Abstract
Arachidonic acid (AA) is the precursor to leukotrienes (LT), potent mediators of the inflammatory response. In the 35+ years since cysteinyl-LTs were reported to mediate antigen-induced constriction of bronchi in tissue from asthma patients, numerous cellular responses evoked by the LTs, such as chemoattraction and G protein-coupled receptor (GPCR) activation, have been elucidated and revealed a potential for 5-lipoxygenase (5-LOX) as a promising drug target that goes beyond asthma. We describe herein early work identifying 5-LOX as the key enzyme that initiates LT biosynthesis and the discovery of its membrane-embedded helper protein required to execute the two-step reaction that transforms AA to the progenitor leukotriene A4 (LTA4). 5-LOX must traffic to the nuclear membrane to interact with its partner and undergo a conformational change so that AA can enter the active site. Additionally, the enzyme must retain the hydroperoxy-reaction intermediate for its final transformation to LTA4. Each of these steps provide a unique target for inhibition. Next, we describe the recent structures of GPCRs that recognize metabolites of the 5-LOX pathway and thus provide target alternatives. We also highlight the role of 5-LOX in the biosynthesis of anti-inflammatory lipid mediators (LM), the so-called specialized pro-resolving mediators (SPM). The involvement of 5-LOX in the biosynthesis of LM with opposing functions undoubtedly complicates the continuing search for 5-LOX inhibitors as therapeutic leads. Finally, we address the recent discovery of how some allosteric 5-LOX inhibitors promote oxygenation at the 12/15 carbon on AA to generate mediators that resolve, rather than promote, inflammation.
1. Introduction
The moniker leukotriene (LT) was coined to describe arachidonic acid (AA)-derived compounds (originally identified as the “slow-reacting substance of anaphylaxis”) isolated from leukocytes that exhibit UV spectra indicative of a conjugated triene1,2. Dahlen and co-workers reported that LTs induced the constriction of human bronchi; while histamine was known to produce a similar contraction, leukotriene C4/leukotriene D4 (LTC4/LTD4) was 1000 times more potent3. Within a few years of this report, the enzyme that initiates the biosynthesis of LT was purified and identified as a lipoxygenase (LOX) that introduced a hydroperoxy moiety at C5 of AA to generate 5-HydroPeroxyEicosaTetraEnoic acid, 5-HPETE). However, this LOX was unlike other lipoxygenases that had been characterized, as it was subsequently shown that 5-HPETE was only an intermediate and that a second reaction catalyzed by the enzyme transformed the hydroperoxide into the epoxide LTA44 (Fig. 1). As its downstream products induced the constriction of bronchi and thus suggested a link between LTs and asthma, 5-LOX became a drug-target. After roughly 40 years and despite substantial drug-discovery effort, there is only one compound on the market that is a direct inhibitor of 5-LOX, and that one has a poor pharmacological profile (for review see Wenzel et al5). When the crystal structure of a stabilized variant of 5-LOX was first reported there was expectation that it would provide a framework for the development of “novel, purpose-designed inhibitors for the treatment of asthma and for probing leukotriene involvement in cardiovascular disease and cancer6.” These expectations have been unmet, as the structures solved to date are not good models for structure-based drug design (SBDD). In the first reported structure, the active site was obstructed by two plugging aromatic residues7. A subsequent structure of the same variant bound to the competitive inhibitor nordihydroguaiaretic acid (NDGA) further complicated matters. This structure revealed an active site that had been “blown open,” with the peptide regions of the shielding helices disordered8.
Figure 1. Molecular structure of 5-LOX metabolites.
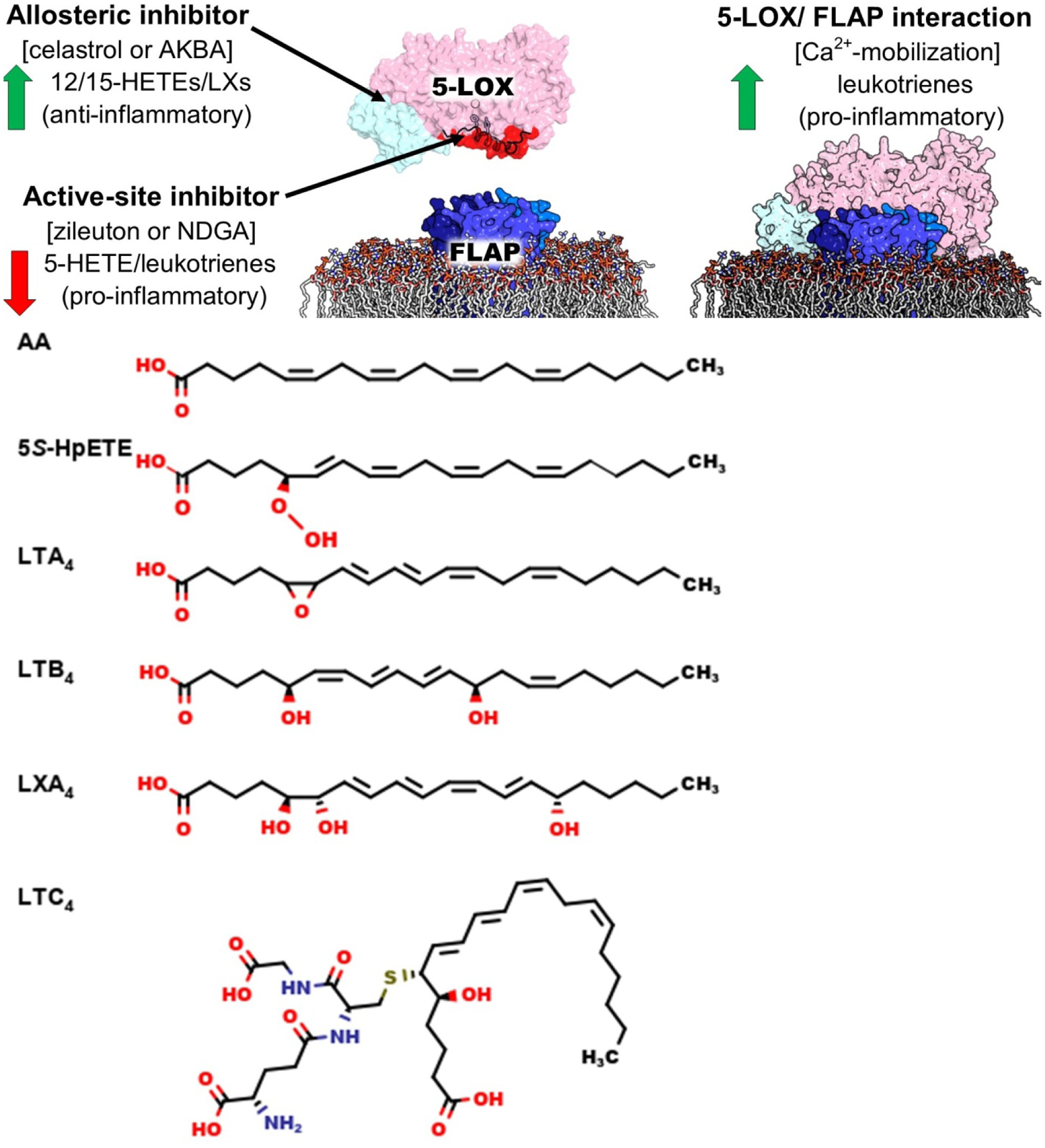
5-LOX transforms AA to 5S-HpETE and then to LTA4. LTA4 can be further modified to either LTB4 or LTC4. The triple hydroxylation of AA to LXA4 is catalyzed by 5-LOX together with 12/15-LOXs, but might also be produced by 5-LOX alone in the presence of noncompetitive inhibitors that alter the regio-specificity of the enzyme.
In contrast, antagonists of LTC4/LTD4 such as montelukast, which compete for binding to a G protein-coupled receptor (GPCR), have proven to be highly effective in treating allergic rhinitis and for long-term control of asthma. (Other strategies have focused efforts at inhibiting leukotriene A4 hydrolase or leukotriene C4 synthase9–11, but this work is beyond the scope of this review.) Given the success of GPCR antagonists one might ask “is 5-LOX still a drug target?” The biosynthetic pathway initiated by 5-LOX is complex, and the downstream products of the 5-LOX reaction have biological roles beyond inducers of smooth muscle cell contraction. Over the years a broader appreciation for the role of 5-LOX-derived LM in chronic inflammation, and the various disorders that result from an unchecked inflammatory response, e.g., cancer, osteoarthritis, neuroinflammation, and cardiovascular disease, suggest other therapeutic roles for 5-LOX inhibitors12–15. Moreover, our increased understanding of 5-LOX at the structural, biochemical and cellular levels provides new avenues for the design of lead compounds with high specificity that exploit features unique to 5-LOX16,17. However, while targeting 5-LOX to inhibit the production of pro-inflammatory LTs is a reasonable strategy, 5-LOX also contributes to the generation of the lipoxins (LX) and other specialized pro-resolving mediators (SPM) that are anti-inflammatory in nature, for review see Serhan18(Fig. 1). Thus, now the question of 5-LOX inhibition becomes more nuanced and challenging - can we limit the production of LTs without a concomitant decrease in pro-resolving products of the 5-LOX pathway?
2. Antagonizing LT production at the source
There are several challenges to the development of highly specific 5-LOX inhibitors. (1) 5-LOX is an iron enzyme, and while iron chelators or redox inhibitors might be potent in vitro, the ubiquity of iron enzymes in vivo complicates this inhibition strategy. (2) Humans express several LOXs, and the sequence identity among isoforms is only around 40%. However, the core active site is highly conserved, making isoform specificity challenging. In addition, (3) the fatty acid substrate does not have a 3-dimensional pattern of H-bond donors and acceptors that might be readily exploited to engineer specificity; indeed, the flexible hydrophobic substrate must rely on complementary van der Waals interactions to be accurately positioned at the catalytic machinery. Nevertheless, 5-LOX has an expanded repertoire of molecular interactions relative to other LOXs, and these interactions may provide paths toward inhibitor specificity. 5-LOX must accommodate both the substrate and the hydroperoxy intermediate in the active site, while other lipoxygenases release their hydroperoxy products. Moreover, the enzyme interacts with a partner protein to ensure completion of the two-step reaction, the nuclear membrane-embedded protein 5-lipoxygenase-activating protein (FLAP)19.
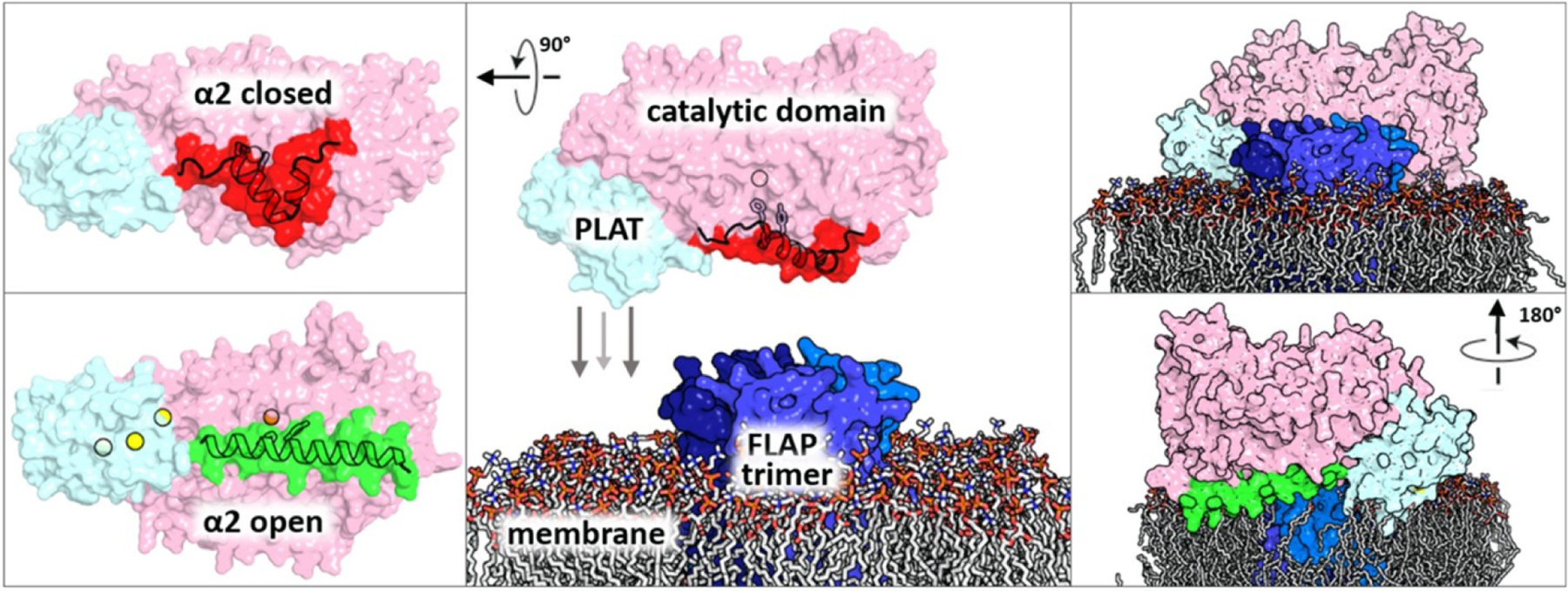
LOXs are widespread in nature, and humans express six distinct isoforms20,21. The animal enzymes are composed of two domains, an amino terminal β-barrel domain, which in 5-LOX confers Ca2+-dependent membrane binding,22,23 and a larger, mostly helical catalytic domain that harbors iron (Fe2+ when resting, Fe3+ when activated) held in place by three invariant histidine side chains and the free C-terminal main-chain carboxyl of an invariant Ile. Helices that appear to help define the active site are an “arched” helix that bends over the active site and contributes an invariant Leu to clamp the substrate pentadiene for attack and helix-α2, which in most LOX structures rims that active site, but in 5-LOX provides “corking” residues that block access to the catalytic machinery (Fig. 2).
