SUMMARY
Low-protein diets promote metabolic health in humans and rodents. Despite evidence that sex and genetic background are key factors in the response to diet, most protein intake studies examine only a single strain and sex of mice. Using multiple strains and both sexes of mice, we find that improvements in metabolic health in response to reduced dietary protein strongly depend on sex and strain. While some phenotypes were conserved across strains and sexes, including increased glucose tolerance and energy expenditure, we observed high variability in adiposity, insulin sensitivity, and circulating hormones. Using a multi-omics approach, we identified mega-clusters of differentially expressed hepatic genes, metabolites, and lipids associated with each phenotype, providing molecular insight into the differential response to protein restriction. Our results highlight the importance of sex and genetic background in the response to dietary protein level, and the potential importance of a personalized medicine approach to dietary interventions.
Graphical abstract
In brief
Green et al. find that restricting dietary protein promotes metabolic health in mice but that the specific benefits observed are dependent upon sex, strain, the level of restriction, and age. Using a multi-omics approach, the authors gain molecular insight into the physiological effects of dietary protein and find that the role of the hormone FGF21 depends upon both sex and strain.
INTRODUCTION
Dietary protein plays a critical role in the regulation of metabolic health and lifespan in multiple diverse species, including rodents and humans (Levine et al., 2014; Mair et al., 2005; Solon-Biet et al., 2014). While current dietary advice recommends eating ~10% calories from protein, eating 10%–35% calories from protein is believed to be acceptable (Institute of Medicine, 2005). Many researchers and medical professionals support consuming more protein (Rodriguez, 2015), though this position is by no means unanimous (Richter et al., 2019). This message is reinforced by the popularization of high-protein fad diets, which promise speedy weight loss.
Some short-term studies have found that high protein intake improves glucose control in humans (Dong et al., 2013; Gannon et al., 2003; Seino et al., 1983). However, long-term epidemiological studies suggest that high protein intake is associated with an increased risk of diabetes, cardiovascular disease, and mortality in humans (Halbesma et al., 2009; Lagiou et al., 2007; Linn et al., 2000; Sluijs et al., 2010). A recent randomized clinical trial found that a low-protein (LP) diet improves metabolic health in humans, reducing weight and adiposity and improving glycemic control, and similar positive metabolic effects of an LP diet are observed in rodents (Fontana et al., 2016; Cummings et al., 2018; Laeger et al., 2014; Maida et al., 2016).
The benefits of an LP diet may be due in part to increased energy expenditure (EE); an LP diet results in reduced weight gain or weight loss despite increased food intake (Laeger et al., 2014). These effects have been linked to fibroblast growth factor 21 (FGF21), a hormone secreted in response to nutrient stress that initiates adaptive changes in metabolism and feeding and is induced by LP diets in mice, rats, and humans (Fontana et al., 2016; Hill et al., 2019; Laeger et al., 2014). Increased thermogenesis via the FGF21-uncoupling protein 1 (UCP1) axis mediates changes in weight and glycemic control (Feldmann et al., 2009; Hill et al., 2017), and C57BL/6 males lacking Fgf21 show no change in EE, weight, or food intake on an LP diet (Laeger et al., 2014).
Calorie restriction (CR) is one of the most well-studied nutritional interventions and robustly extends the lifespan and health span of model organisms and mammals (Green et al., 2022). The effects of CR on the health and longevity of mice vary due to genetic background (strain) and sex (Liao et al., 2010; Mitchell et al., 2016; Rikke et al., 2010). Many compounds have also been shown to have sexually dimorphic effects on mouse lifespan (Miller et al., 2014; Strong et al., 2008, 2016). Studies in Drosophila and other insects have observed sex-dependent effects of dietary protein on metabolism and lifespan (Camus et al., 2019; Jensen et al., 2015; Magwere et al., 2004; Maklakov et al., 2008).
This led us to consider that sex and genetic background might affect the metabolic and molecular response of mice to different levels of dietary protein. Here, we comprehensively determined the physiological, metabolic, and molecular impact of dietary protein on young male and female C57BL/6J (C57) and DBA/ 2J (DBA) inbred mice, young genetically heterogeneous male and female HET3 (HET) mice, aged male and female C57 mice, and male and female C57 mice lacking Fgf21. We selected these strains due to the widespread use of C57 mice for previous studies, the reputation of DBA mice as being less metabolically responsive, and the heterogeneous genetic makeup of HET mice, which better mimic the genetic diversity of humans and have been extensively used in aging studies (Cheng et al., 2019).
Mice were fed one of three isocaloric diets with varying levels of protein. We found that many of the phenotypes we measured–such as body weight, EE, and glucose tolerance– correlated with protein intake across sexes and strains. However, significant changes in these responses were, for the most part, induced only by the lowest protein diet, suggesting that there may be a ‘‘threshold’’ of protein intake as recently suggested (Wu et al., 2021). We observed the most apparent and significant effects in C57BL/6J males, confirming our hypothesis that the effects of dietary protein depend on both genetic background and sex.
Some effects of an LP diet were strongly dependent upon sex and strain, including blood levels of FGF21 and changes in adiposity and insulin sensitivity. Our molecular analyses demonstrate that the physiological and metabolic changes we observe are underpinned by diverse strain- and sex-dependent molecular changes. We identified gene-metabolite-lipid mega-clusters that correlate with key phenotypes and found that with an LP diet there was a strong relationship between EE, lipid storage, and ketone bodies, as well as between glycemic control and amino acid metabolism. Surprisingly, we found that blood FGF21 levels and expression did not cluster tightly with many metabolic and molecular phenotypes; using Fgf21–/– mice, we determined that the effects of FGF21 are sex specific. Finally, we found that age blunts the positive effects of an LP diet on insulin sensitivity in males and EE in both males and females.
We conclude that many aspects of the response to a reduced protein diet in mice depend on both sex and genetic background. While a personalized medicine approach to alleviate age-associated diseases such as diabetes and obesity is vital, consuming less protein may be an easy and translatable way to promote and restore metabolic health, even when started at older ages.
RESULTS
The effect of dietary protein level on weight and body composition is dependent on sex and genetic background
We placed 10-week-old C57BL/6J (C57), DBA/2J (DBA), and HET3 (HET) male and female mice on either a Control (21% calories from protein), a medium protein (MP, 14% calories from protein), or an LP (7% calories from protein) diet (Figure 1A). All three diets were isocaloric, as calories from fat were kept at 20%, and carbohydrates were used to replace calories from protein (Table S1).
Figure 1. An LP diet has sex- and strain-specific effects on body weight and composition.
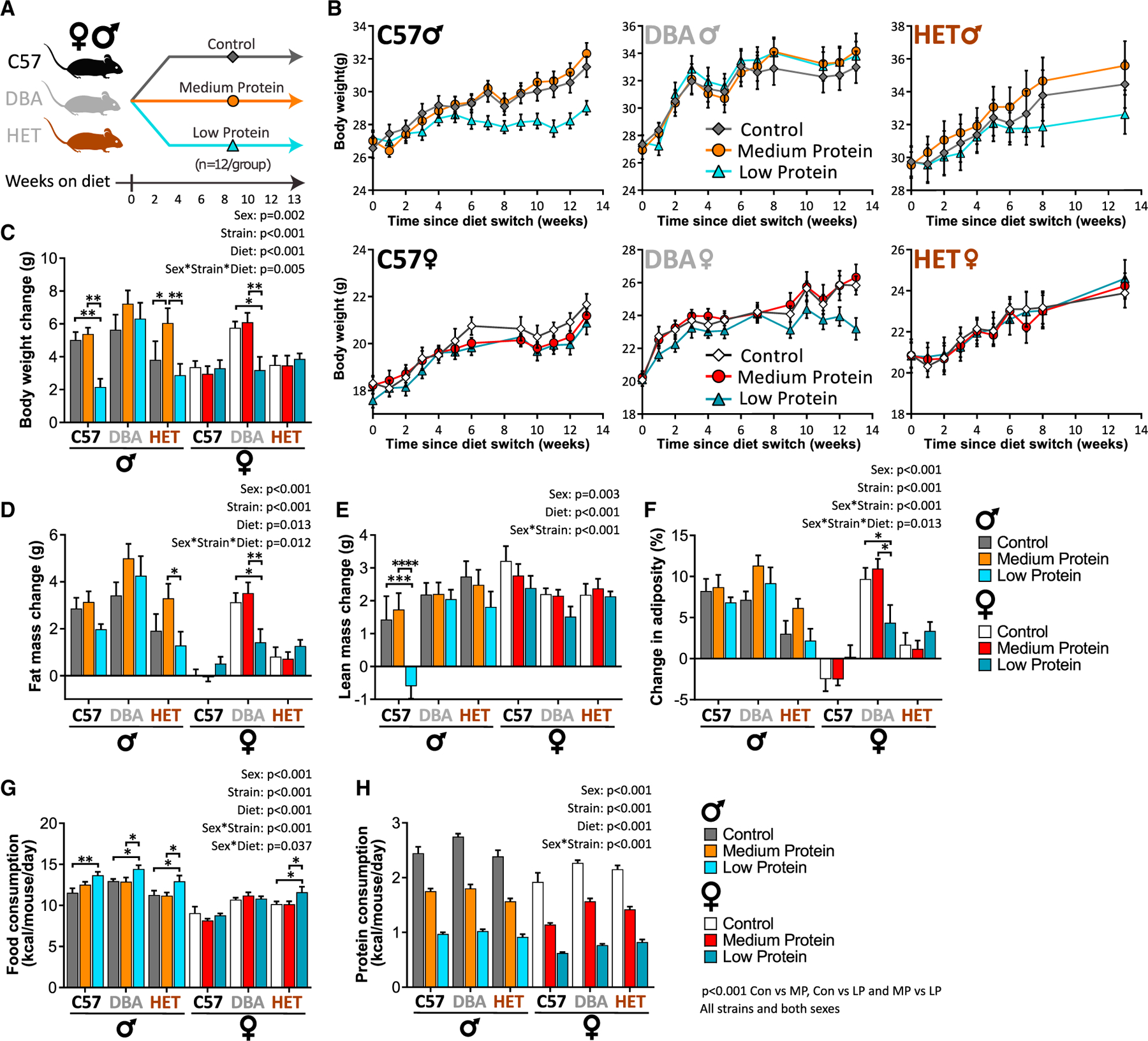
(A) Experimental plan: male and female C57, DBA, and HET mice were fed the indicated diets starting at 10 weeks of age.
(B) Weight.
(C–F) The change in weight (C), fat mass (D), lean mass (E), and adiposity (F) over the course of the experiment.
(G) Food consumption was measured in home cages after 9 weeks on diet.
(H) Calculated protein intake.
(B–H) n = 11–12 mice/group; exact n in Table S7.
(C–H) Three-way ANOVA between sex, strain, and diet groups with post hoc Benjamini-Hochberg (BH) corrected test for pairwise comparisons, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. p values for the overall effect of sex, strain, diet, and the interactions represent the significant p values from the three-way ANOVA. Data are represented as mean ± SEM.
All mice gained weight over time. As expected, LP-fed male C57 mice gained significantly less weight than Control and MP-fed counterparts; similarly, LP-fed HET males and DBA females gained less weight than their Control or MP-fed counterparts (Figures 1B and 1C). In contrast, the weight gain of C57 females, HET females, and DBA males was not significantly altered by reduced protein diets (Figures 1B and 1C).
Over the course of the study, we observed significant effects of sex and strain on fat mass gain (Figure 1D) and an overall significant effect of sex on lean mass gain (Figure 1E). LP-fed C57 males lost 0.5 g of lean mass over 13 weeks (Figure 1E), and LP-fed DBA females gained significantly less fat mass than their Control- and MP-fed counterparts (Figure 1D). Adiposity increased over time in all groups except C57 females (Table S2A). While diet did not significantly affect adiposity, three-way ANOVA indicated a significant effect of both sex and strain on adiposity, and a significant interaction between all three (Figure 1F). DBA males and females had the greatest final adiposity, irrespective of diet (Table S2A). The reduced weight gain of some LP-fed groups was not due to reduced caloric intake; male mice of all strains and female HET mice ate more when fed an LP diet (Figure 1G). Despite the increased food consumption of some groups, both the MP and LP groups were protein restricted relative to Control-fed mice (Figure 1H).
Dietary protein-mediated improvements in glycemic control depend on sex and strain
To examine the effect of dietary protein level on glucose homeostasis, we performed glucose (GTT) (Figure 2A), insulin (ITT) (Figure S1), and alanine tolerance tests (ATT) (Figure S2A), and collected fasting blood to perform a homeostasis model assessment (HOMA) (Figures 2B–2E, S2B, and S2C). Glucose tolerance was significantly improved in LP-fed C57 mice relative to Control- and MP-fed mice (Figure 2A). HET males showed a graded response to protein intake, with improved glucose tolerance on the MP diet, and further improvements on LP. In contrast, dietary protein level did not affect the glucose tolerance of DBA mice or HET females. Fasting blood glucose (FBG) followed a similar pattern, with decreases in C57 mice and HET males (Figure 2B).
Figure 2. Sex- and strain-dependent effects of dietary protein on glucose homeostasis and energy expenditure.
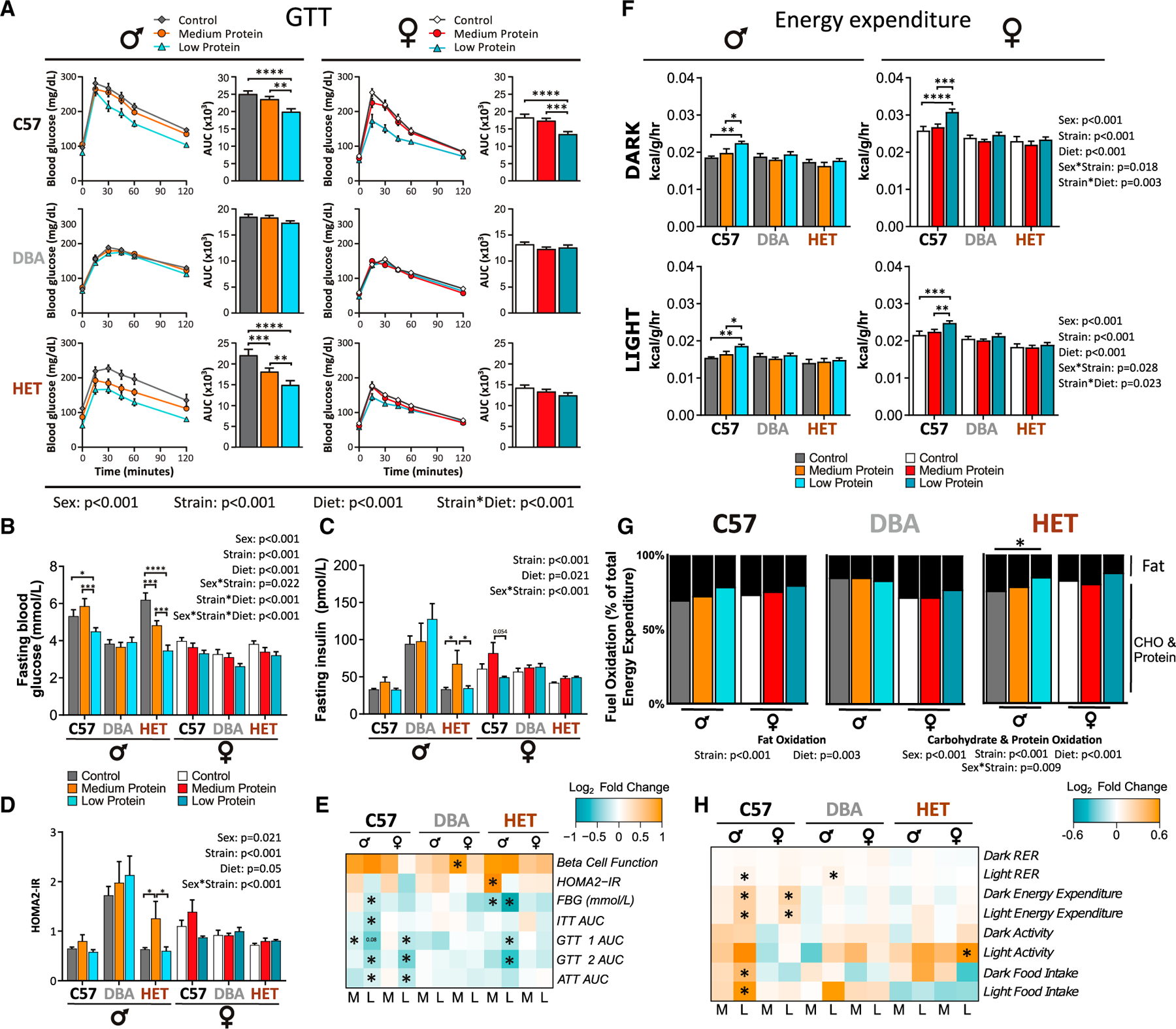
(A) Glucose tolerance test (GTT) after 12 weeks on diet.
