ABSTRACT
Eukaryotic cells use post-translational modifications to diversify and dynamically coordinate the function and properties of protein networks within various cellular processes. For example, the process of autophagy strongly depends on the balanced action of kinases and phosphatases. Highly conserved from the budding yeast Saccharomyces cerevisiae to humans, autophagy is a tightly regulated self-degradation process that is crucial for survival, stress adaptation, maintenance of cellular and organismal homeostasis, and cell differentiation and development. Many studies have emphasized the importance of kinases and phosphatases in the regulation of autophagy and identified many of the core autophagy proteins as their direct targets. In this review, we summarize the current knowledge on kinases and phosphatases acting on the core autophagy machinery and discuss the relevance of phosphoregulation for the overall process of autophagy.
KEYWORDS: Autophagosome, macroautophagy, PAS, phagophore, posttranslational modification
Introduction
Eukaryotic cells maintain their physiological complexity by employing fine-tuned regulatory networks that orchestrate multi-layered inter- and intraconnected pathways. Post-translational modifications represent one essential regulatory mechanism by which cells modulate and diversify the function of biological molecules and dynamically coordinate their signaling networks. In higher eukaryotes, aberrant post-translational modifications are linked to several developmental disorders but also severe pathologies, including neurodegeneration, inflammatory, cardiac, and autoimmune diseases as well as cancer [1–3].
Post-translational modifications can be both reversible and irreversible. Phosphorylation is one type of reversible post-translational modification that is implicated in a broad spectrum of cellular processes and physiological states. The addition of phosphate groups onto proteins creates a new chemical entity, diversifying the chemical nature of the protein surface. The modified protein might interact differently with substrates and binding partners, and/or adopt a new conformation with altered activity. In eukaryotic cells, the phosphorylation status of a biological molecule reflects the balanced action of a kinase and its counteracting phosphatase [2–4]. Kinases and phosphatases are therefore key regulators of cellular functions. They act as molecular switches to navigate and regulate different biological processes by influencing the activity, function and localization of proteins, lipids, and small molecules within almost all cellular pathways.
Biochemically, kinases are enzymes that modulate the transfer of a γ-phosphate moiety from high energy phosphate-donors, usually ATP and more rarely GTP, to a target substrate. The kinome of eukaryotes can be divided into four main superfamilies depending on the substrate molecule that is phosphorylated: proteins, lipids, carbohydrates, and metabolites, such as nucleotides, riboflavin, creatine or dihydroxyacetone [5–8].
Phosphorylation by kinases can be reverted by a class of enzymes known as phosphatases [9]. Phosphatases carry out dephosphorylation via a hydrolysis reaction wherein a water molecule is used to release the phosphate group from the substrate, leaving behind an alcohol derivative. The activity, subcellular localization and substrate specificity of phosphatases are controlled by interactions with regulatory subunits. Thus, phosphatases are often hetero-multimeric complexes consisting of a catalytic subunit and scaffold protein and/or regulatory subunits [10]. The activity of phosphatases can also be regulated by “docking interactions” between different amino acid motifs within the phosphatase and its substrate [11]. Phosphatases are generally classified into six super-families based on the catalytic domain homology: protein tyrosine phosphatases, protein phosphatases (PPP), protein phosphatase, Mg2+- or Mn2+-dependent (PP2C/PPM), lipid phosphatases, haloacid dehalogenases and nudix hydrolases [12]. Among these, the PPP super-family comprises the following complexes: protein phosphatase 1 (PPP1), protein phosphatase 2 (PPP2), protein phosphatase 3 (PPP3/PP2B), protein phosphatase 4 (PPP4), protein phosphatase 5 (PPP5), protein phosphatase 6 (PPP6) and protein phosphatase with EF-hand (PPEF/PPP7) [13]. Protein phosphatases can also be classified into four sub-types based on their substrates: Ser/Thr phosphatases, Tyr phosphatases, His phosphatases and dual-specificity phosphatases (DSP) [10,12]. Although protein kinases can phosphorylate uncommon amino acids such as Lys, Arg, Asp, Cys and Glu, the identity of the phosphatases that eventually revert these modification remains elusive [4,14].
The role of kinases and phosphatases in human health has been a subject of great scientific interest. Phosphorylation and dephosphorylation of macromolecules are tightly regulated in the process of macroautophagy, hereafter autophagy. Autophagy is a highly conserved intracellular transport pathway by which cytoplasmic material is targeted to the lytic compartment, the vacuole in yeast and the lysosome in mammals for degradation and recycling [15]. This cytoplasmic material can include unnecessary or dysfunctional proteins, superfluous or damaged organelles, and harmful pathogens. Proper autophagy function is important for many biological processes, including embryonic development, cellular differentiation and adaptation to changing environmental conditions [16]. In addition, dysregulation of autophagy exacerbates a plethora of clinical conditions such as neurodegenerative disorders, cancer, obesity and infectious diseases [17–19].
Autophagy can be either a nonselective or a selective process [20]. Nonselective autophagy is primarily induced as a response to starvation and serves in the turnover of bulk cytoplasm in an apparent random manner. The target of rapamycin complex 1 (TORC1) kinase complex is the master nutrient sensor of the cell. TORC1 activity is regulated by nutrient levels, in particular amino acids [21]. In the presence of nutrients, TORC1 is active and phosphorylates numerous substrates, including components of the autophagy machinery, thereby inhibiting the progression of autophagy. Upon starvation, TORC1 is inhibited, which is paralleled by the dephosphorylation of these proteins and autophagy induction. Selective autophagy is not under the control of TORC1, but requires receptor proteins, also known as autophagy receptors, which recognize cargo specifically, therefore acting in a non-stochastic manner [20].
Although nonselective and selective autophagy differ in the mode of cargo selection, both processes utilize the same core machinery, and cargo destined for degradation are sequestered into double-membrane vesicles known as autophagosomes [15,20]. Autophagosomes eventually deliver their cargo into the vacuole/lysosome by fusing with the limiting membrane of these organelles. Autophagosome formation is driven by the concerted action of a group of proteins collectively called Atg (autophagy-related) proteins [15], and around 20 of them comprise the core Atg machinery. Many of these Atg proteins are functionally and structurally conserved among eukaryotes [22,23]. They are classified into six functional modules: the Atg1/ULK (unc-51 like autophagy activating kinase) complex, the phosphatidylinositol 3-kinase (PtdIns3K) complex I, the Atg9-positive vesicles, the Atg2-Atg18/ATG2-WIPI (WD repeat domain, phosphoinositide interacting) protein complex, the Atg12 conjugation system and the Atg8 conjugation system [15].
In yeast, autophagosomes are formed in close vicinity to the vacuole and the endoplasmic reticulum (ER) [24–27], whereas in mammals, they appear to be generated throughout the cytoplasm but adjacent to specific subdomains of the ER [28–30]. Autophagosome formation requires the assembly of a distinct multi-component structure called the phagophore assembly site (PAS), which is anchored onto the vacuole in yeast [31]. The PAS is a highly dynamic entity and evolves from the hierarchical recruitment of core Atg proteins [15,32]. Once assembled, the PAS gives rise to a cisterna known as the phagophore [15]. The phagophore expands through the acquisition of lipids and the fusion of its extremities leads to the formation of the autophagosome, and the concomitant sequestration of the cargo destined for degradation. Autophagosome biogenesis can be divided into five discrete steps: 1) initiation, which is characterized by the formation of the PAS and then a phagophore; 2) phagophore expansion and closure, which leads to the generation of an autophagosome; 3) autophagosome maturation, which is characterized by the release of Atg proteins from the complete autophagosome and the acquisition of the fusion machinery; 4) autophagosome fusion with a lysosome/vacuole; and 5) lysosomal/vacuolar degradation of the autophagosomal cargo (Figure 1).
Figure 1.