Figure 2. Overlay of inhibitors and substrate in LOX structures.
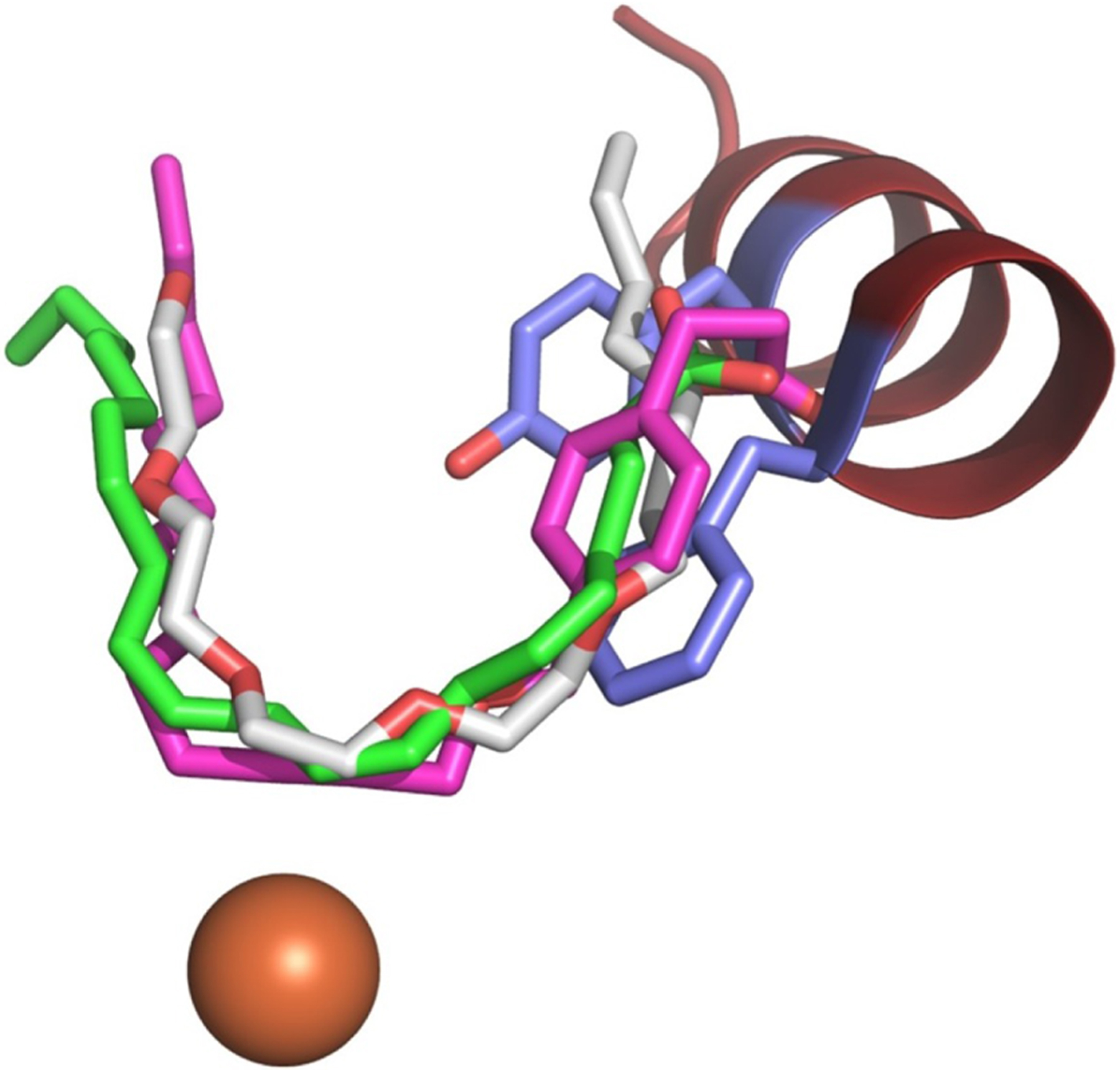
The catalytic iron (orange, sphere) sits at the base of the U-shaped channel defined by superimposed co-structures of 8R-LOX (4QWT) with AA (green C, sticks), 12-LOX (3RDE) with OPP (magenta C, sticks), and 15-LOX-2 (4NRE) with substrate mimetic (gray C, sticks). The Phe/Tyr (blue C, sticks) of helix-α2 (maroon, ribbon) from 5-LOX (3O8Y) plug the entrance to the U-shaped channel.
Mammalian LOXs share a common substrate, and each is named for the distinct oxygenated product it generates. The polyunsaturated fatty acid, AA, has three pentadienes and while 5-LOX positions C7 for attack, 15-lipoxygenase (15-LOX) attacks at C13 and 12-lipoxygenase (12-LOX) at C10. A robust model for how LOXs position AA in the active site24 to generate a distinct HPETE isomer can be proposed by integrating the information from the structures of (1) a bacterial LOX in complex with phospholipid25, (2) the catalytic domain of 12-LOX in complex with the isoform-specific inhibitor 4-(2-oxapentadeca-4-yne)phenylpropanoic acid (OPP)26, (3) 15-lipoxygenase-2 (15-LOX-2) with a substrate mimic27 and finally, (4) an anaerobic structure of the complex of an 8R-lipoxygenase (8R-LOX) with AA28. These structures reveal a U-shaped cavity with invariant Leu and Ile side chains to position one of three substrate pentadienes for attack. As one can see from Fig. 2, the AA revealed in the 8R-LOX structure superimposes with the 12-LOX and 15-LOX-2 inhibitors. The substrate in the 8R-enzyme is positioned in a deep cavity with C10 at the catalytic iron. Can 5-LOX adopt a conformation with a fully defined U-shaped channel? As illustrated in Fig. 2, the Phe/Tyr from the Stable-5-LOX structure overlap with the inhibitors/substrate in the U-shaped cavity.
The first step in a LOX reaction is the abstraction of a hydrogen from the central carbon of a pentadiene. This step is followed by oxygenation at C(+/−2) of the substrate. Oxygenation is antarafacial to the site of H-abstraction, and an O2 pocket, defined by an invariant Leu and an Ala/Gly “switch” is clearly visible in the reported crystal structures to date29. This suite of available structures also reveals the core active site and confirms that cavity depth contributes to defining which pentadiene is centered for attack by the catalytic machinery. In addition, a wealth of experimental data indicate that LOXs can differ with respect to which “end” of the substrate is innermost30. The “inverse entry” model (into the common U-shaped cavity) nicely explains the relationship between 15-LOX-2 and 5-LOX, which share 42% sequence identity. “Tail first” entry of AA positions C13 and “head-first” entry C7 to generate the 15-S- and 5-S-HPETEs, respectively, as opposite faces of the substrate align at the catalytic center. Thus 5-LOX and 15-LOX-2 channels are of comparable depth, but opposite with respect to AA orientation17. When considering how the substrate sits in the active site, the most unique aspect of substrate binding of a given LOX is the 3-dimensional relationship between the position of the O2 pocket and substrate carboxyl, the relationship is defined by depth and direction of substrate entry.
One caveat with envisioning a 5-LOX: substrate model is that the active site in the crystal structure of 5-LOX (in the absence of inhibitor) is not accessible to solvent. This is not the case for two other “unoccupied” animal LOX structures reported (PDB: 2FNQ, 3FG1) or for the structures reported in the presence of inhibitors (4NRE, 2P0M, 3RDE). Some sort of conformational change must occur in 5-LOX before the AA can slide into position. If one assumes that an open-cavity conformation, as observed in 15-LOX-2 or 15-LOX-1, is available to 5-LOX, two amino acids that block site access, namely Phe-177 and Tyr-181, would reorient to be surface exposed (Fig. 2). Mutation of these amino acids impair nuclear membrane localization of the enzyme in human embryonic kidney (HEK293) cells16, a tantalizing observation that might suggest that membrane-binding is coupled to the opening of 5-LOX. In addition, the presence of FLAP was able to “rescue” these 5-LOX mutants, which display a diminished capacity for LTA4 biosynthesis: their co-expression with FLAP restored wild-type levels of products formed in HEK293 cells. This latter observation is consistent with a physical protein–protein interaction, beyond co-localization, of 5-LOX and FLAP. The ability of FLAP to “rescue” 5-LOX mutants in cellulo suggests that a 5-LOX-FLAP interaction either completes the 5-LOX active site or promotes restoration of a native-like conformation.
It is tempting to speculate that FLAP shields the 5-LOX active site to minimize the escape of the intermediate16. The substrate to intermediate reaction involves a shift in the pattern of double bonds: in place of the cis double bond between C5 and C6 there is a trans double bond between C6 and C7. Yet unlike in other lipoxygenase this structural change may not result in the HPETE being ejected from the active site. Instead, C10 of the intermediate is positioned for a second H-abstraction. How does 5-LOX maintain the intermediate for the second attack? While it is possible that the intermediate is released and simply re-binds, in both cases 5-LOX must be able to accommodate two distinct substrate conformations.
3. Inhibiting the handoff of substrate from FLAP to 5-LOX
Early efforts to develop 5-LOX inhibitors led to the discovery of FLAP. Scientists at Merck reported that the indolealkanoic acid MK-886 inhibited LT formation in intact neutrophils, and that the mechanism of inhibition was through binding to a transmembrane protein, for review31. This inhibition was not observed in cell-free assays, and MK-866 had no effect on the activity of purified 5-LOX32. FLAP is a trimeric protein of 18 kD protomers and belongs to a super-family now known as the MAPEGs (Membrane-Associated Proteins in Eicosanoid and Glutathione metabolism)33. The subsequent enzyme in the LT pathway, LTC4 synthetase, is a MAPEG as well, which conjugates glutathione to LTA4 to produce LTC4.
The first structure of FLAP was published in 2007 at a modest resolution of 4.0 Å34. The homotrimeric structure revealed the inhibitor MK-591 (a derivative of MK-886 carrying a quinoline moiety) bound in a hydrophobic pocket formed by neighboring protomers, with all three grooves in the trimer occupied. Mutagenesis studies have shown that the AA-binding site and the inhibitor binding sites partially overlap35. More than 10 years later higher-resolution structures of FLAP, bound to MK-866 and to the quinoline DG-031 (2.4 and 2.6 Å), were published36. The significantly improved resolution for these complexes allowed for the detailed placement of side-chains, inhibitors, and the water molecules that mediate interactions with the protein. A SBDD approach to generate inhibitors targeting both FLAP and microsomal prostaglandin E2 synthase-1 (mPGES-1, another MAPEG member) was initiated based on the inhibitor-bound structures of both proteins. Inhibitors to FLAP (MK-591, MK-866, and DG-031) deeply embed orthogonally to the trimeric protein core near the phosphate-exposed region of FLAP. In contrast, DG-031 binds in a shallow groove parallel to the helical bundle of mPGES-136. This orientation of the inhibitor extends lengthwise from the lipid headgroups to the hydrophobic tail region of the membrane. Additionally, the LTC4 synthase structure first published in 2007 has a similar placement for the detergent dodecylmaltoside (DDM), bound superficially in the shallow interprotomer groove37. Although the trimeric four-helix bundle structure is conserved between MAPEGs, inhibitor binding modes can vary among members. We utilized MemProtMD, a database of molecular dynamic (MD) simulations of lipids around membrane-embedded proteins, to visualize the different binding locations of inhibitors to MAPEGs with respect to the bilayer38 (Fig. 3). The results lead us to suggest that AA likely adopts the latter DDM-like conformation: with the ligand parallel to the helical bundle and carboxylate extending towards the cytosol and poised for uptake by 5-LOX. But the question remains: how does 5-LOX receive AA from FLAP?
