Abstract
Modified nucleotides in tRNA are critical components of the translation apparatus but their importance in the process of translational regulation had until recently been greatly overlooked. Two breakthroughs have recently allowed to more fully understand the importance of tRNA modifications in bacterial physiology. One is the identification of the full set of tRNA modifications genes in model organisms such as Escherichia coli K12. The second is the improvement of available analytical tools to monitor tRNA modifications patterns. The role of tRNA modifications vary greatly with the specific modification within a given tRNA and with the organism studied. Their absence or reduction can lead to cell death, pleiotropic phenotypes or have no apparent visible effect. By linking translation through their decoding functions to metabolism through their biosynthetic pathways, tRNA modifications are emerging as important components of the bacterial regulatory toolbox.
Keywords: Decoding efficiency, post-transcriptional regulation, translation
The resurgence of tRNA modifications as key players in bacterial physiology.
As the central adaptors between mRNA molecules and elongating peptides, transfer RNAs (tRNAs) are at the heart of the translation machinery and must therefore interact very specifically with a great diversity of molecules in the cell (such as ribosomes, elongation factors, mRNAs, aminoacyl-tRNA synthetases) [1]. This is made possible by the diversification of tRNA structures by modifications of both the sugar and base moieties [2]. Specific tRNA modifications play key roles in the accuracy and efficiency of decoding, act as determinants or anti-determinants for proteins of the translation apparatus or toxins and can also be critical for monitoring tRNA integrity and stability as quality control checkpoints [2,3]. The Anti-codon Stem Loop (ASL) portion of the tRNA molecule is the most modified particularly the nucleosides that interacts with the third base of the codon or the wobble position (position 34) and the ones that are located just before the first base of the codon (position 37)(Fig. 1). These modifications can have critical roles expanding or restricting the decoding properties of a given tRNA molecules [4]. Positions outside the ASL usually have more structural roles [3] but have also shown to influence the decoding properties [5].
Figure 1. Full set of tRNA modification genes in the model gram-negative E. coli K12 MG1655.
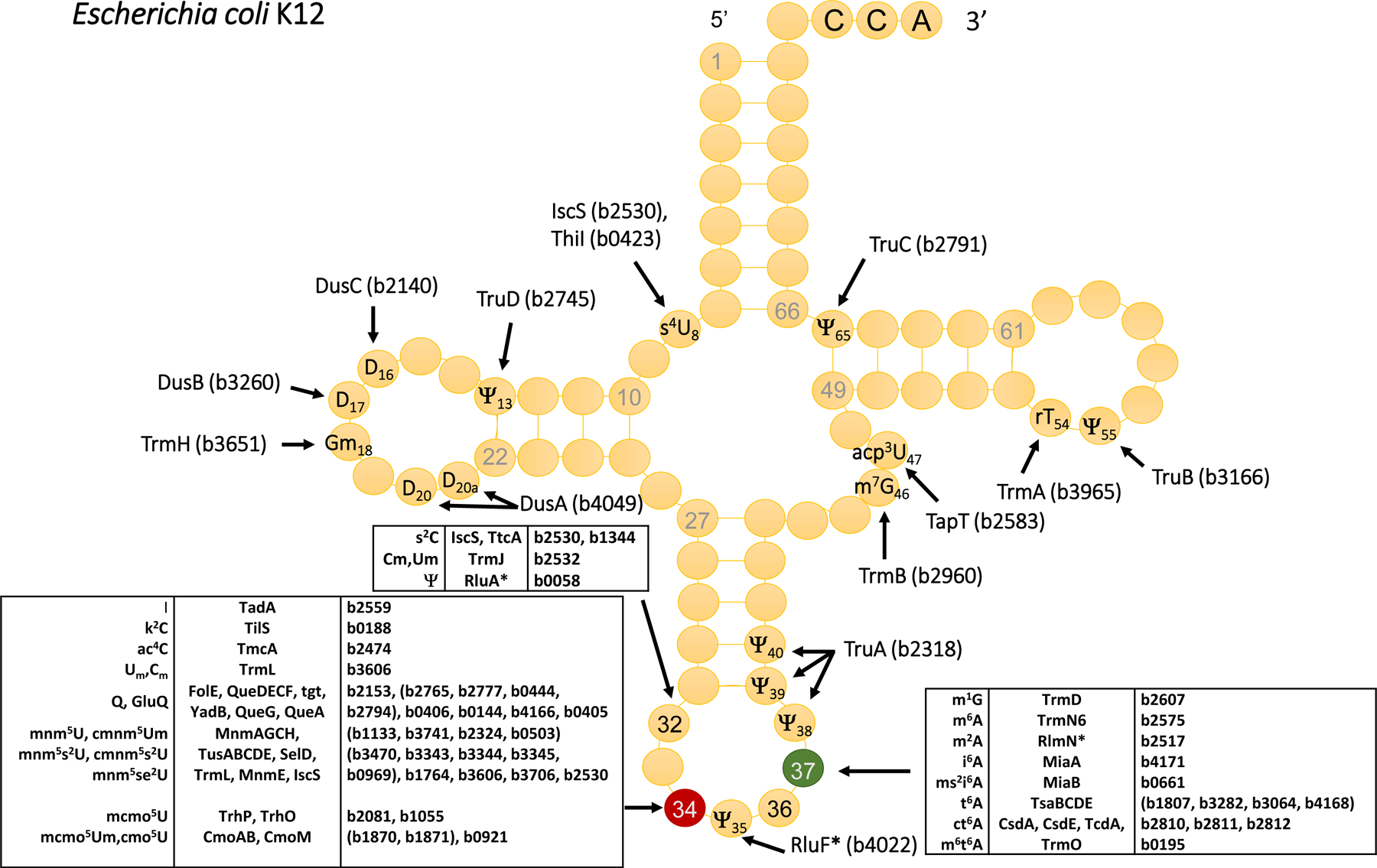
The references for the functional role of every gene can be found in UniProt release 2020_04 (August 12, 2020) [114] using the gene locus tags as input.
Even if most tRNA modifications were discovered ~50 years ago [6], it is only recently that the full appreciation of their importance in biology in general and in bacterial physiology in particular has more broadly emerged. This is mostly because the full set of tRNA modification genes has only been recently identified in a few model organisms such as Escherichia coli (Fig. 1) and yeast [7] and because novel analytical techniques have led to more accurate and sensitive quantification methods [8].
This review will focus on recent developments revealing the emerging role of tRNA modifications as regulatory molecules in bacteria, evaluating tRNA modifications enzymes as possible antibacterial targets and reviewing newly discovered roles for bacterial tRNA modifications in the process.
Completeness of the bacterial tRNA modification landscape, a work in progress.
The first known tRNA modification, pseudouridine (Ψ), was initially identified in yeast and then shortly after in bacteria [9]. The following 25 years can be considered as the first golden period in the tRNA modification field (reviewed in [6]). The majority of the known tRNA modifications were discovered and around half the genes involved in their synthesis identified, mainly in the Gram-negative model Escherichia coli K12 and Salmonella typhimurium LT2 [10]. This pioneer work laid out most of the concepts on the importance of tRNA modifications for bacterial physiology but these remained known to only a few specialists for a long time, or at least until 2005 [10–12]. Thanks to the development of whole genome sequencing technology, bioanalytical tools, and DNA recombinant technology, the last 15 years have witnessed a second golden era for tRNA modifications. First, many of the cases of “missing” genes were solved. Nowadays, the complete sets of genes coding for tRNA modification enzymes are available for the gram-negative E. coli K12 and the gram-positive Mycoplasma capricolum ([13,14] and Fig. 1 & 2A). It took several decades to finalize E. coli, but the recent discoveries of the genes of the t6A37, acp3U47 and cmo5U34 pathways have translated in discovering novel enzymes families and chemistries (see Fig. 1 legend for access to references ), allowed the identification of disease genes in humans [15–17], reinforcing Jacques Monod’s statement on “what is true in E. coli is true in the elephant” and the importance of bacterial model organisms in fundamental research [18]. Second, improvements in mass-spectrometry and the arrival of whole genome sequencing based detection methods now allow to capture the identity and location of tRNA modifications in more organisms (Fig. 3). This information is essential as, for the moment, predicting the exact nature and positions of all tRNA modifications in a given organism just based on the presence/absence of homologs of known modification enzymes remains problematic. In our experience, orthology-based methods can correctly predict ~70–80% of the tRNA modifications (V. de Crécy-Lagard, unpublished data). Indeed, new modifications are constantly being discovered as more diverse bacteria are analyzed. For example, though Vibrio cholerae is taxonomically closely related to E. coli, a totally new modification, acetylated acp3U (acacp3U), was recently identified in this organism [19]. Also, cases of changes in target specificity can make direct annotations transfers between homologs perilous. For example, the synthesis of m5U54 in tRNA and m5U1939 in 23S RNA are catalyzed by predicted orthologs in B. subtilis and Mycoplasma capricolum respectively [20]. Likewise, non-orthologous replacements make homology-based predictions not totally reliable. For examples, synthesis of m5U54, xo5U34 and ac4C34 are catalyzed by non-orthologous enzymes in B. subtilis and E. coli [21–23]. Thus, the presence/absence of a known modification gene does not always warrant the presence/absence of the corresponding modification at given position of tRNA as many factors influence these modifications events [24].