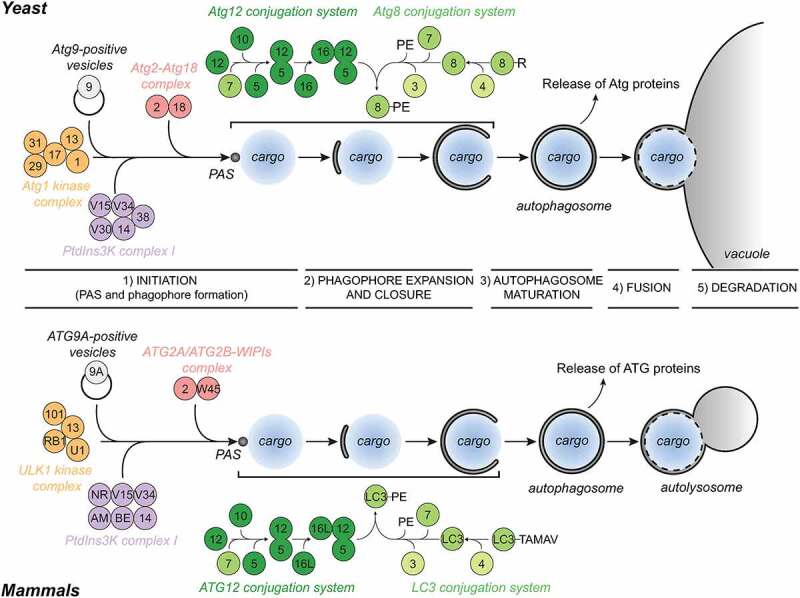
Schematic of the mechanism of autophagy in yeast and mammalian cells. Schematics of the five discrete steps that characterize the process of autophagy, and the Atg modules involved. For details see main text. All yeast and mammalian Atg proteins are indicated with their respective numbers. AM, AMBRA1; BE, BECN1; NR, NRBF2; RB1, RB1CC1; U1, ULK1; V15, Vps15/PIK3R4; V30, Vps30; V34, Vps34/PIK3C3; W45, WDR45/WIPI4. Black arrows indicate the assembly hierarchy of the Atg machinery and the progression of autophagy.
Growing evidence has revealed that the fine-tuned regulation of autophagy is coordinated by kinases and phosphatases. Regardless of the type of autophagy, PAS assembly, autophagosome formation and delivery of the cargo to the lytic compartment are highly regulated by phosphorylation. Many Atg proteins have been found to be phosphorylated, and several kinases and phosphatases have been implicated in the different steps of autophagy. In this review, we focus on the kinases and phosphatases that are reported to act directly on the core Atg machinery and compare the yeast enzymes with their mammalian counterparts (Figure 2).
Figure 2.
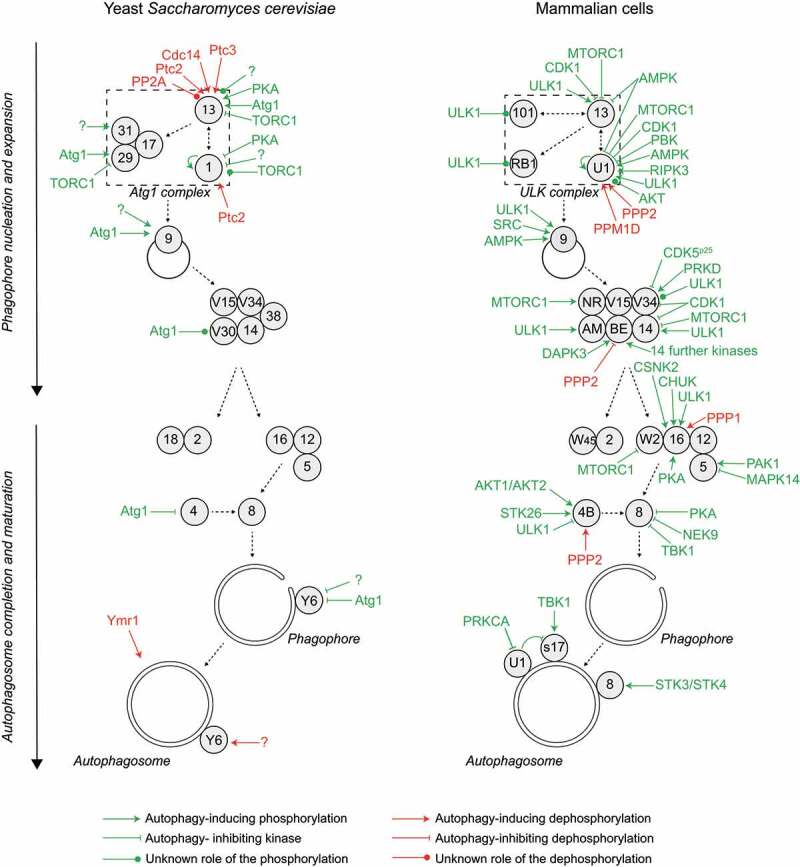
Autophagy progression is fine-tuned by multiple phosphorylation and dephosphorylation events regulating the core Atg machinery. Autophagy is initiated by the formation of a phagophore assembly site (PAS), where the hierarchical assembly of the indicated proteins drives the process of autophagosome formation, characterized by the formation of a phagophore and its expansion. Many of these Atg proteins are subject to phosphoregulation, as detailed in the text. This overview shows the yeast (left panel) and mammalian (right panel) kinases and phosphatases acting on the autophagy pathway. For simplicity, the effect of the kinase or phosphatase on the pathway, rather than the effect on the specific target protein, is indicated. All yeast and mammalian Atg proteins are indicated with their respective numbers. AM, AMBRA1; BE, BECN1; RB1, RB1CC1; NR, NRBF2; S17, STX17; U1, ULK1; V15, Vps15/PIK3R4; V30, Vps30; V34, Vps34/PIK3C3; W2, WIPI2; W45, WDR45/WIPI4; Y6, Ykt6. Note that BECN1 is regulated by numerous kinases, including AKT1, AMPK, BCR-ABL (an oncogenic fusion protein between BCR [BCR activator of RhoGEF and GTPase] and ABL1 [ABL proto-oncogene 1, non-receptor tyrosine kinase], which is found in most patients with chronic myelogenous leukemia), CAMK2, CSNK1G2, DAPK1, DAPK2, EGFR (epidermal growth factor receptor), PTK2 (protein tyrosine kinase 2), MAPKAPK2 (MAPK activated protein kinase 2), MAPKAPK3, PGK1 (phosphoglycerate kinase 1), ROCK1 (Rho associated coiled-coil containing protein kinase 1), ULK1 and STK4, details are shown in Figure 3. Color code: green, kinase; red, phosphatase. Black dashed arrows indicate the assembly hierarchy of the Atg machinery and the progression of autophagy.
Phosphoregulation of Atg1/ULK kinases during autophagosome formation initiation
The Atg1 kinase complex in yeast, conserved with the ULK kinase complex in mammals [33], is central and essential for autophagy initiation under all conditions.
Yeast Atg1 belongs to the family of serine/threonine kinases, and is part of the core Atg machinery [33]. The presence of Atg1 is essential for the correct assembly of the PAS, as well as its maturation and subsequent progression of autophagy [32]. Therefore, Atg1 is viewed as a key regulator of autophagy, acting early during initiation of the process. During selective autophagy, the Atg1 kinase complex assembles on the cargo recruited in proximity of the vacuole, resulting in local clustering and activation of the Atg1 kinase [34]. In bulk autophagy, however, a specific cargo serving as an assembly platform is missing. Instead, Atg1 assembles into a pentameric complex with Atg13, Atg17, Atg29 and Atg31 [35–38]. Several of these complexes further interact with each other via multiple Atg13-Atg17 interactions, resulting in a higher-order oligomeric structure that constitutes the early PAS for bulk autophagy [37]. This clustering of the Atg1 complex both in bulk and selective autophagy is the basis of Atg1 kinase activation, leading to the auto-phosphorylation of Thr226 and Ser230, which are present in a regulatory amino acid loop that is necessary for kinase activation (Table 1) [39,40]. This self-activation event leads to the recruitment of additional Atg proteins and the subsequent maturation of the PAS into a site where the phagophore can form (Figure 2). It was recently proposed that the early PAS also behaves as a liquid-like condensate, where the Atg1 complex undergoes phase separation to form a liquid droplet [41]. In this scenario, phase separation of the Atg1 complex facilitates kinase activation by increasing the collision frequency between single Atg1 molecules [41].