Figure 3. FLAP embedded in the membrane.
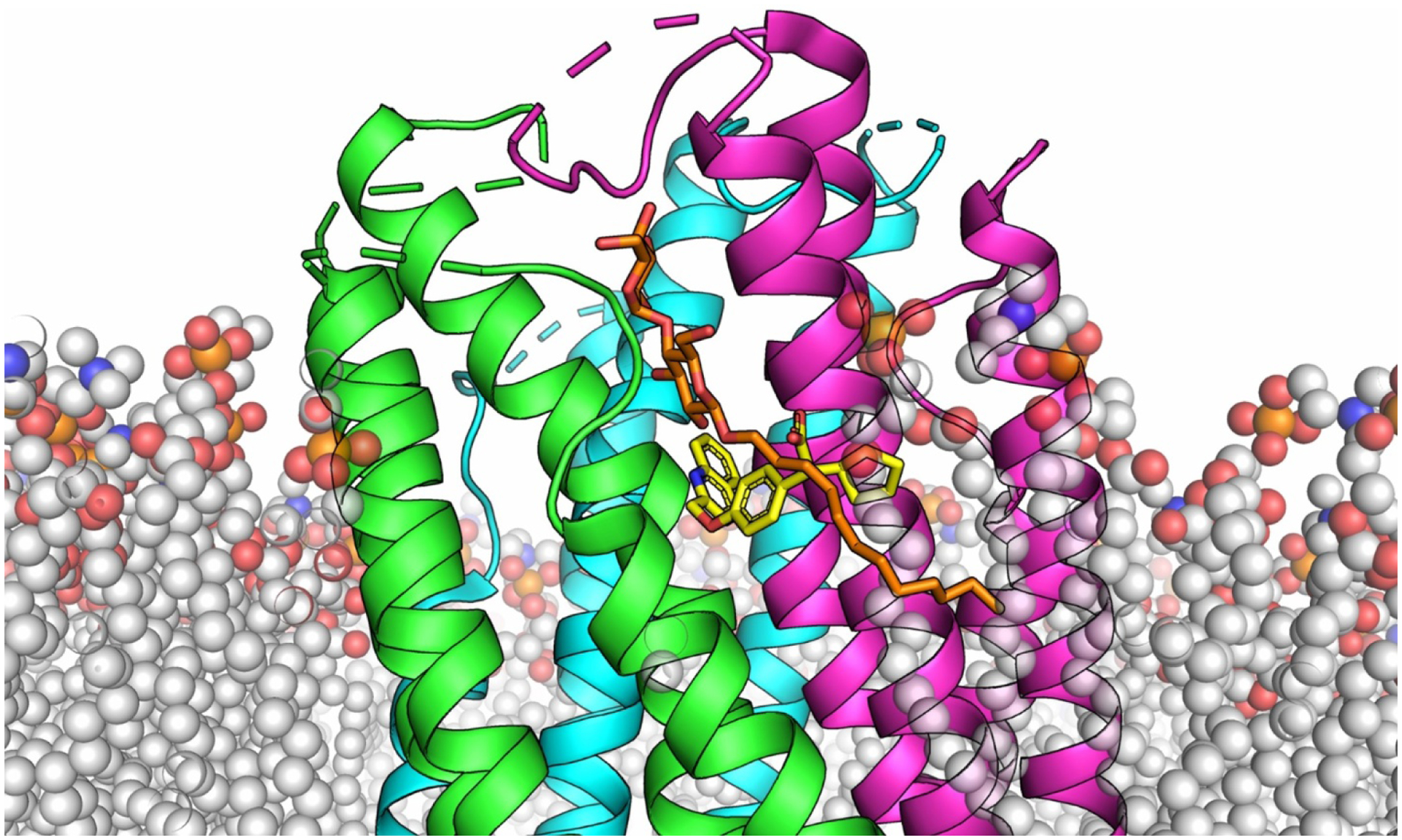
The three protomers of FLAP are shown as cartoons and colored differently for each chain (green, cyan, magenta). The phospholipids are shown as transparent spheres (C (grey), O (red), N (blue) P (orange)). The inhibitor DG-031 (yellow, C) is shown as sticks and inserts perpendicular to the helical bundle near the phosphate region of the bilayer. DDM (orange, C, sticks) superimposed from the LTC4 synthase structure runs parallel to the helical bundle and extends past the interface to the cytoplasm. We predict that AA adopts a similar binding mode as DDM.
4. Modeling the interaction of 5-LOX and FLAP at the membrane
Co-localization of FLAP and 5-LOX at the nuclear membrane is well established, but a physical interaction between the partners in vitro is only beginning to be detailed39. The ability of FLAP to “rescue” 5-LOX mutants in cellulo, a decreased 5-LOX-FLAP co-localization in the presence of FLAP inhibitors, and previous work with fluorescence lifetime imaging microscopy (FLIM) and crosslinking40–42 are consistent with a protein-protein interaction. In addition, Kumar et al. (2020) have demonstrated that AA is sufficient for 5-LOX to bind FLAP incorporated into nanodiscs. However, their low-resolution electron microscopy (EM) structure, which is supported by Mass Spec data, cannot define the molecular details of the protein-protein interface. We propose a model of the 5-LOX-FLAP complex that combines structures of 5-LOX and FLAP, which have been embedded in the lipid bilayer according to Orientations of Proteins in Membranes (OPM), and were manually docked for efficient AA transfer.
The design of the complex requires 5-LOX to be anchored to the membrane by loops in its β-barrel domain, which have been shown to confer membrane binding, and the AA-binding sites of FLAP and 5-LOX to align for optimal substrate transfer22,23. In order to design the model, we generated a wild-type version of 5-LOX from the Stable-5-LOX structure by adding back in silico the membrane-binding loops of the Polycystin-1, Lipoxygenase, Alpha-Toxin (PLAT) domain that were removed for enhanced solubility and crystallizability. We used the WT 8R-LOX (~40% sequence identity) structure (2FNQ), which shares these loops as a template to allow for more accurate loop placement for our WT 5-LOX. Next, we modeled an “open” conformation of Stable-5-LOX by forcing helix-α2 to the conformation observed in 8R-LOX with AA (4QWT) and 15-LOX-2 with a substrate mimetic (4NRE). This “opened” WT 5-LOX was docked to the membrane by the Positioning of Proteins in Membrane (PPM 2.0 server) from the OPM database43,44. Aliphatic and aromatic residues from the membrane-binding loops were calculated to penetrate the hydrophobic bilayer. Surprisingly, residues near the C-terminus of helix-α2 of the catalytic domain, which are distal to the active site, were also predicted to insert in the membrane. Once again, we employed the MD-simulated version of FLAP embedded in a 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) lipid bilayer by MemProtMD to approximate the native nuclear membrane where 5-LOX would bind. The overall logic that went into construction of this presented model in Fig. 4 is as follows: (1) The entrance to the 5-LOX active site should lie close to the AA site in FLAP. While protein-AA complex structures are not available for either FLAP or 5-LOX, excellent models for these sites can be extrapolated from known structures of homologues. As mentioned above, we suspect that the AA site of FLAP is positioned similarly to the detergent DDM in the homologous LTC4 synthase structure (at 1.8 Å) where it runs parallel to the helix bundle past the MK-591 binding site towards the cytosolic face of the protein. The helices of FLAP extend ~15 Å above the bilayer into the cytosol allowing for an ample protein-protein interface both above and within the membrane plane. For the AA binding site of 5-LOX, the structure of 8R-LOX+AA served as the best model. In 5-LOX, AA should have the inverse orientation due to regio- and stereo- specificity rules, and we positioned it at a depth consistent with attack at C728. A second constraint that must be satisfied is that the putative 5-LOX membrane-insertion loops should have access to the bilayer. Hence, the use of PPM server of OPM as detailed. The resulting model (Fig. 5) displays all the important features mentioned above: (1) the substrate, which is bound with its “tail” innermost in membrane-bound FLAP, can slide directly into the 5-LOX active site “head-first” so that the carboxyl is innermost, as is required for product stereochemistry. This distance from the AA bound to FLAP to the substrate access portal is less than 10 Å, as shown. And (2) the interacting surfaces of 5-LOX and FLAP as docked in the membrane are remarkably complementary to allow for substantial protein-protein interactions between the helices and loops of FLAP that extend into the cytoplasm with the elements of helix-α2 and the arched helix from 5-LOX. The legitimacy of the model awaits experimental validation, but the prediction combines the available structural, computational, and experimental data. The docked model should allow one to generate new hypotheses to be tested.
Figure 4. 5-LOX docked to membrane and FLAP.
(middle) The catalytic domain of 5-LOX (pink, surface) is shown with helix-α2 (red, cartoon) in the closed conformation with the Phe/Tyr (sticks, outlined) plugging the substrate access portal to the iron (orange, sphere). The (PLAT) domain (cyan, surface) initiates binding to the membrane upon Ca2+ binding. (top left) 5-LOX rotated to better illustrate closed helix-α2. The viewing angle is from the membrane plane. (bottom left) 5-LOX with Ca2+ bound (yellow, sphere) and helix-α2 elongated (green, cartoon) to an open conformation. (top right) 5-LOX in open conformation docked to membrane by Positioning of Proteins in Membrane (PPM) 2.0 server with putative AA-binding site of FLAP near substrate access portal of 5-LOX. (bottom right) Complex rotated to illustrate how both the membrane-binding loops of the PLAT domain along with helix-α2 inserted into the bilayer.
Figure 5. Transfer of AA from FLAP to 5-LOX.
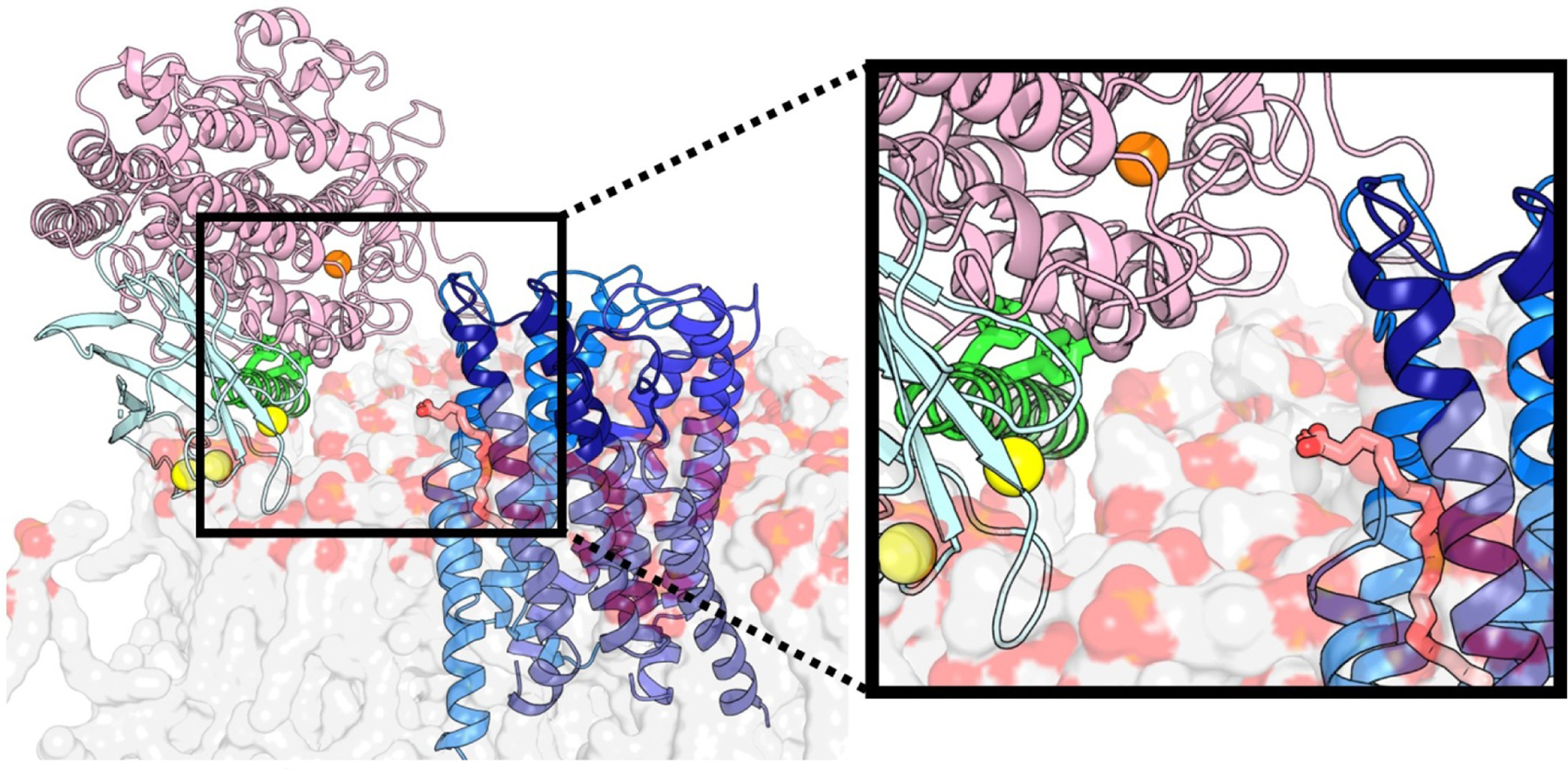
The FLAP trimer (blue, cartoon) is embedded in the membrane (grey/red, transparent surface). AA (salmon, sticks) between the protomers of FLAP is poised for uptake from 5-LOX (catalytic domain pink, cartoon). Access to the catalytic iron (orange, sphere) is controlled by the Phe/Tyr plug (green, sticks) of helix-α2 (green, cartoon). Ca2+ (yellow, sphere) bound to the PLAT domain (cyan, cartoon) is the main driver of 5-LOX to the membrane.