Figure 2. Minimal tRNA modification sets.
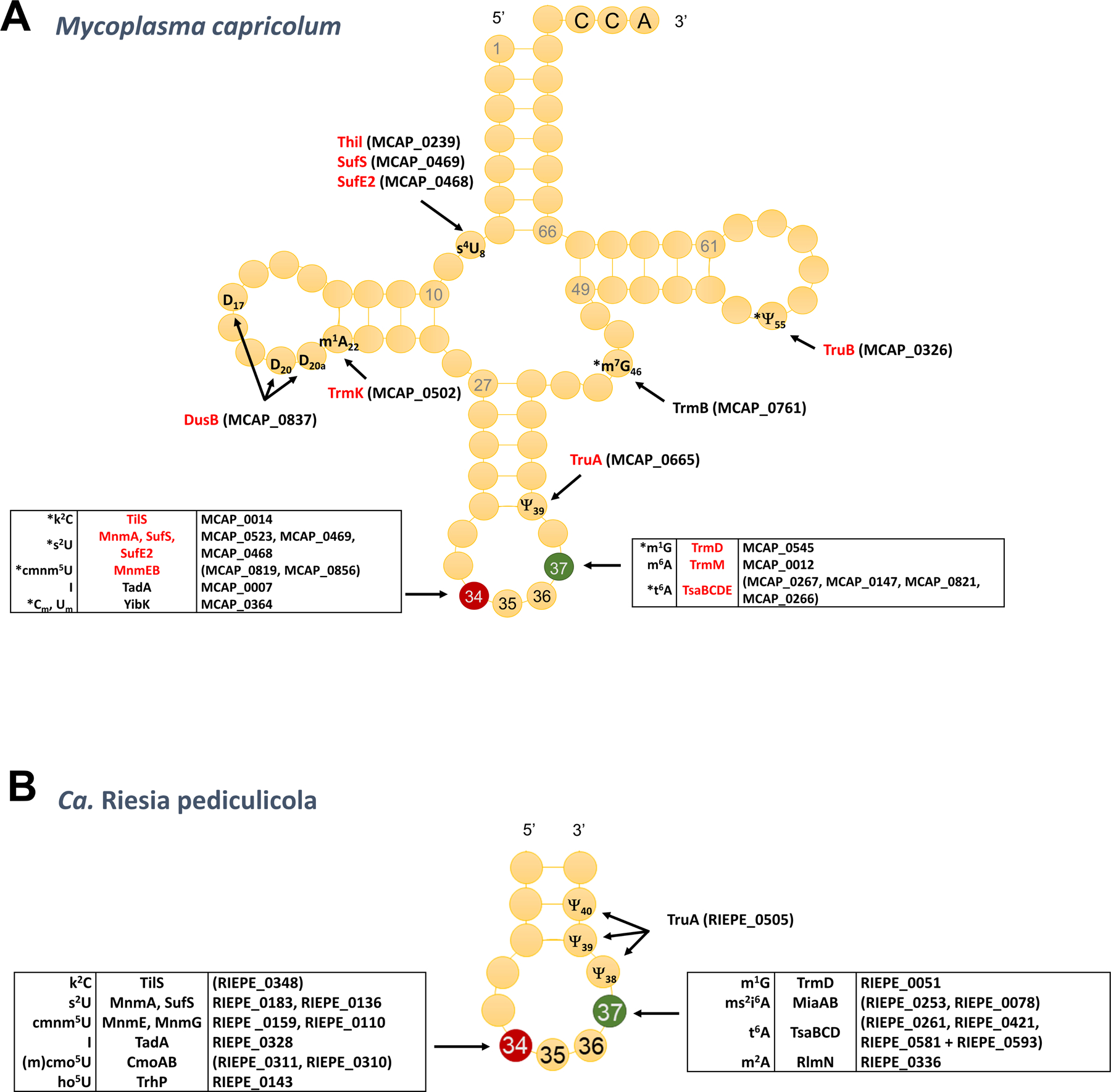
(A) Full set of tRNA modifications genes in the minimal gram-positive Mycoplasma capricolum, in red are the ones conserved in Mycoplasma JCVI-Syn3 (see Table S2 for gene list) and are starred (*) the ones considered that are most resistant to gene loss in Molllicute evolution [33]; (B) Predicted tRNA modifications in Ca. Riesia pediculicola and corresponding gene, updated from [41].
Figure 3. Current pipelines to identify tRNA modifications.
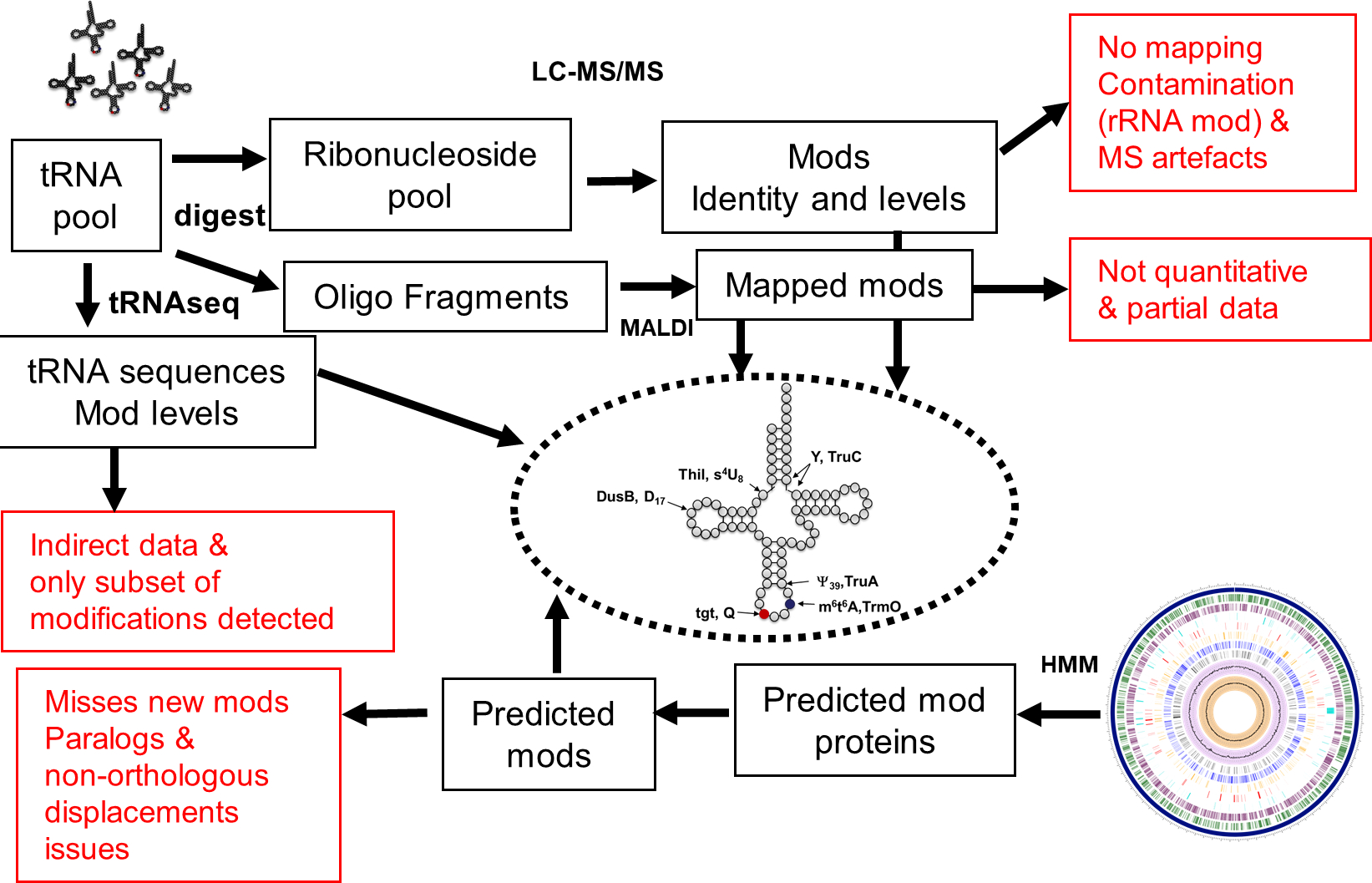
Summary of methods currently used to identify and map tRNA modifications that are tRNA-seq, mass spectrometry (LC-MS/MS or MALDI-based), and comparative genomics (using Hidden Markov Models or HMM to predict the presence/absence of tRNA modification). The major issues with each method are listed in red.