Table 1.
Characterized phosphosites on the yeast Atg machinery
|
Atg protein and phosphosites |
Function |
Kinase Phosphatase |
References |
|
| Atg1 | ||||
| Ser34 | Kinase activity decrease | ? | ? | [52] |
| Thr226 | Autoactivation, autophagy | Atg1 | Ptc2 | [39,41,110] |
| Ser230 | Autoactivation, autophagy | Atg1 | ? | [39] |
| Ser508 | PAS localization and autophagy inhibition | PKA | ? | [51] |
| Ser515 | PAS localization and autophagy inhibition | PKA | ? | [51] |
| Ser518 | ? | TORC1 | ? | [53] |
| Atg4 | ||||
| Ser307 | Atg4 proteolytic activity and autophagy inhibition | Atg1 | ? | [158] |
| Atg9 | ||||
| Ser19 | Phagophore formation induction | Atg1 | ? | [142] |
| Ser122 | Enhancement of the interaction With Atg23 and Atg27, and autophagy | ? | ? | [116] |
| Ser657 | Phagophore formation induction | Atg1 | ? | [142] |
| Ser802 | Phagophore formation induction | Atg1 | ? | [142] |
| Ser831 | Phagophore formation induction | Atg1 | ? | [142] |
| Ser948 | Phagophore formation induction | Atg1 | ? | [142] |
| Ser969 | Phagophore formation induction | Atg1 | ? | [142] |
| Atg13 | ||||
| Ser344 | PAS localization and autophagy | PKA | ? | [68] |
| Ser348 | Mutated in Atg13[8SA], possibly inhibiting autophagy | ? | (Cdc14) | [65,97] |
| Ser428 | Atg17 binding and autophagy inhibition | TORC1 Atg1 | Ptc2 Ptc2 | [41,53] [67] |
| Ser429 | Atg17 binding and autophagy inhibition | TORC1 Atg1 | Ptc2 Ptc2 | [41] [67] |
| Ser437 | PAS localization and autophagy inhibition, mutated in Atg13[8SA] | PKA | (Ptc2, Ptc3) | [65,68,110] |
| Ser438 | Mutated in Atg13[8SA], possibly inhibiting autophagy | ? | (Ptc2, Ptc3) | [65,110] |
| Ser496 | Autophagy inhibition, mutated in Atg13[8SA] | Atg1 | (Ptc2, Ptc3) | [53,65,110] |
| Ser535 | Mutated in Atg13[8SA], Possibly inhibiting autophagy | Atg1 | (Cdc14) | [53,65,97] |
| Ser541 | Mutated in Atg13[8SA], possibly inhibiting autophagy | ? | (Cdc14) | [65,97] |
| Ser581 | PAS localization and autophagy inhibition | PKA | ? | [68] |
| Ser637 | ? | TORC1 | ? | [53] |
| Ser644 | TORC1 | ? | [53] | |
| Ser646 | Mutated in Atg13[8SA], Possibly inhibiting autophagy | TORC1 | (Cdc14) | [53,65,97] |
| Ser649 | Mutated in Atg13[8SA], possibly inhibiting autophagy | ? | (Ptc2, Ptc3) | [65,110] |
| Atg29 | ||||
| Ser106 | Autophagy induction | Atg1 | ? | [53] |
| Thr115 | Autophagy inhibition | TORC1 | ? | [53] |
| Ser129 | Autophagy induction | Atg1 | ? | [53] |
| Ser197 | Release of self-inhibition, autophagy induction | ? | ? | [73] |
| Ser199 | Release of self-inhibition autophagy induction | ? | ? | [73] |
| Ser201 | Release of self-inhibition | Atg1 | ? | [53,73] |
| Atg31 | ||||
| Ser174 | Positive regulation of Atg9 trafficking and autophagy | ? | ? | [75] |
| Vps30 | ||||
| Ser85 | ? | Atg1 | ? | [53] |
| Ykt6 | ||||
| Thr158 | Autophagosome-vacuole fusion inhibition | ? | ? | [182,183] |
| Ser182 | Autophagosome-vacuole fusion inhibition | Atg1 | ? | [182,183] |
| Ser183 | Autophagosome-vacuole fusion inhibition | Atg1 | ? | [182,183] |
Additional phosphosites on the core Atg machinery modified by Atg1 and TORC1 have been systematically mapped in vivo and in vitro [53], but their relevance for autophagy progression remains to be established.
Putative enzymes catalyzing a given dephosphorylation are in parentheses.
ULK1 and ULK2 are the orthologs of Atg1 in more complex eukaryotes [42]. Similar to yeast Atg1, ULK1 and ULK2 form a multimeric kinase complex with ATG13, RB1CC1/FIP200 (RB1 inducible coiled-coil 1), and ATG101 (Figure 2), which are also essential for autophagy [43–46]. Similar to its yeast homolog, ULK1 harbors an autophosphorylation site in the regulatory amino acid loop, at Thr180 (Table 2), and it is also activated by local clustering during selective mitophagy [47,48]. Two additional autophosphorylation site have been described on murine ULK1 (Ser1043 and Ser1047, which correspond to Ser1042 and Thr1046 in the human protein) [49]. Phosphorylation of these residues enhances human ULK1 interaction with CUL3 (cullin 3) E3 ligase via KLHL20 (kelch like family member 20), promoting ULK1 ubiquitination and degradation [50].
Table 2.