We have focused our model on the interaction of 5-LOX with FLAP, but it should be noted that 5-LOX has also been reported to have a stabilizing partner protein, which is soluble: coactosin-like protein (CLP)45. CLP, a 16 kD protein, has been shown to augment 5-LOX activity in cell-free assays46. Esser et al. have identified Trp 102 of the 5-LOX PLAT domain as essential for its interaction with CLP47. In support of a role for CLP in 5-LOX nuclear localization, Allain et al. reported that W102A-5-LOX expressed in HEK293 cells fails to traffic to the nucleus as compared to the wild-type enzyme48. While CLP could indeed also participate in the 5-LOX-FLAP interaction, it is not essential for LT synthesis49. Thus, we have constructed a minimalist model, lacking CLP. Notably in the model, 5-LOX-Trp102 lies along the membrane-binding face of 5-LOX, precluding a simultaneous interaction with CLP.
5. Leukotriene seven transmembrane receptors (7TMs)
Once leukotriene B4 (LTB4) or LTC4 is generated inside the cell what happens? The export of these LTs has been demonstrated (at least for LTC4) to utilize an active transport mechanism50. While a LTB4 active transport system has not been identified, LTC4 is exported to the extracellular space by the ATP-dependent transporter Multidrug Resistance-associated Protein 1 (MRP1)51. The precursor LTA4 is also exported from the cell through binding to the heterotetramer S100A8/A9, which acts as a molecular chaperone. LTA4 has a half-life of 3 seconds at 37 °C at pH 7.4 due to the highly reactive conjugated triene epoxide, which is subject to non-enzymatic hydrolysis1. LTA4 is predominantly produced in neutrophils, with over 50% released to the extracellular space where it can bind serum albumin. This transport supports production of LTB4 and LTC4 transcellularly by cells that do not possess 5-LOX activity, such as platelets, smooth muscle cells, endothelial cells and red blood cells52. Once the LT products have exited the cell, they function in an autocrine and/or paracrine function to escalate the inflammatory response. These LM bind to specific G protein-coupled receptors on the cell surface and transduce signals that trigger changes intracellularly.
The largest superfamily of membrane proteins is that of the GPCRs. These proteins signal across the plasma membrane when they bind a variety of extracellular stimuli such as neurotransmitters, peptides, hormones, ions, and lipids (for review of lipid GPCRs see Audet and Stevens53). The family is the most targeted class of proteins for pharmaceutical development; approximately 34% of the current small molecule, FDA-approved drugs target GPCRs (for a comprehensive review of FDA-approved drugs and molecular drug targets see Santos et al54). GPCRs have very “druggable” pockets positioned on the outside of the cell, obviating the need for the ligand to cross the plasma membrane. The receptors consist of an extracellular N-terminal domain followed by a region of seven transmembrane helices (7TM). An intracellular C-terminal domain may include an amphipathic helix-8 parallel to the plasma membrane. Three extracellular loops in Rhodopsin-like or class A GPCRs are generally responsible for forming a partial lid over the orthosteric pocket and aid in recognition of the ligand. The bound ligand stabilizes specific receptor conformations of the 7TM core and a repositioning of the three intracellular loops. A stabilized conformation dictates coupling to specific protein partners on the cytoplasmic side of the membrane to initiate intracellular signaling cascades. Receptors often have basal or constitutive activity as they can adopt conformations that couple to G proteins in the absence of agonist. An inverse agonist lowers this basal activity while a neutral antagonist does not lower constitutive activity but blocks binding of other ligands. Partial and full agonist bind and cause an increase in efficacy over basal activity by stabilizing a more productive conformation for coupling to heterotrimeric G proteins, resulting in an increase or full signaling efficacy, respectively. Sodium ions appear to be negative allosteric modulators of many class A GPCRs. Na+ binds tightly in the intramembranous region of the helix bundle via an associated water cluster with the conserved Asp2.50 (Ballesteros–Weinstein residue numbering) and stabilizes the ligand free state of the receptor55.
LTs and the other LM generated in the 5-LOX pathway bind to specific GPCRs classified into four main groups. The dihydroxy LT receptors recognize LTB4, with the leukotriene B4 receptor 1 (BLT1R) expressed exclusively in leukocytes and deemed the major physiological player for inflammation and the leukotriene B4 receptor 2 (BLT2R) expressed more widely56. BLT1R binds LTB4 with 2 log units greater affinity than that of BLT2R; various hydroxyeicosanoids and 12(S)-hydroxyheptadecatrienoic acid are higher affinity ligands to BLT2R57. Both receptors signal mainly through the Gi/O family of transducers that provoke adenylate cyclase inhibition or phospholipase C (PLC) stimulation through coupling to GQ/G11 family. There are two cysteinyl-LT receptors, designated CysLT1R and CysLT2R, which bind to the glutathione-conjugates of LTA4, that is, LTC4, LTD4 or LTE4. Cys-LT receptors preferentially bind LTD4 over LTC4 > LTE4. CysLT1R and CysLT2R mainly couple to the GQ/G11 family and promote PLC stimulation that result in phosphatidylinositol turnover and Ca2+ mobilization. Next, we will discuss the more recently discovered and delineated receptors. The formyl peptide receptor 2 (FPR2/ALX) binds the trihydroxy-LM LXA4 with low nanomolar affinity. This receptor also recognizes the peptidyl ligand WKYMVm, which at subnanomolar concentrations induces a full agonist response equivalent to that of LXA4. The FPR2/ALX receptor, like some other chemoattractant/chemokine receptors, utilizes a CD38-dependent cyclic ADP-ribose for Ca2+ flux and chemotaxis primarily through the Gi/O family transduction mechanisms, with secondary transduction mechanism through the GQ/G11 family. We will not discuss in detail the fourth class that includes the oxoeicosanoid receptor 1 (OXER1). We are also including purinergic (P2Y12), G protein-coupled receptor (GPR17), (GPR99), (GPR32) and chemerin receptor 1 (ChemR23) in the fourth class, with the first three receptors recognizing the cysteinyl-LTs and the last two receptors binding the anti-inflammatory lipoxygenase metabolites of ω3-polyunsaturated fatty acids, namely the SPMs. For a comprehensive listing of the leukotriene, lipoxin and oxoeicosanoid receptors see Chemical Reviews “Leukotriene Receptors” from 2011 and for the most up-to-date reference, see the International Union of Basic and Clinical Pharmacology Review (IUPAR) website7. We will focus on the BLT1R, CystLT1R, and CystLT2R receptors in some depth due to their clinical relevance to disease states and recent crystal structures.
5.1. BLT1 Receptor
The BLT1R binds to LTB4 with sub-nanomolar affinity, with a Kd of 0.15 nM in COS-7 cells as determined by [3H]LTB4. This dihydroxylated LM is the most potent chemoattractant in humans and stimulates neutrophil recruitment to extravascular sites to initiate the early immune response58. Recently, the enantioselectivity of BLT1R was demonstrated using the 5R,12S- enantiomer of the bioactive eutomer 5S,12R-LTB4. The latter induced Ca2+ mobilization and chemotaxis in neutrophils by 830- to >3100-fold more efficient compared to the 5R,12S-configurated distomer59. Humans express BLT1R almost solely in inflammatory cells, e.g., macrophages, neutrophils, eosinophils, dendritic cells, T-cells and B-cells. LTB4 signals mostly in a pertussis toxin (PTX) sensitive manner via the Gαi-proteins, but in certain cell types, signaling is not PTX-sensitive; other coupling mechanisms include Ca2+ mobilization and phospholipase C activation. The BLT1R signaling pathway activates the mitogen-activated protein kinases (MAPKs). The C-terminal tail of BLT1R can be phosphorylated at Thr308 to uncouple from G protein recognition and promote desensitization to the stimulus60. Phosphorylation yields a receptor β-arrestin complex that interacts with clathrin and eventually leads to receptor internalization. Five BLT1R drugs have entered phase 2 clinical trials for the treatment of rheumatoid arthritis, cystic fibrosis, and non-small cell lung cancer, among other pathologies, for review see61.
The structure of BLT1R bound to the antagonist BIIL260 was solved by X-ray crystallography to 3.7 Å resolution with diffraction data from crystals generated with the lipidic-cubic phase method (5X33)62. More recently, the structure of BLT1R bound to selective antagonist MK-D-046 (2.88 Å) was released to the protein data bank and published (7K15)63.The first structure revealed a receptor in the inactive state with the benzamidine moiety of BIIL260 bound innermost in the highly conserved Na+ binding site. Asp2.50 is the conserved amino acid inside the seven-transmembrane domain that positions the Na+-centered water cluster that inactivates the receptor. Even at this modest resolution, the highly hydrophobic antagonist is fully refined with 3 of the 4 benzene rings of BIIL260 inside the orthosteric binding site forming edge-to-π interactions with phenyl groups and CH-to-π interactions with branch chained amino acids (Fig. 6e/f). The innermost benzene of BIIL260 forms hydrophobic interactions with the helix bundle, with the amidine creating a salt-link with Asp2.50 of the Na+ site. Mutational and docking studies hint that LTB4 might share most of the binding site that BIIL260 occupies. Entrance to the orthosteric binding site appears to occur through a vestibule open to the extracellular surface, see Fig. 6f. The extracellular loop 2 of BLT1R shares the common β-hairpin motif of related GPCRs. Many other lipid-binding GPCR structures, including cannabinoid receptor 1 (CB1), lysophospholipid S1P receptor (S1P1R), and GPR40, revealed a vestibule inaccessible via the extracellular surface, an indication that access to the binding site is likely achieved by lateral diffusion in the plane of the membrane, as was rigorously detailed for the human melatonin receptor 1 (MT1R)64. MK-D-046 from the higher resolution structure occupies similar positions at the top of the orthosteric pocket as BIIL260 while the Na+- Asp2.50 salt-link is maintained in the bottom of the pocket. The 7TM region is identical in the two BLT1R structures, even though different fusion proteins (lysozyme for 5X33 and flavodoxin for 7K15) were engineered into intracellular loop 3 to facilitate crystallization. Now that the BLT1R structure has been solved and the molecular determinants for antagonist binding have been mapped out, will new more favorable compounds to aid in the treatment of inflammatory disorders be developed? One might also ask if similar strategies to obtain crystal structures of BLT1R might also work for the BLT2R receptor.
Figure 6. Leukotriene GPCR structures.
