conceived the experiments and managed the resources. All authors have read and agreed to the published version of the manuscript.
Supplementary data
Figures S1 and S2 and Tables S1 to S3 are available as Supplementary data at JAC Online.
References
- 1. Pappas PG, Kauffman CA, Andes DR. et al. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 2015; 62: e1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oñate JM, Rivas P, Pallares C. et al. Colombian consensus on the diagnosis, treatment, and prevention of Candida Spp. disease in children and adults. Infectio 2019; 23: 271. [Google Scholar]
- 3. Perlin DS. Echinocandin resistance in Candida. Clin Infect Dis 2015; 61 Suppl 6: S612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Healey K, Perlin D.. Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. J Fungi 2018; 4: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garcia-Effron G, Park S, Perlin DS.. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob Agents Chemother 2009; 53: 112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Effron G, Lee S, Park S. et al. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Chemother 2009; 53: 3690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katiyar SK, Alastruey-Izquierdo A, Healey KR. et al. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother 2012; 56: 6304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia-Effron G, Katiyar SK, Park S. et al. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob Agents Chemother 2008; 52: 2305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singh-Babak SD, Babak T, Diezmann S. et al. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog 2012; 8: e1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cowen LE, Steinbach WJ.. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot Cell 2008; 7: 747–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ceballos Garzon A, Amado D, Robert E. et al. Impact of calmodulin inhibition by fluphenazine on susceptibility, biofilm formation and pathogenicity of caspofungin-resistant Candida glabrata. J Antimicrob Chemother 2020; 75: 1187–93. [DOI] [PubMed] [Google Scholar]
- 12. Shapiro RS, Robbins N, Cowen LE.. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 2011; 75: 213–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LaFayette SL, Collins C, Zaas AK. et al. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog 2010; 6: e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reinoso-Martín C, Schüller C, Schuetzer-Muehlbauer M. et al. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot Cell 2003; 2: 1200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miyazaki T, Izumikawa K, Nagayoshi Y. et al. Functional characterization of the regulators of calcineurin in Candida glabrata. FEMS Yeast Res 2011; 11: 621–30. [DOI] [PubMed] [Google Scholar]
- 16. Juvvadi PR, Lee SC, Heitman J. et al. Calcineurin in fungal virulence and drug resistance: prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach. Virulence 2017; 8: 186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beom JY, Jung JA, Lee K-T. et al. Biosynthesis of nonimmunosuppressive FK506 analogues with antifungal activity. J Nat Prod 2019; 82: 2078–86. [DOI] [PubMed] [Google Scholar]
- 18. Breuder T, Hemenway CS, Movva NR. et al. Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc Natl Acad Sci USA 1994; 91: 5372–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoehamer CF, Cummings ED, Hilliard GM. et al. Changes in the proteome of Candida albicans in response to azole, polyene, and echinocandin antifungal agents. Antimicrob Agents Chemother 2010; 54: 1655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bruneau JM, Maillet I, Tagat E. et al. Drug induced proteome changes in Candida albicans: comparison of the effect of β(1,3) glucan synthase inhibitors and two triazoles, fluconazole and itraconazole. Proteomics 2003; 3: 325–36. [DOI] [PubMed] [Google Scholar]