Characterized phosphosites on the human Atg machinery
|
Atg protein and phosphosites |
Function |
Kinase |
Phosphatase |
References |
| AMBRA1 | ||||
| Ser465 | ? | ULK1 | ? | [72] |
| Ser635 | ? | ULK1 | ? | [72] |
| ATG4B | ||||
| Ser34 | ATG4B proteolytic activity | AKT1 | ? | [160,161] |
| stimulation | AKT2 | ? | [160,161] | |
| Ser316 | ATG4B proteolytic activity and autophagy inhibition | ULK1 | PPP2/PP2A | [159] |
| Ser383 | ATG4B proteolytic activity stimulation | STK26 | ? | [162] |
| Ser383 | ATG4B proteolytic activity stimulation | ? | ? | [163] |
| Ser392 | ATG4B proteolytic activity stimulation | ? | ? | [163] |
| ATG5 | ||||
| Thr75 | Autophagy inhibition | MAPK14 | ? | [149] |
| Thr101 | Stability, binding to ATG16L1 autophagy enhancement | PAK1 | ? | [149] |
| ATG9A | ||||
| Tyr8 | ATG9A trafficking and autophagy induction | SRC | ? | [117] |
| Ser14 | ATG9A trafficking and autophagy induction | ULK1 | ? | [117] |
| Ser761 | YWHAZ binding and | AMPK | ? | [118] |
| autophagy induction | ULK1 | ? | [118] | |
| ATG13 | ||||
| Ser44 | Mitotic autophagy inhibition | CDK1 | ? | [62] |
| Ser225 [Ser224*] | ULK1 activity inhibition | AMPK | ? | [70] |
| Mitotic autophagy inhibition | CDK1 | ? | [61,62] | |
| Ser259 [Ser258*] | ULK1 activity inhibition | MTORC1 | ? | [70] |
| Mitotic autophagy inhibition | CDK1 | ? | [61] | |
| Thr332 | Mitotic autophagy inhibition | CDK1 | ? | [62] |
| Thr342 | Mitotic autophagy inhibition | CDK1 | ? | [62] |
| Ser355 [Ser318***] | Mitophagy upregulation | ULK1 | ? | [71] |
| ATG14 | ||||
| Ser3 | PtdIns3K and autophagy inhibition | MTORC1 | ? | [125] |
| Ser29 | PtdIns3K and autophagy inhibition | ULK1 | ? | [123,124,206,207] |
| Ser223 | PtdIns3K and autophagy inhibition | MTORC1 | ? | [125] |
| Thr233 | PtdIns3K and autophagy inhibition | MTORC1 | ? | [125] |
| Ser383 | PtdIns3K and autophagy inhibition | MTORC1 | ? | [125] |
| Ser440 | PtdIns3K and autophagy inhibition | MTORC1 | ? | [125] |
| Mitotic autophagy inhibition | CDK1 | ? | [61] | |
| ATG16L1 | ||||
| Ser139 | Binding to ATG12–ATG5 and autophagy enhancement | CSNK2 | PPP1 | [148] |
| Ser278 | Regulating ATG16L1 stability and autophagy induction | CHUK | ? | [145] |
| Regulating ATG16L1 stability and autophagy induction | ULK1 | ? | [146] | |
| Ser269 | ATG16L1 degradation induc tion and utophagy inhibition | PRKA/PKA | ? | [147] |
| Ser287 [Ser268***] | ATG16L1 degradation induction and autophagy inhibition | PRKA/PKA | ? | [147] |
| ATG101 | ||||
| Ser11 | ? | ULK1 | ? | [72] |
| Ser203 | ? | ULK1 | ? | [72] |
| BECN1 | ||||
| Ser15 [Ser14*] | PtdIns3K activation, PRKN interaction | ULK1 | ? | [72,208,209] |
| Ser30 | PtdIns3K activation | ULK1 | ? | [72,207,209] |
| PtdIns3K activation | PGK1 | ? | [210] | |
| Ser90 | Autophagy induction | DAPK3 | PPP2/PP2A | [129] |
| MAPKAPK2 | ? | [211] | ||
| MAPKAPK3 | ? | [211] | ||
| CAMK2 | ? | [212] | ||
| Ser93 | PtdIns3K activation | AMPK | ? | [213] |
| Ser96 | Autophagy induction | ULK1 | ? | [72] |
| PtdIns3K activation | AMPK | ? | [213] | |
| Thr108 | BCL2-BCL2L1 interaction | STK4 | ? | [214] |
| Thr119 | BCL2-BCL2L1 interaction | DAPK | ? | [215,216] |
| BCL2-BCL2L1 interaction | ROCK1 | ? | [217] | |
| Tyr229 | Progression of autophagy | EGFR | ? | [218] |
| Tyr233 | ATG14 interaction. | PTK2 | ? | [219] |
| PtdIns3K complex formation | EGFR | ? | [218] | |
| Autophagy regulation | BCR-ABL | ? | [220] | |
| Ser234 | Regulation of function | AKT1 | ? | [221] |
| Ser279 | ? | ULK1 | ? | [72] |
| Ser295 | Regulation of function | AKT1 | ? | [221] |
| Tyr352 | Autophagy regulation | BCR-ABL | ? | [220] |
| Autophagy regulation | EGFR | ? | [218] | |
| Ser377 | ? | ULK1 | ? | [72] |
| Thr388 | Dimerization, association with PtdIns3K complex | AMPK | ? | [222] |
| Ser409 | RUBCN interaction | CSNK1G2 | ? | [223] |
| GABARAPL2 | ||||
| Ser87 | ATG4 protein binding inhibition | TBK1 | ? | [151] |
| Ser88 | ATG4 protein binding inhibition | TBK1 | ? | [151] |
| MAP1LC3A | ||||
| Ser12 | Autophagosome association and autophagy inhibition | PRKA/PKA | ? | [152] |
| MAP1LC3B | ||||
| Ser12 | Autophagosome recruitment and autophagy inhibition | PRKA/PKA | ? | [152] |
| Thr50 | Fusion with lysosomes and autophagy enhancement | STK3 | ? | [153,154] |
| Fusion with lysosomes and autophagy enhancement | STK4 | ? | [153,154] | |
| SQSTM1 degradation block | NEK9 | ? | [153] | |
| MAP1LC3C | ||||
| Ser93 | ATG4 protein binding inhibition | TBK1 | ? | [151] |
| Ser99 | ATG4 protein binding inhibition | TBK1 | ? | [151] |
| NRBF2 | ||||
| Ser113 | PtdIns3K assembly and activity inhibition | MTORC1 | ? | [136] |
| Ser120 | PtdIns3K assembly and activity inhibition | MTORC1 | ? | [136] |
| PIK3C3 | ||||
| Ser249 | ? | ULK1 | ? | [72] |
| Thr159 | Interaction with BECN1 reduction and | CDK1 | ? | [127] |
| autophagy inhibition | CDK5 p25 | ? | [127] | |
| Thr677 | PtdIns3K activity and autophagy induction - | PRKD1 | ? | [128] |
| RB1CC1 | ||||
| Ser943 | ? | ULK1 | ? | [72] |
| Ser986 | ? | ULK1 | ? | [72] |
| Ser1323 | ? | ULK1 | ? | [72] |
| STX17 | ||||
| Ser202 | PAS association and autophagy enhancement | TBK1 | ? | [185] |
| ULK1 | ||||
| Thr180 | Autoactivation, autophagy Induction | ULK1 | ? | [48] |
| Ser317 | Autoactivation, autophagy induction | AMPK | ? | [58] |
| Ser423 | Association with STX17, auto-phagosome-lysosome fusion and autophagic flux inhibition | PRKCA | ? | [184] |
| Ser467 | Autophagy/mitophagy induction AMPK | AMPK | 59? | [59] |
| Ser469 | Autophagy inhibition | PBK | ? | [63] |
| Ser495 | Autophagy inhibition | PBK | ? | [63] |
| Ser533 | Autophagy inhibition | PBK | ? | [63] |
| Ser556 [Ser555*] | Autophagy/mitophagy induction AMPK | AMPK | 59,60,64? | 59,60,64 |
| Thr575 [Thr574*] | Autophagy/mitophagy induction AMPK | AMPK | 59? | [59] |
| Ser623 [Ser622*] | Mitotic autophagy inhibition | CDK1 | ? | [62] |
| Thr636 [Thr635*] | Mitotic autophagy inhibition | CDK1 | ? | [62] |
| Ser638 [Ser637*] | Autophagy inhibition | AMPK | PPP2/PP2A | [54,55,59,60] |
| PPM1D | [56,57] | |||
| MTORC1 | [54] | |||
| Thr654 [Thr653*] | Mitotic autophagy inhibition | CDK1 | ? | [62] |
| Thr660 [Thr659*] | Autophagy induction | AMPK | ? | [60] |
| Ser747 [Ser746*] | Genotoxic stress-induced Autophagy stimulation | RIPK3 | ? | [57] |
| Ser758 [Ser757*] | Inhibition of AMPK-ULK1 interaction, autophagy inhibition | MTORC1 | ? | [54] |
| Mitotic autophagy inhibition | CDK1 | ? | [61] | |
| Ser775 [Ser774*] | Insulin-regulated site, unknown effect on ULK1 activity | AKT | ? | [64] |
| Ser777** | Autoactivation, autophagy induction | AMPK | ? | [58] |
| Ser1042 [Ser1043*] | KLHL20-CUL3 E3 ligase binding and ULK1 turnover enhancement | ULK1 | ? | [49,50] |
| Thr1046 [Ser1047*] | KLHL20-CUL3 E3 ligase binding and ULK1 turnover enhancement | ULK1 | ? | [49,50] |
| WIPI2 | ||||
| Ser395 | HUWE1 binding, autophagy inhibition | MTORC1 | ? | [140] |
Sites in square brackets indicate sites mapped on a different isoform or in a different organism: *corresponding residue in the murine protein; **in the murine protein only, residue not conserved in the human protein; *** originally reported amino acid position, but on a different isoform.