(B) Fasting blood glucose (FBG) during GTT.
(C and D) Fasting insulin (C) was determined and HOMA2-IR (D) was calculated after 6 weeks on diet.
(E) Log2 fold changes of the indicated glycemic control measures for MP- (M) and LP-fed (L) mice relative to Control-fed mice. B cell function (%B) calculated using HOMA2, GTT1 was conducted after 3 weeks on diet, GTT2 at 12 weeks, insulin tolerance test (ITT) at 4 weeks, alanine tolerance test (ATT) at 5 weeks, FBG from GTT2.
(F) Energy expenditure (EE).
(G) Fuel oxidation for fats and carbohydrates and protein was calculated from EE and respiratory exchange ratio (RER). Colored bars represent the proportion of energy (in kcal) from protein and carbohydrate and black bars indicate the proportion of energy from fat oxidation.
(H) Log2 fold changes of metabolic chamber data, including EE/BW, food intake, RER, and spontaneous activity.
(A–H) Exact n for each experiment in Table S7.
(A–D, F, and G) Three-way ANOVA between sex, strain, and diet groups with post hoc BH corrected test for pairwise comparisons, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. p values for the overall effect of sex, strain, diet, and the interactions represent the significant p values from the three-way ANOVA. (E and H) *p < 0.05, BH-corrected pairwise comparison for each phenotype for MP or LP diet compared with Control-fed mice of the same sex and strain. Data are represented as mean ± SEM.
Only LP-fed C57 males showed improved insulin sensitivity during an ITT (Figure S1), and no groups showed significant improvements in fasting insulin (Figure 2C). Glucose tolerance is most directly responsive to hepatic insulin sensitivity and pancreatic b cell function. Consistent with LP diets improving suppression of hepatic gluconeogenesis, we saw significantly improved alanine tolerance (Fernandes and Blom, 1974; Mutel et al., 2011) in LP-fed C57 mice and HET males (Figure S2A).
We used FBG and insulin data (Figure 2C) to determine insulin resistance and fasting b cell function using the HOMA2 method (Geloneze et al., 2009; Mather, 2009; Song et al., 2016). Consistent with our ITT data, we found that insulin sensitivity was not significantly altered by MP or LP diets (Figures 2D and S2B), which may reflect that the Control-fed mice are young, lean, and healthy. However, pancreatic b cell function (HOMA2 %B) was improved significantly in LP-fed C57 males, with a similar trend in HET males (p = 0.13) (Figure S2C). These changes may indicate that the improved glucose tolerance of LP-fed male C57 and HET mice may be due to a combination of improved suppression of hepatic glucose production, as indicated by the improved alanine tolerance, as well as improved b cell function and insulin sensitivity (Figures S1, S2A, and S2C).
LP diet feeding generally led to improvements in glucose homeostasis across sexes and strains and was not associated with negative effects (Figure 2E). The effect size and statistical significance varied dramatically with sex and strain and was strongest in C57 male and female mice and HET males.
The relationship between changes in energy expenditure and protein intake are sex and strain dependent
LP diets promote increased EE, which is thought to contribute to both weight loss and improved glycemic control (Hill et al., 2017; Laeger et al., 2014). We found an overall effect of sex and strain on EE, with females having substantially higher EE than males, and HET mice tending to have the lowest EE of the three strains examined. Both male and female LP-fed C57 mice had significantly increased EE relative to mice on the Control or MP diets (Figure 2F). Neither the MP nor LP diets altered EE in HET or DBA mice (Figure 2F), which was surprising as male HET mice had both decreased weight gain and improved glycemic control (Figures 1B and 2A). These results suggest that although EE correlates with metabolic improvements in C57 mice, increased EE may not be required for an LP diet to improve metabolic health.
In C57 males, protein intake correlated positively with body weight change and negatively with EE, adiposity, and levels of FGF21 (Figure S3; Table S2B). While a similar set of phenotypes correlated with protein intake in C57 females, EE did not correlate with changes in body composition. EE also did not correlate with changes in body composition in HET females, except for a small positive correlation with adiposity (Figure S3; Table S2B). In DBA mice, EE was negatively correlated with change in body weight, fat, and adiposity, and in DBA females EE was also negatively correlated with hepatic Fgf21 expression (Figure S3; Table S2B). In sharp contrast to C57 mice, in DBA males none of these parameters correlated with protein intake. However, all of these parameters correlated with protein intake in DBA females, indicating a distinct sexually dimorphic response of EE to protein intake in DBA mice (Figure S3; Table S2B). Despite the increased EE of C57 mice, activity was largely unchanged; only LP-fed HET females had increased activity, during the light phase only (Figure S4A).
The respiratory exchange ratio (RER) is a measure of metabolic substrate usage (Bruss et al., 2010; Farinatti et al., 2016). As we substituted carbohydrates for protein in the MP and LP diets, we anticipated that RER would increase. Using a three-way ANOVA, we found that strain and diet played a significant role in the response of RER, as did the interaction between sex and strain. RER increased in LP-fed mice, reaching statistical significance in male C57 and DBA mice during the light phase (Figure S4B). Only male HET mice showed a significant change in the proportion of energy derived from either fat or carbohydrate/protein (Bruss et al., 2010; Lusk, 1924) (Figure 2G). However, the three-way ANOVA indicated a significant overall effect of strain and diet on fat oxidation, and of sex, strain, and diet on carbohydrate/protein oxidation (Figure 2G). No changes in substrate utilization were significant in females (Figure 2G). Reducing dietary protein affected RER and EE similarly across all strains and sexes, but the effect size and significance depended upon on genetic background. In contrast, we observed strain- and sex-dependent effects of dietary protein on activity level and food intake. The effects of an LP diet on parameters of energy balance are most striking in C57 mice and muted in both DBA and HET mice (Figure 2H).
Correlation of phenotypic data with protein intake and multivariate analysis indicates conserved global changes by LP diets
We used multivariate analysis to comprehensively identify strain- and sex-dependent responses to reduced dietary protein. We determined the calories from protein eaten by each individual mouse and correlated it with 31 phenotypic measurements obtained from each animal. We used these correlations in a heatmap and performed hierarchical clustering on the strength and direction of each correlation (Figure 3A; Table S2C).
Figure 3. Correlation analysis identifies strain- and sex-dependent and independent physiological and metabolic responses to LP diet.
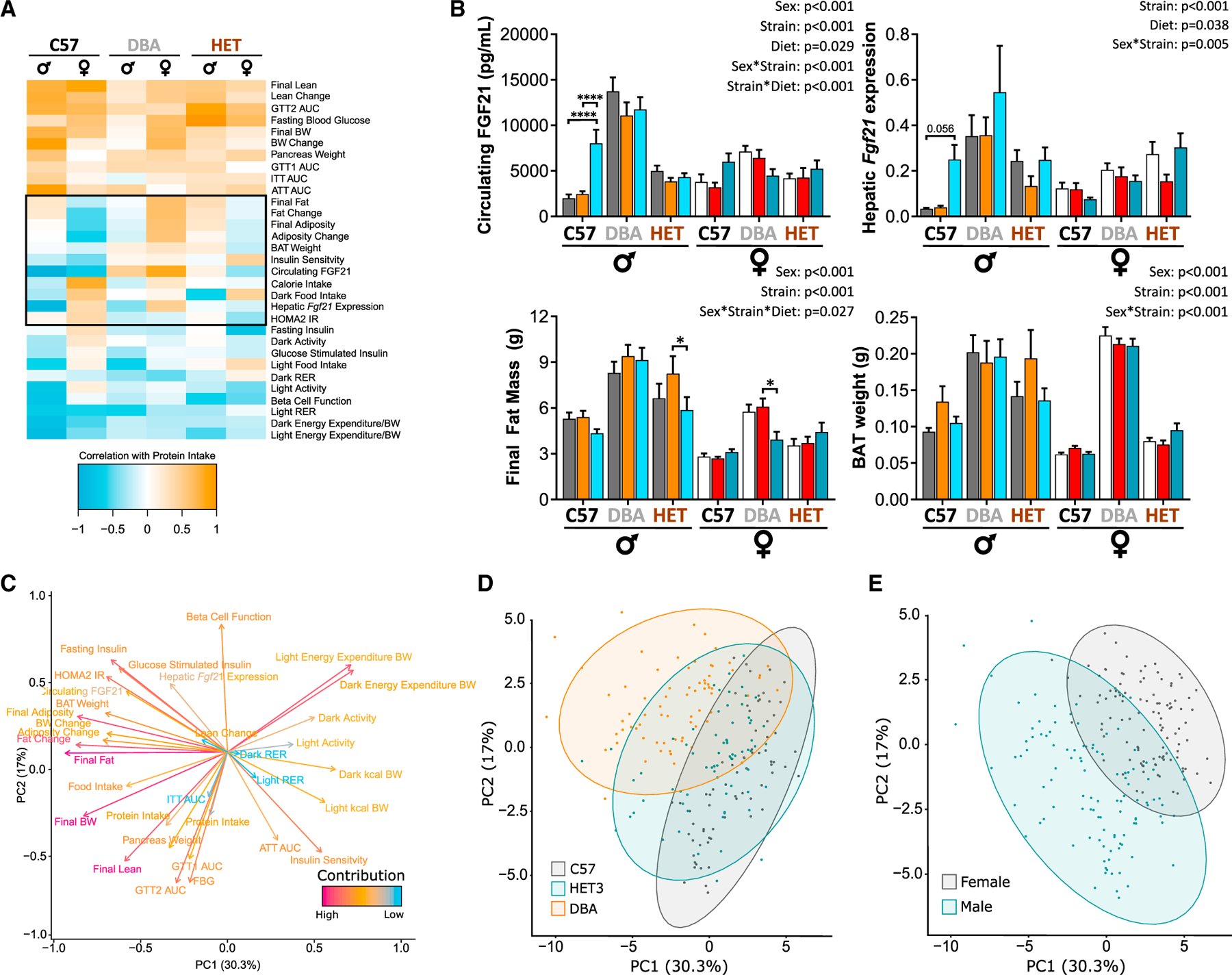
(A) Phenotypic measurements correlated with consumption of protein (kcals) in each mouse (Pearson’s correlation) and clustered (hierarchical clustering). Phenotypic measurements that do not cluster as well appear in the middle of the correlation plot (black outline).
(B) Selected phenotypic measurements within this black box were plotted.
(C–E) PCA on phenotypic measurements from each individual mouse.
(C) Phenotypic measurements were visualized: positively correlated variables point to the same side of the plot; negatively correlated variables point to opposite sides of the plot. Length and color of arrows indicate contribution to the principal components.
(D and E) Individual mice visualized by (D) strain and (E) sex indicating separation between the groups along PC1 and PC2.
(A–E) n for each measurement is detailed in Table S7.
(B) Three-way ANOVA between sex, strain, and diet groups with post hoc BH-adjusted test for pairwise comparisons; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. p values for the overall effect of sex, strain, diet, and the interactions represent the significant p values from the three-way ANOVA. Data are represented as mean ± SEM.
High protein intake was positively correlated with glucose intolerance (GTT area under the curve [AUC]), FBG, and body weight across all strains and sexes. High protein intake correlated negatively with EE, RER, activity, and β cell function across both sexes and most strains. In the middle of the plot (black outline, Figure 3A) are correlations that do not cluster well because they vary in a sex- and strain-dependent manner. These phenotypes include the effect of dietary protein on blood FGF21, hepatic Fgf21 expression, final fat mass, brown adipose tissue (BAT) weight (Figure 3B), insulin sensitivity (Figure S2), calorie intake (Figure 1G), and change in adiposity (Figure 1F).
To determine which phenotypes contribute the most to differences between groups, we performed a principal component analysis (PCA) with the phenotypic data. We visualized the contribution of each phenotype to the variation seen between mice (Figure 3C) and found that protein intake, lean mass, FBG, and GTT AUC correlate together, and that these also correlated with pancreas weight and to a lesser extent ITT AUC. Additionally, EE, activity, and RER grouped together (Figure 3C). Intriguingly, many adiposity-related phenotypes were correlated with circulating FGF21, including BAT weight and fasting insulin (Figure 3C).
Both genetic background (Figure 3D) and sex (Figure 3E) influenced the phenotypic response to dietary protein level. There was limited overlap between C57 and DBA mice, whereas HET mice, which contain both C57 and DBA alleles, overlapped both C57 and DBA mice (Figure 3D). We saw large differences between sexes, with greater variability in males, reflecting the pronounced differences in the response to an LP diet between strains (Figure 3E). The greatest contributors to PC1, which seems to be more involved in differences in sex (x axis in Figures 3C–3E), were final fat mass, final body weight, dark and light EE, final adiposity, and fat mass change (Table S2D). The greatest contributors to PC2, which seems to be more related to differences in strain (y axis in Figures 3C–3E), were related to glucoregulatory control, including insulin sensitivity, β cell function, fasting insulin, FBG, lean mass, GTT AUC, and HOMA2 IR.
Molecular responses to an LP diet are sex and strain dependent
Due to the central role of the liver in metabolic homeostasis, in particular glucose metabolism, and the sex- and strain-dependent effect of dietary protein on glucose tolerance and hepatic Fgf21 expression, we conducted a detailed molecular analysis of the liver. We performed transcriptomic, metabolomic, and lipidomic analyses of the livers of C57 and DBA male and female mice after 13 weeks on either a Control, MP, or LP diet.