Only 226 individual tRNA sequences (with modification information) from 19 different bacteria are available in the Modomics database [25] (Date February 4 2020). Hence the mapping of tRNA modifications along the bacterial tree of life is still very sparse (Fig. 4 and Table S1). Expanding this knowledge is critical for the field to progress. Recent studies that combine mass-spectrometry and tRNA-Seq methods to detect and map tRNA modifications [19,26–28] suggest that gathering such information on phylogenetically diverse bacteria is getting easier and could be more systematically pursued as shown in this recent analysis of modifications of microbiome bacteria [29]. Combining several types of analytical methods with predictions of modifications genes is required to provide an accurate depiction of the final pattern of modifications (both the nature and positions) in a given organism (Fig. 3).
Figure 4. Poor coverage of bacterial species with sequenced tRNAs.
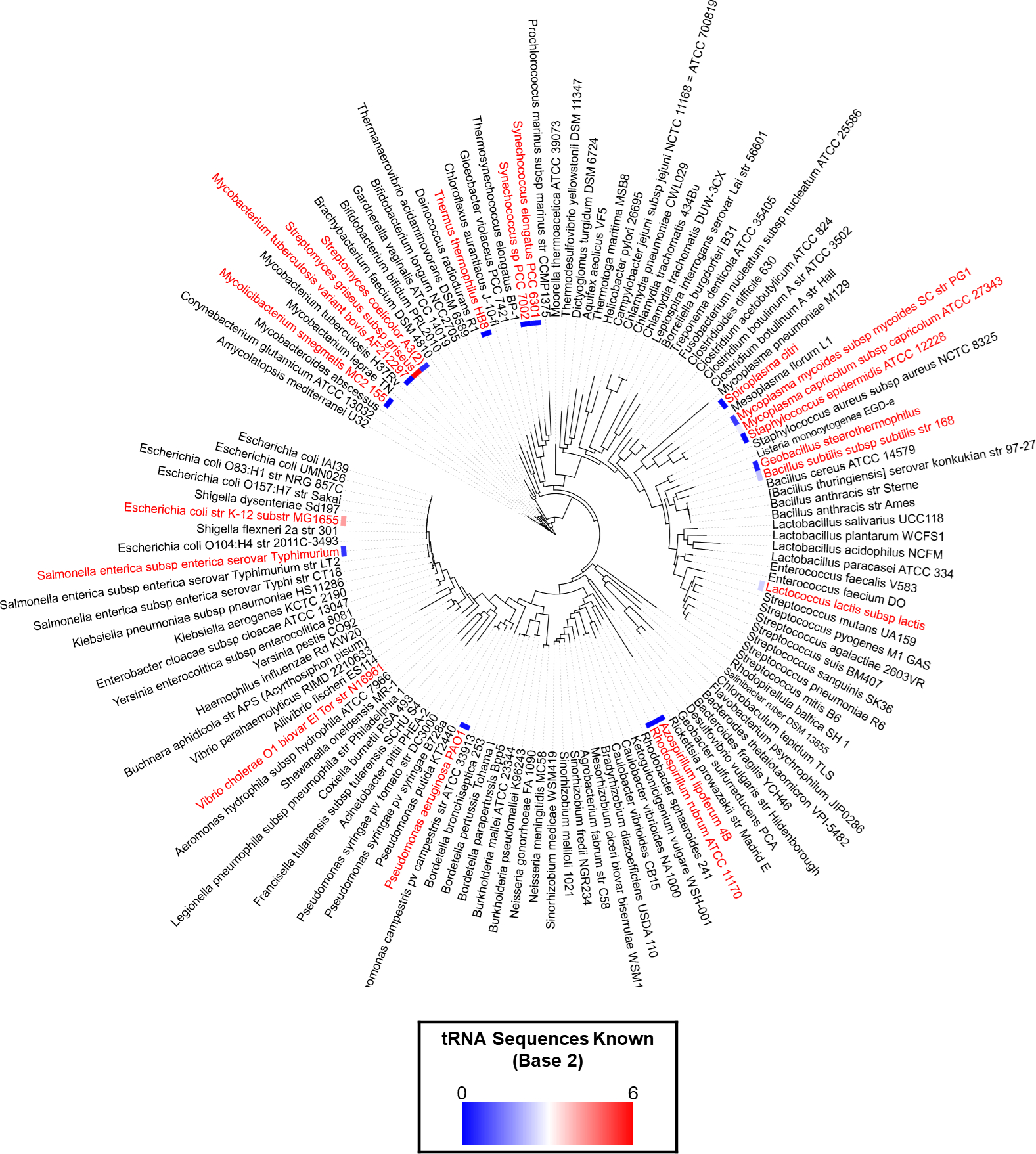
Organisms in red have available sequence data for tRNAs. It can vary from the full set to just a few The number of known tRNA sequences for each species was transformed into log2 and is represented by a color gradient to the left of each species name. The color gradient range goes from 0 (blue) to 6 (red)., The full data is available in Table S1. The species tree was created in iTol [115] using the list of 120 reference bacteria of the Patric database [116] merged with the organisms that had tRNA sequenced data to give a total of 131 species in the trees.
Finally, modification levels are not static and can vary with physiological conditions [12,26]. The recent improvements in analytical tools allow to better track these fluctuations even at the level of the tRNA sequence and in certain cases link them to physiological roles as discussed in the last section of this review.