Many of the listed sites need further rigorous validation to be defined as target sites of the kinases and phosphatases. Additional phosphorylation sites not indicated in this table have been mapped; however, the responsible kinases and/or their biological relevance remain mostly unknown.
Several Atg1/ULK complex subunits are phosphoregulated (Table 1, 2, and Figure 2). Atg1 itself is phosphoregulated by several other kinases, which fine-tune its activity and assembly into the pentameric complex. One of the kinases that acts on Atg1 is the cAMP-dependent protein kinase complex (PKA). PKA, which senses extracellular nutrients, phosphorylates Ser508 and Ser515 of Atg1 (Table 1) [51] to influence Atg1 localization. When phosphorylated, Atg1 recruitment to the PAS is blocked, thus inhibiting autophagy progression when nutrients are available. Intriguingly, substitution of these residues with non-phosphorylatable Ser-to-Ala rescues the localization defect induced by an overexpression of PKA, although autophagosome biogenesis is still impaired [51]. Atg1 is also phosphorylated on its N-terminus, at Ser34, which diminishes its kinase activity [52]. The kinase responsible for this modification, however, remains unknown.
Several sites on Atg1 have been reported to be phosphorylated by TORC1, among these is Ser518, which is found in vivo and is directly targeted by TORC1 also in vitro (Table 1) [53]. MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1) directly targets and phosphorylates ULK1 at Ser638 and Ser758 (corresponding to Ser637 and Ser757 in the murine protein; Table 2) [54]. These modifications regulate the interaction between ULK1 and another kinase, AMP-activated protein kinase (AMPK) [54]. AMPK activity maintains phosphorylation of Ser638, thereby inhibiting autophagy induction [54]. Interestingly, this phosphorylation site has been reported to be dephosphorylated by two phosphatases: PPP2 and PPM1D (protein phosphatase 1D magnesium-dependent, delta isoform; see below) [55–57]. The targeting of Ser638 by different kinases and different phosphatases suggests that it represents an important regulatory residue of ULK1.
Under glucose starvation, AMPK phosphorylates ULK1 at Ser317 and Ser777 (the latter is present only in mouse ULK1), which promotes ULK1 activation and autophagy progression (Table 2) [58]. Phosphorylation of Ser758 by MTORC1 counteracts the AMPK activation of ULK1 by disrupting binding between ULK1 and AMPK. Further AMPK-dependent phosphorylation of ULK1 at residues Ser467, Ser556, Thr575 and Thr660 (Ser467, Ser555, Thr574 and Thr659 in mouse ULK1) positively regulate mitophagy and starvation-induced autophagy [59,60], although one of these sites, Ser556, has also been reported to play an inhibitory role [60]. Thus, although MTORC1-dependent phosphorylations have so far always been reported to inhibit ULK1 function, ULK1 phosphorylation by AMPK appears to both activate and inhibit autophagy. This apparent discrepancy could stem from the different modes of autophagy induction, such as different factors involved and/or different nutrient situations.
One of the ULK1 sites targeted by MTORC1, Ser758, can also be phosphorylated by CDK1 (cyclin dependent kinase 1) in mitosis, which results in an inhibition of autophagy induction during this cell cycle stage [61]. Additional CDK1 phosphorylation sites on ULK1 that inhibit autophagy during mitosis include Ser623, Thr636 and Thr654 (corresponding to Ser622, Thr635 and Thr653 in the murine ULK1) [62]. Altogether, these findings suggest that different kinases can target the same residue depending on the cellular situation.
Additionally, direct phosphorylation of ULK1 at Ser469, Ser495 and Ser533 by PBK (PDZ binding kinase) inhibited autophagy induction by reducing ULK1 stability, complex assembly, and kinase activation. In contrast, mutation of these sites to non-phosphorylatable alanine residues led to enhanced autophagy induction and promoted apoptosis [63]. Moreover, phosphorylation on ULK1 at Ser747 (Ser746 in mouse ULK1), catalyzed by the RIPK3 (receptor-interacting serine-threonine kinase 3), was found to be required for genotoxic stress-induced unconventional autophagy, but not for conventional bulk autophagy [57]. ULK1 phosphorylation by RIPK3 requires the prior dephosphorylation of Ser638 by the phosphatase PPM1D [56,57]. Finally, AKT1 (AKT serine/threonine kinase 1) phosphorylation site has been identified at Ser774 in mouse ULK1 (corresponding to Ser775 in the human homolog), which is increased upon insulin stimulation (Table 2) [64]. Whether this phosphorylation plays a regulatory role has not been addressed yet. Taken together, ULK1 is a major hub of phosphoregulation, targeted by several different kinases and phosphatases (Figure 3A).
Figure 3.
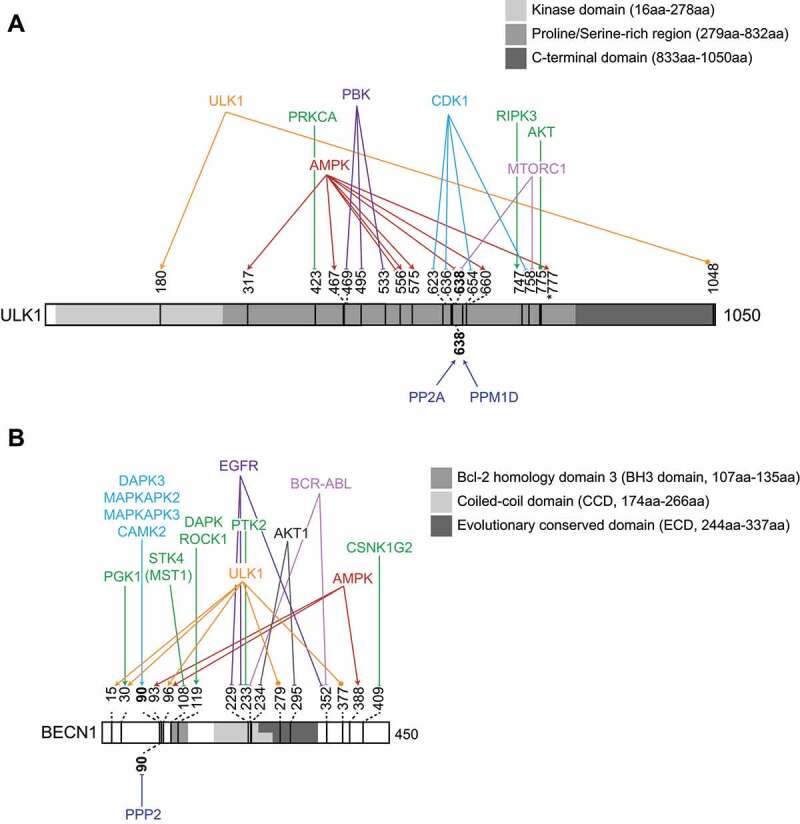
ULK1 and BECN1 are central hubs for the phosphoregulation of autophagy progression. Schematic representation of ULK1 (A) and BECN1 (B), depicting the characterized phosphosites, and the identified kinases and phosphatases that modify them. Pointed arrowheads highlight modifications that positively regulate autophagy, flat arrowheads indicate the ones inhibiting autophagy, and arrows ending in a circle indicate that the mode of regulation is unknwon. Described domains are indicated. Note that different annotations for domains in BECN1 exist; the shown domains are obtained from [203–205].
