Abstract
Background/Aim: Glioblastoma (GBM) is one of the deadliest human cancers responding very poorly to therapy. Although the central nervous system has been traditionally considered an immunologically privileged site with an enhanced immune response, GBM appears to benefit from this immunosuppressive milieu. Immunomodulatory molecules play an important role in immune tumor-host interactions. Non-classical human leukocyte antigens (HLA) class Ib molecules HLA-E, HLA-F, and HLA-G have been previously described to be involved in protecting semi-allogeneic fetal allografts from the maternal immune response and in transplant tolerance as well as tumoral immune escape. Unfortunately, their role in GBM remains poorly understood. Our study, therefore, aimed to characterize the relationship between the expression of these molecules in GBM on the transcriptional level and clinico-pathological and molecular features of GBM as well as the effect of ionizing radiation. Materials and Methods: We performed the analysis of HLA-E, HLA-F, and HLA-G mRNA expression in 69 GBM tissue samples and 21 non-tumor brain tissue samples (controls) by reverse transcription polymerase chain reaction. Furthermore, two primary GBM cell cultures had been irradiated to identify the effect of ionizing radiation on the expression of non-classical HLA molecules. Results: Analyses revealed that both HLA-E and HLA-F are significantly up-regulated in GBM samples. Subsequent survival analysis showed a significant association between low expression of HLA-E and shorter survival of GBM patients. The dysregulated expression of both molecules was also observed between patients with methylated and unmethylated O-6-methylguanine-DNA methyltransferase (MGMT) promoter. Finally, we showed that ionizing radiation increased HLA-E expression level in GBM cells in vitro. Conclusion: HLA-E and HLA-F play an important role in GBM biology and could be used as diagnostic biomarkers, and in the case of HLA-E also as a prognostic biomarker.
Keywords: Glioblastoma, non-classical human leukocyte antigens, HLA-E, HLA-F, ionizing radiation, prognosis
Glioblastoma (GBM) is one of the most aggressive primary brain tumors with a very poor prognosis. The current treatment approach involves surgery, if possible, followed by radiotherapy with a total dose of 60 Gy and concomitant chemotherapy with the alkylating agent temozolomide (TMZ). Sometimes, adjuvant chemotherapy with TMZ is given as monotherapy, which together with concomitant chemoradiotherapy significantly increase median overall survival (OS) of patients to 12-15 months (1-4). Nevertheless, GBM progression and recurrence eventually manifest due to several factors. One possible explanation is related to the immunologically privileged status of the brain, made possible by the existence of the blood-brain barrier, which may be breached in pathological conditions leading to recruitment of leukocytes (5). Alterations of immunomodulatory molecules, such as human leukocyte antigens (HLA), may then lead to changes in immune interactions between the tumor and host organism contributing to the immune escape of tumors (6).
HLA molecules are coded by closely linked polymorphic genes and comprise the so-called major histocompatibility complex (MHC), which has a large impact on cell recognition and immunological defense (7). MHC is further divided into 3 classes (I-III). Class I consists of classical and non-classical HLA molecules, which differ in the rate of polymorphism and tissue distribution. While classical HLA class Ia molecules are comprised of HLA-A, HLA-B, and HLA-C, which are highly polymorphic and widely expressed in most tissues, non-classical HLA class Ib molecules are represented by HLA-E, HLA-F, and HLA-G. These relatively conserved molecules are generally characterized by a more restricted tissue distribution. Their expression was originally observed in placental trophoblasts and fetal tissue under physiological conditions, where they have a fundamental role in maternal immune response/tolerance during pregnancy (8-10).
The mechanism by which HLA-E exerts its function directly involves receptors cluster of differentiation 94/natural killer group 2 (CD94/NKG2) expressed on the surface of natural killer (NK) cells and a subset of cluster of differentiation 8-positive (CD8+) T lymphocytes (11-13). HLA-E binds a subset of peptides, derived either from the leader sequences of other MHC class I molecules, namely HLA-A, HLA-B, HLA-C, and HLA-G, or exogenous viral proteins (14,15). Like the processing of HLA class Ia molecules, the binding is facilitated by the peptide loading complex, which includes transmembrane glycoprotein tapasin and a transporter associated with antigen processing 1 or 2 (TAP1/TAP2). HLA-E is then transported partially in a B-Cell receptor-associated protein 31 (Bap31)-dependent manner from the endoplasmic reticulum lumen to the cell surface where it presents bound signal peptides to NK cells by interacting with NKG2 receptors A-E (16,17). All NKG2 receptors dimerize with CD94 except for NKG2D, which associates with DNA polymerase III subunit tau (DNAX)-activating protein of 10KDa (DAP10) or DNAX-activating protein of 12KDa (DAP12) (18). While the interaction of HLA-E and CD94/NKG2C or CD94/NKG2E results in an activating effect on NK-mediated cell lysis, leading to the destruction of the HLA-E expressing cell, the interaction with CD94/NKG2A or CD94/NKG2B receptor has an inhibitory effect on NK cells. Moreover, the affinity of HLA-E for CD94/NKG2A has been reported to be 6- to 8-times higher than the affinity for CD94/NKG2C. Therefore, these interactions allow for the dynamic control of cytotoxicity (13,19-21).
The role of HLA-F is still not well understood. It is the smallest of the HLA class I molecules and a predominantly empty and intracellular protein acting as a ligand for TAP (22) and specialized receptors expressed by immune cells, such as immunoglobulin(Ig)-like transcript 2 (ILT2), ILT4 (23), killer cell Ig Like receptor, three Ig domains and long cytoplasmic tail 2 (KIR3DL2), killer cell Ig like receptor, two Ig domains and short cytoplasmic tail 4 (KIR2DS4), and killer cell Ig like receptor, three Ig domains and short cytoplasmic tail 1 (KIR3DS1) (24-26). HLA-F was also reported to act as a chaperone in activated lymphocytes in the absence of signal peptides, stabilizing the heavy chain of other HLA class I molecules in the form of open conformers and escorting them to the cell surface, where they participate in cross-presentation of exogenous antigens (27,28).
HLA-G has been first observed on the surface of extravillous cytotrophoblast cells in the placenta, hinting at its role in maternal immune response/tolerance. Compared to other HLA class Ib molecules, HLA-G is highly polymorphic, with several of the alleles connected to recurrent pregnancy loss and pre-eclampsia (29-31). Its function might extend beyond the mediation of maternal immune tolerance, considering the fact it was also found in other types of cells and tissues, such as T cells derived from the thymus (32), mesenchymal stem cells (33), and cornea (34).
The aberrant expression of HLA class Ib molecules was observed in various tumors, including GBM, with most studies reporting overexpression of their mRNA and protein molecules in tumors compared to non-tumor tissues (35-40). Moreover, there is evidence of an association between the expression of HLA-E, HLA-F, and HLA-G and the OS length of patients with GBM (35,36,41-43), as well as the impact of irradiation on these molecules in several malignancies (6,36). However, the expression of HLA class Ib molecules in GBM is still not well-characterized. Therefore, the aim of this study was to analyze the expression of HLA-E, HLA-F, and HLA-G in GBM and non-tumor brain tissue samples, characterize the prognostic potential of these molecules, and study the effect of ionizing radiation on the expression of the above molecules in primary GBM cell cultures.