The importance of specific modifications can only be understood in a given context
If some tRNA modifications are found in a very limited set of isoacceptors or bacteria, such as m5 s2U only found in thermophiles [30], many are widespread. Defining which modification(s) in a given tRNA is/are more important, eventually “essential”, for life, is a difficult task. First, for most modifications involved in decoding, one cannot separate the modifications from the context of the tRNA sequences as there is a clear co-evolution of the two. For example, lysidine (k2C) is a modification at the wobble C position of tRNAIleCAU only found in Bacteria (see [31] for recent review). When modified this tRNA can decode AUA codons but without the k2C modification, it will decode AUG codon leaving the minor Ile codon without a cognate tRNA. Hence, the lysidine synthesis gene is essential and very conserved throughout the bacterial kingdom. One exception is Mycoplasma mobile where the loss of the lysidine synthase gene co-occurs with the mutation in the tRNAIle anticodon from CAU to UAU and possibly other mutation(s) in the decoding site of the ribosome (as discussed in [32] and [33]). Other examples of co-evolution of the tRNA sets and modification genes are detailed in the recent analysis by Diwan and Agashe [34]. Second, both for modifications required for accurate decoding or for correct tRNA folding, different organisms will use different solutions for the same problem. For example, ribothymidine at position 54 (m5U54), that is critical for tRNA stability in bacteria, is replaced by m1Ψ54 in many Archaea [35]. The two modifications fulfill similar roles but are introduced by totally different enzymes. Third, the requirements for modifications are going to be extremely dependent on environmental factors such as temperature or salinity that are known to greatly influence tRNA structure [3]. For example, hyperthermophiles have unique modifications as mentioned above and modify their tRNA more extensively when grown at higher temperature [3]. With these reservations in place, it is possible to list key functional constraints for specific tRNA isoacceptors groups such as split codon box discrimination or codon/anticodon binding that can be solved by the presence of different modifications in different organisms [4], [36].
Defining the minimal tRNA modification set(s) required for bacterial life
Early studies trying to predict the minimal sets of tRNA modifications required for life that were based on phylogeny. Essentiality or conservation gave very different results [37–39]. These ranged from three to nine modifications and these did not always overlap. More recent studies that focused on synthesizing a minimal genome [40] (in red in Fig. 2A and Table S2) or analyzing gene losses in Mollicutes [33] led to minimal set in Gram-positive of around 8 modifications (starred in Fig. 2A). Another strategy to identify minimal tRNA modification sets is to analyze genomes of intracellular symbionts of insects with reduced genomes [18]. The analysis of the human louse endosymbiont Ca. Riesia pediculicola, suggests that organisms can survive with just six modifications in the ASL [41] (Fig. 2B). This number could even be further reduced to the barebone minimum of two modifications (t6A37 and m1G37) in Ca. Spiroplasma holothuricola a gut endosymbiont of Zygothuria oxysclera [42]. Though this prediction requires experimental validation, it must have been made possible by concomitant changes in the anticodon of tRNAs decoding split boxes [42]. In summary, if most extant bacteria that live in competitive environments contain 30–50 distinct modifications in tRNAs to maintain an efficient and accurate translation, very few modifications are conserved in all bacteria and most can be lost in certain conditions, with the exception of t6A37 and m1G37. Both these modifications are universal but because of their essentiality in bacteria and because their synthesis machineries are different between bacteria and eukarya [2], they are also the two tRNA modification pathways that can be targeted for the development of antibacterial compounds.
Development of tRNA modifications enzymes as antibacterial targets.
Not many modifications are essential for bacterial growth, even fewer are essential in a wide range of bacteria. For example, tadA, the gene involved in I34 synthesis in tRNAArgICG is essential in E. coli because it allows this tRNA to decode all CGU/C/A codons [31]. Of note, the post-transcriptional process that results in RNA sequences that differ from the DNA sequence from which they are transcribed, such as the case of A to I, is called editing [43]. The tadA gene is not essential in B. subtilis [44] for reasons that are not clear as no tRNAArgUCG gene that would make tadA dispensable is found in this organism [33]. Possibilities include; 1) the presence of an unknown redundant deaminase in B. subtilis; 2) a different decoding context that allows unmodified tRNAArgACG to decode CGU/C/A codons as seen in plant cytosol [45] or Mycoplasma capricolum [46]; or that the essentiality is cause by another function of TadA. For example, in E. coli, this proteins edits the mRNA of toxin/antitoxin systems in E. coli [47]. Even if further work is required to understand the dispensability of tadA in several bacteria [48], the prediction is that compounds such as xanthorrhizol that inhibit TadA [49], might only target a narrow range of organisms.
The three tRNA modifications that are essential in a wide range of bacteria are lysidine (k2C34), m1G37 and t6A37. The lysidine modification is only found in bacterial tRNAs, so we have here an ideal scenario, with an antibacterial target essential in bacteria but absent in humans. A small subset of bacteria has escaped the requirement for lysidine by mutating their tRNAIleCAU to tRNAIleUAU, as discussed above but these are rare cases. As a proof of principle, a few ATP and lysine analogs have been shown to inhibit the lysidination reaction [50] but none seem to have been developed further, maybe because studies have shown that tRNAIleCAU to tRNAIleUAU suppressor mutations allows B. subtilis tilS mutant to survive [51].
The issue of t6A37 essentiality also turned out to be quite complex. This modification was shown to be a positive determinant for tRNA charging by Isoleucyl-tRNA synthetase (IleRS ) from E. coli and not from yeast [52] providing a rational for the essentiality of t6A synthesis genes in many Bacteria such as E. coli, Staphylococcus aureus or Pseudomonas aeruginosa [53] but not in S. cerevisiae [54]. However, t6A-deficient strains can be constructed by deleting the synthesis genes in other bacterial species such as Deinococcus radiodurans, Synechocystis PCC6803 or Streptococcus mutans [52,55], and the reasons why t6A is not essential in these species are still not clear. That said, the essentiality of t6A in many pathogens make its synthesis pathway an attractive target particularly as two of the proteins of the t6A synthesis complex, TsaB and TsaE, are specific to Bacteria [53] and inhibitors for the ATPase activity of TsaE have already been identified [56].
The most advanced target is TrmD, the enzyme involved in generating m1G37 and member of the COG0336 family [57]. The trmD gene has been shown to be essential in over 23 bacteria including many human pathogens [58]. Because this methylase family is not orthologous to the one that catalyzes the same reaction in eukaryotes (Trm5, COG2520, while TrmD belongs to COG0336) [57], it was identified as an antibacterial target early on [59], and studies combining structure function and medicinal chemistry to develop leads have followed [60–62], suggesting that drug developments programs are in progress. The recent discovery that the absence of TrmD affects the expression of drug efflux proteins make this antibacterial target all the more compelling [63].
tRNA modifications defects can lead to pleiotropic phenotypes possibly caused by protein aggregation
Multiple studies on the role of different ASL modifications in yeast and mammals (such as t6A37, xms2U34 or Queuosine34 (Q34)), clearly show that their absence leads to protein homeostasis defects and to increased levels of protein aggregation that give rise to pleiotropic phenotypes [64–66]. In Bacteria, elimination of the same three ASL modifications also give rise to pleiotropic phenotypes. Indeed, deletion of mnmE and mnmG/gidA genes have been shown to have pleiotropic effects on growth, cell division and virulence in a wide range of pathogenic bacteria both gram-positive and gram-negative (see [67] and discussion of [68] for reviews). These genes encode proteins that form a complex involved in the formation of mnm5U34 but this has been experimentally validated in only a few species such as E. coli, B. subtilis [69] and Salmonella [70]. The molecular mechanisms underlying these pleiotropic phenotypes are far from understood as the expression of hundreds of proteins are affected. Only in one case it was shown that the translation of a specific virulence protein, cytotoxic necrotizing factor 1, is affected [68], but it is not known if this is a direct or indirect effect.
Deleting the genes involved in Q synthesis also gives raise to diverse sets of phenotypes: reduced viability in stationary phase in E. coli [71]; sensitivity to oxidative stress in Streptococcus thermophilus [72] or defects in virulence in Shigella flexneri [73,74] There again, the molecular basis for all these phenotypes remain unknown.