Phosphoregulation of the Atg1/ULK kinase complex subunits during autophagosome formation initiation
Atg13 is another direct target of TORC1 under nutrient-rich conditions in yeast (Figure 2 and Table 1) [65]. When TORC1 is inhibited, Atg13 becomes rapidly dephosphorylated, leading to bulk autophagy induction [36]. More than 40 phosphorylation sites on Atg13 have been mapped, and some of these sites have been verified as direct TORC1 or Atg1 targets in vitro [53,65,66]. Ser428 and Ser429, which are direct targets of both TORC1 and Atg1 [41,53,67], appear to be of regulatory importance, as these residues are involved in Atg17 binding upon starvation (Table 1). Phosphorylation of Ser496 also negatively regulates autophagy induction and the dephosphorylation of several further Ser residues in the Atg1 binding region of Atg13, which is key in mediating Atg1-Atg13 interaction [66]. Atg13 is also a target of PKA [68]. PKA-mediated phosphorylation of Ser344, Ser437 and Ser581 regulates the association of Atg13 with the PAS (Table 1). The Atg13-Atg17 interaction is affected by phosphorylation of these sites as well, probably by the modification of Ser437 in particular, as this residue is located next to the Atg17-interacting region in Atg13 [66]. Direct TORC1 phosphorylation sites on Atg13 include Ser428, Ser637, Ser644 and Ser646 [53]. A mutant of Atg13, Atg13[8SA], which potentially lacks phosphorylation at eight serine residues (Ser348, Ser437, Ser438, Ser496, Ser535, Ser541, Ser646 and Ser649), has been reported to bypass the negative TORC1 regulation (Table 1) [65]. Recently, these sites were shown to be phosphorylated in vivo [53], however, only Ser646 was found to be a TORC1 target both in vitro and in vivo, and Ser496 and Ser535 are targeted by Atg1. How these phosphorylation sites contribute to autophagy inhibition mechanistically and how TORC1 and Atg1 cooperate in this process remain to be examined.
Similar to its yeast homolog, mammalian ATG13 undergoes dramatic changes in its phosphorylation status upon autophagy induction, but in contrast to yeast Atg13, this does not appear to influence ATG13 binding affinity to ULK1 and ATG101 [69]. ATG13 is a target of AMPK and MTORC1, which phosphorylate murine Ser224 (Ser225 in human ATG13) and Ser258 (Ser259 in human ATG13), respectively, and negatively regulate autophagy [46,70]. Both these sites, as well as Ser44, Thr332 and Thr342, are targeted by CDK1 during mitotic autophagy regulation [61,62]. CDK1 phosphorylation has been suggested to negatively regulate mitotic autophagy. ATG13 is also a target of ULK1, which phosphorylates its Ser355 (corresponding to Ser318 in isoform 2). This phosphorylation influences mitophagy function positively by mediating ATG13 localization to mitochondria, but it does not affect bulk autophagy, indicating that ULK1 might phosphorylate ATG13 at different locations in the cell, depending on the type of autophagy that needs to be executed [71]. Other ULK1 phosphosites on ATG13 have been reported, but their role is still not understood [43,54,62,72].
Yeast Atg29, which is part of the Atg17-Atg31-Atg29 ternary complex (Figure 2), also shows differential phosphorylation depending on the nutrient availability [53,73]. The function of Atg29 is regulated by an inhibitory peptide located at its extreme C-terminus. Upon autophagy induction, the C-terminus becomes phosphorylated, inducing a conformational change that releases the inhibition by this peptide and thus allows autophagy to progress [73]. Three residues, Ser197, Ser199 and Ser201, appear to be particularly important for this regulation, but it remains to be determined whether these sites are phosphorylated in vivo (Table 1) [73]. Only Ser201 phosphorylation has been confirmed in cultured cells in vivo and appears to be directly targeted by Atg1 [53]. Additional phosphorylation sites on Atg29 have been mapped. In addition to Ser201, Ser106 and Ser129 are Atg1 targets, whereas Thr115 appears to be directly modified by TORC1 (Table 1) [53]. Mutation of the three Ser residues to non-phosphorylatable Ala, or of the Thr115 to a phosphomimicking Glu, results in reduced bulk autophagy [53]. Phosphorylation of Atg29 also appears to regulate its binding to Atg11 and subsequently the targeting of Atg17-Atg29-Atg31 to the vacuole under nutrient starvation, but which phosphorylation sites are responsible for these events has not been analyzed [73]. These findings indicate that, in contrast to what has been previously suggested, Atg11 may contribute to the efficiency of bulk autophagy under nitrogen-starvation conditions, similarly to its role during glucose deprivation-induced autophagy [74]. Several phosphorylation sites have been identified on Atg31, and Ser174 in particular plays an important role in promoting the autophagic flux because it positively regulates Atg9 trafficking (Table 1) [75]. The kinase responsible for Ser174 modification, as well as the role of the phosphorylated sites, remain unknown.
Similar to yeast Atg11 and Atg17, RB1CC1 acts as a scaffold protein to recruit the Atg machinery in mammals. It directly interacts with ATG13 and is considered the functional homolog of yeast Atg11 and Atg17 [43,44,46,76–79]. Similar to Atg11, RB1CC1 binds autophagy receptors, thus linking the ULK1 complex to the autophagosomal cargo [80–83]. ULK1 was shown to phosphorylate RB1CC1 on Ser943, Ser986 and Ser1323 (Table 2) [46,72]. The importance of this phosphoregulation and how it affects autophagy, however, is not known yet. While no phosphorylation has been reported on Atg17 to date, Atg11 contains direct Atg1 target sites, but their relevance remains unexplored [53].
The mammalian ULK complex does not contain Atg29 and Atg31 homologs but rather contains a unique subunit, ATG101, which shows no functional or structural homology to any yeast protein (Figure 2). Loss of ATG101 results in the aberrant recruitment of downstream factors necessary for phagophore expansion, leading to a defect in autophagosome biogenesis [43,69,84]. Two ULK1 consensus phosphorylation sites are present in human ATG101 (Ser11 and Ser203), but whether these sites are important for autophagy progression remains unknown [72].
Protein phosphatases regulating the Atg1/ULK kinase complex
A major class of phosphatases counteracting TORC1 function and the early steps of autophagy is the serine/threonine-protein phosphatase PP2A (PP2A) family of phosphatase holoenzymes. PP2A phosphatases are ubiquitous, and they are involved in multiple cellular functions [85–87]. In yeast, PP2A phosphatases consist of heterotrimeric complexes composed of 1) the scaffold A subunit, Tpd3 (tRNA processing deficient 3), 2) a regulatory B subunit, Cdc55 (cell division control 55) or Rts1 (rox three suppressor 1), and 3) a catalytic C subunit, Pph21 (protein phosphatase 21) or Pph22 [88]. Although Pph21 and Pph22 have redundant roles, the function of these two phosphatases differs based on their interaction with their regulatory subunits and their substrates [89].
Approximately 2% of Pph21 and Pph22 form a heterodimer with Tap42 (two A phosphatase associated protein 42), one of the targets of the TORC1 kinase complex [89,90]. Phosphorylated Tap42 competes with Cdc5 and Rts1 to bind the PP2A phosphatase catalytic subunit C. Upon nutrient depletion, TORC1 becomes inactivated and Tap42 is dephosphorylated, triggering its dissociation from Pph21 and Pph22. This event allows the binding of Cdc55 and Rts1, along with Tpd3, to Pph21 and Pph22, leading to the formation of active Cdc55-Tpd3- and Rts1-Tpd3-containing PP2A phosphatase complexes [90,91].
Recently, it was shown that yeast PP2A phosphatases play a role in dephosphorylating Atg13 (Figure 2) [92]. Although pph21∆, pph22∆, cdc55∆ and rts1∆ single knockout cells do not show any defects in autophagy, the double deletion pph21∆ pph22∆ displays severely impaired autophagy [92]. A similar phenotype is observed in the double knockout strain cdc55∆ rts1∆. Additional investigations showed that the autophagy defect observed in these knockout mutants was due to inefficient dephosphorylation of Atg13. Another report reached the opposite conclusion [93] that PP2A phosphatases negatively regulate starvation-induced autophagy. In the latter report, overexpression of the phosphatase-dead Pph22H186N variant or the inactive pph22-172 allele did not block autophagy induction. Conversely, overexpression of either PPH21 or PPH22 leads to impaired autophagy [93]. Further analyses are needed to clarify how PP2A phosphatases regulate autophagy.