At the transcriptional level, there were significant differences in the response to an LP diet relative to sex and strain (Table S3A). There were profound differences in the number of genes altered by LP in each strain, e.g., the expression of almost 2,000 genes was significantly altered by an LP diet versus Control diet in C57 males, whereas only 24 genes were altered by LP in DBA males (Table S3B, adjusted p < 0.05). While C57 females showed fewer altered genes than C57 males, DBA females had many more genes altered by an LP diet than DBA males. There was a clear impact of restriction level, as only 40 genes, all in C57 males, were significantly differentially expressed in MP-fed mice (Table S3B, adjusted p < 0.05).
We enriched significantly differentially expressed genes (unadjusted p < 0.05) using KEGG analysis to identify specific pathways altered by MP and LP diets in each strain and sex (Figures 4A and 4B; Table S3C). In LP-fed C57 mice, arachidonic acid metabolism, fatty acid degradation, and PPAR signaling pathways were all increased (Figure 4B), which may reflect an overall shift in metabolism to greater fatty acid oxidation, which is also seen in CR (Bruss et al., 2010). This may relate to the increased circulating FGF21 levels we see in both C57 males and females (Figure 3B), as FGF21 promotes hepatic fatty acid oxidation through PPARα (Badman et al., 2007). Pathways decreased in LP-fed C57 males and females fell into three camps: amino acid and glucose metabolism, cholesterol metabolism, and the immune system (Figure 4B). Interestingly, one downregulated pathway shared between males and females, glycine, serine, and threonine metabolism, has been implicated as a key metabolic hub associated with lifespan (Aon et al., 2020). Uniquely upregulated in DBA females were oxidative phosphorylation, the TCA cycle, thermogenesis, and several amino acid pathways, while in DBA males the lysosome, synaptic vesicle cycle, and sphingolipid metabolism were upregulated. In C57 and DBA males, nicotinamide as well as alanine, aspartate, and glutamate metabolisms were downregulated (Table S3C). This further supports the idea that the molecular mechanisms underpinning the response to an LP diet is both sex and strain dependent and that it involves a shift in fuel utilization.
Figure 4. Changes in hepatic transcriptional and lipid pathways with LP diet are both sex and strain dependent.
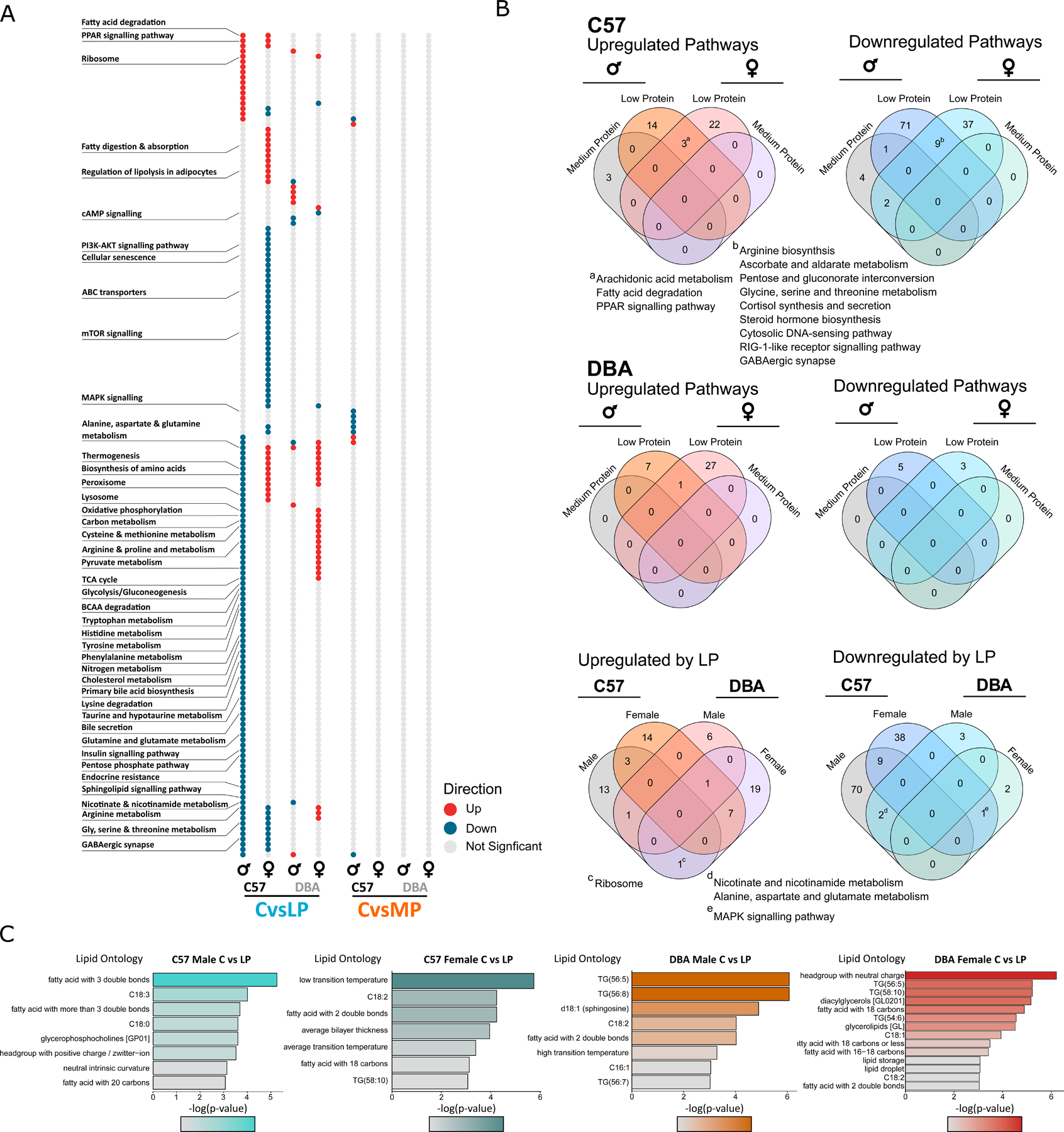
(A and B) Transcriptional differences between LP- or MP-fed mice relative to Control-fed mice.
(A) Significantly up- and downregulated pathways for each sex, diet, and strain were determined using KEGG enrichment, n = 6 per group, p < 0.05. Pathways of interest are highlighted.
(B) Venn diagram of overlap between significant pathways, with pathways of interest highlighted.
(C) Lipid enrichment analysis between Control and LP-fed mice of each sex and strain (n = 6 per group, p < 0.05).
We saw sex- and strain- dependent changes at the lipid level, with the most in LP-fed C57 males and a few significantly altered lipids in female C57s and male and female DBAs (Tables S4A and S4B). Using lipid ontology (LION) enrichment on significantly altered lipids (unadjusted p < 0.05), we found distinct lipid pathways altered in each sex and strain (Figure 4C; Table S4C). In C57 males, highly unsaturated fatty acids and ceramides were most significantly altered on an LP diet, whereas in females an LP diet appeared to impact the lipid bilayer (Figure 4C). In DBAs, there were changes in highly unsaturated triacylglycerols and ceramides, and in males changes in sphingosines (Figure 4C).
Sex- and strain-dependent changes were also observed in the hepatic metabolome (Figure 5; Table S5A). While a similar number of metabolites were significantly altered in male and female C57 mice, after adjustment for the false discovery rate, very few metabolites were altered in DBAs (Table S5B). Differentially altered metabolites (unadjusted p < 0.05) were enriched for KEGG pathways using Metaboanalyst (Figure 5A; Table S5C). As with the hepatic transcriptome, the greatest number of altered pathways by LP was in C57 males (Figure 5A). Arginine biosynthesis, which is also involved in production of urea, was significantly altered in all groups, and most strongly upregulated in C57 females.
Figure 5. Changes in hepatic metabolic pathways with LP diet are both sex and strain dependent.
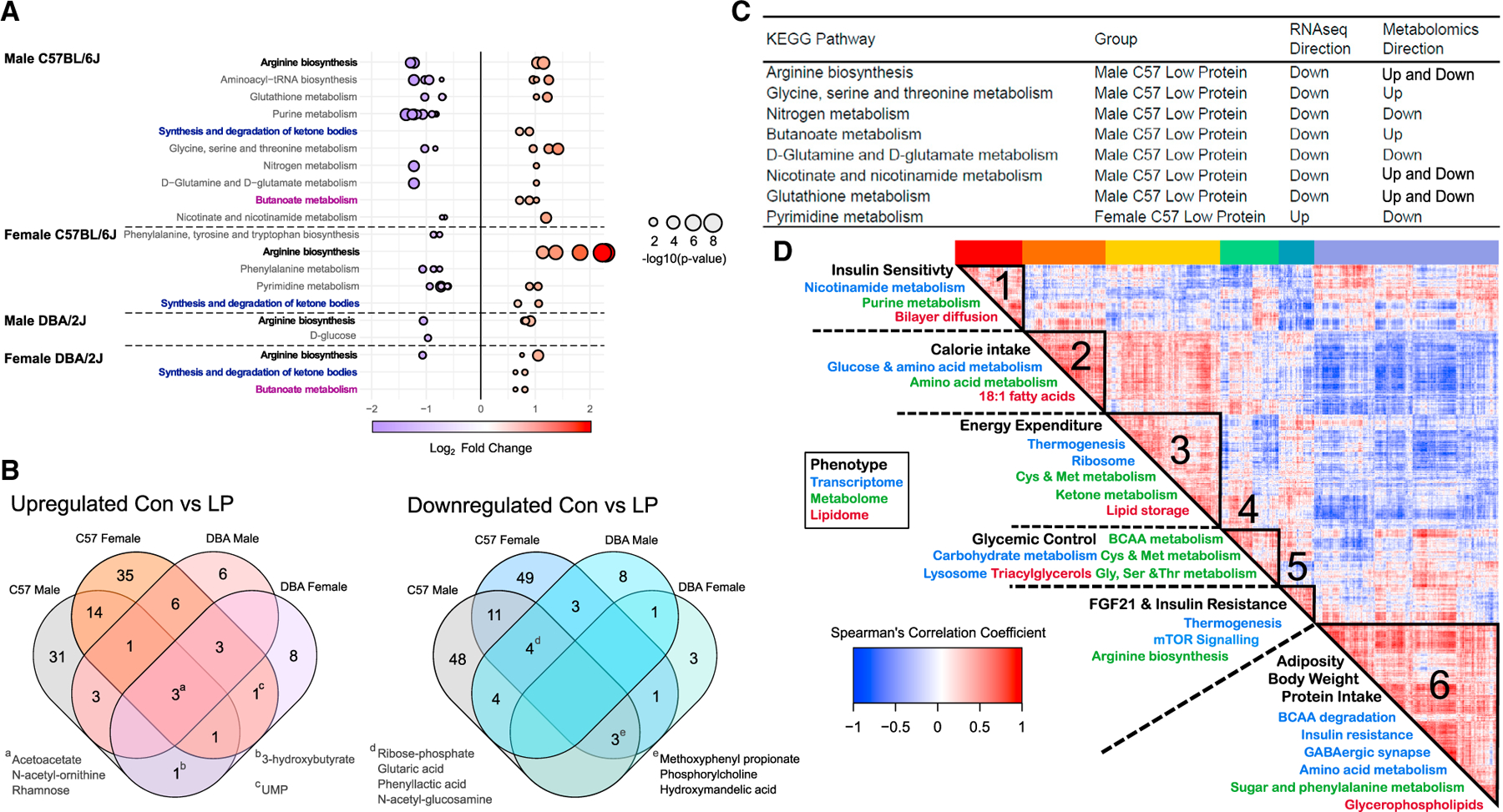
(A) Significantly up- and downregulated pathways were determined using KEGG enrichment of metabolites (n = 6 per group, unadjusted p < 0.05). Dots represent individual metabolites and color indicates log2 fold change. Matching pathways between groups are highlighted in bold with corresponding colors.
(B) Venn diagram of overlap of significant pathways, with pathways of interest highlighted.
(C) KEGG pathways present in both the transcriptional and metabolomics analyses in Control diet versus LP diet (Tables S3C and S5C) and the overall direction of change.
(D) LP diets produce changes across phenotypes that correlate with genes, metabolites, and lipids. Spearman’s rank order correlation matrix of a total of 831 significant observations, including phenotypic (black), transcriptomic (blue), metabolomics (green), and lipidomic (red) changes between Control and LP diet groups. Hierarchical clustering identified 6 mega-clusters (outlined in black; Table S5E). Enriched pathways listed in Table S5F; phenotypes and pathways of interest in each cluster are highlighted.
The synthesis and degradation of ketone bodies were significantly upregulated in C57s and female DBAs, with butanoate (butyric acid) metabolism also increased in male C57 and female DBA mice. This may indicate a shift toward ketogenesis, which is interesting as ketogenic diets are typically low in both protein and carbohydrates (Roberts et al., 2017), and the LP diet we used here has abundant carbohydrates. FGF21 may play a role, as it is induced by an LP (9.5% of calories from protein) ketogenic diet and is a key regulator of ketogenesis (Badman et al., 2007). Isoleucine restricted C57 males likewise have elevated FGF21 and increased ketones despite high levels of carbohydrate (Yu et al., 2021).
Three metabolites were significantly altered in both sexes and strains: a b-keto acid, acetoacetate; a substrate of nitrogen metabolism, N-acetyl-ornithine; and the deoxy-sugar, rhamnose. Interestingly, the ketone, 3-hydroxybutyrate, was significantly upregulated in C57 males and DBA females only (Figure 5B). Only 14% of the total metabolites significantly altered on an LP diet in the hepatic metabolome (unadjusted p < 0.05) appeared in both C57 and DBA mice (male or female). One metabolite was unique to DBA mice, arginosuccinic acid, an intermediate of the urea cycle, which was downregulated (Table S5D). In male C57 and DBA mice tetradecanoylcarnitine, O-phosphoethanolamine and tetradecadienyl-L-carnitine were upregulated and glycylproline, allantoin, D-glucose and glycerol 3-phosphate were downregulated, which may indicate a shift in energy metabolism on an LP diet (Table S5D).
We identified pathways that were enriched in KEGG for both the hepatic transcriptome and metabolome (Figure 5C). The most overlap was found in LP-fed C57 males; arginine biosynthesis, D-glutamine and D-glutamate metabolism, nitrogen metabolism, nicotinate and nicotinamide metabolism, and glutathione metabolism were broadly downregulated in the liver. In some cases, the transcriptome and metabolome did not agree as to the direction of change; this effect has been seen previously and may indicate that the transcriptional downregulation of catabolic genes is leading to an accumulation of metabolites (Neinast et al., 2019). This pattern is observed in glycine, serine, and threonine metabolism as well as butanoate metabolism. Only one overlapping pathway, pyrimidine metabolism, was observed in C57 females, and no overlapping pathways were observed in DBA males or females.