In organisms like E. coli or Staphylococcus aureus where t6A37 is essential, limiting expression of the t6A genes leads to cell division defects [75,76], increased protein glycation [77] and induction of the stringent response [78]. Even in organisms that can survive without t6A, its absence leads to diverse and pleiotropic phenotypes. The t6A-deficient Deinococcus radiodurans strains are more sensitive to mitomycin C [79], while in Streptococcus mutans the same t6A-deficient mutants are compromised in biofilm formation, and more sensitive than the wild-type to low pH and oxidative stress [52,55,80]. Growth defects and induction of the stringent response are observed in B. subtilis when t6A is absent [81–83]. Deleting t6A genes in Synechocystis PCC6803 affects salt tolerance, altered pigmentation, and cyanophycin accumulation [52,84]. No specific mistranslated protein have been identified in any of these cases, but proteomic analysis in D. radiodurans does suggest protein homeostasis is affected as t6A deficient strains overexpress express chaperones such as GroEL and ClpB [52].
tRNA modifications particularly of the ASL are one of the factors that affect translation kinetics, so any perturbation on these modification levels leads to amino acids misincorporation and misfolding [85]. The consequences are an imbalance in protein homeostasis, an idea first put forward by Nedialkova and Leidel in yeast [86] that could explain many of the pleiotropic phenotypes caused by ASL modification defects in bacteria and will require further experimental validation.
tRNA modifications provide both targets and protection from ribonucleases
It is well established in eukaryotes that tRNA modifications act as quality control signals and tRNAs that lack modifications are degraded by specific machineries (see introduction of [87] for review). tRNA modifications can also be determinants for toxins and nucleases that generate tRNA fragments with regulatory roles under stress [88]. Recent studies suggest tRNA modifications play similar roles in bacteria, but the field is much more advanced in eukaryotes. The first studies showing the importance of modifications (mainly ψ55 and m7G47) on the tRNA structure and integrity were performed in thermophilic bacteria, where they seem to also have regulatory roles allowing Thermus thermophilus to grow at different temperatures [89]. More recently it was shown that absence s4U8 in V. cholerae leads to total tRNA degradation by the tRNA degradosome particularly if other modifications such as ψ55 are also missing [87]. Like in eukaryotes tRNA modifications can be determinants for bacterial toxins as shown for the PrrC toxin that requires the presence of the t6A modification to cleave its target tRNA [55].
Regulatory mechanisms mediated by tRNA modifications.
As we extensively discussed in a previous review [90], the expression of specific genes can be affected by the levels of a given modification (see Box 1 for the definition of Modification Tunable Transcripts or MoTTs). MoTTs can be directly or indirectly part of regulatory loops and the number of examples is steadily increasing (Fig. 5 and Table 1), allowing general themes to emerge. First, the cases of regulation by MoTTs seem to occur for genes that are already regulatory hubs with multiple layers of control such as RpoS or KatA/KatB [91]. Second, regulatory systems that rely on translation speed such as regulation by attenuation or translational coupling seem to be very sensitive to the presence of modification in the decoding tRNAs as already reported by the pioneer studies of Bruce Ames [92] and Charles Yanofsky [93]. Third, the number of “sensor codons” that are affected by the level of the regulated modification can vary greatly from a unique Ser codon [94], to a total reprogramming of the proteome in the hypoxic response in M. smegmatis [26] similar to the reprogramming seen in eukaryotes in stress or cancer [95].
Box 1: What are Modification Tunable Transcripts (MoTTs)?
The term was coined by Dedon and Begley [96] and can be summarized as transcripts that will be translated with different efficiencies in response to the levels of a specific modification. MoTTs can identified by analyzing codon biases and looking specifically for genes enriched for the codon(s) that is/are decoded by the modified tRNA(s). For example, in Mycobacterium bovis BCG, the choice of the Thr-codon at a specific position will determine if it is decoded by a tRNA that is modified or not by cmo5U [26]. Translation of transcripts enriched in the modification-dependent codon (in this case Thr-ACG) will be more sensitive to changes in cmo5U than transcripts enriched in the modification independent codons (in this case Thr-ACC). Combining codon usage bias analysis with proteomics analysis often required to identify MoTTs. Some MoTTs cannot be identified with codon bias analyses if all isoacceptors tRNAs used are modified. In these cases, stretches of codons (such as the in the MgtL example [107]) or possibly specific di or tri-codon environment [113] can make the translation efficiency of the MoTT very sensitive to the presence/absence of the modification.
Figure 5.
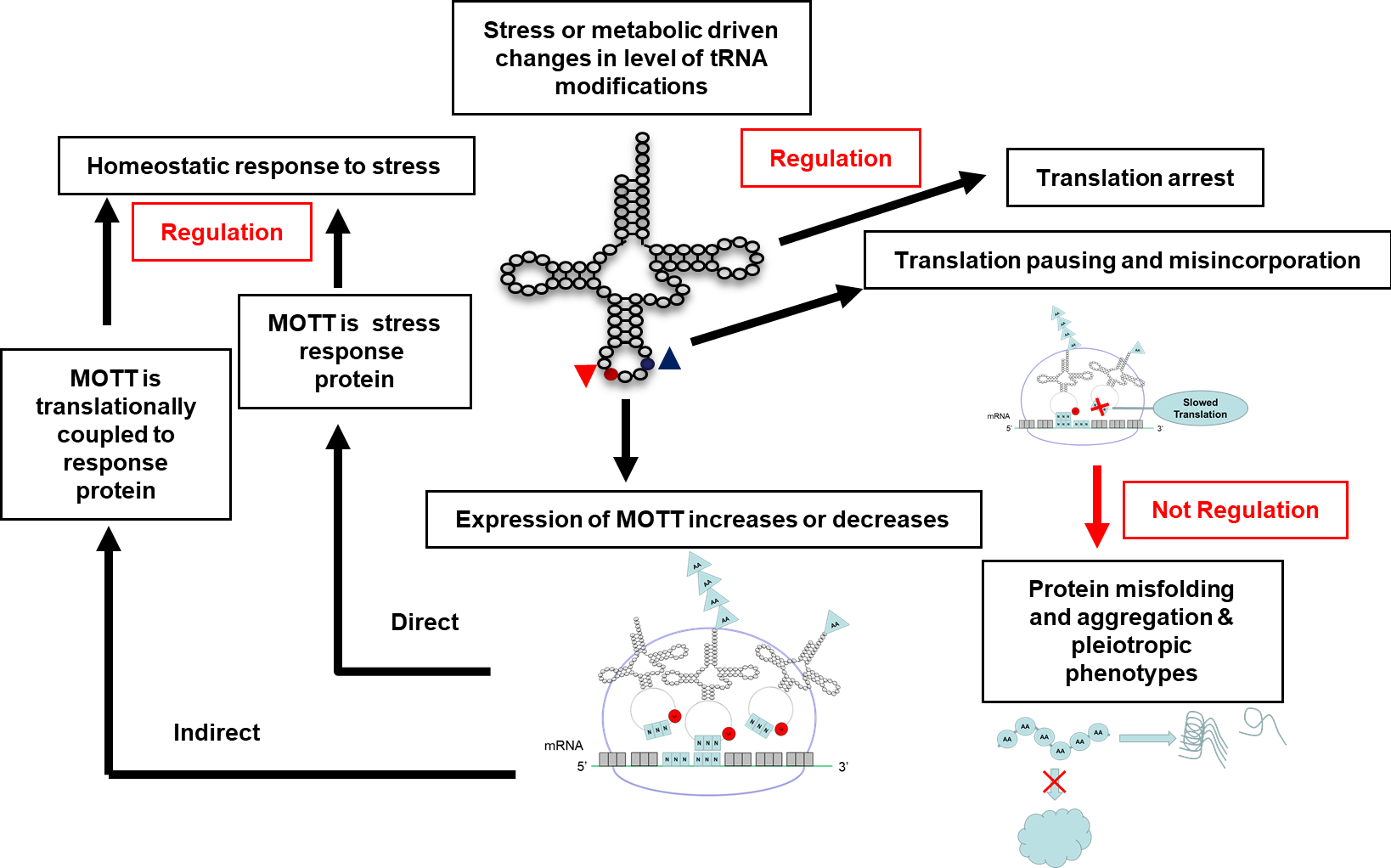
Possible regulatory consequences of changes in tRNA modification levels that affect decoding efficiency.
Table 1.