PP2A phosphatases are highly conserved from yeast to humans and share 74% amino acid similarity. Similar to yeast PP2A, human PPP2 are heterotrimeric complexes composed of PPP2R1A (protein phosphatase 2 scaffold subunit Aalpha) or PPP2R1B (protein phosphatase 2 scaffold subunit Abeta), one of 15 regulatory B subunits that comprise 4 subfamilies namely: PPP2R2 (protein phosphatase 2 regulatory subunit B), PPP2R5 (protein phosphatase 2 regulatory subunit B’), PPP2R3 (protein phosphatase 2 regulatory subunit B”) and STRN (striatin), and one catalytic C subunit PPP2CA (protein phosphatase 2 catalytic subunit alpha) or PPP2CB (protein phosphatase 2 catalytic subunit beta) [85,94,95].
Under starvation conditions, the regulatory subunit PPP2R2A binds to the catalytic subunit PPP2CA or PPP2CB, leading to the release of the PPP2 inhibitor IGBP1 (immunoglobulin binding protein 1), a mammalian homolog of Tap42 [55]. The active PPP2R2A-containing PPP2 complex promotes autophagy by specifically dephosphorylating Ser637 of mouse ULK1 (corresponding to Ser638 in human ULK1), a MTORC1 and AMPK kinase target site (Table 2) [54,55,59]. Intriguingly, MTORC1 also phosphorylates ULK1 at Ser757, but the PPP2R2A-containing PPP2 complex is unable to dephosphorylate this site [55]. This observation highlights the possibility that other phosphatases may regulate autophagy induction by dephosphorylation of ULK1.
As the yeast PP2A mutant pph21∆ pph22∆ did not show complete Atg13 dephosphorylation [92], this observation led to an investigation aimed at identifying the other phosphatase(s) involved. The tyrosine protein phosphatase Cdc14 was identified to cooperate in this dephosphorylation event. Cdc14 can dephosphorylate both phospho-Ser/Thr and phospho-Tyr residues and has been extensively studied for its role in cell cycle control [96]. In the absence of Cdc14 phosphatase activity, bulk autophagy is blocked, which is paralleled by hyper-phosphorylated Atg13 [97]. Conversely, hyperactivation of Cdc14 triggers autophagy under growing conditions [97], further supporting a positive role of Cdc14 function in autophagy induction. Ser348, Ser535, Ser541 and Ser646 within Atg13, which are phosphorylated and are mutated in the Atg13[8SA] variant, match the phosphorylation consensus of Cdc14 (Table 1) [97,98]. Cdc14 function in autophagy, however, appears not to be restricted to Atg13 dephosphorylation, as expression of ATG8 and ATG13 [97] also depends on the function of Cdc14, indicating that this phosphatase also participates in the transcriptional regulation of autophagy. Cdc14 has two mammalian homologs, CDC14A and CDC14B, which also regulate the cell cycle [99]. The role of human CDC14 phosphatases in mammalian autophagy, however, remains unknown. As the structure and function of Cdc14 phosphatase are conserved from yeast to humans [100], it is plausible that the human homologs possess a similar role.
In addition to PP2A and Cdc14, the PP2C are a third class of phosphatases acting on Atg1/ULK1 and Atg13. PP2C phosphatases form a class of phosphatases that comprises the yeast phosphatase two C (Ptc) family, which consists of seven members with a highly conserved catalytic domain [101–104]. PP2C phosphatases are monomeric and do not possess regulatory subunits; their activity is regulated by binding to Mn2+ or Mg2+ [10]. By dephosphorylating different substrates, PP2C orchestrate the function of a myriad of proteins, including those involved in the high-osmolarity glycerol pathway [105,106], mitogen-activated protein kinase (MAPK) signaling cascade [106–108], the unfolded protein response [108] and the DNA damage checkpoint [109].
Two of the yeast PP2C family phosphatases, the functionally redundant Ptc2 and Ptc3, play a key role in enhancing autophagy in yeast by directly acting on the Atg1 kinase complex (Figure 2). In particular, rapamycin- or DNA damage-induced autophagy is blocked in the ptc2∆ ptc3∆ double mutant, which also displays hyperphosphorylated Atg13 both in the presence of nutrients and upon rapamycin treatment [110]. Moreover, the autophagy defect of the ptc2∆ ptc3∆ strain is bypassed by overexpression of Atg13[8SA], which cannot be phosphorylated at 8 different residues (described above) [110]. In contrast, expressing Atg13[8SA] from the endogenous ATG13 promoter in ptc2∆ ptc3∆ cells only partially rescued the autophagy impairment, suggesting that Atg13 has additional phosphorylation sites that are targeted by Ptc2 and Ptc3. The authors of this study proposed that Atg13 (S429), between the mutated residues of Atg13[8SA], is probably the site targeted by Ptc2 and Ptc3, as its dephosphorylation is crucial for the Atg13-Atg17 interaction and normal progression of autophagy under starvation conditions [66]. In vitro, in addition to Atg1 (Thr226), Ptc2 dephosphorylates Atg13 (Ser428 and Ser429), promoting PAS organization by triggering the formation of phase-separated liquid droplets consisting of the Atg1 kinase complex (Table 1) [41].
In humans, 16 different genes encode 22 different PPM isozymes, which also arise from alternate splicing [111]. PPM1D-dependent dephosphorylation of mouse ULK1 at Ser637 (corresponding to Ser638 in humans) is reported to be necessary for genotoxin-induced autophagy in mouse embryonic fibroblasts (Table 2) [56]. In particular, mouse ppm1d−/- primary thymocytes showed reduced dephosphorylation of ULK1 at Ser637 and impaired autophagic flux after X-ray irradiation [56]. It should be noted that dephosphorylation of human ULK1 at Ser638 is necessary for starvation-induced autophagy [54]. Thus, an appealing hypothesis is that human PPM1D regulates ULK1 in a similar way in mice.
Phosphoregulation during phagophore nucleation: Atg9/ATG9A
The formation and subsequent activation of the Atg1/ULK kinase complex in the early stages of autophagosome biogenesis are essential for correct phagophore nucleation and its expansion into an autophagosome. Particularly, both Atg1 and ULK1 phosphorylate and thus regulate many core Atg proteins via a defined consensus amino acid sequence (Figure 2) [72,112].
One such core Atg protein that undergoes Atg1/ULK-dependent phosphoregulation is Atg9. Atg9 is the only conserved multi-spanning membrane protein within the core Atg machinery and is present in cytoplasmic vesicles and tubules, which are believed to be involved in the nucleation of the phagophore [113,114]. Yeast Atg9 has been identified as a direct substrate of Atg1 [112]. Peptide arrays combined with quantitative in vivo confirmation revealed six bona fide Atg1 target sites on Atg9 (Ser19, Ser657, Ser802, Ser831, Ser948 and Ser969, see Table 1), which are required for the efficient recruitment of Atg8 and Atg18 to the PAS, and for subsequent formation of the phagophore [112]. Although mutation of these phosphorylation sites did not affect Atg9 vesicle trafficking, abrogation of Atg1 kinase activity resulted in accumulation of Atg9 at the PAS, suggesting that Atg1 kinase regulates Atg9 trafficking by phosphorylating one or more targets [115]. Ser122 of Atg9 is also phosphorylated and this modification is important for Atg9 interactions with both Atg23 and Atg27 and for its trafficking to the PAS [116]. However, mutation of this site into an alanine results in only a slight reduction in autophagy. The kinase responsible for Ser122 phosphorylation remains unknown [116]. In higher eukaryotes, ATG9A trafficking is regulated by the SRC kinase, which directly phosphorylates ATG9A at Tyr8, promoting autophagy [117]. In response to starvation, ULK1 phosphorylates ATG9A at Ser14, and this modification redirects ATG9A vesicles from the trans-Golgi network to endosomes for autophagy initiation (Table 2) [117]. ATG9A is also phosphorylated in its C-terminus on Ser761 by AMPK and ULK1, enhancing its binding to YWHAZ/14-3-3 zeta and thereby promoting autophagosome formation [118].