Materials and Methods
Patient sample cohort. The retrospective multi-institutional (University Hospital Brno, University Hospital Ostrava, St. Anne’s University Hospital Brno) cohort study included 69 patients with histopathologically confirmed primary GBM and 21 non-tumor tissues, with the latter serving as controls. The study was approved by the local Ethics Committee and all enrolled patients signed the informed consent form. The clinico-pathological and molecular characteristics of GBM patients are summarized in Table I. Non-tumor brain tissues were obtained via therapeutic resections in patients with pharmacoresistant, intractable epilepsy and only brain tissue lacking evidence of dysplastic changes from the non-dominant temporal lobe was used. Forty-one GBM patients underwent adjuvant concomitant chemoradiotherapy according to the Stupp protocol. In summary, 60 Gy of fractionated radiotherapy were administered at the primary site of the tumor, followed by 42 cycles of TMZ chemotherapy. A subset of patients was further indicated to adjuvant TMZ monotherapy.
Table I. Clinico-pathological and molecular characteristics of the GBM patient cohort.
WHO: World Health Organization; MGMT: O-6-methylguanine-DNA methyltransferase; IDH1R132: the residue R132 of the isocitrate dehydrogenase 1 gene.
Primary cell cultures. Two primary GBM cell cultures were derived from fresh tumor tissue samples obtained from GBM patients who underwent surgical resection at the Department of Neurosurgery of the University Hospital Brno. The fresh tissue sample was enzymatically dissociated with the Papain Dissociation System (Worthington Biochemical Corporation, Lakewood, NJ, USA) for 20 min at 37˚C and then processed to a single cell suspension according to the manufacturer’s instructions. Single cell suspension was seeded into a 25 cm2 tissue culture flask (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum, 2 mM GlutaMAX, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 1% non-essential amino acids (all Thermo Fisher Scientific, Waltham, MA, USA). After 1-3 weeks, adherent cells, which covered more than 2/3 of the culture flask, were passaged using Trypsin-ethylenediaminetetraacetic acid (Trypsin-EDTA) solution (Sigma-Aldrich, St. Louis, MO, USA).
Tissue sample preparation and nucleic acid extraction. All tissue samples were frozen and stored at –80˚C in RNA stabilization solution RNAlater (Thermo Fisher Scientific). Samples were later homogenized with 1.4 mm ceramic beads and total RNA enriched for small RNAs was isolated using mirVana miRNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The concentration of extracted RNA was measured using ultraviolet-visible spectrophotometry in NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). RNA integrity was assessed with both electrophoresis in 1% agarose gel and capillary electrophoresis in 2200 TapeStation using the RNA ScreenTape System (Agilent Technologies, Santa Clara, CA, USA).
Irradiation of primary cell cultures. Based on similar methodology used in our previous study (44), two primary GBM cell cultures (GBM1 and GBM2) were grown to approximately 60% confluence, irradiated with 2 Gy (Cs-137 γ-radiation, 2 Gy/min), and upon reaching 90% confluence, subcultured into new culture flasks. This procedure was repeated at regular intervals up to 32 Gy, when the dose was raised to 4 Gy and administered up to a total dose of 40 Gy, rendering the cell lines radioresistant in the process (GBM1-R, GBM2-R). Parental cells were cultured in the same conditions without irradiation treatment and used as controls (GBM1-C, GBM2-C). Both pairs of primary cell lines then received a radiation dose of 0, 5 or 10 Gy and were cultured and lysed after 24 h and 72 h. The whole experiment was performed in biological triplicate with each also performed in technical triplicate. RNA from both radioresistant and control cell cultures was isolated from cell lysates using mirVana miRNA Isolation Kit according to the manufacturer's instructions and the concentration was measured using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific).
Quantification of HLA mRNA expression levels by reverse transcription polymerase chain reaction (RT-qPCR). High-Capacity cDNA Reverse Transcription Kit was used for cDNA synthesis according to the manufacturer’s instructions (Thermo Fisher Scientific). Real-time PCR was performed using LightCycler 480 Instrument II (Roche, Basel, Switzerland) and TaqMan Gene Expression Master Mix together with specific gene expression assays for HLA-E (Hs03045171_m1), HLA-F (Hs04185703_gH), HLA-G (Hs00365950_g1, Hs03045108_m1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Hs02786624_g1), and peptidylprolyl isomerase A (PPIA) (Hs99999904_m1) (Thermo Fisher Scientific), with the latter two serving as normalizers, which were chosen based on the past experience and current literature. All PCR reactions were performed in technical duplicates or monoplicates when measuring expression in fresh tissue samples and lysates from primary line cell irradiation experiment, respectively. The crossing point cycle (Cp) data were determined using the Second Derivative Maximum Method. When measuring expression levels in fresh frozen tissue samples, prior to normalization, Cp values were first corrected using an interplate control to minimize interplate variability. To compare the expression of HLA-E and HLA-F mRNAs, the expression levels were normalized using the 2−ΔCp method. ΔCP values were calculated according to the following formula: ΔCP=AvgCP(HLA)-AvgCP(normalizer), where AvgCP(HLA) and AvgCP(normalizer) are average values calculated from CP values obtained from technical replicates, with the former corresponding to HLA-E or HLA-F gene expression and the latter to the normalizer gene expression. PPIA or the combination of PPIA and GAPDH (an average Cp value) was used as a normalizer for relative quantification of HLA expression in fresh tissue samples and primary cell lines, respectively. In the case of the irradiation experiment, the average normalized expression value was then calculated from 3 biological replicate values which were, in turn, calculated from the technical replicate values of normalized HLA expression. Finally, the values on the y-axes in corresponding graphs were calculated as the ratios of normalized expression values at 72 h to 24 h after exposure to irradiation to a particular dose. Prism 8 software (GraphPad Software, San Diego, CA, USA) was used for the following statistical evaluation, including non-parametric Mann-Whitney U-test, Spearman correlation, receiver operating characteristic (ROC) analysis, Kaplan-Meier survival analysis, and linear regression line fitting. A p-value of 0.05 was selected as a threshold of significance. In the case of ROC and survival analysis, OS was defined as the time elapsed from the beginning of the treatment, i.e., surgical resection, until the patient’s death or the censoring date. Progression-free survival (PFS) was similarly defined as the time elapsed from the beginning of the treatment until the date of GBM progression. Patients were divided into the short-survival and long-survival groups based on the 12-month OS and 6-month PFS cut-offs.