Example of regulations by tRNA modifications in Bacteria
| Organism | Stress | Mod | tRNA | Mod Enzyme | Target protein/codon | Mechanism |
|---|---|---|---|---|---|---|
| Escherichia coli K12 | Low iron | ms2i6A37 | Ser-UGA | Fe/S dependent Enzyme MiaB Ser-UCA |
U ofSer-UCA | Low iron, leads to reduced MiaB activity and lower modification of tRNASer- and to poor translation of uof and fur encoding the negative regulator of the low iron response (reviewed in [90]) |
| ? | i6A37 | Leu-UAA | MiaA | RpoS. IraP,Hfq Leu-UUX |
MiaA, TrmL and TusA identifies as MOTTs but mechanisms that affects modification levels unknown [104,105] | |
| ? | C/U34m | Leu-C/UAA | TrmL | RpoS Leu-UUX |
[105] | |
| ? | s2U34 | Leu-UAA | TusA | RpoS Leu-UUX |
[105] | |
| Growth phase | mcmo5U | Pro-UGG | CmoM | ? | The ratio of mcmo5U/cmo5U increases clearly during growth phase but the molecular mechanisms are yet to be discovered [106]. | |
| Salmonella typhimurium LT2 | Low Mg2+ | m1G37 | Pro-C/G/UGG | Mg2+ dependent enzyme TrmD | MgtL, Pro-CCX |
Low Mg2+ leads to reduced TrmD activity and lower modification of tRNAPro that triggers attenuation of the MgtL leader peptide allowing expression of the coupled mgtA transporter gene [107] |
| Pseudomonas aeruginosa PAO1 | H2O2 | m7G47 | Asp-GUC Phe-GAA |
TrmB | KatA and KatB Phe-UUC Asp-GAC |
Transcription of trmB is increased under oxidative stress by an unknown mechanism leading to increased translation of the katAB genes [108] |
| H2O2 | C/U/Am32 | TrmJ | OxyR ? |
trmJ is essential for a proper expression of katB during H2O2 stress through a modulation of oxyR expression it was not shown that OxyR translation is affected at specific codons [109] | ||
| H2O2 | s2C32 | TtcA | KatA ? |
The expression of TtcA is under the positive control of OxyR. It is postulated but not proven that this leads to increased translation of the catalase gene KatA [110] | ||
| Mycobacterium bovis BCG | Low O2 | cmo5U34 | Thr-UGU | CmoR (BCG_0224) | DosR, Thr-ACG |
Reprogramming of tRNA and modification pool switches proteome and allows increased expression of DosR the regulator of the hypoxic response [26]. |
|
Salmonella typhimurium LT2 & E. coli K12 |
UV | s4U | Many | NA | NA | Presence of s4U under UV stress leads to cross-linking of tRNAs and to a translation block similar to stringent response [111,112]. |
We anticipate the list of MoTTs involved in regulation will grow as more modification genes get correctly annotated and as the technological platforms required to identify them, such as quantification of tRNA modification and tRNA transcripts levels, as well as whole cell transcriptome and proteome [96] become more accessible. There are indeed many reports of phenotypes linked to the absence of modifications that are yet to be understood at the molecular level. As discussed above some of these might be caused by general defects of protein homeostasis and not by perturbed regulatory loops. However, we anticipate that the arsenal of “omics” analysis should allow to unearth unknown MoTTs both in recent examples like the discovery that deleting miaB involved in the synthesis of ms2i6A37 influences morphogenesis and moenomycin biosynthesis in Streptomyces ghanaensis ATCC14672 [97], or in studies published over 20 years ago like the report that the miaE mutant of S. typhimurium has growth defects on specific carbon sources suggesting that growth occurs on succinate, fumarate, or malate only if the isopentenyl group of tRNA is hydroxylated [98].
Concluding Remarks and Future Perspectives
It seems we are just at the beginning of grasping the role tRNA modifications play in bacteria. Indeed, most of the published work focuses only on a few model organisms, and even though the molecular role of a given modification on tRNA stability or in codon-anticodon pairing can be very conserved, the consequences of eliminating it can have very different physiological consequences. The most extreme example is t6A37 that is essential in some organisms like E. coli but not required for normal growth in others like Deinococcus radiodurans [52]. Studies of more organisms could reveal additional cases of alternate strategies that allow to fulfil the same function help identify narrow-range antibacterial targets.
One factor that greatly hampers our understanding of the physiological role of tRNA modifications is the lack of data on their nature and positions in most organisms in the bacterial tree illustrated with just the small set of reference organisms shown in Fig. 4. As discussed above, predictions based on genomics analyses are valuable, but many factors make them inaccurate. These would be greatly improved if tRNA modification maps were available for chosen organisms covering the diversity of the bacterial tree. This goal is currently technically challenging as only a few laboratories master the needed mass-spectrometry tools [19] and might require the improvement of sequencing techniques to directly identify tRNA modifications. Recent development of the Nanopore platform suggests that this is possible [99].
Recent technological advances to measure translation speed [100] will also help to understand the role of modifications in modulating translation efficiency at the codon level. This step is critical to better predict why the translation of a specific codon and not another is affected by modification levels and could allow to better predict MoTTs. In general, the added complexity of regulation by translation speed mediated by tRNA modification levels will need to be integrated in regulation models.
Finally, tRNA modifications are only a small part of much larger epitranscriptomics cellular process, with modification of rRNA but also of mRNA and other non-coding RNAs. The field of epitranscriptomic has exploded in recent years mainly because of the discoveries of the regulatory roles of modification of mRNA in eukaryotes [101] and interplays between the tRNA and mRNA modification processes have recently emerged [102]. The field of mRNA modification in bacteria is not as developed but a recent study found that TadA not only edited tRNAs but also mRNA molecules [43], and m6A has been detected in bacterial mRNAs [103], so it would not be surprising that other more interconnections between modifications of tRNAs and of other types of RNA molecules might be discovered in the near future.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (grant no. R01 GM70641 to V.d.C.-L.). We thank Peter C. Dedon, Geoffrey Hutinet and Henri Grosjean for their insightful comments on the manuscript.
Glossary
- Anticodon
Triplet sequence of tRNA that hybridizes with the codon sequence on the mRNA.
- Anticodon Stem Loop (ASL)
Decoding region of the tRNA.
- Editing
A post Translational process that results in RNA sequences that differ from their DNA sequences.
- Epitranscritomic
The biology of RNA modifications and their associated regulatory factors.
- Isoacceptor tRNA
Any transfer RNA species obtainable from a given organism that can be acylated by the same amino acid; they may differ in their anticodons.
- Modification Tunable Transcripts (MoTT)
See Box 1.
- Split codon boxes
In the genetic code table, each codon box contains codons that start with the same two bases but differ by the third base. Hence, each codon box harbors four codons. In split-codon boxes these codons can encode different amino acids usually in 2/2 or 1/3 arrangements.
- Non-orthologous replacements
Proteins of different evolutionary origins that fulfill the exact same function.
- Wobble base
A wobble base pair is a pairing between two nucleotides in RNA molecules that does not follow Watson-Crick base pair rules.