To determine the potential relationships between molecular changes and whole-organism physiology and metabolism, we constructed a correlation matrix using the phenotypic, transcriptomic, metabolomic, and lipidomic data. First, we identified the statistically significant changes in gene expression, metabolite levels, and lipid levels induced by an LP diet in each group (e.g., C57 males); this approach ensures that strain-specific phenotypes unrelated to protein intake did not affect our results. Concatenating all the statistically significant changes from each group with the phenotypic data from each individual animal gave us a total of 831 inputs, including 83 metabolites, 623 transcripts, 79 lipids, and 46 phenotypes. Spearman’s correlation coefficients were then calculated for 831 × 831 = 690,561 pairwise comparisons and then were rendered by hierarchical clustering (based on 1 – correlation coefficient between all molecules), ultimately settling on 6 clusters with an average cluster occupancy of 139 measures (Table S5E).
We enriched genes, metabolites, and lipids for pathways in each cluster and related these to the corresponding cluster phenotypes (Table S5F). While these results are based on correlations and are not necessarily causative, they are consistent with a broad reprogramming of hepatic metabolism by an LP diet, and link specific molecular and metabolic processes in the liver directly to the phenotype. As proof of the validity of this approach, calorie intake on an LP diet correlated with changes in macronutrient metabolism at all -omics levels (Figure 5D, cluster 2).
Hepatic expression of Fgf21 and circulating levels of FGF21 both clustered with thermogenesis, mammalian target of rapamycin (mTOR) signaling, and arginine biosynthesis (Figure 5D, cluster 5). Genes in this cluster were enriched with the regulatory motif for the transcription factor c-Myc, which is reported to be regulated by FGF21 (Liu et al., 2015). FGF21 is essential for LP-induced increases in EE in C57 males, promotes lipid oxidation (Chalvon-Demersay et al., 2019), and has been linked to mTORC1 signaling (Cornu et al., 2014; Gong et al., 2016; Minard et al., 2016). The correlation with arginine biosynthesis may be related to the role of arginine as a regulator of mTORC1 activity (Chantranupong et al., 2016), or due to the effect of arginine on mitochondrial biogenesis and whole-body oxidation (McKnight et al., 2010). Intriguingly, FGF21 did not cluster with EE; instead, dark and light EE clustered with genes and metabolites in which KEGG and GO terms for thermogenesis, oxidative phosphorylation, and ribosome were overrepresented, as well as enrichment for metabolites related to cystine and methionine metabolism (Figure 5D, cluster 3).
Several phenotypes associated with glucose homeostasis correlated with lysosomal gene expression as well as the metabolism of cystine and methionine; glycine, serine, and threonine metabolism; and branched-chain amino acid (BCAA) metabolism (Figure 5D, cluster 4). Both BCAAs and methionine have been linked to the regulation of blood glucose for many years (Felig et al., 1969; Lees et al., 2017; Lynch and Adams, 2014; Yu et al., 2018), but lysosomal gene expression and glycine, serine, and threonine metabolism have not been. Intriguingly, recent work has identified glycine, serine, and threonine metabolism as a key metabolic hub associated with lifespan (Aon et al., 2020); lysosomal signaling has likewise been linked to longevity (Lapierre et al., 2013; Wang et al., 2017).
Adiposity and insulin resistance clustered with genes involved in BCAA degradation (Figure 5D, cluster 6). This connection is logical, as we and others have shown that dietary BCAAs regulate weight, body composition, and insulin resistance (Cummings et al., 2018; Fontana et al., 2016; Jang et al., 2016; Newgard et al., 2009; Yu et al., 2021). This cluster was also strongly associated with phenylalanine metabolism, which is thought to reduce food intake through promoting satiety and improved glucose tolerance (Alamshah et al., 2017).
Fgf21 regulates some of the responses to a low-protein diet in a sex-dependent manner
LP diets induce FGF21, and deletion of either Fgf21, its receptor, or the upstream amino acid sensor Gcn2 attenuates the metabolic impact of an LP diet (Hill et al., 2017, 2019; Laeger et al., 2014, 2016). However, most studies of FGF21 and LP diets were conducted in C57 males, and as described above, we find that the response of FGF21 to an LP diet is strain dependent and sexually dimorphic. While both hepatic Fgf21 expression and FGF21 blood levels cluster with insulin resistance, FGF21 does not cluster with many other LP-induced metabolic phenotypes. This suggested to us that FGF21 may not be directly linked to all the metabolic benefits of an LP diet.
We placed wild-type (WT) and Fgf21–/– (FGF21 KO) mice on a C57 background on Control or LP diets and examined how sex and genotype impacted the metabolic response to an LP diet (Figure 6A). WT males gained less weight on an LP diet than on a Control diet, but deletion of Fgf21 blunted this effect (Figures 6B–6D). The effect of FGF21 on male weight gain was driven by increased lean mass gain in LP-fed FGF21 KO mice, and not by changes in fat accretion or adiposity (Figures 6E–6H). In females, there was no effect of an LP diet on weight gain (Figure 6I). However, loss of Fgf21 reduced lean mass gain and fat accretion regardless of diet (Figures 6J and 6K). There was an overall effect of an LP diet on female adiposity (Figures 6L and 6M).
Figure 6. FGF21 is responsible for some, but not all, metabolic changes on an LP diet.
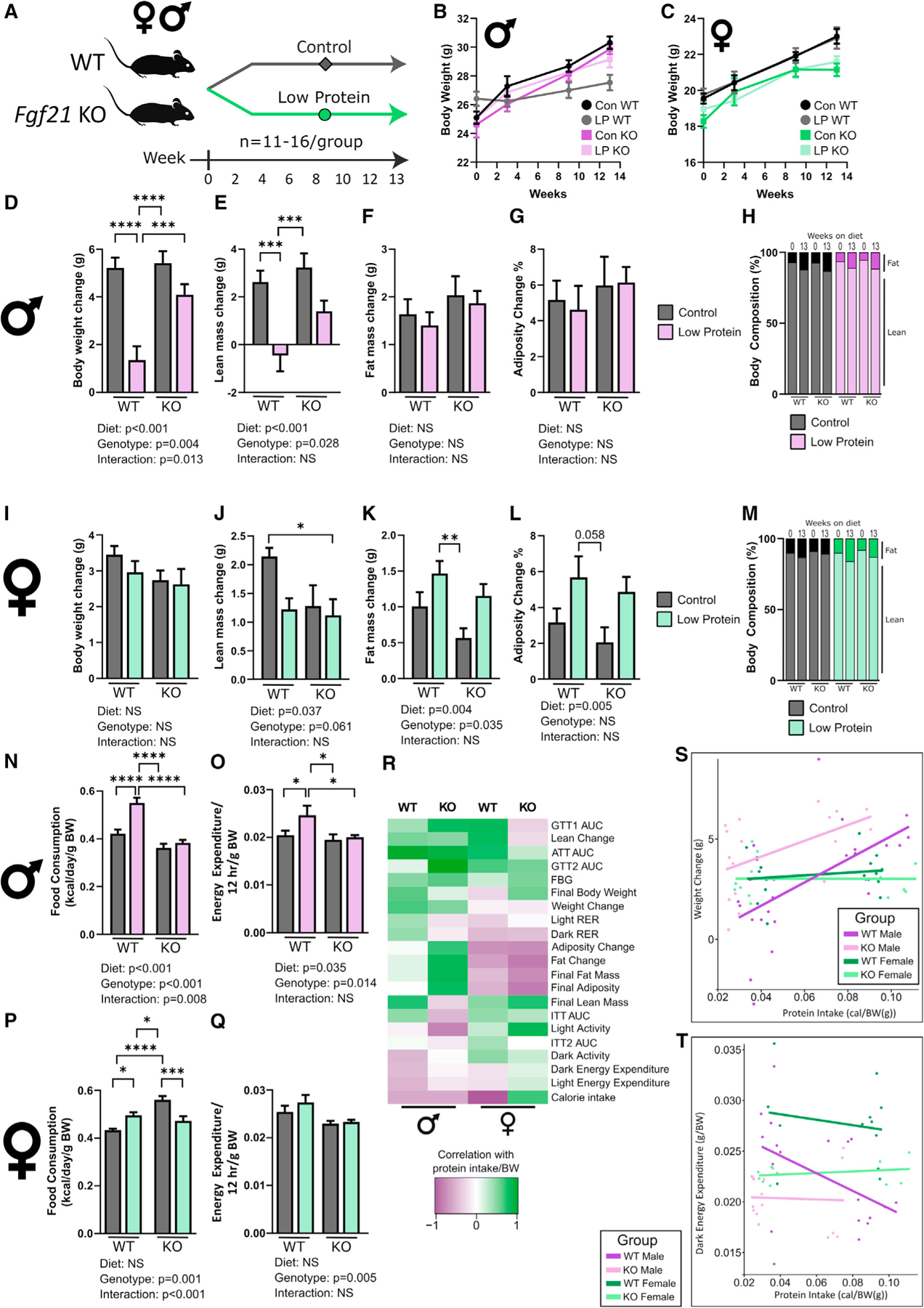
(A) Experimental plan (n for each phenotype in Table S7).
(B and C) Weight of male (B) and female (C) mice.
(D–G) Change in weight (D), lean mass (E), fat mass (F), and adiposity (G).
(H) Body composition was determined at diet start and 13 weeks later.
(I–L) In female WT and Fgf21–/– mice, we measured the change in weight (I), lean mass (J), fat mass (K), and adiposity (L).
(M) Body composition was determined at diet intervention start and 13 weeks later.
(N and O) Food consumption (N) (at 3 weeks) and energy expenditure (EE, dark phase, 7–9 weeks) (O) of males.
(P and Q) Food consumption (P) and EE (dark phase) (Q) of females.
(R) Phenotypes were correlated with protein intake using Pearson’s correlation.
(S and T) Correlations with regression lines calculated from (R) to examine the relationship between sex, genotype, and protein intake with weight change and EE (dark phase); p values in Table S6.
(B–G, I–L, and N–Q) Two-way ANOVA for significant differences between sex and diet and significant interactions with post hoc Sidak test, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Data are represented as mean ± SEM.
FGF21 reportedly mediates LP-induced increases in food consumption and EE (Chalvon-Demersay et al., 2019), and we observed that WT males, but not FGF21 KO males, have increased EE and food consumption when fed an LP diet (Figures 6N and 6O). In females, we did not observe a clear effect on food consumption; food intake in LP-fed WT females was mildly increased, while FGF21 KO females on a Control diet had greater consumption than WT females, which was decreased by LP diet feeding (Figure 6P). Female FGF21 KO mice had lower EE regardless of diet (Figure 6Q).
We performed glucose, insulin, and ATTs (Figures S5A–S5F). As expected, an LP diet improved glucose tolerance in WT mice but as previously reported (Hill et al., 2019), the effect of an LP diet was blocked in FGF21 KO mice (Figures S5A and S5D). FGF21 KO mice of both sexes had decreased insulin sensitivity, regardless of diet (Figures S5B and S5E). Alanine tolerance, which is reflective of hepatic gluconeogenesis from L-alanine (Fernandes and Blom, 1974; Mutel et al., 2011), was improved by an LP diet in both sexes and genotypes (Figures S5C and S5F), suggesting that an LP diet improves hepatic insulin sensitivity via an FGF21-independent mechanism. This agrees with our clustering analysis (Figure 5D) and our recent finding that restriction of isoleucine promotes hepatic insulin sensitivity via a FGF21-independent mechanism (Yu et al., 2021).
We correlated the protein intake of each individual mouse with its phenotypic measurements and performed hierarchical clustering on the correlation coefficients (Figure 6R). Phenotypes such as weight change, lean change, and AUC for ATTs correlated positively with protein intake, while EE and activity levels were negatively correlated; many of these relationships were maintained to some degree in FGF21 KO males, but several were flipped (Figure 6R). In females, the correlation of these phenotypes with protein intake was also flipped in several instances, including AUC for GTTs and ITTs, and calorie intake; additionally, deletion of Fgf21 in females promoted adiposity as protein decreased (Figure 6R). The response of total calories to protein intake flipped in response to genotype, but not sex (Figure S5G).
While the relationship between protein intake and phenotypes such as weight change was sexually dimorphic and FGF21 independent (Figure 6S), others such as EE (Figure 6T) and adiposity (Figure S5H) have a sexually dimorphic response, being dependent upon FGF21 in males, but not females. Some phenotypes, such FBG and ATT AUC, were totally independent of both sex and genotype (Figures S5I and S5J; Table S6). These unique relationships tended to appear in different mega-clusters (Figure 5D).
Aging blunts the sex-dependent metabolic impact of a low-protein diet
While an LP diet has generally favorable effects on glycemic control, weight, and EE in young mice, there is a great need for interventions that can start later in life. Protein requirements are thought to change with age; and aged mice have been shown to maintain a memory of early life nutrition (Hahn et al., 2019; van Dijk et al., 2017).
To determine if LP diets are effective later in life, we placed aged (21–22 months) male and female C57BL/6J.Nia mice from the NIA Aging Rodent Colony on either Control or LP diets (Figure 7A). While aged female mice maintained their weight on an LP diet, aged male mice switched to the LP diet lost weight, primarily due to a significant loss in lean mass, although fat mass also trended downward (Figures 7B–7F). There was a striking effect of sex on both change in fat mass and change in adiposity, in which aged males lose over time and females gain (Figures 7G and 7H).
Figure 7. Aged C57BL/6 male and female mice have improved glycemic control on an LP diet.