Results
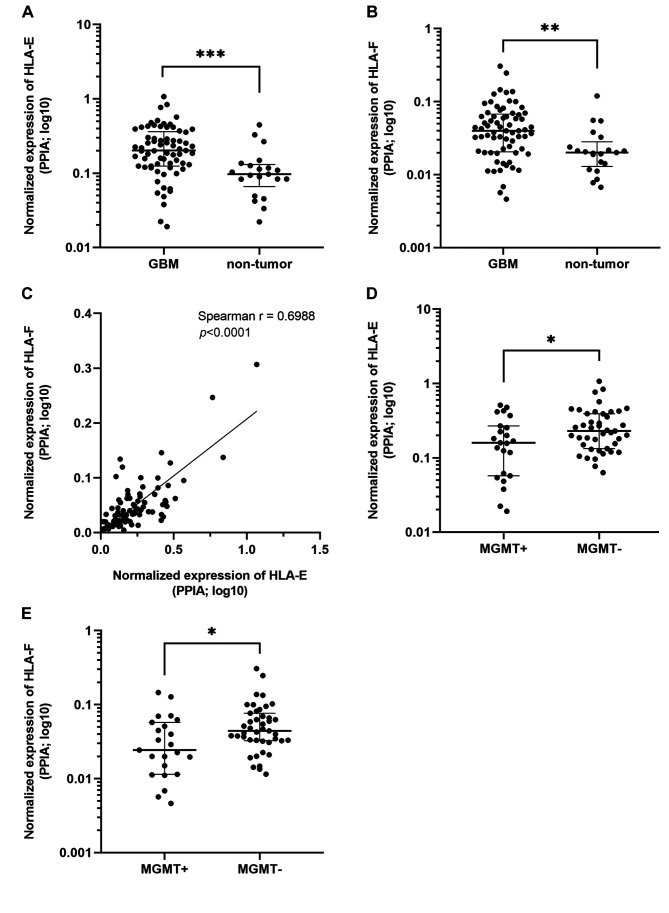
HLA-E and HLA-F are overexpressed in glioblastoma patient tissue samples compared to non-tumor tissue samples. Expression levels of HLA class Ib molecules were analyzed in an independent cohort of 69 primary GBM tissue samples and 21 non-tumor tissue samples serving as controls. The RT-qPCR analysis using TaqMan Gene Expression assays followed by Mann-Whitney analysis confirmed that both HLA-E and HLA-F are up-regulated in GBM samples (FC=2.05, p=0.0001, and FC=2.02, p=0.0014, respectively; Mann-Whitney analysis; Figure 1A and B), while the expression of HLA-G mRNA was not detected. A significant correlation between the expression levels of HLA-E and HLA-F in all samples (Spearman r=0.6988; p<0.0001; Figure 1C) and the dysregulated expression of HLA-E and HLA-F among patients with methylated and unmethylated MGMT promoters (p=0.0477 and p=0.0172, respectively; Figure 1D and E) were also observed. No significant dysregulation of HLA-E and HLA-F was observed among patients with the wild-type and mutated isocitrate dehydrogenase 1 (IDH1) gene (p=0.8555 and p=0.7640, respectively).
Figure 1. Results from the Mann-Whitney U-test and correlation analyses comparing mRNA levels of (A) HLA-E in glioblastoma (GBM) and nontumor tissues (p=0.0001); (B) HLA-F in GBM and non-tumor tissues (p=0.0014); (C) HLA-E and HLA-F in all tissue samples (Spearman r=0.6988; p<0.0001); (D) HLA-E in GBM samples with methylated and unmethylated O-6-methylguanine-DNA methyltransferase (MGMT) promoter (MGMT+ and MGMT-, respectively; p=0.0477); (E) HLA-F in MGMT+ and MGMT- GBM samples (p=0.0172).
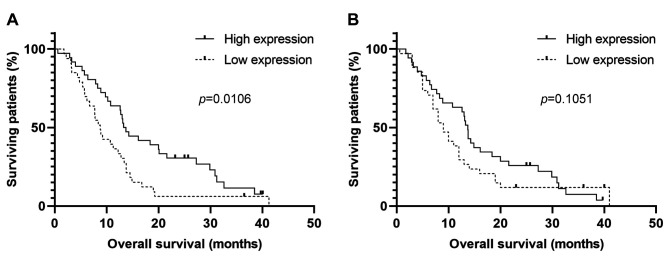
The expression of HLA-E is associated with overall survival of glioblastoma patients. ROC analysis of HLA-E and HLA-F expression in the cohort of GBM patients who underwent complete therapy according to the Stupp protocol (radiotherapy 60 Gy, 42 cycles of TMZ, and if possible, adjuvant therapy with TMZ) revealed that HLA-E was able to distinguish patient survival over 12 months with 65.71% sensitivity and 61.76% specificity. Significant association of HLA-E expression and OS has been subsequently confirmed by Kaplan-Meier analysis (p=0.0106; Gehan-Breslow-Wilcoxon test). Specifically, lower expression of HLA-E has been significantly associated with a poorer survival (Figure 2A). On the other hand, HLA-F tissue expression levels showed a similar trend but not a significant association with OS in GBM patients (p=0.1051; Gehan-Breslow-Wilcoxon test; Figure 2B). Finally, no significant association of HLA-E and HLA-F expression was observed in connection to PFS.
Figure 2. Kaplan-Meier survival analysis of (A) HLA-E and (B) HLA-F expression in GBM patients. Comparison of survival curves was performed using the Gehan-Breslow-Wilcoxon test.
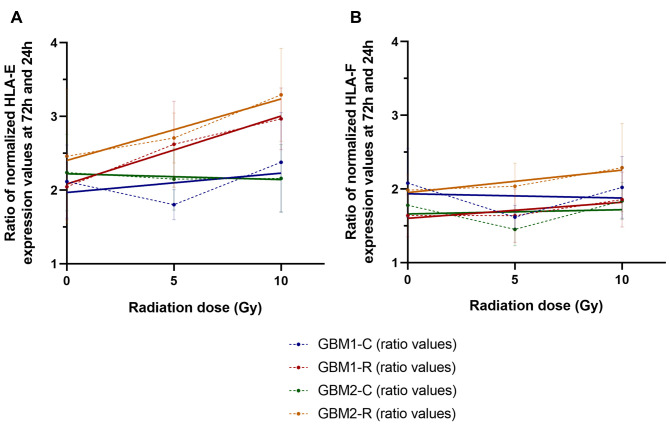
Ionizing radiation elevates expression levels of HLA-E in glioblastoma cells in vitro. We also performed in vitro analysis of the effect of ionizing radiation on the expression of HLA-E and HLA-F in two tissue-derived primary GBM cell cultures. Each culture was divided into two pairs, with one being regularly irradiated to a total dose of 40 Gy, eventually becoming radioresistant, and the other used as an untreated, radiosensitive control. The radioresistant and control cell lines then received a dose of 0, 5 or 10 Gy and the HLA-E and HLA-F expression was measured at 24 h and 72 h after exposure to radiation. The results demonstrated that compared to controls, both radioresistant cell lines were characterized by a visibly greater radiation dose-dependent change in HLA-E expression between 24 h and 72 h after exposure to radiation (Figure 3A). No significant dose-dependent change in HLA-F expression was observed between 24 h and 72 h in any of the paired primary cell lines (Figure 3B).
Figure 3. Results from reverse transcription polymerase chain reaction (RT-qPCR) comparing the changes in mRNA levels of (A) HLA-E and (B) HLA-F in 2 radioresistant (GBM1-R, GBM2-R) and 2 radiosensitive parental (GBM1-C, GBM2-C; control) primary GBM cell cultures from 24 h to 72 h after receiving a dose of 0, 5 or 10 Gy. Average values (ratios of normalized expression values at 72 h to values at 24 h) are denoted by points connected with a dashed line. Linear regression (trend) lines are shown to demonstrate the trend of dose-dependent changes in HLA-E and HLA-F expression.