Abbreviations not explained in text
- m1G
1-methylguanosine
- m1Ψ
1-methylpseudouridine
- m1U
1-methyluracil
- m5 s2U
5-methyl-2-thiouridine
- mnm5U
5-methylaminomethyluridine
- m7G
7-methylguanosine
- acacp3U
acetylated 3-(3-amino-3-carboxypropyl)uridine
- I
inosine
- k2C
lysidine
- ac4C
N4-acetylcytidine
- t6A
N6-threonylcarbamoyladenosine
- Ψ
pseudouridine
- Q
queuosine
- m5U
5-methyluridine
- xo5U
5-hydroxyuridine derivatives
- cmo5U
uridine 5-oxyacetic acid
- Am
2′-O-methyladenosine
- Cm
2′-O-methylcytidine
- Um
2′-O-methyluridine
References
- 1.Shepherd J and Ibba M (2015) Bacterial transfer RNAs. FEMS Microbiol. Rev 39, 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El Yacoubi B et al. (2012) Biosynthesis and function of posttranscriptional modifications of Transfer RNAs. Annu. Rev. Genet 46, 69–95 [DOI] [PubMed] [Google Scholar]
- 3.Lorenz C et al. (2017) tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosjean H et al. (2010) Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett. 584, 252. [DOI] [PubMed] [Google Scholar]
- 5.Cochella L and Green R (2005) An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308, 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosjean H (2005) Modification and editing of RNA: Historical overview and important facts to remember. In Fine-Tuning of RNA Functions by Modification and Editing 12pp. 1–22 [Google Scholar]
- 7.Phizicky EM and Hopper AK (2010) tRNA biology charges to the front. Genes Dev. 24, 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helm M and Motorin Y (2017) Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet 18, 275–291 [DOI] [PubMed] [Google Scholar]
- 9.Cohn WE (1960) bjork GR 1. J. Biol. Chem 235, 1488–1498 [PubMed] [Google Scholar]
- 10.Björk GR et al. (1987) Transfer RNA modification. Annu Rev Biochem 56, 263–87 [DOI] [PubMed] [Google Scholar]
- 11.Nishimura S and Watanabe K (2006) The discovery of modified nucleosides from the early days to the present: A personal perspective. J. Biosci 31, 465–475 [DOI] [PubMed] [Google Scholar]
- 12.Persson BC (1993) Modification of tRNA as a regulatory device. Mol. Microbiol 8, 1011–1016 [DOI] [PubMed] [Google Scholar]
- 13.Björk GR and Hagervall TG (2014) Transfer RNA modification: presence, synthesis, and function. EcoSal Plus, DOI: 10.1128/ecosalplus.ESP-0007-2013 [DOI] [PubMed] [Google Scholar]
- 14.de Crécy-Lagard V et al. (2007) Comparative RNomics and Modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB Life 59, 634–58 [DOI] [PubMed] [Google Scholar]
- 15.Edvardson S et al. (2017) tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet 25, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun DA et al. (2017) Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet 49, 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrondel C et al. (2019) Defects in t(6)A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun 10, 3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmer C (2008) Microcosm: E. coli and the new science of life, New York: Pantheon. [Google Scholar]
- 19.Kimura S et al. (2020) Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat Chem Biol (in press) DOI: 10.1101/723049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lartigue C et al. (2014) The flavoprotein Mcap0476 (RlmFO) catalyzes m5U1939 modification in Mycoplasma capricolum 23S rRNA. Nucleic Acids Res. 42, 8073–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urbonavicius J et al. (2005) Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria--evolutionary implications. Nucleic Acids Res. 33, 3955–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu H et al. (2018) Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine. Nucleic Acids Res. 46, 9160–9169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taniguchi T et al. (2018) Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat. Chem. Biol 14, 1010–1020 [DOI] [PubMed] [Google Scholar]
- 24.Barraud P and Tisné C (2019) To be or not to be modified: Miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB Life 71, 1126–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boccaletto P et al. (2018) MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chionh YH et al. (2016) tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun 7, 13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puri P et al. (2014) Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol. Microbiol 93, 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grobe S et al. (2019) Identification and quantification of (t)RNA modifications in Pseudomonas aeruginosa by Liquid Chromatography–Tandem Mass Spectrometry. ChemBioChem 20, 1430–1437 [DOI] [PubMed] [Google Scholar]
- 29.Schwartz MH et al. (2018) Microbiome characterization by high-throughput transfer RNA sequencing and modification analysis. Nat. Commun 9, 5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hori H et al. (2018) Transfer RNA modification enzymes from thermophiles and their modified nucleosides in tRNA. Microorganisms 6, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson EM and Alexander RW (2019) Bacterial wobble modifications of NNA-decoding tRNAs. IUBMB Life 71, 1158–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniguchi T et al. (2013) Decoding system for the AUA codon by tRNAIle with the UAU anticodon in Mycoplasma mobile. Nucleic Acids Res 41, 2621–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grosjean H et al. (2014) Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet. 10, e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diwan GD and Agashe D (2018) Wobbling forth and drifting back: the evolutionary history and impact of bacterial tRNA modifications. Mol. Biol. Evol 35, 2046–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterjee K et al. (2012) The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA 18, 421–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grosjean H and Westhof E (2016) An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 44, 8020–8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forster AC and Church GM (2006) Towards synthesis of a minimal cell. Mol. Syst. Biol 2, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cermakian N and Cedergren R (1998) Modified nucleosides always were: an evolutionary model. In Modification and Editing of RNA (Grosjean H and Benne R, eds), pp. 535–541, ASM Press [Google Scholar]
- 39.Ouzounis CA et al. (2006) A minimal estimate for the gene content of the last universal common ancestor--exobiology from a terrestrial perspective. Res. Microbiol 157, 57. [DOI] [PubMed] [Google Scholar]
- 40.Hutchison CA et al. (2016) Design and synthesis of a minimal bacterial genome. Science 351, aad6253. [DOI] [PubMed] [Google Scholar]
- 41.de Crécy-Lagard V et al. (2012) Decoding in Candidatus Riesia pediculicola, close to a minimal tRNA modification set? Trends cell Mol. Biol 7, 11–34 [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y et al. (2018) Unique tRNA gene profile suggests paucity of nucleotide modifications in anticodons of a deep-sea symbiotic Spiroplasma. Nucleic Acids Res. 46, 2197–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bar-Yaacov D et al. (2018) RNA editing in bacteria: occurrence, regulation and significance. RNA Biol. 15, 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koo B-M et al. (2017) Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 4, 291–305.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aldinger CA et al. (2012) The absence of A-to-I editing in the anticodon of plant cytoplasmic tRNA (Arg) ACG demands a relaxation of the wobble decoding rules. RNA Biol. 9, 1239–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokobori S et al. (2013) Life without tRNAArg-adenosine deaminase TadA: evolutionary consequences of decoding the four CGN codons as arginine in Mycoplasmas and other Mollicutes. Nucleic Acids Res. 41, 6531–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bar-Yaacov D et al. (2017) RNA editing in bacteria recodes multiple proteins and regulates an evolutionarily conserved toxin-antitoxin system. Genome Res. 27, 1696–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W-H et al. (2017) OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 45, D940–D944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yogiara et al. (2015) Escherichia coli ASKA clone library harboring tRNA-specific adenosine deaminase (tadA) reveals resistance towards xanthorrhizol. Molecules 20, 16290–16305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salowe SP et al. (2009) The catalytic flexibility of tRNAIle-lysidine synthetase can generate alternative tRNA substrates for isoleucyl-tRNA synthetase. J. Biol. Chem 284, 9656–9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fabret C et al. (2011) Life without the essential bacterial tRNAIle2–lysidine synthetase TilS: a case of tRNA gene recruitment in Bacillus subtilis. Mol. Microbiol. 80, 1062–1074 [DOI] [PubMed] [Google Scholar]
- 52.Thiaville PC et al. (2015) Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol. Microbiol 98, 1199–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiaville PC et al. (2014) Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol. 11, 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Yacoubi B et al. (2009) The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 37, 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bacusmo JM et al. (2017) The t(6)A modification acts as a positive determinant for the anticodon nuclease PrrC, and is distinctively nonessential in Streptococcus mutans. RNA Biol. 15, 508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lerner CG et al. (2007) From bacterial genomes to novel antibacterial agents: discovery, characterization, and antibacterial activity of compounds that bind to HI0065 (YjeE) from Haemophilus influenzae. Chem. Biol. Drug Des 69, 395–404 [DOI] [PubMed] [Google Scholar]
- 57.Goto-Ito S et al. (2017) Trm5 and TrmD: two enzymes from distinct origins catalyze the identical tRNA modification, m1G37. Biomolecules 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo H et al. (2014) DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 42, D574–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hill PJ et al. (2013) Selective inhibitors of bacterial t-RNA-(N1G37) methyltransferase (TrmD) that demonstrate novel ordering of the lid domain. J. Med. Chem 56, 7278–7288 [DOI] [PubMed] [Google Scholar]