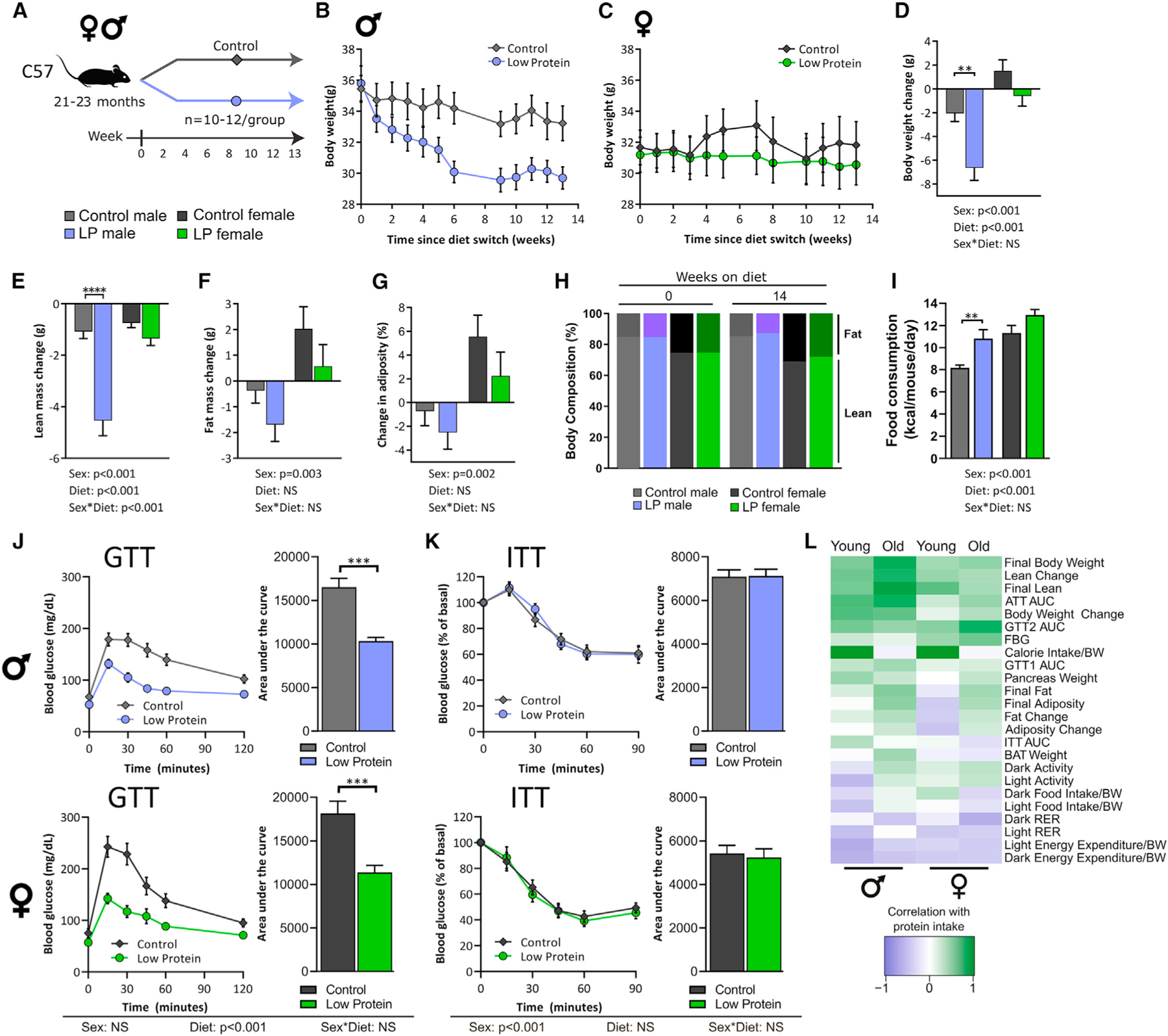
(A) Experimental plan.
(B and C) Weight of male (B) and female (C) mice.
(D–G) Change in weight (D), fat mass (E), lean mass (F), and adiposity (G).
(H) Body composition was determined at diet intervention start and 14 weeks later.
(I) Food consumption.
(J and K) GTT (J) and ITT (K) after 9 and 10 weeks on diet, respectively.
(L) Phenotypes were correlated with protein intake using Pearson’s correlation and compared with young C57 mouse data (shown in Figures 1 and 2), Y, young; O, old.
(B–K) n for each phenotype in Table S7. Two-way ANOVA between sex and diet with post hoc Sidak test for pairwise comparisons, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Data are represented as mean ± SEM.
Despite losing weight, aged LP-fed males had higher food intake (Figure 7I), similar to what we observed in young C57s (Figure 1H). Surprisingly, an LP diet did not increase EE in aged mice (Figure S6A), whereas a significant increase was seen in LP-fed young mice (Figure 2F). In both sexes RER was increased in the dark phase with LP feeding (Figure S6B), but as with their young counterparts, little change was seen in activity (Figure S6C). This may account for the increase in adiposity seen in aged females fed an LP diet but does not explain the weight loss seen in males.
Both aged males and females fed the LP diet had improved glucose tolerance (Figure 7J). In contrast to young males, LP-fed aged males do not have improved insulin sensitivity (Figure 7K). We correlated phenotypic parameters with level of protein in the diet and compared this with the results previously determined in young C57 mice. In general, young and aged mice had similar phenotypic responses to an LP diet (Figure 7L). However, in young C57 females, fat mass and adiposity correlate negatively with protein intake, while the opposite is true in young male C57 mice. In aged C57 female mice, the response of these parameters to an LP diet is blunted, with old female mice responding more similarly to aged males (Figure 7L).
DISCUSSION
‘‘A calorie is not just a calorie,’’ and the macronutrient composition of the diet may be just as important in determining health as how many calories are consumed. An important role for dietary protein in human metabolic health has been suggested by both retrospective and prospective cohort studies (Lagiou et al., 2007; Levine et al., 2014; Sluijs et al., 2010; Vergnaud et al., 2013), and we recently demonstrated that protein restriction (PR) improves human metabolic health (Fontana et al., 2016).
Several groups have shown that dietary protein is an important regulator of metabolic health in mice (Fontana et al., 2016; Laeger et al., 2014; Solon-Biet et al., 2014, 2015). However, the majority of these studies have characterized the response to dietary protein exclusively in C57BL/6J males, and recent work has highlighted that sex and genetic background impact the response to nutritional interventions (Green and Lamming, 2021). The high interpersonal variability of humans in response to diets has been attributed in part to genetic variation (Bashiardes et al., 2018; Zeevi et al., 2015).
Here, we have performed the first comprehensive analysis of the impact of sex and strain on the response to dietary levels of protein. Broadly, we have found that there are strain- and sex-dependent effects of an LP diet. In the strains we examined, the metabolic response to an LP diet is sexually dimorphic, with both C57 and HET males responding more strongly than their female counterparts; this is flipped in DBA mice, with DBA females responding more strongly than DBA males.
The response to dietary protein was not linear, and in combination with the work of others suggests that there is a threshold at which PR no longer exerts its effects (Wu et al., 2021). This is particularly apparent in C57 mice, with the phenotypes and liver transcriptome of mice fed a MP diet (14% protein) being virtually indistinguishable from those of mice fed a Control diet (21% protein). This threshold may be strain dependent, as a MP diet did improve the glucose tolerance of HET males and also decreased weight gain and increased food intake. LP-fed HET males did not increase their EE but did alter their fuel source utilization, which in humans has been linked to weight change without changes in EE (Zurlo et al., 1990).
Across all strains and sexes, lean mass, GTT AUC, ITT AUC, FBG, and body weight all generally increased with increasing protein in the diet. In contrast, RER, EE, and activity levels decreased as dietary protein intake increased across strains and sexes. This indicates that despite the genetic variation between these strains of mice, dietary protein has many broadly conserved negative effects on the metabolic health of mice—just as it does in humans, where higher consumption of dietary protein is linked to diabetes.
A number of phenotypes, including blood FGF21, hepatic Fgf21 expression, fat mass, adiposity, insulin sensitivity, and fasting insulin, had a more varied response to dietary protein level in different strains and sexes. LP diets increased FGF21 only in C57 mice, and only male C57 mice had increased hepatic Fgf21 expression on an LP diet. Our finding is in agreement with recent work showing reduced induction of FGF21 by an LP diet in intact females as compared with males or ovariectomized females (Larson et al., 2017). DBA and HET mice, including those sexes that had improved metabolic health on an LP diet, did not show increased levels or expression of FGF21 on LP. Indeed, across strains and sexes, our phenotypic correlation and multiomics analyses in Figures 3 and 5 find that although FGF21 level and hepatic expression correlate with fasting insulin, β cell function, and HOMA2-IR, and cluster with thermogeneic genes and metabolites, many other phenotypes we observed do not correlate as strongly with FGF21. Interestingly, FGF21 did cluster with mTOR signaling (Figure 5D), which supports previous work indicating that FGF21 modulates mTORC1 activity (Gong et al., 2016). Biosynthesis of arginine, a key agonist of mTORC1, was also found in this cluster (Chantranupong et al., 2016; Wang et al., 2015).
We examined the effect of Fgf21 deletion on the response to dietary protein in C57 mice of both sexes. In males, our findings confirmed an important role for FGF21 in the metabolic response to an LP diet, including the LP-induced increase in food intake and EE (Hill et al., 2019; Laeger et al., 2014). FGF21 is not as strongly induced by an LP diet in C57 females, and our study of female Fgf21–/– mice yielded some surprising results. We identified FGF21-dependent and independent responses to an LP diet in both sexes. Female Fgf21–/– mice maintained a lower body mass regardless of diet, possibly due to slightly lower adiposity, and lean mass gain was blunted in Control-fed female Fgf21–/– mice (Figures 6C and 6J). The well-characterized FGF21-dependent increase in EE and food consumption in LP-fed mice was observed only in males (Figures 6O and 6Q), and loss of Fgf21 completely ablated the relationship between EE and protein intake in males, but not females (Figure 6T).
We conclude that while FGF21 plays an important role in the metabolic response to an LP diet, some effects of an LP diet, particularly in females, are FGF21 independent. Thus, the specific role of FGF21 in many LP-diet-induced phenotypes varies by sex. Investigation of the role of FGF21 in both sexes and in genetic backgrounds other than C57 mice will likely be highly useful in understanding the role of FGF21 in the response to an LP diet, particularly with regards to phenotypes that do not cluster with FGF21, including EE, hepatic gluconeogenesis (glucose and alanine tolerance, and FBG) and ketogenesis, and adiposity.
EE clustered with genes and metabolites in which KEGG and GO terms for thermogenesis, oxidative phosphorylation, and ribosome were overrepresented (Figure 5D, cluster 3). We also found an enrichment for metabolites related to cysteine and methionine metabolism, and ketone metabolism. While methionine restriction (MR) has metabolic benefits similar to those of an LP diet, the effects examined here do not result from MR. All our diets were supplemented with additional methionine as is common for rodent diets (Reeves et al., 1993); the 7% LP diet used here contained a total of 1.9 g/kg methionine, significantly greater than the levels typically used in MR studies (Hasek et al., 2010; Miller et al., 2005; Wanders et al., 2017; Yu et al., 2018). Further, hepatic levels of methionine were not significantly altered by LP feeding in C57BL/6J males and DBA/2J mice of either sex. Finally, many effects of MR are blocked by the presence of cysteine (Wanders et al., 2016), which is present in our diets.
FGF21 has been identified as a key regulator of ketogenesis (Badman et al., 2007; Domouzoglou and Maratos-Flier, 2011). The increased synthesis of ketones in LP-fed mice suggests that these may be an important fuel source despite the high levels of dietary carbohydrate. Ketogenesis is regulated by hepatic mTORC1 (Sengupta et al., 2010), and LP-fed mice have reduced mTORC1 activity (Lamming and Bar-Peled, 2019; Lamming et al., 2015; Solon-Biet et al., 2014). mTORC1 regulates numerous cellular processes, including translation and ribosome biogenesis, processes that are enriched in cluster 3. The role of ketogenesis, which has positive effects on health span and aging in mice and the metabolic benefits of LP diets may be an important area for future research (Roberts et al., 2017).
Other changes that relate to health may provide clues for future research. Insulin sensitivity, a hallmark of metabolic health was strongly related to lysine and arginine transport, purine metabolism, nicotinamide metabolism, and lipid bilayer diffusion in the liver. Nicotinamide and purine metabolism are key factors in NAD+ metabolism, which has been linked to metabolic health in both mice and humans (Banks et al., 2008; Connell et al., 2019). The regulation of hepatic NAD+ metabolism may contribute to the effects of protein on hepatic insulin sensitivity. Intake of L-arginine has been associated with the salvage of purines (such as adenosine) and is thought to be involved in salvaging these molecules for ATP regeneration (Kocic et al., 2012). As dysregulation of ATP generation and inefficient nutrient oxidation in the mitochondria may be a central cause of insulin resistance (Petersen et al., 2004; Ritov et al., 2005), this may indicate that reorganization of amino acid metabolic pathways on an LP diet may contribute to improvements in insulin sensitivity.
Finding strategies to combat or reverse diabetes and obesity is of the utmost importance as the population ages. Mid-life interventions are likely to be of use to a much broader population than interventions that are effective only early in life. While the impact of an LP diet in aged mice was somewhat blunted, an LP diet still promoted metabolic health in both sexes. While we and others have observed that LP diets started early in life can extend the lifespan and health span of male mice (Richardson et al., 2021; Solon-Biet et al., 2014), we find that late-life initiation of an LP diet leads to a loss of lean mass in males. Lean mass loss in humans is associated with sarcopenia and frailty, and increasing–not decreasing–dietary protein has been investigated as a way of combating sarcopenia and frailty the elderly (Beasley et al., 2013; Paddon-Jones and Rasmussen, 2009; Paddon-Jones et al., 2008). A recent retrospective analysis of age-specific mortality in mice found that lower protein consumption was associated with decreased age-specific mortality up to middle age, but that higher protein consumption was associated with decreased mortality in later life (Senior et al., 2019). Thus, there may be drawbacks to late-life initiation or continuation of LP diets, at least in certain sexes and age groups.
Limitations of study
Limitations of our work include the relatively short length of the studies; the use of lean, young, and healthy mice; and the limiting of our aged and FGF21 KO studies to C57 mice. Our molecular analysis was limited to the liver, and while this is the first organ to be exposed to absorbed nutrients, dietary protein has direct effects on multiple tissues. Our mega-cluster analysis linking molecular and phenotypic data are based on correlations, and while the broad strokes of our analysis make biological sense and are backed up by our follow-up studies on Fgf21, additional research will be required to determine which of the molecular changes we have identified are causative, particularly the relationship between FGF21, ketogenesis, and changes in metabolism.
Our diets are based on whey rather than casein; however, LP studies conducted with either protein source have found broadly similar effects (Fontana et al., 2016; Laeger et al., 2014; Maida et al., 2016, 2017). To keep diets isocaloric, the removal of dietary protein was balanced by addition of carbohydrates, and while changes in protein likely drive the metabolic responses to an LP diet, the protein to carbohydrate ratio, the type of dietary carbohydrate consumed, and diet-induced changes to the microbiome may also play a role (Hu et al., 2018; Solon-Biet et al., 2014, 2015, 2016; Speakman et al., 2016; Wali et al., 2021).
Our results suggest that while an LP diet across the genetically heterogeneous human population is likely to be broadly beneficial, as we and others have observed (Fontana et al., 2016; Levine et al., 2014; Sluijs et al., 2010), only a subset of individuals will have a strongly favorable response. Identifying genomic regions and alleles that control the metabolic response to an LP diet will therefore not only help unlock new molecular mechanisms to promote metabolic health but will enable a ‘‘food as medicine’’ approach to prevent and treat diabetes and obesity. Understanding how sex and genetic background dictate the response to nutritional interventions is key to translating these interventions to the clinic. In addition, disentangling these differences will help us to better understand the pathways which mediate the benefits of an LP diet, with a view to producing personalized nutritional interventions based on genetic background.
STAR☆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dudley W. Lamming (dlamming@medicine.wisc.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The accession number for the hepatic RNAseq gene expression data reported in this paper can be obtained from Gene Expression Omnibus (GEO) (GEO: GSE181301).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse information
All procedures were performed in conformance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee of the William S. Middleton Memorial Veterans Hospital (Madison, WI, USA).
Young female and male C57BL/6J (#000664) DBA/2J (#000671) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) at nine weeks of age, acclimatized on chow diet Purina 5001 (Purina Mills, Richmond, IN, USA) for one week before experiment start, and housed 2 per cage. HET3 mice are the F2 progeny of (BALB/cJ x C57BL/6J ) mothers and (C3H/HeJ x DBA/2J) fathers; female BALB/cJ (#000651), male C57BL/6J (#000664), female C3H/HeJ (#000659) and male DBA/2J (#000671) were obtained from The Jackson Laboratory and bred to produce heterogeneous HET3 F2 mice. Mice were acclimatized on chow diet Purina 5001 for one week before experiment start and housed 2–3 per cage. All mice were maintained at a temperature of approximately 22°C, and health checks were completed on all mice daily.