Discussion
The function of non-classical HLA Ib molecules HLA-E, HLA-G, and lately HLA-F has been extensively studied, both in physiological and pathological conditions. Since they are well-known to be expressed on the surface of placental extravillous trophoblasts, these immunomodulating molecules were originally considered to only have a limited role in providing maternal immune tolerance to a semi-allogeneic fetus (8-10,45). Their expression was also eventually observed in normal non-fetal tissues. For example, HLA-E expression was detected in B and T lymphocytes, natural killer cells (NK), macrophages, and endothelial cells (46), while HLA-G proteins were observed in thymic epithelial cells and cells of erythropoietic lineage from bone marrow or the endothelium (47). HLA-F has been detected in a number of diverse tissues and cell lines, including liver, skin, and bladder cells and the surface of the monocyte cell line (48,49). However, for several decades, minimal attention has been given to further exploration of their role and relevance. Under pathological conditions, expression of both HLA-E and HLA-F was detected in various malignancies in tumor cells in vivo (50-52) and cell lines in vitro (22,50,53). Unfortunately, the relationship between these immunomodulating molecules and their function in GBM remains poorly understood.
Similarly to our previously reported findings (36), our current study was performed on a larger independent patient sample cohort showing increased expression of HLA-E in GBM samples compared to non-tumor tissue samples. This agreed with various studies that detected the overexpression of this molecule in both solid and hematopoietic malignancies (11,38-41,43,54-60). HLA-E is known to have the ability to bind to receptors CD94/NKG2A or CD94/NKG2B (inhibitory effect) and CD94/NKG2C or CD94/NKG2E (activating effect), expressed on the surface of NK cells and cytotoxic lymphocytes with 6- to 8-times higher affinity for the CD94/NKG2A inhibitory receptor compared to that for CD94/NKG2C activating receptor (20,21), but roughly the same as that for the CD94/NKG2E activating receptor (20). Deactivation of NK cells or cytotoxic lymphocytes through checkpoint inhibitor CD94/NKG2A appears to be one of many tumor mechanisms used for deceiving the immune system’s surveillance (61), which could explain why many published studies link high expression of HLA-E in tumor cells with poor prognosis of patients (40,56,62,63). It also needs to be noted that several other studies did not find any association between OS and the expression of HLA-E in renal cancer (38), rectal cancer (57), and GBM (39,58).
Nevertheless, Benevolo et al. associated high expression of HLA-E in colorectal carcinoma with favorable prognosis. In their study, high expression of HLA-E correlated significantly with high expression of its preferential ligand donor HLA-A with the presence of lymphoid cell infiltrates. These results hint at HLA-E favoring activating immune responses to colorectal carcinoma. Authors also suggested that tumor cells entertain extensive negotiation with the immune system until a compromise between recognition and escape is reached (64). Spaans et al. came to a similar conclusion in their study of the HLA-E expression in three most common histopathological types of cervical carcinoma where they observed a positive correlation of HLA-E expression with OS (59). Finally, John et al. identified HLA-E as one of the 14 genes with prognostic potential, whose higher expression in stage III melanomas is associated with a better outcome (65). Albeit seemingly counterintuitive to previously mentioned studies associating higher HLA-E expression with worse prognosis, these studies support our own results from the previous (36) and the current studies, showing a positive correlation between HLA-E expression and length of survival.
There is also evidence for the binding of specific molecules blocking the NKG2A inhibitory receptor and the existence of cells lacking this receptor, which may cause higher cytotoxicity against HLA-E expressing tumor cells through NKG2C, NKG2D, or NKG2E activating receptors (11,66,67). Since the central nervous system has been traditionally considered an immunologically privileged site (5) and roles of HLA class Ib molecules in relationship with GBM pathophysiology and antitumor response still remain unclear, there is a possibility that the distinct tumor microenvironment and the presence of specific molecules therein could also play role in involvement of activating receptors, and thus, in the better prognosis of patients with higher expression of HLA-E. For example, Michaëlsson et al. identified chaperonin heat shock protein 60 (hsp60) as a ligand donor for HLA-E during conditions of cellular stress, giving rise to a complex which cannot be efficiently recognized by CD94/NKG2A inhibitory receptors (68).
In our study, we also observed significantly higher expression of HLA-F in GBM tissue samples compared to non-tumor tissue samples. These findings correspond to the results of several similar already published studies (35,37,41). As for the relationship between the expression of HLA-F by malignant cells and its prognostic significance, our results did not show any significant association with OS despite having a trend similar to that of HLA-E expression. This contrasts with several recent studies focused on HLA-F expression in various cancers, including gastric cancer, stage II breast cancer, and gliomas, which reported a negative correlation of HLA-F expression with patient survival (35,51,56). However, the study by Chen et al. was specifically focused on low-grade gliomas (35), which have a different molecular background compared to most GBMs. With respect to the number of enrolled patients, there is a large difference between this study and ours, so it is possible that the analysis of a higher number of patient samples would reveal a significant, albeit possibly positive correlation of HLA-F expression with OS. On the other hand, in contrast to other HLA class Ib molecules, HLA-F (previously known as HLA-5.4) contains only 5 of the 10 highly conserved amino acids needed for antigen recognition (69). Also of note is the fact that ER export of HLA-F is dependent on the structure of its cytoplasmic tail (70), which is significantly shorter in one of its alternatively spliced isoforms due to exon 7 exclusion during transcription (69). It has been, thus, suggested that due to the differences in its structure, HLA-F may exert a different biological function from that of other HLA class I molecules (22). This alternative role might perhaps not be associated with overall prognosis. However, such a claim would require further investigation in the future studies.
Radiotherapy is an integral part of comprehensive care for cancer patients, including GBM patients. To identify the effect of ionizing radiation on the expression of studied HLA molecules, we hereby provide results for 2 paired primary cell cultures of GBM, each pair consisting of 1 radioresistant and 1 radio-sensitive control cell line. Compared to controls, we observed a visibly greater radiation dose-dependent change in HLA-E expression in radioresistant GBM cells between 24 h and 72 h after the exposure to radiation. No visible dose-dependent change in HLA-F expression was observed in the case of both paired primary cell lines. So far, to our knowledge, no data have been reported on the potential association of HLA-F expression and radiation, and thus, our study provides the first insight into this topic.
To this day, there are only a handful of studies reporting on the effect of ionizing radiation on HLA-E expression. Riederer et al. observed up-regulated levels of HLA-E in macrovascular endothelial cells (ECs) after the exposure of cells to a sublethal gamma radiation dose of 4 Gy (71). A study by Pereira et al. focused on inducing senescence in primary human fibroblasts by exposing the cells to a total dose of 10 Gy at a rate of 5 Gy/min, which had a significant impact on HLA-E up-regulation, but not HLA-G and MHC class Ia molecules (72). Finally, Michelin et al. reported HLA-E up-regulation in melanoma cells after a single dose of 10-20 Gy of gamma-radiation (73). Our results showed up-regulation of HLA-E expression induced by a dose of 5 or 10 Gy. We, therefore, conclude that moderately small doses of ionizing radiation may lead to up-regulation of HLA-E expression both in physiological and pathological conditions.