- 60.Zhong W et al. (2019) Thienopyrimidinone derivatives that inhibit bacterial tRNA (guanine37-N(1))-methyltransferase (TrmD) by restructuring the active site with a tyrosine-flipping mechanism. J. Med. Chem 62, 7788–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jaroensuk J et al. (2019) Crystal structure and catalytic mechanism of the essential m(1)G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa. RNA 25, 1481–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong W et al. (2019) Targeting the bacterial epitranscriptome for antibiotic development: discovery of novel tRNA-(N(1)G37) methyltransferase (TrmD) inhibitors. ACS Infect. Dis 5, 326–335 [DOI] [PubMed] [Google Scholar]
- 63.Masuda I et al. (2019) tRNA methylation Is a global determinant of bacterial multi-drug resistance. Cell Syst. 8, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schaffrath R and Leidel SA (2017) Wobble uridine modifications–a reason to live, a reason to die?! RNA Biol. 14, 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pollo-Oliveira L et al. (2020) Loss of Elongator- and KEOPS-dependent tRNA modifications leads to severe growth phenotypes and protein aggregation in yeast. Biomolecules 10, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuorto F et al. (2018) Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 37, e99777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shippy CD and Fadl AA (2014) tRNA modification enzymes GidA and MnmE: potential role in virulence of bacterial pathogens. Int.l J. of Mol. Sci 15, 18267–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu H and Kim KS (2012) mRNA context dependent regulation of cytotoxic necrotizing factor 1 translation by GidA, a tRNA modification enzyme in Escherichia coli. Gene 491, 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moukadiri I et al. (2018) Bacillus subtilis exhibits MnmC-like tRNA modification activities. RNA Biol. 15, 1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shippy DC et al. (2013) Virulence characteristics of Salmonella following deletion of genes encoding the tRNA modification enzymes GidA and MnmE. Microb. Pathog 57, 1–9 [DOI] [PubMed] [Google Scholar]
- 71.Noguchi S et al. (1982) Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J. Biol. Chem 257, 6544–6550 [PubMed] [Google Scholar]
- 72.Thibessard A et al. (2004) Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl Env. Microbiol 70, 2220–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durand J et al. (1994) vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of E. coli K-12. J Bacteriol 176, 4627–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gradler U et al. (2001) A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase. J Mol Biol 306, 455–67. [DOI] [PubMed] [Google Scholar]
- 75.Handford JI et al. (2009) Conserved network of proteins essential for bacterial viability. J. Bacteriol 191, 4732–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng L et al. (2007) Conditional mutation of an essential putative glycoprotease eliminates autolysis in Staphylococcus aureus. J. Bacteriol 189, 2734–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katz C et al. (2010) The ubiquitous conserved glycopeptidase Gcp prevents accumulation of toxic glycated proteins. MBio 1, e00195–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bergmiller T et al. (2011) Single-cell time-lapse analysis of depletion of the universally conserved essential protein YgjD. BMC Microbiol. 11, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Onodera T et al. (2013) Deinococcus radiodurans YgjD and YeaZ are involved in the repair of DNA cross-links. Extremophiles 17, 171–179 [DOI] [PubMed] [Google Scholar]
- 80.Bitoun JP et al. (2014) Deficiency of BrpB causes major defects in cell division, stress responses and biofilm formation by Streptococcus mutans. Microbiology 160, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karst JC et al. (2009) The ATPase activity of an “essential” Bacillus subtilis enzyme, YdiB, is required for its cellular function and is modulated by oligomerization. Microbiology 155, 944–956 [DOI] [PubMed] [Google Scholar]
- 82.Benoist C et al. (2015) Constitutive stringent response restores viability of Bacillus subtilis lacking structural maintenance of chromosome protein. PLoS One 10, e0142308–e0142308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ogura M et al. (2019) Bacillus subtilis YlxR, which is involved in glucose-responsive metabolic changes, regulates expression of tsaD for protein quality control of pyruvate dehydrogenase. Front. Microbiol 10, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zuther E et al. (1998) Mutation of a gene encoding a putative glycoprotease leads to reduced salt tolerance, altered pigmentation, and cyanophycin accumulation in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 180, 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stein KC and Frydman J (2019) The stop-and-go traffic regulating protein biogenesis: How translation kinetics controls proteostasis. J. Biol. Chem 294, 2076–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nedialkova DD and Leidel SA (2015) Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kimura S and Waldor MK (2019) The RNA degradosome promotes tRNA quality control through clearance of hypomodified tRNA. Proc. Natl. Acad. Sci. U. S. A 116, 1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chanfreau GF (2017) Impact of RNA modifications and RNA-modifying enzymes on eukaryotic ribonucleases. In The Enzymes 41pp. 299–329 [DOI] [PubMed] [Google Scholar]
- 89.Hori H (2019) Regulatory factors for tRNA modifications in extreme-thermophilic bacterium Thermus thermophilus. Front. Genet 10, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pollo-Oliveira L and de Crécy-Lagard V (2019) Can protein expression be regulated by modulation of tRNA Modification profiles? Biochemistry 58, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gottesman S (2019) Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem 294, 11685–11700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lewis JA and Ames BN (1972) Histidine regulation in Salmonella typhimurium. XI. The percentage of transfer RNAHis charged in vivo and its relation to the repression of the histidine operon. J. Mol. Biol 66, 131–42 [DOI] [PubMed] [Google Scholar]
- 93.Gollnick P and Yanofsky C (1990) tRNA(Trp) translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J. Bacteriol 172, 3100–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Veĉerek B et al. (2007) Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 26, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Advani VM and Ivanov P (2019) Translational control under stress: reshaping the translatome. BioEssays 41, 1900009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Endres L et al. (2015) Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 12, 603–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sehin Y et al. (2019) Gene ssfg_01967 (miaB) for tRNA modification influences morphogenesis and moenomycin biosynthesis in Streptomyces ghanaensis ATCC14672. Microbiology 165, 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Persson BC et al. (1998) The ms2io6A37 modification of tRNA in Salmonella typhimurium regulates growth on citric acid cycle intermediates. J. Bacteriol 180, 3144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith AM et al. (2019) Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One 14, e0216709–e0216709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao J et al. (2019) Translatomics: the global view of translation. Int. J. Mol. Sci 20, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frye M et al. (2016) RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet 17, 365–372 [DOI] [PubMed] [Google Scholar]
- 102.Ontiveros RJ et al. (2020) Coordination of mRNA and tRNA methylations by TRMT10A. Proc. Natl. Acad. Sci 117, 7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deng X et al. (2015) Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 43, 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aubee JI et al. (2016) The i6A37 tRNA modification is essential for proper decoding of UUX-Leucine codons during rpoS and iraP translation. RNA 22, 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aubee JI et al. (2017) TrmL and TusA are necessary for rpoS and MiaA is required for HFQ expression in Escherichia coli. Biomolecules 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sakai Y et al. (2016) Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 44, 509–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hou Y-M et al. (2019) Codon-specific translation by m1G37 methylation of tRNA. Front. Genet 9, 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thongdee N et al. (2019) TrmB, a tRNA m7G46 methyltransferase, plays a role in hydrogen peroxide resistance and positively modulates the translation of katA and katB mRNAs in Pseudomonas aeruginosa. Nucleic Acids Res. 47, 9271–9281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jaroensuk J et al. (2016) Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa. Nucleic Acids Res. 44, 10834–10848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Romsang A et al. (2018) Pseudomonas aeruginosa ttcA encoding tRNA-thiolating protein requires an iron-sulfur cluster to participate in hydrogen peroxide-mediated stress protection and pathogenicity. Sci. Rep 8, 11882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomas G and Favre A (1975) 4-thiouridine as the target for near-ultraviolet light induced growth delay in Escherichia coli. Biochem. Biophys. Res. Commun 66, 1454–1461 [DOI] [PubMed] [Google Scholar]
- 112.Kramer GF et al. (1988) Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and ApppGpp as components of an adaptive response. J Bacteriol 170, 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chevance FFV and Hughes KT (2017) Case for the genetic code as a triplet of triplets. Proc. Natl. Acad. Sci 114, 4745–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Letunic I and Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 [DOI] [PubMed] [Google Scholar]
- 116.Wattam AR et al. (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