Aged C57BL/6J.NIA mice maintained with ad libitum access to NIH 31 diet were obtained from the NIA Aged Rodent Colony and then group housed locally with ad libitum access to Purina 5001 diet prior to experiment start.
To generate FGF21 KO mice, we crossed CMV-Cre mice (Schwenk et al., 1995) from the Jackson Laboratory (006054) with mice expressing a floxed allele of FGF21 (Potthoff et al., 2009) from The Jackson Laboratory (022361), then crossed with C57BL/6J mice to remove CMV-Cre; genotyping was performed using PCR, primers can be found in the key resources table. PCR Protocol: Stage 1, 95C°, 1:30; Stage 2 (x8) 95°C, 0:30, step down, 68°C, 1:00; Stage 3 (x28) 95°C, 0:30, 61.5°C, 0:30, 72°C, 1:00; Stage 4 72°C, 5:00, hold at 4°C. PCR was performed using DreamTaq DNA Polymerase (Thermo Fisher Scientific, EP0705) according to insert instructions.
At the start of the experiment, mice were randomized to receive either the 21% (TD.180161), 14% (TD.180141), or 7% (TD.10192) protein diet; all diets were obtained from Envigo. Dietary protein was primarily derived from whey, with a minor amount derived from corn, and supplemental L-methionine added, as is common for whey or casein based diets as they have low levels of sulfur amino acids (The Journal of Nutrition, 1977; Reeves, 1997; Reeves et al., 1993). Within the diet series, calories from protein were replaced by calories from carbohydrates, while calories from fat were held fixed at 20%, making the diets isocaloric (3.6 Kcal/g). Full diet descriptions, compositions and item numbers are provided in Table S1. The randomization of mice was performed at the cage level to ensure that all groups had approximately the same initial starting weight and body composition. The number of animals in each group used for each experiment is listed in Table S7. Mice were housed in a SPF mouse facility in static microisolator cages, except when temporarily housed in a Columbus Instruments Oxymax/CLAMS metabolic chamber system. Mice were housed under a 12:12 h light/dark cycle with free access to food and water, except where noted in the procedures below.
METHOD DETAILS
In vivo procedures
Glucose, insulin, and alanine tolerance tests were performed by fasting the mice overnight for 16 hours and then injecting glucose (1 g kg−1), insulin (0.75 U kg−1) or alanine (2 g kg−1) intraperitoneally (i.p.) (Bellantuono et al., 2020; Yu et al., 2018). For glucose stimulated insulin secretion, 2 g kg−1 was used. Glucose measurements were taken using a Bayer Contour blood glucose meter (Bayer, Leverkusen, Germany) and test strips. Mouse body composition was determined using an EchoMRI Body Composition Analyzer (EchoMRI, Houston, TX, USA). For assay of multiple metabolic parameters [O2, CO2, food consumption, respiratory exchange ratio (RER), energy expenditure] and activity tracking, mice were acclimated to housing in a Oxymax/CLAMS metabolic chamber system (Columbus Instruments) for ~24 h and data from a continuous 24 h period was then recorded and analyzed. Food consumption in home cages was measured by moving mice to clean cages, filling the hopper with a measured quantity of fresh diet in the morning and measuring the remainder in the morning 3–6 days later. The amount was adjusted for the number of mice per cage, the number of days that passed and the relative weights of the mice (i.e., heavier mice were credited with a larger relative portion of the food intake). Mice were euthanized by cervical dislocation after an overnight (16h) fast and tissues for molecular analysis were flash-frozen in liquid nitrogen or fixed and prepared as described below.
Assays and kits
Blood for fasting insulin and FGF21 was obtained following an overnight fast. Plasma insulin was quantified using a ultra-sensitive mouse insulin ELISA kit (90080), from Crystal Chem (Elk Grove Village, IL, USA). Blood FGF21 levels were assayed by a mouse/ rat FGF-21 quantikine ELISA kit (MF2100) from R&D Systems (Minneapolis, MN, USA).
Quantitative PCR
Liver was extracted with Trireagent (Sigma, St Louis, MO, USA). Then, 1 µg of RNA was used to generate cDNA (Superscript III; Invitrogen, Carlsbad, CA, USA). Oligo dT primers and primers for real-time PCR were obtained from Integrated DNA Technologies (IDT, Coralville, IA, USA). Reactions were run on a StepOne Plus machine (Applied Biosystems, Foster City, CA, USA) with Sybr Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA). Actin was used to normalize the results from gene-specific reactions. Primer sequences can be found in the key resources table.
Transcriptomic analysis
RNA was extracted from liver as previously described (Cummings et al., 2018). The concentration and purity of RNA was determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and RNA was diluted to 100–400 ng/µl for sequencing. The RNA was then submitted to the University of Wisconsin-Madison Biotechnology Center Gene Expression Center & DNA Sequencing Facility, and RNA quality was assayed using an Agilent RNA NanoChip. RNA libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation protocol (Illumina, San Diego, CA) with 250ng of mRNA, and cleanup was done using RNA Clean beads (lot #17225200). Reads were aligned to the mouse (Mus musculus) with genome-build GRCm38.p5 of accession NCBI:GCA_000001635.7 and expected counts were generated with ensembl gene IDs (Zerbino et al., 2018).
Analysis of significantly differentially expressed genes (DEGs) was completed in R version 3.4.3 (R Core Team, 2017) using edgeR (Robinson et al., 2010) and limma (Ritchie et al., 2015). Gene names were converted to gene symbol and Entrez ID formats using the mygene package. Initially, 51,912 transcripts were identified across all Genes with zero counts per million (cpm) in six or more individuals were removed, leaving 17,575 transcripts. To reduce the impact of external factors not of biological interest that may affect expression, data was normalized to ensure the expression distributions of each sample are within a similar range. We normalized using the trimmed mean of M-values (TMM), which scales to library size. Heteroscedascity was accounted for using the voom function, DEGs were identified using an empirical Bayes moderated linear model, and log coefficients and Benjamini-Hochberg (BH) adjusted p-values were generated for each comparison of interest (Benjamini and Hochberg, 1995). DEGs were used to identify enriched pathways, both Gene Ontology (for Biological Processes) and KEGG enriched pathways were determined for each contrast. All genes, log2 fold-changes and corresponding unadjusted and Benjamini-Hochberg adjusted p-values can be found in Table S3A.
Metabolomic analysis
Metabolite extraction
To extract metabolites from the liver, snap-frozen liver samples were ground at liquid nitrogen temperature with a Cryomill (Retsch, Newtown, PA). The resulting liver powder (~20 mg) was weighed and then metabolites were extracted by adding −20°C acetonitrile:methanol:water (40:40:20 v/v) extraction solution, vortexed, and centrifuged at 16,000 x g for 10 min at 4°C. The volume of the extraction solution (µL) was 40x the weight of liver (mg) to make an extract of 25 mg tissue per mL solvent. The supernatant was collected for liquid chromatography-mass spectrometry (LC-MS) analysis.
Metabolite measurement by LC-MS
A quadrupole-orbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific, San Jose, CA) operating in negative or positive ion mode was coupled to hydrophilic interaction chromatography via electrospray ionization and used to scan from m/z 70 to 1000 at 1 Hz and 140,000 resolution. LC separation was on a XBridge BEH Amide column (2.1 mm x 150 mm, 2.5 µm particle size, 130 Å pore size; Waters, Milford, MA) using a gradient of solvent A (20 mM ammonium acetate, 20 mM ammonium hydroxide in 95:5 water: acetonitrile, pH 9.45) and solvent B (acetonitrile). Flow rate was 150 µL/min. The LC gradient was: 0 min, 85% B; 2 min, 85% B; 3 min, 80% B; 5 min, 80% B; 6 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 16 min, 25% B; 18 min, 0% B; 23 min, 0% B; 24 min, 85% B; 30 min, 85% B. Autosampler temperature was 5°C, and injection volume was 3 µL. Data were analyzed using the MAVEN software.
Metabolite intensities were normalized and rescaled to mean=0 and standard deviation=1. To detect significantly differentially expressed metabolites between treatment groups, an empirical Bayes moderated linear model was fitted to each metabolite. The empirical Bayes approach shrinks the estimated sample variances by borrowing information from across metabolites. Fold changes were estimated using contrasts between each control group and its LP counterpart (e.g. Male C57 Control vs Male C57 LP). P-values for each comparison were adjusted using the Benjamini-Hochberg (BH) procedure using a FDR of 5% (Benjamini and Hochberg, 1995). Pathway analysis was conducted using unadjusted p-values significant for each contrast (p<0.05), and enriched using KEGG in Metaboanalyst (version 4.0), significant pathways can be seen in Table S5C. All genes, log2 fold-changes and corresponding unadjusted and Benjamini-Hochberg adjusted p-values can be found in Table S5A.
Lipidomics analysis
Approximately 10mg of liver tissue was transferred to labeled bead-mill tubes (1.4 mm, MoBio Cat# 13113–50) where lipids were extracted in a solution of 250 µL PBS, 225 µL MeOH containing internal standards (Avanti SPLASH LipidoMix (Lot#12) at 10 µL per sample) and 750 µL MTBE (methyl tert-butyl ether). The sample was homogenized in five 30 s cycle using the TissueLyzer followed by a rest on ice for 15 minutes. After centrifugation at 16,000 g for 5 minutes at 4 °C, 500 µL of the upper phases were collected and evaporated to dryness under a gentle nitrogen stream at room temperature. Lipid samples are reconstituted in 150 µL IPA and transferred to an LC-MS vial with insert (Agilent 5182–0554 and 5183–2086) for analysis. Concurrently run are a process blank sample and pooled quality control (QC) sample. The pooled QC was prepared by taking equal volumes (~10 µL) from each sample after final resuspension.
Lipid extracts were separated on a Waters Acquity UPLC CSH C18 1.7 µm 2.1 × 100 mm column maintained at 65 °C connected to an Agilent HiP 1290 Sampler, Agilent 1290 Infinity pump, equipped with an Agilent 1290 Flex Cube and Agilent 6546 Accurate Mass Q-TOF dual ESI mass spectrometer. For positive mode, the source gas temperature was set to 225 °C, with a gas flow of 11 L/min and a nebulizer pressure of 50 psig. VCap voltage was set at 3500 V, fragmentor at 110 V, skimmer at 85 V and Octopole RF peak at 750 V. For negative mode, the source gas temperature was set to 325 °C, with a drying gas flow of 12 L/min and a nebulizer pressure of 30 psig. VCap voltage was set at 3000 V, fragmentor at 125 V, skimmer at 75 V and Octopole RF peak at 750 V. Reference masses in positive mode (m/z 121.0509 and 922.0098) were infused with nebulizer pressure at 2 psig, in negative mode reference masses (m/z 1033.988, 966.0007, 112.9856 and 68.9958) were infused with a nebulizer pressure at 5 psig. Samples were analyzed in a randomized order in both positive and negative ionization modes in separate experiments acquiring with the scan range m/z 100 – 1700. Mobile phase A consists of ACN:H2O (60:40 v/v) in 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consists of IPA:ACN:H2O (90:9:1 v/v) in 10 mM ammonium formate and 0.1% formic acid. The chromatography gradient for both positive and negative modes starts at 15% mobile phase B then increases to 30% B over 2.4 min, it then increases to 48% B from 2.4 – 3.0 min, then increases to 82% B from 3 – 13.2 min, then increases to 99% B from 13.2 – 13.8 min where it’s help until 15.4 min and then returned to the initial conditioned and equilibrated for 4 min. Flow is 0.5 mL/min throughout, injection volume is 5 µL for positive and 7 µL negative mode. Tandem mass spectrometry is conducted using the same LC gradient at collision energies of 20 V and 40 V. QC samples (n=6) and blanks (n=4) were injected throughout the sample queue and ensure the reliability of acquired lipidomics data. Results from LC-MS experiments were collected using Agilent Mass Hunter (MH) Workstation and analyzed using the software packages MH Qual, MH Quant, Lipid Annotator (Agilent Technologies) to prepare the data set. Pathway enrichment analysis of the lipidomics data was conducted using the Lipid Ontology (LION) enrichment analysis web application (Molenaar et al., 2019). All lipids, log2 fold-changes and corresponding unadjusted and Benjamini-Hochberg adjusted p-values can be found in Table S4B.
Integrative analyses
Four data types (metabolomics, transcriptomics, lipidomics, phenotypic outcomes) were obtained from experiments with three factors (strain (C57, DBA), sex, and diet (LP or control)) from 41 mice (five C57 female mice fed Control diet, six C57 female mice fed LP diet, five C57 male mice fed Control diet, five C57 male mice fed LP diet, six DBA female mice fed Control diet, three DBA female mice fed LP diet, five DBA male mice fed Control diet, and six DBA male mice fed LP diet). The data consisted of 364 metabolites, 17575 probes, 343 lipids, and 46 phenotypes.
To identify molecules of interest in each of the -omics datasets, significantly differentially expressed molecules between Control and Low Protein Groups were identified using an empirical Baye’s moderated linear model. The Benjamini-Hochberg method was applied to control false discovery rate, selecting those with adjusted p-value < 0.05 (Benjamini and Hochberg 1995). Metabolomics, transcriptomics, and lipidomics data were preprocessed, log2 transformed, z-scale normalized across molecules and samples for each data type individually. Phenotypic outcome data were similarly z-scale normalized just across phenotypes. The data consisted of 831 inputs across individual mice which had no missing data points including 83 metabolites, 623 transcripts, 79 lipids, and 46 phenotypes.
To integrate the data, all four datatypes were concatenated for each comparison. Correlations were performed between the 18,328 data points using Spearman’s rank (831 × 831 = 690,561 correlations). Complete hierarchical clustering was used to reorder molecules based on 1 – Spearman correlation between all molecules. The number of clusters were determined by silhouette scores (Rousseeuw, 1987). However, for the diet cluster, the silhouette curve was atypical, so the number of clusters were determined based on the 6 clusters observed in the heatmap and the consistencies of pathways and phenotypes within clusters; heatmaps and correlation plots were generated for all comparisons.
All analyses were performed in R (v. 3.6.1) using gplots (Warnes, 2005) (v. 3.0.3), cluster (Mächler et al., 2012) (v. 2.1.0), and limma (Ritchie et al., 2015) (v. 3.42.2). BiomaRt (Durinck et al., 2005) was used to convert ensembl gene ids to entrez gene ids. For each cluster, the over representation of KEGG (Kanehisa et al., 2017) pathways from genes were determined using kegga from limma package.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Most statistical analyses were conducted using Prism, version 8 (GraphPad Software, San Diego, CA, USA) and R (version 4.1.0). Tests involving multiple factors were analyzed by either a three-way analysis of variance (ANOVA) with Sex, Strain and Diet as categorical variables, followed by Benjamini-Hochberg post hoc testing for pairwise comparisons or by two-way ANOVA with Sex and Diet or Sex and Genotype as categorical variables followed by a Tukey–Kramer post hoc test for multiple comparisons. Data distribution was assumed to be normal but was not formally tested. Statistical parameters can be found in the figures, figure legends, and Table S7. Transcriptomics and metabolomics data were analyzed using R (version 3.3.1). All correlations where depicted were produced using Pearson’s correlation (except for the integrative mega-clusters plot) and PCA plots were produced by imputing missing data and scaling the data using the R package ‘‘missMDA’’ using the PCA analysis and plots were generated using the R package ‘‘factoextra’’ (Kassambara and Mundt, 2020; Lê et al., 2008).