Up-regulated HLA-E expression could be related to the radiation-induced expression of interferon gamma (IFN-γ), a cytokine and the only member of a type II class of interferons. It has the ability to up-regulate the expression of HLA class I molecules (43,74,75), including HLA-E (76-79), on the surface of various normal and malignant cell types and was also reported to rapidly induce expression of TAP1 in HeLa cells via activation of signal transducer and activator of transcription 1α (Stat1α) homodimers, known as γ-activated factor (GAF), which bind to the gamma activating sequence (GAS) element in the TAP1 promoter (80). IFN-γ is physiologically secreted by infiltrating immune cells, including natural killer cells and CD8+ T lymphocytes (81-83), further promoting their proliferation (84,85). Moreover, the presence of the CD8+ T cell infiltrate in GBM has been associated with longer survival (86) and the decrease of IFN-γ was, in turn, reported to confer increased resistance of GBM cells to cytotoxicity (87). One explanation for the positive correlation of HLA-E expression with OS reported in our current study could be that the further up-regulation of initially higher basal HLA-E levels via IFN-γ or other radiation-induced factors may lead to an eventual oversaturation of NKG2A inhibitory receptors in vivo, shifting the balance towards immune activating signals.
To our knowledge, there is no study describing the relationship between HLA-E or HLA-F expression and chemotherapy. However, a study by Davidson et al. evaluated the expression of HLA-G in malignant effusions in ovarian carcinoma prior to and after chemotherapy. In light of their results, authors speculate that reduced expression of HLA-G in effusions obtained during or following chemotherapy and its correlation with improved survival could be related to preferential susceptibility of HLA-G expressing cells to the administered therapy (88). In our current study, we report significant dysregulation of both HLA-E and HLA-F between patients with a different status of MGMT promoter methylation, which is a predictive biomarker of GBM associated with TMZ. In terms of TMZ as the main chemotherapy agent for GBM, we cannot rule out its possible influence either. Therefore, animal models of GBM would be useful for a future study of the possible effect of concomitant therapy.
One limitation of our current study is the limited number of fresh-frozen tissue samples compared to some of the referenced studies with larger sample cohorts, which could have an impact on the results. However, similarly to our previous study (36), we demonstrated a positive correlation between HLA-E expression and OS of patients with GBM, this time in an independent larger sample cohort.
To conclude, we speculate that the expression of HLA-E by neoplastic cells may represent a coincidental selective pro-host advantage related to a better response to subsequent therapeutic modalities. However, the role of immunomodulatory non-classical HLA-E and HLA-F molecules in the antitumor response and development of GBM is still unknown. Whether the unexpected positive correlation of HLA-E expression with survival is related to the shift in balance between the activating and inhibitory signals to NK cells caused by the administered therapeutic modalities, leading to a favored occupancy of the activating receptors by this molecule is unclear, and this question remains a subject for further studies.
Conflicts of Interest
The Authors declare that they have no conflicts of interest regarding this study.
Authors’ Contributions
Conceptualization of the study: L.K. and J.S. Data curation: M.V., M.Hen. and V.V. Funding acquisition: L.K. Investigation: T.H., A.K., F.Si. and M.V. Methodology: T.H., O.S. and J.S. Project administration: J.S. Providing resources: R.J., M.Her., M.S. and R.L. Supervision of the study: R.J., M.Her., M.S., R.L. and O.S. Data validation: T.K., F.So. and V.K. Data visualization: A.K., F.Si. and M.V. Writing of the original draft: T.H., M.R. and J.S. Review and editing of the original draft: M.V., T.K. and P.F.
Acknowledgements
This work was financially supported by the Ministry of Health of the Czech Republic, grant no. 17-32758A, and conceptual development of research organizations (Masaryk Memorial Cancer Institute, 00209805; and University Hospital Brno, 65269705). We acknowledge the Core Facility Genomics CEITEC MU, which was supported by the NCMG research infrastructure (LM2018132 funded by MEYS CR), for their support in obtaining scientific data presented in this paper.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups , National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin. 2020;70(4):299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, Di Meco F, Lieberman F, Zhu JJ, Stragliotto G, Tran D, Brem S, Hottinger A, Kirson ED, Lavy-Shahaf G, Weinberg U, Kim CY, Paek SH, Nicholas G, Bruna J, Hirte H, Weller M, Palti Y, Hegi ME, Ram Z. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318(23):2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 5.Pachter JS, de Vries HE, Fabry Z. The blood-brain barrier and its role in immune privilege in the central nervous system. J Neuropathol Exp Neurol. 2003;62(6):593–604. doi: 10.1093/jnen/62.6.593. [DOI] [PubMed] [Google Scholar]
- 6.Gallegos CE, Michelin S, Dubner D, Carosella ED. Immunomodulation of classical and non-classical HLA molecules by ionizing radiation. Cell Immunol. 2016;303:16–23. doi: 10.1016/j.cellimm.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Balner H. The major histocompatibility complex of primates: evolutionary aspects and comparative histogenetics. Philos Trans R Soc Lond B Biol Sci. 1981;292(1057):109–119. doi: 10.1098/rstb.1981.0019. [DOI] [PubMed] [Google Scholar]
- 8.King A, Allan DS, Bowen M, Powis SJ, Joseph S, Verma S, Hiby SE, McMichael AJ, Loke YW, Braud VM. HLA-E is expressed on trophoblast and interacts with CD94/NKG2 receptors on decidual NK cells. Eur J Immunol. 2000;30(6):1623–1631. doi: 10.1002/1521-4141(200006)30:6<1623::AID-IMMU1623>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248(4952):220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 10.Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H, Geraghty DE. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J Immunol. 2003;171(3):1376–1384. doi: 10.4049/jimmunol.171.3.1376. [DOI] [PubMed] [Google Scholar]
- 11.Wischhusen J, Friese MA, Mittelbronn M, Meyermann R, Weller M. HLA-E protects glioma cells from NKG2D-mediated immune responses in vitro: implications for immune escape in vivo. J Neuropathol Exp Neurol. 2005;64(6):523–528. doi: 10.1093/jnen/64.6.523. [DOI] [PubMed] [Google Scholar]