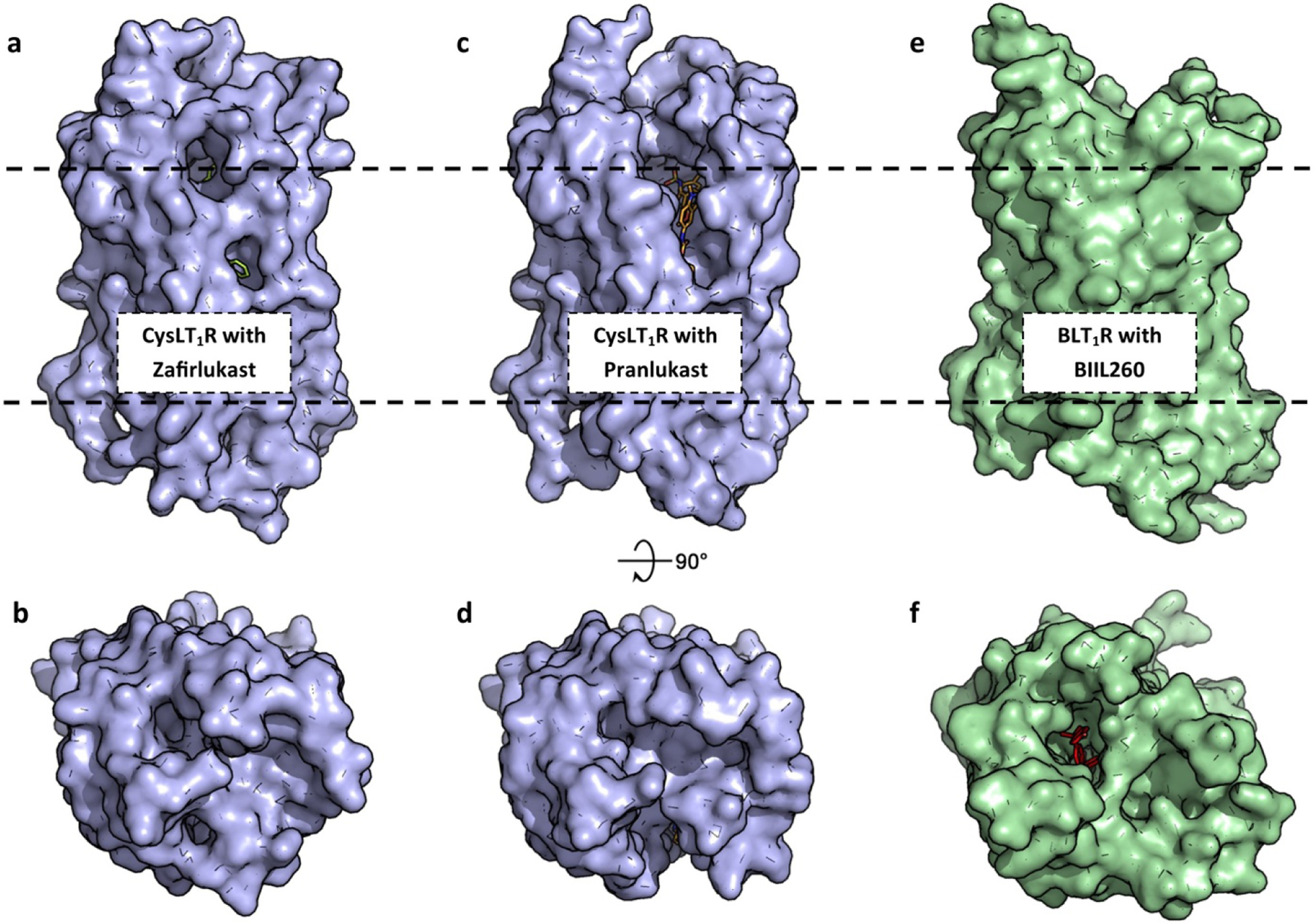
(a) Zafirlukast (green, sticks) is bound in the orthosteric pocket of CysLT1R (blue, surface, 6RZ5). Ligands to CysLT1R are predicted to enter laterally near the hydrophobic portion of bilayer (black, dashes). (b) The extracellular loops of CysLT1R fully encapsulate the orthosteric pocket. (c,d) Pranlukast (orange, sticks) bound to CysLT1R (6RZ4). (e,f) BIIL260 (magenta, sticks) and other orthosteric ligands are predicted to access BLT1R (green, surface) from the extracellular side (5X33).
5.2. CysLT1 and CysLT2 receptor
Slow-reacting substance of anaphylaxis (SRS-A) was first discovered in the late 1930s as a substance released when the perfused lungs of guinea pigs were treated with cobra venom. While histamine was known to cause a reversible and rapid contraction of smooth muscle, SRS-A caused a slow and sustained contraction. In the 1970s, scientists at a company called Fisons discovered an analogue of chromoglycate, FPL-55712, that antagonized the action of SRS-A65. Later that decade future Nobel Laureate Bengt Samuelsson proposed the structure of SRS-A as a cysteine-containing peptide which carries a tetraene epoxide2. SRS-A was later shown to be a mixture of the cysteine-containing peptide-lipid conjugates of LTC4, LTD4, and LTE4, which differ by successive modifications by a transpeptidase and dipeptidase. The CysLT receptors are broadly expressed in most types of leukocytes and most importantly in lung smooth muscle cells where antagonism of the CysLT1R is used for the treatment of allergic rhinitis and asthma. There are two FDA-approved drugs that target CysLT1R; montelukast (Singulair) and zafirlukast (Accolate), as well as the widely used drug in Japan, pranlukast (Onon). Annual prescriptions of montelukast are >30 million in the USA. However, the FDA has updated and added a boxed warning for use of montelukast as post-marketing reports have indicated neuropsychiatric adverse events. The current interpretation is that benefit-risk considerations support the continued prescription of montelukast for asthma, but not for the treatment of allergic rhinitis66. Additionally, responses to the various “lukast” drugs vary greatly, especially among children for the treatment of asthma. At this point, the design of new drugs that target the CysLT receptors appears to be a prudent strategy. We will now explore in detail the structures of the two CysLT receptors and how they compare with other lipid-binding GPCRs.
The structures of the CysLT receptors were published separately in 2019 by the Cherezov group67,68. These receptors have a sequence identity of 36% and share the classic 7TM helices, but differ in the canonical amphipathic helix 8 resolved in only the CysLT2R structure. They differ in the linker region between TM7 and helix 8, with CysLT1R having a very flexible GG8.48 motif, while CysLT2R has a GE8.48 in a similar position. Interestingly, the disease mutation of GS8.48 of CysLT1R increases signaling efficacy, likely due to improved stability of helix 8, which is known to help regulate G protein and β-arrestin coupling. Overall, the orthosteric pockets of the two receptors are highly similar, and only differ by conservative substitutions. For example, residues K371.31 and H2847.32 from CysLT2R are residues that make key contacts with selective antagonists and if replaced with the CysLT1R residues of R371.31 and Q2847.32, the CysLT2R had decreased potency to LTD4, as well as inhibition by antagonists. A more detailed structure-activity relationship of the CysLT receptors was produced by docking compounds that are highly selective to either CysLT receptor to delineate the molecular differences in binding modes between the two GPCRs. The ligands are predicted to access the orthosteric pocket for both CysLT receptors through lateral diffusion from the membrane between TM4 and TM5, rather than from the extracellular space as observed for BLT1R (Fig 5a/c). One mode of selectivity to the different CysLT receptors is conferred by a narrow cleft opening to the lipid membrane where ligand is accessed. CysLT1R has a F1504.52, while in CysLT2R the replacement F4.52L lessens this restriction and allows larger hydrophobic compounds to enter the orthosteric site. At the top of the pocket, CysLT1R has an Y261.26 that participates in hydrogen bonds with an oxo-pentanoic-acid moiety in the most selective antagonists whereas CysLT2R has an F411.35. Altogether, these high-resolution crystal structures provide an excellent framework for understanding mechanisms of ligand selection and subtype selectivity. They also provide robust models to guide a structure-based drug design approach to new therapeutics.
6. SPMs in the 5-LOX world: birth of the lipoxins & active resolution
Only few years after the discovery of the crucial role of 5-LOX in the biosynthesis of the pro-inflammatory LTs, a critical enzymatic function of 5-LOX became obvious in the formation of anti-inflammatory LXs69,70. These tri-hydroxylated AA metabolites were found to possess a tetraene structure with hydroxy moieties in 5,6,15(S)- (LXA4) or in 5,14,15(S)- (LXB4) position. It was found that both a 12/15-LOX and the 5-LOX act in conjunction for LX biosynthesis, where 12/15-LOXs cause oxygenation of C15 to produce 15-HETE which is then oxygenated by 5-LOX at C5 to generate the 5,6 epoxide that is converted by hydrolases to the 5,6,15 or the 5,14,15-dihydroxy structures. For LX formation, 5-LOX and the 12/15-LOX may occur either in one single cell such as in macrophages, monocytes or eosinophils or in different cell types that express either 5-LOX (e.g. neutrophils) or 12/15-LOX (e.g. platelets, epithelial cells), producing LXs by transcellular biosynthetic routes, for review see71. Notably, 15-LOX-1 is capable of catalyzing all enzymatic steps in LXA4 biosynthesis, without the need of 5-LOX72. More recent studies revealed the existence of epimers of the two LXs, i.e. 15-epi-LXA4 and 15-epi-LXB4, where the hydroxy moiety in 15-position is R-configured, also called aspirin-triggered LX (AT-LX) because acetylated cyclooxygenase-2 (COX-2) upon aspirin treatment generates 15(R)-HpETE as precursor for conversion by 5-LOX73.
Most intriguing was the finding that LXs are anti-inflammatory mediators, in sharp contrast to pro-inflammatory LTs, which can now be explained by divergent receptor interactions. LXs act at their own specific GPCRs, e.g., FPR2/ALX or other LXA4-specific receptors to evoke anti-inflammatory responses, interfere with LTB4 functions at the BLT1R, antagonize CysLT1R and CysLT2R to block pro-inflammatory responses, or interact with intracellular targets, e.g. protein kinase C, for review74. In this way, LXs reduce vascular permeability, stop excessive neutrophil infiltration and activation, stimulate macrophages for efferocytosis of apoptotic neutrophils, and are potent chemoattractants of monocytes required for wound healing. Therefore, LXs were proposed to counteract many biological functions of LTs and to represent beneficial LM that terminate inflammation and promote its resolution without acting as immunosuppressants74.
Today, LXs are known to belong to the superfamily of SPM. These are LM that terminate and resolve inflammatory processes, prevent tissue damage, support healing and promote the return to tissue homeostasis18. Besides LXs, the SPM comprise ω3-PUFA (i.e., eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA) and docosapentaenoic acid (DPA)-derived resolvins (RV), protectins (PD), and maresins (MaR) which limit neutrophil influx and activation, but activate macrophages for efferocytosis of apoptotic neutrophils and cellular debris, bacterial killing, and phagocytosis of bacteria. Thus, SPM support the relief of inflammation with beneficial functions in host defense, pain, organ protection, wound healing and tissue repair. Importantly and in analogy to LX biosynthesis, 5-LOX was reported to catalyze reactions involved in the transformations of EPA (20:5) and DHA (22:6) towards so-called E-series and D-series RVs, while other SPMs such as DHA-derived PDs (10,17-dihydroxylated) and MaR (7,14- or 13,14-dihydroxylated) are generated in a 5-LOX-independent fashion mainly by 12/15-LOXs. For generation of E-series RV, liberated EPA is converted to18-HEPE, seemingly via cytochrome P450 enzymes or COX-2, and then oxygenated by 5-LOX and/or 12/15-LOXs (in 5,12 or 17 position). For the formation of D-series RVs liberated DHA is first oxygenated at C17 by 15-LOXs and in a second step at C5 or C7 by 5-LOX. Moreover, in analogy to CysLTs that are formed from LTA4 by conjugation with glutathione, also SPM from their epoxide immediate form can be converted to glutathione-conjugates (called conjugates of tissue regeneration, CTRs) that promote tissue repair and regeneration. For details in SPM structures and biosynthesis, the interested reader is referred to recent reviews by Chiang & Serhan75 and Serhan & Levy76.
In accordance with its function in LX and RV biosynthesis, it is obvious that 5-LOX cannot be considered exclusively as pro-inflammatory enzyme producing LTs, and this issue may prove its role as drug target rather difficult. Thus, pharmacological inhibition of 5-LOX may not only suppress LT formation but could also prevent beneficial LX and RV formation. In fact, in human 5-LOX/15-LOX-1-expressing M2 macrophages that produce LTs and a broad spectrum of SPM upon bacterial challenge, the direct 5-LOX inhibitor zileuton inhibited the generation of the classical 5-LOX products LTB4 and 5-HETE/5HEPE but also of RVs77. Only a few studies addressing the actions of 5-LOX inhibitors have considered SPM in the cellular analysis among generated 5-LOX-derived products78–81. Moreover, in experimental animal disease models, 5-LOX inhibitors like zileuton are commonly studied with focus on prevention of (mainly acute) LT-mediated inflammatory processes but modulation of the subsequent resolution phase is rarely considered. Therefore, even though 5-LOX inhibitors can effectively block pro-inflammatory reactions how they affect SPM formation and thus inflammation resolution is largely unknown. Likely reasons for neglecting SPM are on one hand the low abundance of these LM that are extremely potent with bioactions in the picomolar range. On the other hand, the commonly used simple cell-based assay systems optimized for studying classical 5-LOX activity related to LTs (e.g., ionophore A23187-activated primary leukocytes or leukocytic cell lines) are unsuitable and exhibit only very low or even no capacities for SPM formation. We found that human monocyte-derived macrophages with M2-like phenotype that are activated with relevant pathogenic bacteria (e.g., S. aureus or E. coli) represent suitable models that produce a broad spectrum of LM from endogenous substrates including PG, LTs and substantial SPM members77,82,83. Besides the low abundance, the complex stereochemistry (e.g., trans/cis double bonds, R/S-configuration of hydroxyls) of SPM further complicates the separation and detection by commonly used bioanalytical methods employed to determine LTs. Novel UPLC-MS-MS methods with accurate and sophisticated sample work-up have now advanced SPM bioanalytics, allowing consideration of SPM formation as part of modulation of 5-LOX77,84,85.