- 21. Fernández-Arenas E, Cabezón V, Bermejo C. et al. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics 2007; 6: 460–78. [DOI] [PubMed] [Google Scholar]
- 22. Vaz C, Reales-Calderon JA, Pitarch A. et al. Enrichment of ATP binding proteins unveils proteomic alterations in human macrophage cell death, inflammatory response, and protein synthesis after interaction with Candida albicans. J Proteome Res 2019; 18: 2139–59. [DOI] [PubMed] [Google Scholar]
- 23. Reales-Calderón JA, Vaz C, Monteoliva L. et al. Candida albicans modifies the protein composition and size distribution of THP-1 macrophage-derived extracellular vesicles. J Proteome Res 2017; 16: 87–105. [DOI] [PubMed] [Google Scholar]
- 24. Gil-Bona A, Reales-Calderon JA, Parra-Giraldo CM. et al. The cell wall protein Ecm33 of Candida albicans is involved in chronological life span, morphogenesis, cell wall regeneration, stress tolerance, and host-cell interaction. Front Microbiol 2016; 7: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jayampath Seneviratne C, Wang Y, Jin L. et al. Proteomics of drug resistance in Candida glabrata biofilms. Proteomics 2010; 10: 1444–54. [DOI] [PubMed] [Google Scholar]
- 26. Gómez-Molero E, de Boer AD, Dekker HL. et al. Proteomic analysis of hyperadhesive Candida glabrata clinical isolates reveals a core wall proteome and differential incorporation of adhesins. FEMS Yeast Res 2015; 15: 1–10. [DOI] [PubMed] [Google Scholar]
- 27. Pais P, Costa C, Pires C. et al. Membrane proteome-wide response to the antifungal drug clotrimazole in Candida glabrata: role of the transcription factor CgPdr1 and the Drug:H+ antiporters CgTpo1-1 and CgTpo1-2. Mol Cell Proteomics 2016; 15: 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pais P, Pires C, Costa C. et al. Membrane proteomics analysis of the Candida glabrata response to 5-flucytosine: unveiling the role and regulation of the drug efflux transporters CgFlr1 and CgFlr2. Front Microbiol 2016; 7: 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vargas-Casanova Y, Carlos Villamil Poveda J, Jenny Rivera-Monroy Z. et al. Palindromic peptide LfcinB (21-25)Pal exhibited antifungal activity against multidrug-resistant candida. ChemistrySelect 2020; 5: 7236–42. [Google Scholar]
- 30. Chen S, Zhou Y, Chen Y. et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018; 34: i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Waterhouse AM, Procter JB, Martin DMA. et al. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009; 25: 1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altschul SF, Gish W, Miller W. et al. Basic local alignment search tool. J Mol Biol 1990; 215: 403–10. [DOI] [PubMed] [Google Scholar]
- 34. Notredame C, Higgins DG, Heringa J.. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 2000; 302: 205–17. [DOI] [PubMed] [Google Scholar]
- 35. Källberg M, Wang H, Wang S. et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 2012; 7: 1511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Z, Zhao F, Peng J. et al. Protein 8-class secondary structure prediction using conditional neural fields. Proteomics 2011; 11: 3786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adamczak R, Porollo A, Meller J.. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins 2004; 56: 753–67. [DOI] [PubMed] [Google Scholar]
- 38. Adamczak R, Porollo A, Meller J.. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins 2005; 59: 467–75. [DOI] [PubMed] [Google Scholar]
- 39. Wagner M, Adamczak R, Porollo A. et al. Linear regression models for solvent accessibility prediction in proteins. J Comput Biol 2005; 12: 355–69. [DOI] [PubMed] [Google Scholar]
- 40. Parollo A, Adamczak R, Wagner M. et al. Maximum Feasibility Approach for Consensus Classifiers: Applications to Protein Structure Prediction. 2004. https://folding.cchmc.org/publications/ciras2003_jmeller.pdf.