- 12.Romagnani C, Pietra G, Falco M, Millo E, Mazzarino P, Biassoni R, Moretta A, Moretta L, Mingari MC. Identification of HLA-E-specific alloreactive T lymphocytes: a cell subset that undergoes preferential expansion in mixed lymphocyte culture and displays a broad cytolytic activity against allogeneic cells. Proc Natl Acad Sci USA. 2002;99(17):11328–11333. doi: 10.1073/pnas.172369799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braud VM, Allan DS, O’Callaghan CA, Söderström K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 14.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160(10):4951–4960. [PubMed] [Google Scholar]
- 15.Braud VM, Allan DS, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. 1998;8(1):1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 16.Ladasky JJ, Boyle S, Seth M, Li H, Pentcheva T, Abe F, Steinberg SJ, Edidin M. Bap31 enhances the endoplasmic reticulum export and quality control of human class I MHC molecules. J Immunol. 2006;177(9):6172–6181. doi: 10.4049/jimmunol.177.9.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5(2):103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 18.Gilfillan S, Ho EL, Cella M, Yokoyama WM, Colonna M. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol. 2002;3(12):1150–1155. doi: 10.1038/ni857. [DOI] [PubMed] [Google Scholar]
- 19.Iwaszko M, Bogunia-Kubik K. Clinical significance of the HLA-E and CD94/NKG2 interaction. Arch Immunol Ther Exp (Warsz) 2011;59(5):353–367. doi: 10.1007/s00005-011-0137-y. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser BK, Barahmand-Pour F, Paulsene W, Medley S, Geraghty DE, Strong RK. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174(5):2878–2884. doi: 10.4049/jimmunol.174.5.2878. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan LC, Clements CS, Rossjohn J, Brooks AG. The major histocompatibility complex class Ib molecule HLA-E at the interface between innate and adaptive immunity. Tissue Antigens. 2008;72(5):415–424. doi: 10.1111/j.1399-0039.2008.01138.x. [DOI] [PubMed] [Google Scholar]
- 22.Wainwright SD, Biro PA, Holmes CH. HLA-F is a predominantly empty, intracellular, TAP-associated MHC class Ib protein with a restricted expression pattern. J Immunol. 2000;164(1):319–328. doi: 10.4049/jimmunol.164.1.319. [DOI] [PubMed] [Google Scholar]
- 23.Lepin EJ, Bastin JM, Allan DS, Roncador G, Braud VM, Mason DY, van der Merwe PA, McMichael AJ, Bell JI, Powis SH, O’Callaghan CA. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur J Immunol. 2000;30(12):3552–3561. doi: 10.1002/1521-4141(200012)30:12<3552::AID-IMMU3552>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 24.Burian A, Wang KL, Finton KA, Lee N, Ishitani A, Strong RK, Geraghty DE. HLA-F and MHC-I open conformers bind natural killer cell Ig-like receptor KIR3DS1. PLoS One. 2016;11(9):e0163297. doi: 10.1371/journal.pone.0163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Beltran WF, Hölzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR, Rucevic M, Lamothe-Molina PA, Pertel T, Kim TE, Dugan H, Alter G, Dechanet-Merville J, Jost S, Carrington M, Altfeld M. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat Immunol. 2016;17(9):1067–1074. doi: 10.1038/ni.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol. 2013;191(7):3553–3562. doi: 10.4049/jimmunol.1300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F complex without peptide binds to MHC class I protein in the open conformer form. J Immunol. 2010;184(11):6199–6208. doi: 10.4049/jimmunol.1000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodridge JP, Lee N, Burian A, Pyo CW, Tykodi SS, Warren EH, Yee C, Riddell SR, Geraghty DE. HLA-F and MHC-I open conformers cooperate in a MHC-I antigen cross-presentation pathway. J Immunol. 2013;191(4):1567–1577. doi: 10.4049/jimmunol.1300080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldman-Wohl DS, Ariel I, Greenfield C, Hochner-Celnikier D, Cross J, Fisher S, Yagel S. Lack of human leukocyte antigen-G expression in extravillous trophoblasts is associated with pre-eclampsia. Mol Hum Reprod. 2000;6(1):88–95. doi: 10.1093/molehr/6.1.88. [DOI] [PubMed] [Google Scholar]
- 30.Ober C, Aldrich CL, Chervoneva I, Billstrand C, Rahimov F, Gray HL, Hyslop T. Variation in the HLA-G promoter region influences miscarriage rates. Am J Hum Genet. 2003;72(6):1425–1435. doi: 10.1086/375501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldman-Wohl DS, Ariel I, Greenfield C, Hanoch J, Yagel S. HLA-G expression in extravillous trophoblasts is an intrinsic property of cell differentiation: a lesson learned from ectopic pregnancies. Mol Hum Reprod. 2000;6(6):535–540. doi: 10.1093/molehr/6.6.535. [DOI] [PubMed] [Google Scholar]
- 32.Feger U, Tolosa E, Huang YH, Waschbisch A, Biedermann T, Melms A, Wiendl H. HLA-G expression defines a novel regulatory T-cell subset present in human peripheral blood and sites of inflammation. Blood. 2007;110(2):568–577. doi: 10.1182/blood-2006-11-057125. [DOI] [PubMed] [Google Scholar]
- 33.Nasef A, Mathieu N, Chapel A, Frick J, François S, Mazurier C, Boutarfa A, Bouchet S, Gorin NC, Thierry D, Fouillard L. Immunosuppressive effects of mesenchymal stem cells: involvement of HLA-G. Transplantation. 2007;84(2):231–237. doi: 10.1097/01.tp.0000267918.07906.08. [DOI] [PubMed] [Google Scholar]
- 34.Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in human cornea, an immune-privileged tissue. Hum Immunol. 2003;64(11):1039–1044. doi: 10.1016/j.humimm.2003.08.346. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Sun N, Li R, Sang X, Li X, Zhao J, Han J, Yang J, Ikezoe T. Targeting HLA-F suppresses the proliferation of glioma cells via a reduction in hexokinase 2-dependent glycolysis. Int J Biol Sci. 2021;17(5):1263–1276. doi: 10.7150/ijbs.56357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kren L, Slaby O, Muckova K, Lzicarova E, Sova M, Vybihal V, Svoboda T, Fadrus P, Lakomy R, Vanhara P, Krenova Z, Sterba J, Smrcka M, Michalek J. Expression of immune-modulatory molecules HLA-G and HLA-E by tumor cells in glioblastomas: an unexpected prognostic significance. Neuropathology. 2011;31(2):129–134. doi: 10.1111/j.1440-1789.2010.01149.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin A, Zhang X, Ruan YY, Wang Q, Zhou WJ, Yan WH. HLA-F expression is a prognostic factor in patients with non-small-cell lung cancer. Lung Cancer. 2011;74(3):504–509. doi: 10.1016/j.lungcan.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Seliger B, Jasinski-Bergner S, Quandt D, Stoehr C, Bukur J, Wach S, Legal W, Taubert H, Wullich B, Hartmann A. HLA-E expression and its clinical relevance in human renal cell carcinoma. Oncotarget. 2016;7(41):67360–67372. doi: 10.18632/oncotarget.11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Z, Liang J, Wang Z, Li A, Fan X, Jiang T. HLA-E expression in diffuse glioma: relationship with clinicopathological features and patient survival. BMC Neurol. 2020;20(1):59. doi: 10.1186/s12883-020-01640-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhen ZJ, Ling JY, Cai Y, Luo WB, He YJ. Impact of HLA-E gene polymorphism on HLA-E expression in tumor cells and prognosis in patients with stage III colorectal cancer. Med Oncol. 2013;30(1):482. doi: 10.1007/s12032-013-0482-2. [DOI] [PubMed] [Google Scholar]