Supplementary Material
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Human insulin | Eli Lilly | NDC 0002–8215-17 (Humulin R U-100) |
| TRI reagent | Sigma | T9494 |
| SYBR Green | Thermo Fisher Scientific | 4309155 |
| DreamTaqDNA Polymerase | Thermo Fisher Scientific | EP0705 |
| Superscript III Reverse Transcriptase | Thermo Fisher Scientific | 18080085 |
| Critical commercial assays | ||
| Ultra-sensitive mouse insulin ELISA | Crystal Chem | Cat# 90080 |
| Mouse/Rat FGF-21 Quantikine ELISA Kit | R&D Systems | Cat# MF2100 |
| Deposited data | ||
| Raw and analyzed RNAseq data | This paper | GEO: GSE181301; Table S5 |
| Lipidomics | This paper | Table S9 |
| Metabolomics | This paper | Table S11 |
| Experimental models: Organisms/strains | ||
| Mouse strain: C57BL6/J (Male and Female) | The Jackson Laboratory | Cat# JAX:000664; RRID: IMSR_JAX:000664 |
| Mouse strain: DBA2/J (Male and Female) | The Jackson Laboratory | Cat# JAX: 000671; RRID: IMSR_JAX:000671 |
| Mouse strain: UM-HET3 (Male and Female) | This paper | F2 progeny of (BALB/cJ x C57BL/6J) mothers and (C3H/HeJ x DBA/2J) fathers |
| Mouse strain: BALB/cJ (Female) | The Jackson Laboratory | Cat# JAX: 000651; RRID: IMSR_JAX:000651 |
| Mouse strain: C3H/HeJ (Female) | The Jackson Laboratory | Cat# JAX: 000659; RRID: IMSR_JAX:000659 |
| Mouse strain: C57BL6/J.Nia (Male and Female) | NIA Aged Rodent Colony | C57BL/6J.Nia |
| Mouse strain: Fgf21loxP/loxP | The Jackson Laboratory (Schwenk et al., 1995), (Potthoff et al., 2009) | Cat# JAX:022361; RRID: IMSR_JAX:022361 |
| Mouse strain:C57BL/6J; Fgf21Δ/Δ | Lamming Laboratory (Yu et al., 2021) | N/A |
| Oligonucleotides | ||
| Mouse FGF21 genotyping WT F: ACCCCCTGAGCATGGTAGA |
The Jackson Laboratory | 51425, https://www.jax.org/Protocol?stockNumber=033846&protocolID=37478 |
| Mouse FGF21 genotyping mutant F: CAGACCAAGGAGCACAGACC |
The Jackson Laboratory | 51434, https://www.jax.org/Protocol?stockNumber=033846&protocolID=37478 |
| Mouse FGF21 genotyping common R: GCAGAGGCAAGTGATTTTGA |
The Jackson Laboratory | 51435, https://www.jax.org/Protocol?stockNumber=033846&protocolID=37478 |
| Mouse Cre genotyping common F: GAACCTGATGGACATGTTCAGG |
(Yu et al., 2019) | N/A |
| Mouse Cre genotyping common R: AGTGCGTTCGAACGCTAGAGCCTGT |
(Yu et al., 2019) | N/A |
| Actb: F: ACCTTCTACAATGAGCTGCG | (Fontana et al., 2016) | N/A |
| Actb: R: CTGGATGGCTACGTACATGG | (Fontana et al., 2016) | N/A |
| Fgf21: F: CAAATCCTGGGTGTCAAAGC | (Fontana et al., 2016) | N/A |
| Fgf21: R: CATGGGCTTCAGACTGGTAC | (Fontana et al., 2016) | N/A |
| Software and algorithms | ||
| edgeR package | (Robinson et al., 2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GraphPad Prism | GraphPad | http://www.graphpad.com/scientific-software/prism |
| MetaboAnalyst | (Chong et al., 2019) | https://www.metaboanalyst.ca/ |
| R (Version 3.4.3) | N/A | https://www.r-project.org/ |
| MyGene package | N/A | https://www.bioconductor.org/packages/release/bioc/html/mygene.html |
| Factoextra package | (Kassambara and Mundt, 2020) | https://cran.r-project.org/web/packages/factoextra/index.html |
| missMDA package | N/A | https://cran.r-project.org/web/packages/missMDA/index.html |
| LION/web | (Molenaar et al., 2019) | http://www.lipidontology.com/ |
| Limma package | (Ritchie et al., 2015) | https://bioconductor.org/packages/release/bioc/html/limma.html |
| Other | ||
| Normal Chow | Purina | Cat# 5001 |
| Mouse diets (See Table S1) | This paper | N/A |
Highlights.
Protein restriction (PR) promotes metabolic health in mice
The benefits of PR are influenced by sex, strain, and the degree of restriction
The role of FGF21 in the effects of PR is sex and strain dependent
PR improves metabolic health in aged mice
ACKNOWLEDGMENTS
We thank all members of the Lamming lab for their feedback, R. Jain for assistance with lipidomics, Dr. Herfel (Envigo) for assistance with diets, and Drs. Jay Mitchell and Michael MacArthur for insightful discussions. The Lamming lab is supported in part by the NIA (AG056771, AG062328, and AG061635), the NIDDK (DK125859), and startup funds from UW-Madison. C.L.G. is supported in part by Dalio Philanthropies and is a Glenn Foundation for Medical Research Postdoctoral Fellow. H.H.P. is supported in part by F31AG066311. C.J. was a postdoctoral fellow of the American Diabetes Association (1–17-PDF-076). N.E.R. was supported in part by training grant T32AG000213. D.Y. is supported in part by a fellowship from the American Heart Association (17PRE33410983). M.E.M. is supported in part by a Supplement to Promote Diversity in Health-Related Research (R01AG062328–03S1). R.B. is supported in part by training grant T32DK007665. J.R. is supported in part by DP1DK113643 and metabolomics work supported by P30DK019525. Lipidomics work was supported by a pilot grant to J.S. from the Diabetes Research Center at Washington University, P30DK020579, and a UW BIRCWH Scholars Program award to J.S. (K12HD101368). I.O. is supported by UWCCC support grant P30 CA014520 and Wisconsin Head and Neck Cancer SPORE CEP P50DE026787. Support was also provided by the UW-Madison OVCRGE with funding from the Wisconsin Alumni Research Foundation. This work was supported in part by the US Department of Veterans Affairs (I01-BX004031), and this work was supported using facilities and resources from the William S. Middleton Memorial Veterans Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work does not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cmet.2021.12.018.
DECLARATION OF INTERESTS
D.W.L. has received funding from and is a scientific advisory board member of Aeovian Pharmaceuticals, which seeks to develop novel, selective mTOR inhibitors for the treatment of various diseases.
REFERENCES
- Alamshah A, Spreckley E, Norton M, Kinsey-Jones JS, Amin A, Ramgulam A, Cao Y, Johnson R, Saleh K, Akalestou E, et al. (2017). L-phenylalanine modulates gut hormone release and glucose tolerance, and suppresses food intake through the calcium-sensing receptor in rodents. Int. J. Obes. (Lond) 41, 1693–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aon MA, Bernier M, Mitchell SJ, Di Germanio C, Mattison JA, Ehrlich MR, Colman RJ, Anderson RM, and de Cabo R (2020). Untangling determinants of enhanced health and lifespan through a multi-omics approach in mice. Cell Metab 32, 100–116.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, and Maratos-Flier E (2007). Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab 5, 426–437. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, and Accili D (2008). SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab 8, 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiardes S, Godneva A, Elinav E, and Segal E (2018). Towards utilization of the human genome and microbiome for personalized nutrition. Curr. Opin. Biotechnol 51, 57–63. [DOI] [PubMed] [Google Scholar]
- Beasley JM, Shikany JM, and Thomson CA (2013). The role of dietary protein intake in the prevention of sarcopenia of aging. Nutr. Clin. Pract 28, 684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellantuono I, de Cabo R, Ehninger D, Di Germanio C, Lawrie A, Miller J, Mitchell SJ, Navas-Enamorado I, Potter PK, Tchkonia T, et al. (2020). A toolbox for the longitudinal assessment of healthspan in aging mice. Nat. Protoc 15, 540–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat Methodol 57, 289–300. [Google Scholar]
- Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, and Hellerstein MK (2010). Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am. J. Physiol. Endocrinol. Metab 298, E108–E116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus MF, Piper MD, and Reuter M (2019). Sex-specific transcriptomic responses to changes in the nutritional environment. eLife 8, e47262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalvon-Demersay T, Moro J, Even PC, Chaumontet C, Tomé D, Averous J, Piedcoq J, Gaudichon C, Maurin AC, Fafournoux P, and Azzout-Marniche D (2019). Liver GCN2 controls hepatic FGF21 secretion and modulates whole body postprandial oxidation profile under a low-protein diet. Am. J. Physiol. Endocrinol. Metab 317, E1015–E1021. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, and Sabatini DM (2016). The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CJ, Gelfond JAL, Strong R, and Nelson JF (2019). Genetically heterogeneous mice exhibit a female survival advantage that is age- and site-specific: results from a large multi-site study. Aging Cell 18, e12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Yamamoto M, and Xia J (2019). MetaboAnalystR 2.0: from raw spectra to biological insights. Metabolites 9, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell NJ, Houtkooper RH, and Schrauwen P (2019). NAD+ metabolism as a target for metabolic health: have we found the silver bullet? Diabetologia 62, 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, and Hall MN (2014). Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc. Natl. Acad. Sci. USA 111, 11592–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings NE, Williams EM, Kasza I, Konon EN, Schaid MD, Schmidt BA, Poudel C, Sherman DS, Yu D, Arriola Apelo SI, et al. (2018). Restoration of metabolic health by decreased consumption of branched-chain amino acids. J. Physiol 596, 623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domouzoglou EM, and Maratos-Flier E (2011). Fibroblast growth factor 21 is a metabolic regulator that plays a role in the adaptation to ketosis. Am. J. Clin. Nutr 93, 901S–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JY, Zhang ZL, Wang PY, and Qin LQ (2013). Effects of high-protein diets on body weight, glycaemic control, blood lipids and blood pressure in type 2 diabetes: meta-analysis of randomised controlled trials. Br. J. Nutr 110, 781–789. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Farinatti P, Castinheiras Neto AG, and Amorim PRS (2016). Oxygen consumption and substrate utilization during and after resistance exercises performed with different muscle mass. Int. J. Exerc. Sci 9, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, and Nedergaard J (2009). UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab 9, 203–209. [DOI] [PubMed] [Google Scholar]
- Felig P, Marliss E, and Cahill GF Jr. (1969). Plasma amino acid levels and insulin secretion in obesity. N. Engl. J. Med 281, 811–816. [DOI] [PubMed] [Google Scholar]
- Fernandes J, and Blom W (1974). The intravenous L-alanine tolerance test as a means for testing gluconeogenesis. Pediatr. Res 8, 137. [DOI] [PubMed] [Google Scholar]
- Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, Cava E, Spelta F, Tosti V, Syed FA, et al. (2016). Decreased consumption of branched-chain amino acids improves metabolic health. Cell Rep 16, 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon MC, Nuttall FQ, Saeed A, Jordan K, and Hoover H (2003). An increase in dietary protein improves the blood glucose response in persons with type 2 diabetes. Am. J. Clin. Nutr 78, 734–741. [DOI] [PubMed] [Google Scholar]
- Geloneze B, Vasques AC, Stabe CF, Pareja JC, Rosado LE, Queiroz EC, and Tambascia MA; BRAMS Investigators (2009). HOMA1-IR and HOMA2-IR indexes in identifying insulin resistance and metabolic syndrome: Brazilian Metabolic Syndrome Study (BRAMS). Arq. Bras. Endocrinol. Metabol 53, 281–287. [DOI] [PubMed] [Google Scholar]
- Gong Q, Hu Z, Zhang F, Cui A, Chen X, Jiang H, Gao J, Chen X, Han Y, Liang Q, et al. (2016). Fibroblast growth factor 21 improves hepatic insulin sensitivity by inhibiting mammalian target of rapamycin complex 1 in mice. Hepatology 64, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CL, and Lamming DW (2021). We are more than what we eat. Nat. Metab 3, 1144–1145. [DOI] [PubMed] [Google Scholar]
- Green CL, Lamming DW, and Fontana L (2022). Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol 23, 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn O, Drews LF, Nguyen A, Tatsuta T, Gkioni L, Hendrich O, Zhang Q, Langer T, Pletcher S, Wakelam MJO, et al. (2019). A nutritional memory effect counteracts benefits of dietary restriction in old mice. Nat. Metab 1, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbesma N, Bakker SJ, Jansen DF, Stolk RP, De Zeeuw D, De Jong PE, and Gansevoort RT; PREVEND Study Group (2009). High protein intake associates with cardiovascular events but not with loss of renal function. J. Am. Soc. Nephrol 20, 1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasek BE, Stewart LK, Henagan TM, Boudreau A, Lenard NR, Black C, Shin J, Huypens P, Malloy VL, Plaisance EP, et al. (2010). Dietary methionine restriction enhances metabolic flexibility and increases uncoupled respiration in both fed and fasted states. Am. J. Physiol. Regul. Integr. Comp. Physiol 299, R728–R739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, Laeger T, Albarado DC, McDougal DH, Berthoud HR, Münzberg H, and Morrison CD (2017). Low protein-induced increases in FGF21 drive UCP1-dependent metabolic but not thermoregulatory endpoints. Sci. Rep 7, 8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, Laeger T, Dehner M, Albarado DC, Clarke B, Wanders D, Burke SJ, Collier JJ, Qualls-Creekmore E, Solon-Biet SM, et al. (2019). FGF21 signals protein status to the brain and adaptively regulates food choice and metabolism. Cell Rep 27, 2934–2947.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wang L, Yang D, Li L, Togo J, Wu Y, Liu Q, Li B, Li M, Wang G, et al. (2018). Dietary fat, but not protein or carbohydrate, regulates energy intake and causes adiposity in mice. Cell Metab 28, 415–431.e4. [DOI] [PubMed] [Google Scholar]
- Institute of Medicine (2005). Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids (The National Academies Press; ). [DOI] [PubMed] [Google Scholar]
- Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, et al. (2016). A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat. Med 22, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, McClure C, Priest NK, and Hunt J (2015). Sex-specific effects of protein and carbohydrate intake on reproduction but not lifespan in Drosophila melanogaster. Aging Cell 14, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, and Morishima K (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, and Mundt F (2020). factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.7 https://cran.r-project.org/web/packages/factoextra/index.html.