7. Temporal and spatial considerations in LX/LT biosynthesis
The dual role of 5-LOX in the formation of pro-inflammatory LTs and of anti-inflammatory/pro-resolving SPM raises the question how the biosynthesis of these disparate LM is organized in the cellular context for the divergent biological outcomes. Moreover, it is questionable if a pharmacological concept can be developed at all that allows favorable modulation of LM biosynthesis by interference with the 5-LOX pathway, that is, suppression of LTs without lowering SPM, but rather stimulation of the formation of the latter. In fact, two such concepts have recently emerged that may enable such wishful thinking: a) antagonism of FLAP and b) allosteric modulation/inhibition of 5-LOX leading to a shift of the regio-specificity of 5-LOX to a 12/15-lipoxygenating enzyme.
Fredman et al.86 suggested that the balance of LTs and SPM in leukocytes might be achieved through differential intracellular localization of 5-LOX, where nuclear 5-LOX in proximity to FLAP favors LTB4 formation, and cytoplasmic 5-LOX could favor the biosynthesis of LXA4. They showed that RvD1 promotes nuclear exclusion of 5-LOX by limiting phosphorylation at Ser271 in macrophages and thereby suppresses LTB4 formation but enhances LXA4 biosynthesis86. 5-LOX that is retained in the cytosol of stimulated cells has no access to FLAP and thus the supply of AA to activated 5-LOX for LTA4 formation is excluded. However, such activated cytoplasmic 5-LOX (distant from FLAP) may receive de-novo generated 15-HETE by cytosolic 15-LOX and convert it to LXA4. Such circumstances are conceivable also when FLAP is blocked by antagonists to prevent LT formation from AA, but still permitted to convert other substrates (e.g., 15-HETE, 17-HDHA, 18-HEPE etc.) for SPM formation. Thus, these 15-LOX-formed monohydroxylated precursors might be effectively shuttled to 5-LOX independent of FLAP. Recent reports are indeed in favor of such speculations. For example, in human M2 macrophages activated by pathogenic E. coli, the FLAP inhibitor MK-886 inhibited the formation of classical 5-LOX-derived products while 12/15-LOX products including the SPM RvD5 and MaR1 were elevated77,82. In zymosan-induced murine peritonitis, MK-886 efficiently suppressed LTB4, tr-LTB4 and 5-HETE levels in the peritoneal exudates, while amounts of the SPM LXA4, neuroprotection/protectin D1 (PD1), RvD4 and RvD5 were only moderately impaired80. Similarly, in murine liver injury, the FLAP inhibitor BAY X-1005 blocked cysteinyl-LT formation, but elevated SPM levels87. Others, however, have claimed a role of FLAP in LX and RV formation in human neutrophils where MK-886 suppressed LXA4 and RvD1 under artificial conditions where cells had been challenged with A23187 and exogenous 17-HDHA88 that may circumvent cellular regulation of 5-LOX subcellular redistribution that is governed by phosphorylation89.
Moreover, besides FLAP antagonists, prostaglandin E2 (PGE2) and oxidized phosphatidylcholine inhibited LT formation while enhancing the biosynthesis of LXs. Small molecule 5-LOX inhibitors such as benzenesulfonamide-derivatives81, ginkgolic acid79, 3-O-acetyl-11-keto-β-boswellic acid (AKBA)8, and celastrol78 were found to trigger SPM formation in human neutrophils or macrophages or in inflamed murine peritoneal exudates by different mechanisms. For all these agents, a strong increase in 12-LOX and 15-LOX metabolites, as well as SPM, both in vitro in activated M2 subsets and in vivo in the inflamed murine peritoneum was shown. Also, oxymetazoline, which failed to inhibit 5-LOX product formation, elevated SPM formation90.
The question remains: what mechanisms allow some 5-LOX inhibitors to not only avoid inhibition of LX and RV formation, but promote it? Again, manipulation of 5-LOX subcellular localization and access to FLAP may be causative, as in the case of benzoxanthene lignans that prevented 5-LOX interaction with FLAP at the nuclear membrane of human monocytes80. However, the discovery of an unexpected molecular mode of action of the 5-LOX inhibitor AKBA suggests that 5-LOX, and possibly other human LOXs, possess an allosteric site where appropriate modulators can not only inhibit catalysis but also invoke a shift in the regio-specificity. Similarly, an allosteric site for fatty acids is hypothesized to regulate the C-H activation step in 15-LOX-2 and soybean LOX-1 and impact substrate selectivity, for review see91.
Structural elucidation of human Stable-5-LOX bound to AKBA revealed a deep groove at the interface of the membrane-binding PLAT and the catalytic domains as the binding site for this pentacyclic triterpene that functions as an allosteric modulator of 5-LOX (Fig. 7a)8. AKBA was observed to inhibit formation of LTA4 hydrolysis products and 5-HETE generated by human isolated 5-LOX and 5-LOX expressed in HEK293 cells, as expected. However, AKBA concomitantly elevated the formation of 12-HETE, despite the absence of any 12-LOX or 15-LOX, especially when AA was delivered to 5-LOX by FLAP in a cellular context. Intriguingly, the levels of SPM that require 12/15-lipoxygenation (MaR1, PD1, RvD1, RvD2, and RvD5) were increased by AKBA as well. In contrast, another 5-LOX inhibitor, nordihydroguaiaretic acid (NDGA) bound in the 5-LOX active site and blocked formation of all AA metabolites (Fig. 7b). This latter co-crystal structure left much to be desired, as key helices (the arched helix and helix-α2) that help shape the active site pocket were missing, most likely due to the presence of the of the bulky, inflexible inhibitor. Additional co-crystal structures have been sought for 5-LOX (unpublished data) with a variety of competitive inhibitors with little success. Alternate approaches are required to provide the molecular details needed for SBDD. However, exploitation of this AKBA-binding groove may be a promising approach for isoform-specific modulation of LOX activity due to the targeted inhibition of LTs and the added production of the SPMs offering a novel therapeutic strategy to limit inflammation and promote its resolution.
Figure 7. Small molecule inhibitors bound to 5-LOX.
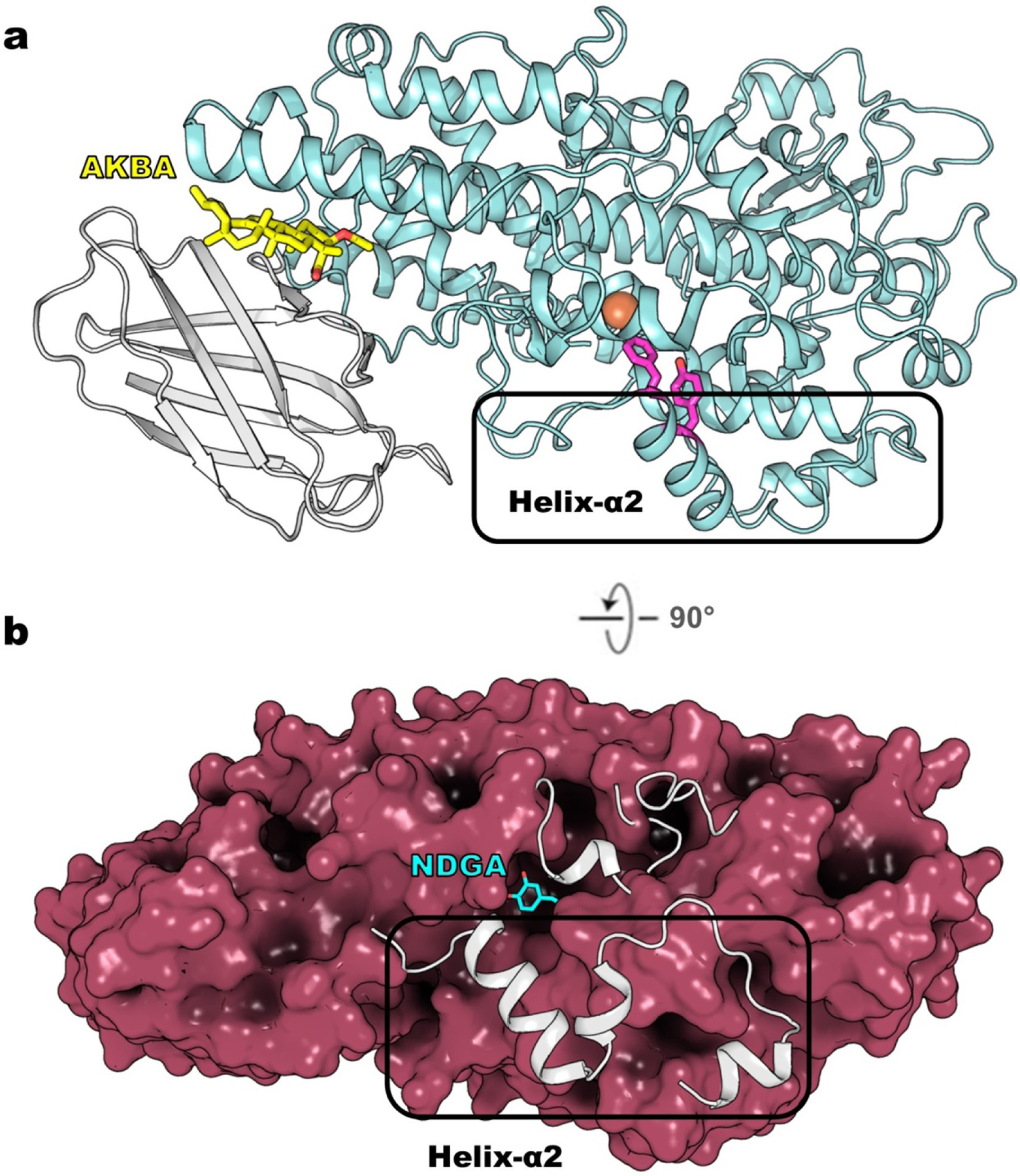
(a) The structure of Stable-5-LOX bound to AKBA (yellow, sticks, 6NCF) revealed a novel allosteric site between PLAT domain (grey, cartoon) and catalytic domain (teal, cartoon). The Phe/Tyr plug (magenta, sticks) of helix-α2 blocks access to the active-site iron (orange, sphere). (b) NDGA (cyan, sticks) bound to Stable-5-LOX (ruby, surface, 6N2W) triggered disordering of key peptide regions that encapsulate the active site, including the arched helix (grey, cartoon above NDGA) and helix-α2 (grey, cartoon that is boxed). The “missing” pieces are the gray helices as observed in the apo Stable-5-LOX (3O8Y).
A similar inter-domain groove as found in 5-LOX is present in 15-LOX-1 and 15-LOX-2 but the access of AKBA is partially restricted and the binding modalities likely differ from that of 5-LOX. Indeed, AKBA failed to inhibit 15-LOX-1 and 15-LOX-2 activities in activated HEK293 cells expressing these LOXs8. Recent unpublished data, however, indicate that AKBA itself evokes LM generation in innate immune cells, but in contrast to typical stimuli like ionophore A32187 or pathogenic E. coli, AKBA almost exclusively induces SPM formation and their 12/15-LOX-derived precursors, especially in M2-like macrophages that strongly express 15-LOX-1. It appears that AKBA directly activates 15-LOX-1 via binding at the inter-domain groove, reflected by AKBA-induced translocation of 15-LOX-1 to a subcellular site in M2 macrophages where PUFA substrate is accessed and converted, independent of Ca2+ and MAPK pathways (unpublished). Besides AKBA, the anti-inflammatory pentacyclic triterpenoid quinone methide celastrol, a natural product from the anti-rheumatic remedy Tripterygium wilfordii, potently inhibited the activity of human isolated 5-LOX (IC50 = 0.19 μM), seemingly acting at the same 5-LOX allosteric site as AKBA78. Indeed, celastrol induced the generation of 12-/15-LOX-derived LM including the SPM RvD5 in human M2 macrophages and strongly impaired zymosan-induced LT formation but at the same time celastrol elevated levels of SPM and related 12-/15-LOX-derived LM in various tissue of mice in vivo. Therefore, it is tempting to speculate that celastrol may act in a similar fashion as AKBA, but not only on 5-LOX but also on 15-LOX-1 by activating this enzyme. Together, these findings with AKBA and celastrol call for consideration of the application of small molecules in inflammatory diseases to push the LM class switch from pro-inflammatory LT to SPM formation in order to promote endogenous programs of inflammation resolution.