- 41.Feng E, Liang T, Wang X, Du J, Tang K, Wang X, Wang F, You G. Correlation of alteration of HLA-F expression and clinical characterization in 593 brain glioma samples. J Neuroinflammation. 2019;16(1):33. doi: 10.1186/s12974-019-1418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J, Shao W, Dorak MT, Li Y, Miike R, Lobashevsky E, Wiencke JK, Wrensch M, Kaslow RA, Cobbs CS. Positive and negative associations of human leukocyte antigen variants with the onset and prognosis of adult glioblastoma multiforme. Cancer Epidemiol Biomarkers Prev. 2005;14(8):2040–2044. doi: 10.1158/1055-9965.EPI-05-0136. [DOI] [PubMed] [Google Scholar]
- 43.Wastowski IJ, Simões RT, Yaghi L, Donadi EA, Pancoto JT, Poras I, Lechapt-Zalcman E, Bernaudin M, Valable S, Carlotti CG Jr, Flajollet S, Jensen SS, Ferrone S, Carosella ED, Kristensen BW, Moreau P. Human leukocyte antigen-G is frequently expressed in glioblastoma and may be induced in vitro by combined 5-aza-2’-deoxycytidine and interferon-γ treatments: results from a multicentric study. Am J Pathol. 2013;182(2):540–552. doi: 10.1016/j.ajpath.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ondracek J, Fadrus P, Sana J, Besse A, Loja T, Vecera M, Radova L, Smrcka M, Slampa P, Slaby O. Global MicroRNA expression profiling identifies unique microRNA pattern of radioresistant glioblastoma cells. Anticancer Res. 2017;37(3):1099–1104. doi: 10.21873/anticanres.11422. [DOI] [PubMed] [Google Scholar]
- 45.Hackmon R, Pinnaduwage L, Zhang J, Lye SJ, Geraghty DE, Dunk CE. Definitive class I human leukocyte antigen expression in gestational placentation: HLA-F, HLA-E, HLA-C, and HLAG in extravillous trophoblast invasion on placentation, pregnancy, and parturition. Am J Reprod Immunol. 2017;77(6) doi: 10.1111/aji.12643. [DOI] [PubMed] [Google Scholar]
- 46.Coupel S, Moreau A, Hamidou M, Horejsi V, Soulillou JP, Charreau B. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood. 2007;109(7):2806–2814. doi: 10.1182/blood-2006-06-030213. [DOI] [PubMed] [Google Scholar]
- 47.Carosella ED, Moreau P, Le Maoult J, Le Discorde M, Dausset J, Rouas-Freiss N. HLA-G molecules: from maternal-fetal tolerance to tissue acceptance. Adv Immunol. 2003;81:199–252. doi: 10.1016/s0065-2776(03)81006-4. [DOI] [PubMed] [Google Scholar]
- 48.Lee N, Geraghty DE. HLA-F surface expression on B cell and monocyte cell lines is partially independent from tapasin and completely independent from TAP. J Immunol. 2003;171(10):5264–5271. doi: 10.4049/jimmunol.171.10.5264. [DOI] [PubMed] [Google Scholar]
- 49.Wei XH, Orr HT. Differential expression of HLA-E, HLA-F, and HLA-G transcripts in human tissue. Hum Immunol. 1990;29(2):131–142. doi: 10.1016/0198-8859(90)90076-2. [DOI] [PubMed] [Google Scholar]
- 50.Derré L, Corvaisier M, Charreau B, Moreau A, Godefroy E, Moreau-Aubry A, Jotereau F, Gervois N. Expression and release of HLA-E by melanoma cells and melanocytes: potential impact on the response of cytotoxic effector cells. J Immunol. 2006;177(5):3100–3107. doi: 10.4049/jimmunol.177.5.3100. [DOI] [PubMed] [Google Scholar]
- 51.Harada A, Ishigami S, Kijima Y, Nakajo A, Arigami T, Kurahara H, Kita Y, Yoshinaka H, Natsugoe S. Clinical implication of human leukocyte antigen (HLA)-F expression in breast cancer. Pathol Int. 2015;65(11):569–574. doi: 10.1111/pin.12343. [DOI] [PubMed] [Google Scholar]
- 52.Wu B, Yang H, Ying S, Lu H, Wang W, Lv J, Xiong H, Hu W. High HLA-F expression is a poor prognosis factor in patients with nasopharyngeal carcinoma. Anal Cell Pathol (Amst) 2018;2018:7691704. doi: 10.1155/2018/7691704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marín R, Ruiz-Cabello F, Pedrinaci S, Méndez R, Jiménez P, Geraghty DE, Garrido F. Analysis of HLA-E expression in human tumors. Immunogenetics. 2003;54(11):767–775. doi: 10.1007/s00251-002-0526-9. [DOI] [PubMed] [Google Scholar]
- 54.Andersson E, Poschke I, Villabona L, Carlson JW, Lundqvist A, Kiessling R, Seliger B, Masucci GV. Non-classical HLA-class I expression in serous ovarian carcinoma: Correlation with the HLA-genotype, tumor infiltrating immune cells and prognosis. Oncoimmunology. 2015;5(1):e1052213. doi: 10.1080/2162402X.2015.1052213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boujelbene N, Ben Yahia H, Babay W, Gadria S, Zemni I, Azaiez H, Dhouioui S, Zidi N, Mchiri R, Mrad K, Ouzari HI, Charfi L, Zidi I. HLA-G, HLA-E, and IDO overexpression predicts a worse survival of Tunisian patients with vulvar squamous cell carcinoma. HLA. 2019;94(1):11–24. doi: 10.1111/tan.13536. [DOI] [PubMed] [Google Scholar]
- 56.Ishigami S, Arigami T, Okumura H, Uchikado Y, Kita Y, Kurahara H, Maemura K, Kijima Y, Ishihara Y, Sasaki K, Uenosono Y, Natsugoe S. Human leukocyte antigen (HLA)-E and HLA-F expression in gastric cancer. Anticancer Res. 2015;35(4):2279–2285. [PubMed] [Google Scholar]
- 57.Reimers MS, Engels CC, Putter H, Morreau H, Liefers GJ, van de Velde CJ, Kuppen PJ. Prognostic value of HLA class I, HLA-E, HLA-G and Tregs in rectal cancer: a retrospective cohort study. BMC Cancer. 2014;14:486. doi: 10.1186/1471-2407-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mittelbronn M, Simon P, Löffler C, Capper D, Bunz B, Harter P, Schlaszus H, Schleich A, Tabatabai G, Goeppert B, Meyermann R, Weller M, Wischhusen J. Elevated HLA-E levels in human glioblastomas but not in grade I to III astrocytomas correlate with infiltrating CD8+ cells. J Neuroimmunol. 2007;189(1-2):50–58. doi: 10.1016/j.jneuroim.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Spaans VM, Peters AA, Fleuren GJ, Jordanova ES. HLA-E expression in cervical adenocarcinomas: association with improved long-term survival. J Transl Med. 2012;10:184. doi: 10.1186/1479-5876-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levy EM, Bianchini M, Von Euw EM, Barrio MM, Bravo AI, Furman D, Domenichini E, Macagno C, Pinsky V, Zucchini C, Valvassori L, Mordoh J. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int J Oncol. 2008;32(3):633–641. [PubMed] [Google Scholar]
- 61.van Hall T, André P, Horowitz A, Ruan DF, Borst L, Zerbib R, Narni-Mancinelli E, van der Burg SH, Vivier E. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J Immunother Cancer. 2019;7(1):263. doi: 10.1186/s40425-019-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun C, Xu J, Huang Q, Huang M, Wen H, Zhang C, Wang J, Song J, Zheng M, Sun H, Wei H, Xiao W, Sun R, Tian Z. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Oncoimmunology. 2016;6(1):e1264562. doi: 10.1080/2162402X.2016.1264562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, Liefers GJ, van den Elsen PJ, van de Velde CJ, Kuppen PJ. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. 2010;185(12):7452–7459. doi: 10.4049/jimmunol.1002629. [DOI] [PubMed] [Google Scholar]