- 41. Krogh A, Larsson B, von Heijne G. et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305: 567–80. [DOI] [PubMed] [Google Scholar]
- 42. Tsirigos KD, Peters C, Shu N. et al. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 2015; 43: W401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hofmann K, Stoffel W.. A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 1993; 374. [Google Scholar]
- 44. Dobson L, Reményi I, Tusnády GE.. CCTOP: a consensus constrained TOPology prediction web server. Nucleic Acids Res 2015; 43: W408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dobson L, Reményi I, Tusnády GE.. The human transmembrane proteome. Biol Direct 2015; 10: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tusnady GE, Simon I.. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001; 17: 849–50. [DOI] [PubMed] [Google Scholar]
- 47. Käll L, Krogh A, Sonnhammer EL.. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 2004; 338: 1027–36. [DOI] [PubMed] [Google Scholar]
- 48. Jones P, Binns D, Chang H-Y. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014; 30: 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Monteoliva L, Martinez-Lopez R, Pitarch A. et al. Quantitative proteome and acidic subproteome profiling of Candida albicans yeast-to-hypha transition. J Proteome Res 2010; 10: 502–17. [DOI] [PubMed] [Google Scholar]
- 50. Sechi S, Chait BT.. Modification of cysteine residues by alkylation. A tool in peptide mapping and protein identification. Anal Chem 1998; 70: 5150–8. [DOI] [PubMed] [Google Scholar]
- 51. Zybailov BL, Florens L, Washburn MP.. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst 2007; 3: 354–60. [DOI] [PubMed] [Google Scholar]
- 52. Perez-Riverol Y, Csordas A, Bai J. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019; 47: D442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deutsch EW, Bandeira N, Sharma V. et al. The ProteomeXchange consortium in 2020: enabling ‘big data’ approaches in proteomics. Nucleic Acids Res 2020; 48: D1145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts—Third Informational Supplement: M27-S3. 2008.
- 55. CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts—Fourth Informational Supplement: M27-S4. 2012.
- 56. Le Pape P, Pagniez F, Abdala-Valencia H.. A new fluorometric method for anti-Leishmania drug screening on axenic amastigotes. Acta Parasitol 2003; 48: 301–5. [Google Scholar]
- 57. Fallon J, Kelly J, Kavanagh K.. Galleria mellonella as a Model for Fungal Pathogenicity Testing. Methods Mol Biol 2012; 845: 469–85. [DOI] [PubMed] [Google Scholar]
- 58. Diekema D, Arbefeville S, Boyken L. et al. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn Microbiol Infect Dis 2012; 73: 45–8. [DOI] [PubMed] [Google Scholar]
- 59. Kumar K, Askari F, Sahu MS. et al. Candida glabrata: a lot more than meets the eye. Microorganisms 2019; 7: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Toda M, Williams SR, Berkow EL. et al. Population-based active surveillance for culture-confirmed candidemia - four sites, United States, 2012-2016. MMWR Surveill Summ 2019; 68: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Whaley SG, Rogers PD.. Azole resistance in Candida glabrata. Curr Infect Dis Rep 2016; 18: 1–10. [DOI] [PubMed] [Google Scholar]
- 62. Alexander BD, Johnson MD, Pfeiffer CD. et al. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 2013; 56: 1724–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zimbeck AJ, Iqbal N, Ahlquist AM. et al. FKS mutations and elevated echinocandin MIC values among Candida glabrata Isolates from U.S. population-based surveillance. Antimicrob Agents Chemother 2010; 54: 5042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu SJ, Chang YL, Chen YL.. Calcineurin signaling: lessons from Candida species. FEMS Yeast Res 2015; 15: 1–7. [DOI] [PubMed] [Google Scholar]
- 65. Kelly J, Kavanagh K.. Proteomic analysis of proteins released from growth-arrested Candida albicans following exposure to caspofungin. Med Mycol 2010; 48: 598–605. [DOI] [PubMed] [Google Scholar]