8. Closing remarks
From the discovery of the “slow reacting substance of anaphylaxis” in the 1930s to recent high-resolution structural studies of enzymes from multiple steps of the LT biosynthetic pathway, to the identification and structures of LT-GPCRs, it is clear that global efforts have contributed to our expanded knowledge of how these autacoids exert their effects. The results of basic research at the molecular level, cell-based assays, whole-animal models, and clinical trials combine to establish the LT pathway as an important contributor to the innate immune response. 5-LOX was originally appreciated as a target for asthma, but LTs are now known to contribute to arthritis92,93, cardiovascular disease15,94, cancer95,96, and the neuropathology of Alzheimer’s disease14,97,98. There have been many chemical tools discovered that can (1) limit the initiation of LT biosynthesis through inhibition of 5-LOX, (2) disrupt the handoff of AA to 5-LOX by FLAP, (3) suppress the bifurcation of the LT pathway by inhibiting LTA4 hydrolase or LTC4 synthase, or (4) antagonize specific LT-GPCRs. The high-resolution structures now available for proteins of the LT pathway can provide excellent models for docking studies with the computational tools available. Moreover, key GPCR structures that remain to be exploited for drug development are now accessible. While a model of the substrate-5-LOX complex still eludes us, new approaches to co-structures are being explored. Lastly, the recently identified allosteric site that lies between the PLAT and catalytic domains of 5-LOX may provide an alternative target and strategy for the generation of LM class-switching compounds to increase the production of pro-resolving LM. Such compounds may not only prevent the pro-inflammatory reactions of LTs, as demonstrated for classical 5-LOX inhibitors like zileuton, but in addition could also support SPM-mediated inflammation resolution and return to homeostasis. Resolution of inflammation is now widely recognized as an active step in the inflammatory process, and tuning the levels of SPMs by modulation of 5-LOX activity could provide a more elegant and nuanced approach to novel therapeutic developments.
Acknowledgements
This work was funded in part by grants to M.E.N. (nos. NIH HL107887 and AHA 16GRNT31000010, and the NIH P50AT002776 seed grant, and the Louisiana Governor’s Biotechnology Initiative) and O.W. (Deutsche Forschungsgemeinschaft (DFG, the German Research Foundation), project no. 316213987, SFB 1278 PolyTarget (project nos. A04 and C02), CRC 1127 ChemBioSys (project no. A04) and Free State of Thuringia and the European Social Fund (2016 FGR 0045).
Abbreviations:
- 12-LOX
12-lipoxygenase
- 15-LOX
15-lipoxygenase
- 15-LOX-2
15-lipoxygenase-2
- 5-HPETE
5(S)-hydroperoxy-6-trans-8,11,14-cis-eicosatetraenoic acid
- 5-LOX
5-lipoxygenase
- 8R-LOX
8R-lipoxygenase
- AA
arachidonic acid
- AKBA
3-O-acetyl-11-keto-β-boswellic acid
- BLT1R
leukotriene B4 receptor 1
- BLT2R
leukotriene B4 receptor 2
- CB1
cannabinoid receptor 1
- ChemR23
chemerin receptor 1
- CLP
coactosin-like protein
- COX-2
cyclooxygenase-2
- CTR
conjugates of tissue regeneration
- CysLT1R
cysteinyl leukotriene receptor 1
- CysLT2R
cysteinyl leukotriene receptor 2
- DDM
dodecylmaltoside
- DHA
docosahexaenoic acid
- DPA
docosapentaenoic acid
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- EM
electron microscopy
- EPA
eicosapentaenoic acid
- FLAP
5-lipoxygenase-activating protein
- FLIM
fluorescence lifetime imaging microscopy
- FPR2/ALX
formyl peptide receptor 2
- GPCR
G protein-coupled receptor
- GPR17, GPR99, GPR32
G protein-coupled receptor 17, 99, 32
- HEK
human embryonic kidney
- LM
lipid mediator
- LOX
lipoxygenase
- LT
leukotriene
- LTA4
leukotriene A4
- LTB4
leukotriene B4
- LTC4
leukotriene C4
- LTD4
leukotriene D4
- LX
lipoxin
- MAPEG
Membrane-Associated Proteins in Eicosanoid and Glutathione metabolism
- MAPK
mitogen-activated protein kinase
- MaR
maresins
- MD
molecular dynamics
- mPGES1
microsomal prostaglandin E2 synthase-1
- MRP1
Multidrug Resistance-associated_Protein 1
- MT1R
human melatonin receptor 1
- NDGA
nordihydroguaiaretic acid
- OPM
Orientations of Proteins in Membranes
- OPP
4-(2-oxapentadeca-4-yne)phenylpropanoic acid
- OXER1
oxoeicosanoid receptor 1
- P2Y12
purinergic
- PD1
neuroprotection/protectin D1
- PD
protectins
- PLAT
Polycystin-1, Lipoxygenase, Alpha-Toxin
- PLC
phospholipase C
- PGE2
prostaglandin E2
- PPM
Positioning of Proteins in Membrane
- PTX
pertussis toxin
- PUFA
polyunsaturated fatty acid
- RV
resolvin
- S1P1R
lysophospholipid S1P receptor
- SBDD
structure-based drug design
- SPM
specialized pro-resolving mediator
- SRS-A
slow-reacting substance of anaphylaxis
- TM
transmembrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interests
The authors declare no competing interests.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
There are no declarations of interest for Nathaniel C. Gilbert, Marcia E. Newcomer, or Oliver Werz.
References
- 1.Borgeat P & Samuelsson B Arachidonic acid metabolism in polymorphonuclear leukocytes: effects of ionophore A23187. Proc Natl Acad Sci U S A 76, 2148–2152 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy RC, Hammarstrom S & Samuelsson B Leukotriene C: a slow-reacting substance from murine mastocytoma cells. Proc Natl Acad Sci U S A 76, 4275–4279 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahlen SE, Hedqvist P, Hammarstrom S & Samuelsson B Leukotrienes are potent constrictors of human bronchi. Nature 288, 484–486 (1980). [DOI] [PubMed] [Google Scholar]
- 4.Shimizu T et al. Characterization of leukotriene A4 synthase from murine mast cells: evidence for its identity to arachidonate 5-lipoxygenase. Proc Natl Acad Sci U S A 83, 4175–4179 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenzel SE & Kamada AK Zileuton: the first 5-lipoxygenase inhibitor for the treatment of asthma. Ann Pharmacother 30, 858–864, doi: 10.1177/106002809603000725 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Funk CD Leukotriene inflammatory mediators meet their match. Sci Transl Med 3, 66ps63, doi: 10.1126/scitranslmed.3002040 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Gilbert NC et al. The structure of human 5-lipoxygenase. Science 331, 217–219, doi: 10.1126/science.1197203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert NC et al. Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products. Nat Chem Biol 16, 783–790, doi: 10.1038/s41589-020-0544-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Numao S et al. Feasibility and physiological relevance of designing highly potent aminopeptidase-sparing leukotriene A4 hydrolase inhibitors. Sci Rep 7, 13591, doi: 10.1038/s41598-017-13490-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munck Af Rosenschöld M et al. Discovery of the Oral Leukotriene C4 Synthase Inhibitor (1S,2S)-2-({5-[(5-Chloro-2,4-difluorophenyl)(2-fluoro-2-methylpropyl)amino]-3-methoxypyrazin-2-yl}carbonyl)cyclopropanecarboxylic Acid (AZD9898) as a New Treatment for Asthma. J Med Chem 62, 7769–7787, doi: 10.1021/acs.jmedchem.9b00555 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Kleinschmidt TK et al. Tandem Benzophenone Amino Pyridines, Potent and Selective Inhibitors of Human Leukotriene C4 Synthase. J Pharmacol Exp Ther 355, 108–116, doi: 10.1124/jpet.115.227157 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Sarveswaran S, Chakraborty D, Chitale D, Sears R & Ghosh J Inhibition of 5-lipoxygenase selectively triggers disruption of c-Myc signaling in prostate cancer cells. J Biol Chem 290, 4994–5006, doi: 10.1074/jbc.M114.599035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta K et al. A double blind, randomized, placebo controlled study of the efficacy and safety of 5-Loxin for treatment of osteoarthritis of the knee. Arthritis Res Ther 10, R85, doi: 10.1186/ar2461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vagnozzi AN, Giannopoulos PF & Praticò D The direct role of 5-lipoxygenase on tau pathology, synaptic integrity and cognition in a mouse model of tauopathy. Transl Psychiatry 7, 1288, doi: 10.1038/s41398-017-0017-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poeckel D & Funk CD The 5-lipoxygenase/leukotriene pathway in preclinical models of cardiovascular disease. Cardiovasc Res 86, 243–253, doi: 10.1093/cvr/cvq016 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Gerstmeier J et al. 5-Lipoxygenase-activating protein rescues activity of 5-lipoxygenase mutations that delay nuclear membrane association and disrupt product formation. FASEB J 30, 1892–1900, doi: 10.1096/fj.201500210R (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitra S, Bartlett SG & Newcomer ME Identification of the Substrate Access Portal of 5-Lipoxygenase. Biochemistry 54, 6333–6342, doi: 10.1021/acs.biochem.5b00930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serhan CN Pro-resolving lipid mediators are leads for resolution physiology. Nature 510, 92–101, doi: 10.1038/nature13479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon RA et al. Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature 343, 282–284 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Funk CD Lipoxygenase pathways as mediators of early inflammatory events in atherosclerosis. Arterioscler Thromb Vasc Biol 26, 1204–1206 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Funk CD, Chen XS, Johnson EN & Zhao L Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediat 68–69, 303–312 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Hammarberg T, Provost P, Persson B & Radmark O The N-terminal domain of 5-lipoxygenase binds calcium and mediates calcium stimulation of enzyme activity. J Biol Chem 275, 38787–38793, doi: 10.1074/jbc.M006136200 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni S, Das S, Funk CD, Murray D & Cho W Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J Biol Chem 277, 13167–13174 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Newcomer ME & Brash AR The structural basis for specificity in lipoxygenase catalysis. Protein science : a publication of the Protein Society 24, 298–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garreta A et al. Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa. Faseb J 27, 4811–4821, doi: 10.1096/fj.13-235952 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu S, Mueser TC, Marnett LJ & Funk MO Jr. Crystal structure of 12-lipoxygenase catalytic-domain-inhibitor complex identifies a substrate-binding channel for catalysis. Structure 20, 1490–1497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobe MJ, Neau DB, Mitchell CE, Bartlett SG & Newcomer ME The structure of human 15-lipoxygenase-2 with a substrate mimic. J Biol Chem 289, 8562–8569, doi: 10.1074/jbc.M113.543777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neau DB et al. Crystal structure of a lipoxygenase in complex with substrate: the arachidonic acid-binding site of 8R-lipoxygenase. J Biol Chem 289, 31905–31913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coffa G & Brash AR A single active site residue directs oxygenation stereospecificity in lipoxygenases: stereocontrol is linked to the position of oxygenation. Proc Natl Acad Sci U S A 101, 15579–15584, doi: 10.1073/pnas.0406727101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider C, Pratt DA, Porter NA & Brash AR Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol 14, 473–488 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young RN Development of novel leukotriene--based anti-asthma drugs: MK-886 and MK-571. Agents Actions Suppl 34, 179–187 (1991). [PubMed] [Google Scholar]
- 32.Gillard J et al. L-663,536 (MK-886) (3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isopropylindol-2-yl]-2,2 - dimethylpropanoic acid), a novel, orally active leukotriene biosynthesis inhibitor. Can J Physiol Pharmacol 67, 456–464 (1989). [DOI] [PubMed] [Google Scholar]
- 33.Jakobsson PJ, Morgenstern R, Mancini J, Ford-Hutchinson A & Persson B Common structural features of MAPEG -- a widespread superfamily of membrane associated proteins with highly divergent functions in eicosanoid and glutathione metabolism. Protein science : a publication of the Protein Society 8, 689–692 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson AD et al. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science 317, 510–512 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Vickers PJ et al. Amino acid residues of 5-lipoxygenase-activating protein critical for the binding of leukotriene biosynthesis inhibitors. J Lipid Mediat 6, 31–42 (1993). [PubMed] [Google Scholar]