- 64.Benevolo M, Mottolese M, Tremante E, Rollo F, Diodoro MG, Ercolani C, Sperduti I, Lo Monaco E, Cosimelli M, Giacomini P. High expression of HLA-E in colorectal carcinoma is associated with a favorable prognosis. J Transl Med. 2011;9:184. doi: 10.1186/1479-5876-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.John T, Black MA, Toro TT, Leader D, Gedye CA, Davis ID, Guilford PJ, Cebon JS. Predicting clinical outcome through molecular profiling in stage III melanoma. Clin Cancer Res. 2008;14(16):5173–5180. doi: 10.1158/1078-0432.CCR-07-4170. [DOI] [PubMed] [Google Scholar]
- 66.Kamiya T, Seow SV, Wong D, Robinson M, Campana D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J Clin Invest. 2019;129(5):2094–2106. doi: 10.1172/JCI123955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lauterbach N, Wieten L, Popeijus HE, Voorter CE, Tilanus MG. HLA-E regulates NKG2C+ natural killer cell function through presentation of a restricted peptide repertoire. Hum Immunol. 2015;76(8):578–586. doi: 10.1016/j.humimm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Michaëlsson J, Teixeira de Matos C, Achour A, Lanier LL, Kärre K, Söderström K. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J Exp Med. 2002;196(11):1403–1414. doi: 10.1084/jem.20020797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geraghty DE, Wei XH, Orr HT, Koller BH. Human leukocyte antigen F (HLA-F). An expressed HLA gene composed of a class I coding sequence linked to a novel transcribed repetitive element. J Exp Med. 1990;171(1):1–18. doi: 10.1084/jem.171.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boyle LH, Gillingham AK, Munro S, Trowsdale J. Selective export of HLA-F by its cytoplasmic tail. J Immunol. 2006;176(11):6464–6472. doi: 10.4049/jimmunol.176.11.6464. [DOI] [PubMed] [Google Scholar]
- 71.Riederer I, Sievert W, Eissner G, Molls M, Multhoff G. Irradiation-induced up-regulation of HLA-E on macrovascular endothelial cells confers protection against killing by activated natural killer cells. PLoS One. 2010;5(12):e15339. doi: 10.1371/journal.pone.0015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pereira BI, Devine OP, Vukmanovic-Stejic M, Chambers ES, Subramanian P, Patel N, Virasami A, Sebire NJ, Kinsler V, Valdovinos A, LeSaux CJ, Passos JF, Antoniou A, Rustin MHA, Campisi J, Akbar AN. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat Commun. 2019;10(1):2387. doi: 10.1038/s41467-019-10335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Michelin S, Gallegos CE, Dubner D, Favier B, Carosella ED. Ionizing radiation modulates the surface expression of human leukocyte antigen-G in a human melanoma cell line. Hum Immunol. 2009;70(12):1010–1015. doi: 10.1016/j.humimm.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 74.Abdel-Wahab Z, Dar MM, Hester D, Vervaert C, Gangavalli R, Barber J, Darrow TL, Seigler HF. Effect of irradiation on cytokine production, MHC antigen expression, and vaccine potential of interleukin-2 and interferon-gamma gene-modified melanoma cells. Cell Immunol. 1996;171(2):246–254. doi: 10.1006/cimm.1996.0200. [DOI] [PubMed] [Google Scholar]
- 75.Cangemi G, Morandi B, D’Agostino A, Peri C, Conte R, Damonte G, Ferlazzo G, Biassoni R, Melioli G. IFN-alpha mediates the up-regulation of HLA class I on melanoma cells without switching proteasome to immunoproteasome. Int Immunol. 2003;15(12):1415–1421. doi: 10.1093/intimm/dxg140. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen S, Beziat V, Dhedin N, Kuentz M, Vernant JP, Debre P, Vieillard V. HLA-E upregulation on IFN-gamma-activated AML blasts impairs CD94/NKG2A-dependent NK cytolysis after haplo-mismatched hematopoietic SCT. Bone Marrow Transplant. 2009;43(9):693–699. doi: 10.1038/bmt.2008.380. [DOI] [PubMed] [Google Scholar]
- 77.Wolpert F, Roth P, Lamszus K, Tabatabai G, Weller M, Eisele G. HLA-E contributes to an immune-inhibitory phenotype of glioblastoma stem-like cells. J Neuroimmunol. 2012;250(1-2):27–34. doi: 10.1016/j.jneuroim.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Malmberg KJ, Levitsky V, Norell H, de Matos CT, Carlsten M, Schedvins K, Rabbani H, Moretta A, Söderström K, Levitskaya J, Kiessling R. IFN-gamma protects short-term ovarian carcinoma cell lines from CTL lysis via a CD94/NKG2A-dependent mechanism. J Clin Invest. 2002;110(10):1515–1523. doi: 10.1172/JCI15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gustafson KS, Ginder GD. Interferon-gamma induction of the human leukocyte antigen-E gene is mediated through binding of a complex containing STAT1alpha to a distinct interferon-gamma-responsive element. J Biol Chem. 1996;271(33):20035–20046. doi: 10.1074/jbc.271.33.20035. [DOI] [PubMed] [Google Scholar]
- 80.Min W, Pober JS, Johnson DR. Interferon induction of TAP1: the phosphatase SHP-1 regulates crossover between the IFN-alpha/beta and the IFN-gamma signal-transduction pathways. Circ Res. 1998;83(8):815–823. doi: 10.1161/01.res.83.8.815. [DOI] [PubMed] [Google Scholar]
- 81.Whitmire JK, Tan JT, Whitton JL. Interferon-gamma acts directly on CD8+ T cells to increase their abundance during virus infection. J Exp Med. 2005;201(7):1053–1059. doi: 10.1084/jem.20041463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 83.Yu J, Wei M, Becknell B, Trotta R, Liu S, Boyd Z, Jaung MS, Blaser BW, Sun J, Benson DM Jr, Mao H, Yokohama A, Bhatt D, Shen L, Davuluri R, Weinstein M, Marcucci G, Caligiuri MA. Pro- and antiinflammatory cytokine signaling: reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity. 2006;24(5):575–590. doi: 10.1016/j.immuni.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 84.Siegel JP. Effects of interferon-gamma on the activation of human T lymphocytes. Cell Immunol. 1988;111(2):461–472. doi: 10.1016/0008-8749(88)90109-8. [DOI] [PubMed] [Google Scholar]
- 85.Weigent DA, Stanton GJ, Johnson HM. Interleukin 2 enhances natural killer cell activity through induction of gamma interferon. Infect Immun. 1983;41(3):992–997. doi: 10.1128/iai.41.3.992-997.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, Parsa AT. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17(11):1381–1385. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kozlowska AK, Tseng HC, Kaur K, Topchyan P, Inagaki A, Bui VT, Kasahara N, Cacalano N, Jewett A. Resistance to cytotoxicity and sustained release of interleukin-6 and interleukin-8 in the presence of decreased interferon-γ after differentiation of glioblastoma by human natural killer cells. Cancer Immunol Immunother. 2016;65(9):1085–1097. doi: 10.1007/s00262-016-1866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davidson B, Elstrand MB, McMaster MT, Berner A, Kurman RJ, Risberg B, Trope CG, Shih IeM. HLA-G expression in effusions is a possible marker of tumor susceptibility to chemotherapy in ovarian carcinoma. Gynecol Oncol. 2005;96(1):42–47. doi: 10.1016/j.ygyno.2004.09.049. [DOI] [PubMed] [Google Scholar]