- Kocic G, Nikolic J, Jevtovic-Stoimenov T, Sokolovic D, Kocic H, Cvetkovic T, Pavlovic D, Cencic A, and Stojanovic D (2012). L-arginine intake effect on adenine nucleotide metabolism in rat parenchymal and reproductive tissues. ScientificWorldJournal 2012, 208239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Henagan TM, Albarado DC, Redman LM, Bray GA, Noland RC, Münzberg H, Hutson SM, Gettys TW, Schwartz MW, and Morrison CD (2014). FGF21 is an endocrine signal of protein restriction. J. Clin. Invest 124, 3913–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Albarado DC, Burke SJ, Trosclair L, Hedgepeth JW, Berthoud HR, Gettys TW, Collier JJ, Münzberg H, and Morrison CD (2016). Metabolic responses to dietary protein restriction require an increase in FGF21 that is delayed by the absence of GCN2. Cell Rep 16, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagiou P, Sandin S, Weiderpass E, Lagiou A, Mucci L, Trichopoulos D, and Adami HO (2007). Low carbohydrate-high protein diet and mortality in a cohort of Swedish women. J. Intern. Med 261, 366–374. [DOI] [PubMed] [Google Scholar]
- Lamming DW, and Bar-Peled L (2019). Lysosome: the metabolic signaling hub. Traffic 20, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Cummings NE, Rastelli AL, Gao F, Cava E, Bertozzi B, Spelta F, Pili R, and Fontana L (2015). Restriction of dietary protein decreases mTORC1 in tumors and somatic tissues of a tumor-bearing mouse xenograft model. Oncotarget 6, 31233–31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. (2013). The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun 4, 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson KR, Russo KA, Fang Y, Mohajerani N, Goodson ML, and Ryan KK (2017). Sex differences in the hormonal and metabolic response to dietary protein dilution. Endocrinology 158, 3477–3487. [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, and Husson F (2008). FactoMineR: an R package for multivariate analysis. J. Stat. Softw 25, 18. [Google Scholar]
- Lees EK, Banks R, Cook C, Hill S, Morrice N, Grant L, Mody N, and Delibegovic M (2017). Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Sci. Rep 7, 9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Suarez JA, Brandhorst S, Balasubramanian P, Cheng CW, Madia F, Fontana L, Mirisola MG, Guevara-Aguirre J, Wan J, et al. (2014). Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab 19, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, and Nelson JF (2010). Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn T, Santosa B, Grönemeyer D, Aygen S, Scholz N, Busch M, and Bretzel RG (2000). Effect of long-term dietary protein intake on glucose metabolism in humans. Diabetologia 43, 1257–1265. [DOI] [PubMed] [Google Scholar]
- Liu HX, Hu Y, French SW, Gonzalez FJ, and Wan YJ (2015). Forced expression of fibroblast growth factor 21 reverses the sustained impairment of liver regeneration in hPPARa(PAC) mice due to dysregulated bile acid synthesis. Oncotarget 6, 9686–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk G (1924). Animal calorimetry. Paper XXIV. Analysis of the oxidation of mixtures of carbohydrate and fat: a correction. J. Biol. Chem 59, 41–42. [Google Scholar]
- Lynch CJ, and Adams SH (2014). Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol 10, 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mächler M, Rousseeuw P, Struyf A, Hubert M, and Hornik K (2012). Cluster: cluster analysis basics and extensions (R package version 2.1.2)
- Magwere T, Chapman T, and Partridge L (2004). Sex differences in the effect of dietary restriction on life span and mortality rates in female and male Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci 59, 3–9. [DOI] [PubMed] [Google Scholar]
- Maida A, Zota A, Sjøberg KA, Schumacher J, Sijmonsma TP, Pfenninger A, Christensen MM, Gantert T, Fuhrmeister J, Rothermel U, et al. (2016). A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J. Clin. Invest 126, 3263–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida A, Chan JSK, Sjøberg KA, Zota A, Schmoll D, Kiens B, Herzig S, and Rose AJ (2017). Repletion of branched chain amino acids reverses mTORC1 signaling but not improved metabolism during dietary protein dilution. Mol. Metab 6, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Piper MD, and Partridge L (2005). Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol 3, e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklakov AA, Simpson SJ, Zajitschek F, Hall MD, Dessmann J, Clissold F, Raubenheimer D, Bonduriansky R, and Brooks RC (2008). Sex-specific fitness effects of nutrient intake on reproduction and lifespan. Curr. Biol 18, 1062–1066. [DOI] [PubMed] [Google Scholar]
- Mather K (2009). Surrogate measures of insulin resistance: of rats, mice, and men. Am. J. Physiol. Endocrinol. Metab 296, E398–E399. [DOI] [PubMed] [Google Scholar]
- McKnight JR, Satterfield MC, Jobgen WS, Smith SB, Spencer TE, Meininger CJ, McNeal CJ, and Wu G (2010). Beneficial effects of L-arginine on reducing obesity: potential mechanisms and important implications for human health. Amino Acids 39, 349–357. [DOI] [PubMed] [Google Scholar]
- Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, and Smith-Wheelock M (2005). Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, Javors MA, Li X, Nadon NL, Nelson JF, et al. (2014). Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell 13, 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard AY, Tan SX, Yang P, Fazakerley DJ, Domanova W, Parker BL, Humphrey SJ, Jothi R, Stöckli J, and James DE (2016). mTORC1 is a major regulatory node in the FGF21 signaling network in adipocytes. Cell Rep 17, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, González-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B, et al. (2016). Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab 23, 1093–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar MR, Jeucken A, Wassenaar TA, van de Lest CHA, Brouwers JF, and Helms JB (2019). LION/web: a web-based ontology enrichment tool for lipidomic data analysis. GigaScience 8, giz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutel E, Gautier-Stein A, Abdul-Wahed A, Amigó-Correig M, Zitoun C, Stefanutti A, Houberdon I, Tourette JA, Mithieux G, and Rajas F (2011). Control of blood glucose in the absence of hepatic glucose production during prolonged fasting in mice: induction of renal and intestinal gluconeogenesis by glucagon. Diabetes 60, 3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neinast MD, Jang C, Hui S, Murashige DS, Chu Q, Morscher RJ, Li X, Zhan L, White E, Anthony TG, et al. (2019). Quantitative analysis of the whole-body metabolic fate of branched-chain amino acids. Cell Metab 29, 417.e4–429.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon-Jones D, and Rasmussen BB (2009). Dietary protein recommendations and the prevention of sarcopenia. Curr. Opin. Clin. Nutr. Metab. Care 12, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon-Jones D, Short KR, Campbell WW, Volpi E, and Wolfe RR (2008). Role of dietary protein in the sarcopenia of aging. Am. J. Clin. Nutr 87, 1562S–1566S. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, and Shulman GI (2004). Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med 350, 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, Mohammadi M, Finck BN, Mangelsdorf DJ, Kliewer SA, and Burgess SC (2009). FGF21 induces PGC-1α and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc. Natl. Acad. Sci. USA 106, 10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017). R: a language and environment for statistical computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Reeves PG (1997). Components of the AIN-93 diets as improvements in the AIN-76A diet. J. Nutr 127 (Suppl 5 ), 838S–841S. [DOI] [PubMed] [Google Scholar]
- Reeves PG, Nielsen FH, and Fahey GC Jr. (1993). AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr 123, 1939–1951. [DOI] [PubMed] [Google Scholar]
- Richardson NE, Konon EN, Schuster HS, Mitchell AT, Boyle C, Rodgers AC, Finke M, Haider LR, Yu D, Flores V, et al. (2021). Lifelong restriction of dietary branched-chain amino acids has sex-specific benefits for frailty and life span in mice. Nat. Aging 1, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Baerlocher K, Bauer JM, Elmadfa I, Heseker H, Leschik-Bonnet E, Stangl G, Volkert D, Stehle P, and German Nutrition Society (DGE). (2019). Revised reference values for the intake of protein. Ann. Nutr. Metab 74, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikke BA, Liao CY, McQueen MB, Nelson JF, and Johnson TE (2010). Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension. Exp. Gerontol 45, 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, and Kelley DE (2005). Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54, 8–14. [DOI] [PubMed] [Google Scholar]
- Roberts MN, Wallace MA, Tomilov AA, Zhou Z, Marcotte GR, Tran D, Perez G, Gutierrez-Casado E, Koike S, Knotts TA, et al. (2017). A ketogenic diet extends longevity and healthspan in adult mice. Cell Metab 26, 539–546.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez NR (2015). Introduction to Protein Summit 2.0: continued exploration of the impact of high-quality protein on optimal health. Am. J. Clin. Nutr 101, 1317S–1319S. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ (1987). Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math 20, 53–65. [Google Scholar]
- Schwenk F, Baron U, and Rajewsky K (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP -flanked gene segments including deletion in germ cells. Nucleic Acids Res 23, 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino Y, Seino S, Ikeda M, Matsukura S, and Imura H (1983). Beneficial effects of high protein diet in treatment of mild diabetes. Hum. Nutr. Appl. Nutr 37, 226–230. [PubMed] [Google Scholar]
- SenGupta S, Peterson TR, Laplante M, Oh S, and Sabatini DM (2010). mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Senior AM, Solon-Biet SM, Cogger VC, Le Couteur DG, Nakagawa S, Raubenheimer D, and Simpson SJ (2019). Dietary macronutrient content, age-specific mortality and lifespan. Proc. Biol. Sci 286, 20190393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluijs I, Beulens JW, van der A DL, Spijkerman AM, Grobbee DE, and van der Schouw YT (2010). Dietary intake of total, animal, and vegetable protein and risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition (EPIC)-NL study. Diabetes Care 33, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. (2014). The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab 19, 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, Mitchell SJ, Coogan SC, Cogger VC, Gokarn R, McMahon AC, Raubenheimer D, de Cabo R, Simpson SJ, and Le Couteur DG (2015). Dietary protein to carbohydrate ratio and caloric restriction: comparing metabolic outcomes in mice. Cell Rep 11, 1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, Cogger VC, Pulpitel T, Heblinski M, Wahl D, McMahon AC, Warren A, Durrant-Whyte J, Walters KA, Krycer JR, et al. (2016). Defining the nutritional and metabolic context of FGF21 using the geometric framework. Cell Metab 24, 555–565. [DOI] [PubMed] [Google Scholar]
- Song YS, Hwang YC, Ahn HY, and Park CY (2016). Comparison of the usefulness of the updated homeostasis model assessment (HOMA2) with the original HOMA1 in the prediction of type 2 diabetes mellitus in Koreans. Diabetes Metab. J 40, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Mitchell SE, and Mazidi M (2016). Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp. Gerontol 86, 28–38. [DOI] [PubMed] [Google Scholar]
- Strong R, Miller RA, Astle CM, Floyd RA, Flurkey K, Hensley KL, Javors MA, Leeuwenburgh C, Nelson JF, Ongini E, et al. (2008). Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell 7, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong R, Miller RA, Antebi A, Astle CM, Bogue M, Denzel MS, Fernandez E, Flurkey K, Hamilton KL, Lamming DW, et al. (2016). Longer lifespan in male mice treated with a weakly estrogenic agonist, an antioxidant, an alpha-glucosidase inhibitor or a Nrf2-inducer. Aging Cell 15, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1977). Report of the American Institute of Nutrition ad hoc Committee on Standards for Nutritional Studies. J. Nutr 107, 1340–1348. [DOI] [PubMed] [Google Scholar]
- van Dijk M, Nagel J, Dijk FJ, Salles J, Verlaan S, Walrand S, van Norren K, and Luiking Y (2017). Sarcopenia in older mice is characterized by a decreased anabolic response to a protein meal. Arch. Gerontol. Geriatr 69, 134–143. [DOI] [PubMed] [Google Scholar]
- Vergnaud AC, Norat T, Mouw T, Romaguera D, May AM, Bueno-de-Mesquita HB, van der A D, Agudo A, Wareham N, Khaw KT, et al. (2013). Macronutrient composition of the diet and prospective weight change in participants of the EPIC-PANACEA study. PLoS One 8, e57300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wali JA, Milner AJ, Luk AWS, Pulpitel TJ, Dodgson T, Facey HJW, Wahl D, Kebede MA, Senior AM, Sullivan MA, et al. (2021). Impact of dietary carbohydrate type and protein-carbohydrate interaction on metabolic health. Nat. Metab 3, 810–828. [DOI] [PubMed] [Google Scholar]
- Wanders D, Stone KP, Forney LA, Cortez CC, Dille KN, Simon J, Xu M, Hotard EC, Nikonorova IA, Pettit AP, et al. (2016). Role of GCN2-independent signaling through a noncanonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes 65, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders D, Forney LA, Stone KP, Burk DH, Pierse A, and Gettys TW (2017). FGF21 mediates the thermogenic and insulin-sensitizing effects of dietary methionine restriction but not its effects on hepatic lipid metabolism. Diabetes 66, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. (2015). Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Niederstrasser H, Douglas PM, Lin R, Jaramillo J, Li Y, Oswald NW, Zhou A, McMillan EA, Mendiratta S, et al. (2017). Small-molecule TFEB pathway agonists that ameliorate metabolic syndrome in mice and extend C. elegans lifespan. Nat. Commun 8, 2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR (2005). gplots: various R programming tools for plotting data
- Wu Y, Li B, Li L, Mitchell SE, Green CL, D’Agostino G, Wang G, Wang L, Li M, Li J, et al. (2021). Very-low-protein diets lead to reduced food intake and weight loss, linked to inhibition of hypothalamic mTOR signaling, in mice. Cell Metab 33, 886–904.e6. [DOI] [PubMed] [Google Scholar]
- Yu D, Yang SE, Miller BR, Wisinski JA, Sherman DS, Brinkman JA, Tomasiewicz JL, Cummings NE, Kimple ME, Cryns VL, et al. (2018). Short-term methionine deprivation improves metabolic health via sexually dimorphic, mTORC1-independent mechanisms. FASEB J 32, 3471–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Tomasiewicz JL, Yang SE, Miller BR, Wakai MH, Sherman DS, Cummings NE, Baar EL, Brinkman JA, Syed FA, et al. (2019). Calorie-restriction-induced insulin sensitivity is mediated by adipose mTORC2 and not required for lifespan extension. Cell Rep 29, 236–248.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Richardson NE, Green CL, Spicer AB, Murphy ME, Flores V, Jang C, Kasza I, Nikodemova M, Wakai MH, et al. (2021). The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. Cell Metab 33, 905–922.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. (2015). Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. (2018). Ensembl 2018. Nucleic Acids Res 46, D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurlo F, Lillioja S, Esposito-Del Puente A, Nyomba BL, Raz I, Saad MF, Swinburn BA, Knowler WC, Bogardus C, and Ravussin E (1990). Low ratio of fat to carbohydrate oxidation as predictor of weight gain: study of 24-h RQ. Am. J. Physiol 259, E650–E657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the hepatic RNAseq gene expression data reported in this paper can be obtained from Gene Expression Omnibus (GEO) (GEO: GSE181301).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