- 36.Ho JD et al. Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP). Biochim Biophys Acta Gen Subj 1865, 129800, doi: 10.1016/j.bbagen.2020.129800 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Martinez Molina D et al. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature 448, 613–616, doi: 10.1038/nature06009 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Newport TD, Sansom MSP & Stansfeld PJ The MemProtMD database: a resource for membrane-embedded protein structures and their lipid interactions. Nucleic Acids Res 47, D390–d397, doi: 10.1093/nar/gky1047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar RB, Purhonen P, Hebert H & Jegerschöld C Arachidonic acid promotes the binding of 5-lipoxygenase on nanodiscs containing 5-lipoxygenase activating protein in the absence of calcium-ions. PLoS One 15, e0228607, doi: 10.1371/journal.pone.0228607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bair AM, Turman MV, Vaine CA, Panettieri RA Jr. & Soberman RJ The nuclear membrane leukotriene synthetic complex is a signal integrator and transducer. Mol Biol Cell 23, 4456–4464, doi: 10.1091/mbc.E12-06-0489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal AK et al. The nuclear membrane organization of leukotriene synthesis. Proc Natl Acad Sci U S A 105, 20434–20439 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandal AK et al. The membrane organization of leukotriene synthesis. Proc Natl Acad Sci U S A 101, 6587–6592 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lomize AL, Pogozheva ID, Lomize MA & Mosberg HI The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct Biol 7, 44, doi: 10.1186/1472-6807-7-44 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lomize MA, Pogozheva ID, Joo H, Mosberg HI & Lomize AL OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40, D370–376, doi: 10.1093/nar/gkr703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Provost P et al. 5-Lipoxygenase interacts with coactosin-like protein. J Biol Chem 276, 16520–16527, doi: 10.1074/jbc.M011205200 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Rakonjac M et al. Coactosin-like protein supports 5-lipoxygenase enzyme activity and up-regulates leukotriene A4 production. Proc Natl Acad Sci U S A (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esser J et al. Coactosin-like protein functions as a stabilizing chaperone for 5-lipoxygenase: role of tryptophan 102. Biochem J 425, 265–274, doi: 10.1042/bj20090856 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Allain EP, Boudreau LH, Flamand N & Surette ME The Intracellular Localisation and Phosphorylation Profile of the Human 5-Lipoxygenase Δ13 Isoform Differs from That of Its Full Length Counterpart. PLoS One 10, e0132607, doi: 10.1371/journal.pone.0132607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basavarajappa D et al. Roles of coactosin-like protein (CLP) and 5-lipoxygenase-activating protein (FLAP) in cellular leukotriene biosynthesis. Proc Natl Acad Sci U S A 111, 11371–11376, doi: 10.1073/pnas.1410983111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam BK, Gagnon L, Austen KF & Soberman RJ The mechanism of leukotriene B4 export from human polymorphonuclear leukocytes. J Biol Chem 265, 13438–13441 (1990). [PubMed] [Google Scholar]
- 51.Sjolinder M, Tornhamre S, Claesson HE, Hydman J & Lindgren J Characterization of a leukotriene C4 export mechanism in human platelets: possible involvement of multidrug resistance-associated protein 1. J Lipid Res 40, 439–446 (1999). [PubMed] [Google Scholar]
- 52.Claesson HE & Haeggström J Human endothelial cells stimulate leukotriene synthesis and convert granulocyte released leukotriene A4 into leukotrienes B4, C4, D4 and E4. Eur J Biochem 173, 93–100, doi: 10.1111/j.1432-1033.1988.tb13971.x (1988). [DOI] [PubMed] [Google Scholar]
- 53.Audet M & Stevens RC Emerging structural biology of lipid G protein-coupled receptors. Protein Sci 28, 292–304, doi: 10.1002/pro.3509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos R et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov 16, 19–34, doi: 10.1038/nrd.2016.230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katritch V et al. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci 39, 233–244, doi: 10.1016/j.tibs.2014.03.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokomizo T, Kato K, Hagiya H, Izumi T & Shimizu T Hydroxyeicosanoids bind to and activate the low affinity leukotriene B4 receptor, BLT2. J Biol Chem 276, 12454–12459, doi: 10.1074/jbc.M011361200 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Okuno T et al. 12(S)-Hydroxyheptadeca-5Z, 8E, 10E-trienoic acid is a natural ligand for leukotriene B4 receptor 2. J Exp Med 205, 759–766, doi: 10.1084/jem.20072329 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokomizo T, Izumi T, Chang K, Takuwa Y & Shimizu T A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature 387, 620–624, doi: 10.1038/42506 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Jagusch H et al. An Alternative Pathway to Leukotriene B(4) Enantiomers Involving a 1,8-Diol-Forming Reaction of an Algal Oxylipin. Org Lett 21, 4667–4670, doi: 10.1021/acs.orglett.9b01554 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Jala VR, Shao WH & Haribabu B Phosphorylation-independent beta-arrestin translocation and internalization of leukotriene B4 receptors. J Biol Chem 280, 4880–4887, doi: 10.1074/jbc.M409821200 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Bhatt L, Roinestad K, Van T & Springman EB Recent advances in clinical development of leukotriene B4 pathway drugs. Semin Immunol 33, 65–73, doi: 10.1016/j.smim.2017.08.007 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Hori T et al. Na(+)-mimicking ligands stabilize the inactive state of leukotriene B(4) receptor BLT1. Nat Chem Biol 14, 262–269, doi: 10.1038/nchembio.2547 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Michaelian N et al. Structural insights on ligand recognition at the human leukotriene B4 receptor 1. Nat Commun 12, 2971, doi: 10.1038/s41467-021-23149-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stauch B et al. Structural basis of ligand recognition at the human MT(1) melatonin receptor. Nature 569, 284–288, doi: 10.1038/s41586-019-1141-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones DG & Kay AB Inhibition of eosinophil chemotaxis by the antagonist of slow reacting substance of anaphylaxis--compound FPL 55712. J Pharm Pharmacol 26, 917–918, doi: 10.1111/j.2042-7158.1974.tb09208.x (1974). [DOI] [PubMed] [Google Scholar]
- 66.Clarridge K, Chin S, Eworuke E & Seymour S A Boxed Warning for Montelukast: The FDA Perspective. J Allergy Clin Immunol Pract, doi: 10.1016/j.jaip.2021.02.057 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Luginina A et al. Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs. Sci Adv 5, eaax2518, doi: 10.1126/sciadv.aax2518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gusach A et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat Commun 10, 5573, doi: 10.1038/s41467-019-13348-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serhan CN, Hamberg M & Samuelsson B Trihydroxytetraenes: a novel series of compounds formed from arachidonic acid in human leukocytes. Biochem Biophys Res Commun 118, 943–949, doi: 10.1016/0006-291x(84)91486-4 (1984). [DOI] [PubMed] [Google Scholar]
- 70.Serhan CN, Hamberg M & Samuelsson B Lipoxins: novel series of biologically active compounds formed from arachidonic acid in human leukocytes. Proc Natl Acad Sci U S A 81, 5335–5339 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Capra V et al. Transcellular biosynthesis of eicosanoid lipid mediators. Biochim Biophys Acta 1851, 377–382, doi: 10.1016/j.bbalip.2014.09.002 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Serhan CN et al. Lipoxin A. Stereochemistry and biosynthesis. J Biol Chem 261, 16340–16345 (1986). [PubMed] [Google Scholar]
- 73.Claria J & Serhan CN Aspirin triggers previously undescribed bioactive eicosanoids by human endothelial cell-leukocyte interactions. Proc Natl Acad Sci U S A 92, 9475–9479 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Serhan CN Lipoxins and aspirin-triggered 15-epi-lipoxins are the first lipid mediators of endogenous anti-inflammation and resolution. Prostaglandins Leukot Essent Fatty Acids 73, 141–162, doi: 10.1016/j.plefa.2005.05.002 (2005). [DOI] [PubMed] [Google Scholar]
- 75.Chiang N & Serhan CN Specialized pro-resolving mediator network: an update on production and actions. Essays Biochem 64, 443–462, doi: 10.1042/ebc20200018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Serhan CN & Levy BD Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. J Clin Invest 128, 2657–2669, doi: 10.1172/jci97943 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Werner M et al. Targeting biosynthetic networks of the proinflammatory and proresolving lipid metabolome. Faseb j 33, 6140–6153, doi: 10.1096/fj.201802509R (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pace S et al. Anti-inflammatory celastrol promotes a switch from leukotriene biosynthesis to formation of specialized pro-resolving lipid mediators. Pharmacol Res 167, 105556, doi: 10.1016/j.phrs.2021.105556 (2021). [DOI] [PubMed] [Google Scholar]
- 79.Gerstmeier J et al. Ginkgolic Acid is a Multi-Target Inhibitor of Key Enzymes in Pro-Inflammatory Lipid Mediator Biosynthesis. Front Pharmacol 10, 797, doi: 10.3389/fphar.2019.00797 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerstmeier J et al. Novel benzoxanthene lignans that favorably modulate lipid mediator biosynthesis: A promising pharmacological strategy for anti-inflammatory therapy. Biochem Pharmacol 165, 263–274, doi: 10.1016/j.bcp.2019.03.003 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Cheung SY et al. Discovery of a benzenesulfonamide-based dual inhibitor of microsomal prostaglandin E(2) synthase-1 and 5-lipoxygenase that favorably modulates lipid mediator biosynthesis in inflammation. Eur J Med Chem 156, 815–830, doi: 10.1016/j.ejmech.2018.07.031 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Werz O et al. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat Commun 9, 59, doi: 10.1038/s41467-017-02538-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jordan PM et al. Staphylococcus aureus-Derived α-Hemolysin Evokes Generation of Specialized Pro-resolving Mediators Promoting Inflammation Resolution. Cell Rep 33, 108247, doi: 10.1016/j.celrep.2020.108247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Colas RA, Shinohara M, Dalli J, Chiang N & Serhan CN Identification and signature profiles for pro-resolving and inflammatory lipid mediators in human tissue. Am J Physiol Cell Physiol 307, C39–54, doi: 10.1152/ajpcell.00024.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hong S et al. Resolvin D1, protectin D1, and related docosahexaenoic acid-derived products: Analysis via electrospray/low energy tandem mass spectrometry based on spectra and fragmentation mechanisms. J Am Soc Mass Spectrom 18, 128–144, doi: 10.1016/j.jasms.2006.09.002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fredman G et al. Resolvin D1 limits 5-lipoxygenase nuclear localization and leukotriene B4 synthesis by inhibiting a calcium-activated kinase pathway. Proc Natl Acad Sci U S A 111, 14530–14535, doi: 10.1073/pnas.1410851111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Titos E et al. Inhibition of 5-lipoxygenase-activating protein abrogates experimental liver injury: role of Kupffer cells. J Leukoc Biol 78, 871–878, doi: 10.1189/jlb.1204747 (2005). [DOI] [PubMed] [Google Scholar]
- 88.Lehmann C et al. Lipoxin and resolvin biosynthesis is dependent on 5-lipoxygenase activating protein. Faseb j 29, 5029–5043, doi: 10.1096/fj.15-275487 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Werz O, Burkert E, Samuelsson B, Radmark O & Steinhilber D Activation of 5-lipoxygenase by cell stress is calcium independent in human polymorphonuclear leukocytes. Blood 99, 1044–1052 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Beck-Speier I, Oswald B, Maier KL, Karg E & Ramseger R Oxymetazoline inhibits and resolves inflammatory reactions in human neutrophils. J Pharmacol Sci 110, 276–284, doi: 10.1254/jphs.09012fp (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Offenbacher AR & Holman TR Fatty Acid Allosteric Regulation of C-H Activation in Plant and Animal Lipoxygenases. Molecules 25, doi: 10.3390/molecules25153374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen M et al. Neutrophil-derived leukotriene B4 is required for inflammatory arthritis. J Exp Med 203, 837–842, doi: 10.1084/jem.20052371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gheorghe KR et al. Expression of 5-lipoxygenase and 15-lipoxygenase in rheumatoid arthritis synovium and effects of intraarticular glucocorticoids. Arthritis Res Ther 11, R83, doi: 10.1186/ar2717 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoxha M et al. Montelukast Use Decreases Cardiovascular Events in Asthmatics. Front Pharmacol 11, 611561, doi: 10.3389/fphar.2020.611561 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao Y, Wang W, Wang Q, Zhang X & Ye L Lipid metabolism enzyme 5-LOX and its metabolite LTB4 are capable of activating transcription factor NF-κB in hepatoma cells. Biochem Biophys Res Commun 418, 647–651, doi: 10.1016/j.bbrc.2012.01.068 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Hennig R et al. 5-Lipoxygenase, a marker for early pancreatic intraepithelial neoplastic lesions. Cancer Res 65, 6011–6016, doi: 10.1158/0008-5472.Can-04-4090 (2005). [DOI] [PubMed] [Google Scholar]
- 97.Valera E, Dargusch R, Maher PA & Schubert D Modulation of 5-lipoxygenase in proteotoxicity and Alzheimer’s disease. J Neurosci 33, 10512–10525, doi: 10.1523/jneurosci.5183-12.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giannopoulos PF et al. 5-lipoxygenase activating protein reduction ameliorates cognitive deficit, synaptic dysfunction, and neuropathology in a mouse model of Alzheimer’s disease. Biol Psychiatry 74, 348–356, doi: 10.1016/j.biopsych.2013.04.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