- 66. Román E, Alonso-Monge R, Miranda A. et al. The Mkk2 MAPKK regulates cell wall biogenesis in cooperation with the Cek1-pathway in Candida albicans. Lenardon MD, ed. PLoS One 2015; 10: e0133476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Alonso-Monge R, Guirao-Abad JP, Sánchez-Fresneda R. et al. The fungicidal action of micafungin is independent on both oxidative stress generation and HOG pathway signaling in Candida albicans. Microorganisms 2020; 8: 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Garcia-Rubio R, Hernandez RY, Clear A. et al. Critical assessment of cell wall integrity factors contributing to in vivo echinocandin tolerance and resistance in Candida glabrata. Front Microbiol 2021; 12: 702779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Singh SD, Robbins N, Zaas AK. et al. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog 2009; 5: e1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Miyazaki T, Inamine T, Yamauchi S. et al. Role of the Slt2 mitogen-activated protein kinase pathway in cell wall integrity and virulence in Candida glabrata. FEMS Yeast Res 2010; 10: 343–52. [DOI] [PubMed] [Google Scholar]
- 71. Vasicova P, Stradalova V, Halada P. et al. Nuclear import of chromatin remodeler Isw1 is mediated by atypical bipartite cNLS and classical import pathway. Traffic 2013; 14: 176–93. [DOI] [PubMed] [Google Scholar]
- 72. Tieg B, Krebber H.. Dbp5 - From nuclear export to translation. Biochim Biophys Acta 2013; 1829: 791–9. [DOI] [PubMed] [Google Scholar]
- 73. Ha SW, Ju D, Xie Y.. Nuclear import factor Srp1 and its associated protein Sts1 couple ribosome-bound nascent polypeptides to proteasomes for cotranslational degradation. J Biol Chem 2014; 289: 2701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen L, Madura K.. Yeast Importin-α (Srp1) performs distinct roles in the import of nuclear proteins and in targeting proteasomes to the nucleus. J Biol Chem 2014; 289: 32339–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mazáň M, Mazáňová K, Farkaš V.. Phenotype analysis of Saccharomyces cerevisiae mutants with deletions in Pir cell wall glycoproteins. Antonie Van Leeuwenhoek 2008; 94: 335–42. [DOI] [PubMed] [Google Scholar]
- 76. De Groot PWJ, Kraneveld EA, Qing YY. et al. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot Cell 2008; 7: 1951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kandasamy R, Vediyappan G, Chaffin WL.. Evidence for the presence of Pir-like proteins in Candida albicans. FEMS Microbiol Lett 2000; 186: 239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Walker LA, Gow NAR, Munro CA.. Elevated chitin content reduces the susceptibility of candida species to caspofungin. Antimicrob Agents Chemother 2013; 57: 146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen Y-L, Konieczka JH, Springer DJ. et al. Convergent evolution of calcineurin pathway roles in thermotolerance and virulence in Candida glabrata. G3 (Bethesda) 2012; 2: 675–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cuéllar-Cruz M, Briones-Martin-Del-Campo M, Cañas-Villamar I. et al. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell 2008; 7: 814–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fonseca E, Silva S, Rodrigues CF. et al. Effects of fluconazole on Candida glabrata biofilms and its relationship with ABC transporter gene expression. Biofouling 2014; 30: 447–57. [DOI] [PubMed] [Google Scholar]
- 82. Rodrigues CF, Rodrigues ME, Henriques M.. Susceptibility of Candida glabrata biofilms to echinocandins: alterations in the matrix composition. Biofouling 2018; 34: 569–78. [DOI] [PubMed] [Google Scholar]
- 83. Park H-S, Chow EWL, Fu C. et al. Calcineurin targets involved in stress survival and fungal virulence. PLoS Pathog 2016; 12: e1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bashir Q, LeMaster DM, Hernández G.. 1H, 13C, 15N chemical shift assignments of the FKBP12 protein from the pathogenic fungi Candida auris and Candida glabrata. Biomol NMR Assign 2020; 14: 105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee Y, Lee KT, Lee SJ. et al. In vitro and in vivo assessment of FK506 analogs as novel antifungal drug candidates. Antimicrob Agents Chemother 2018; 62: e01627-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Malik A, Afaq S, Gamal BE. et al. Molecular docking and pharmacokinetic evaluation of natural compounds as targeted inhibitors against Crz1 protein in Rhizoctonia solani. Bioinformation 2019; 15: 277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bahn Y-S. Exploiting fungal virulence-regulating transcription factors as novel antifungal drug targets. PLoS Pathog 2015; 11: e1004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Omasits U, Ahrens CH, Müller S. et al. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014; 30: 884–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental data are provided in the manuscript and in supplementary files, or are available via ProteomeXchange with identifier PXD021578 (10.6019/PXD021578) and in the NCBI BioProject database with the accession number PRJNA692260.