Phosphoregulation during phagophore nucleation: The PtdIns3K complex I and PtdIns3P
The PtdIns3K complex I generates phosphatidylinositol-3-phosphate (PtdIns3P) on autophagosomal membranes; PtdIns3P is essential for autophagosome biogenesis in both yeast and mammals [119,120]. Under specific conditions, PtdIns3P might be substituted by phosphatidylinositol-5-phosphate, which is synthesized by PIKFYVE (phosphoinositide kinase, FYVE-type zinc finger containing) in mammalian cells (Table 3) [121]. PtdIns3K complex I consists of the phosphatidylinositol 3-kinase Vps34 (vacuole sorting protein 34), the protein kinase Vps15, Vps30/Atg6, Atg14 and Atg38 in yeast, and PIK3C3 (phosphatidylinositol 3-kinase catalytic subunit type 3), PIK3R4 (phosphoinositide-3-kinase regulatory subunit 4), BECN1 (beclin 1), ATG14 and NRBF2 (nuclear receptor binding factor 2) in mammals [119,120].
Table 3.
Phosphoinositides that positively regulate autophagy
| Phosphoinositide | Kinase | Phosphatase | References |
|---|---|---|---|
| Yeast | |||
| PtdIns3P | Vps34 | Ymr1 | [170,224] |
| Inp52 | [170,224] | ||
| Inp53 | [170,224] | ||
| PtdIns4P | Pik1 | Sac1 | [189,191,192,198] |
| Stt4 | Sac1 | [188,191,192,198] | |
| Human | |||
| PtdIns3P | PIK3C3 | MTMR3, MTMR6 | [119,174,175] |
| MTMR14 | |||
| PtdIns4P | PI4K2A | ? | [194,195] |
| PI4KB | SACM1L | [196,198] | |
| PtdIns5P | PIKFYVE | ? | [121] |
Vps30, the yeast BECN1 homolog, is a direct target of Atg1 [53,122], in particular Ser85 [53] (Table 1). This serine is in the poorly conserved N-terminal domain of Vps30, which shows extensive phosphorylation in human BECN1 as well (Table 2). In this region, Ser96 of human BECN1 has been reported to be targeted by ULK1 [72], suggesting that although different residues are modified, Vps30 and BECN1 may be regulated similarly.
The mammalian PtdIns3K complex I appears to be extensively phosphoregulated. Several of its subunits undergo phosphoregulation (Figure 2; Tables 1 and 2). ULK1 and ULK2 phosphorylate ATG14 on Ser29, which enhances PtdIns3K lipid kinase activity [123,124], and MTORC1 negatively regulates ATG14 function via multiple phosphorylation sites (Ser3, Ser223, Thr233, Ser383 and Ser440, see Table 2) [125]. One of these sites, Ser440, is targeted by CDK1 rather than MTORC1 during mitotic autophagy regulation [61]. ULK1 also phosphorylates PIK3C3 at Ser249 [72], as well as the activating molecule in AMBRA1 (autophagy and BECN1 regulator 1) on Ser465 and Ser635 [72]. AMBRA1 has been reported to interact with the PtdIns3K complex via BECN1 and to regulate its association with the ER [126]. PIK3C3 is further phosphorylated on Thr159, which reduces its binding to BECN1 and results in autophagy inhibition. This phosphorylation can be catalyzed by CDK1, but also by the aberrantly activated CDK5p25 variants, which have been associated with neurodegeneration [127]. Moreover, phosphorylation of PIK3C3 (Thr677) by PRKD1 (protein kinase D1) stimulates PtdIns3K activity and autophagy progression [128].
BECN1 is a major hub of phosphoregulation as it is targeted by many kinases, including ULK1, AMPK, DAPK (death associated protein kinase) and many more. The kinase and the corresponding phosphatase have been identified for one phosphorylation site on BECN1 (Ser90; Table 2). Ser90 is phosphorylated by DAPK3 and this modification promotes starvation-induced autophagy [129]. Ser90 is dephosphorylated by PPP2 phosphatases [129]. In particular, the PPP2R2A-containing PPP2 complex is associated with BECN1 in the presence of nutrients, which constitutively dephosphorylates Ser90, inhibiting autophagy. Starvation induces the release of PPP2R2A-containing PPP2 from BECN1, promoting the phosphorylation of Ser90 and consequently autophagy enhancement [129]. This regulation of BECN1 (Ser90), however, appears to be tissue-specific as it is observed in skeletal muscles but not in the heart tissue and liver [129]. Phosphoregulation of BECN1 has been extensively covered in recent reviews [130,131] and will not be discussed in detail here. A summary is provided in Table 2 and Figure 3B.
Atg38 in yeast and NRBF2 in mammals are regulatory components of the autophagy-specific PtdIns3K complex [132–135]. MTORC1-mediated phosphorylation of NRBF2 at Ser113 and Ser120 promotes PtdIns3K complex assembly and activity [136]. Thus, the entire PtdIns3K complex is under heavy phosphoregulation, where phosphorylation both positively and negatively regulates complex assembly and its lipid kinase activity.
PtdIns3P generated by PtdIns3K complex I recruits PtdIns3P-binding proteins such as Atg18 and Atg21 in yeast and their mammalian counterparts, the WIPI proteins, to the site of autophagosome formation [137–139]. WIPI2 stability has been reported to be regulated by phosphorylation, whereby Ser395 modification by MTORC1 promotes an interaction between WIPI2 and the ubiquitin ligase HUWE1 (HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1), thus enhancing its ubiquitination and proteasomal degradation, leading to autophagy inhibition (Table 2) [140]. Phosphorylation-dependent regulation of the yeast protein Atg18 has not been reported to date, although Ser404 is modified by Atg1 in vitro [53]. Atg2, which interacts with Atg18 [141], also contains Atg1 phosphorylation sites [142], but the physiological relevance of these phosphorylations remains unknown.
Phosphoregulation during phagophore expansion
Atg8 protein family members are ubiquitin-like proteins that are essential for the expansion and closure of phagophores. Atg8 proteins are anchored to the phosphatidylethanolamine (PE) present in the autophagosomal membranes through the action of two ubiquitin-like conjugation systems [143]. Yeast Atg16 forms a trimeric complex with the Atg12–Atg5 that oligomerizes and acts as an E3 ligase for the conjugation of Atg8-family proteins to PE [143]. In mammals, the ATG12–ATG5-ATG16L1 complex is recruited to PtdIns3P-positive membranes by WIPI2 [144]. The stability of human ATG16L1 is regulated by phosphorylation of Ser278 [145,146]. ULK1 kinase and CHUK (component of inhibitor of nuclear factor kappa B kinase complex) can phosphorylate this site, although ULK1 appears to be the primary kinase regulating ATG16L1 stability in cultured cells [145,146]. PKA-mediated phosphorylation of ATG16L1 at Ser269 and Ser287 also regulates its stability [147]. Moreover, phosphorylation of ATG16L1 Ser139 by CSNK2 (casein kinase 2) promotes the association of ATG16L1 with the ATG12–ATG5 conjugate (Table 2) and, in turn, autophagy progression [148]. Phospho-Ser139 is dephosphorylated by PPP1 (protein phosphatase 1) [148]. The stability of ATG5 is also phosphoregulated, but by another kinase, the PAK1 (p21 [RAC1]-activated kinase 1). Specifically, phosphorylation of ATG5 (Thr101) by PAK1 protects ATG5 from ubiquitin-dependent degradation and increases the binding of ATG12–ATG5 to ATG16L1, thereby promoting autophagy [149]. In contrast, MAPK14-mediated phosphorylation of Thr75 inhibits ATG5 function [150].
The mammalian Atg8 family proteins MAP1LC3A (microtubule associated protein 1 light chain 3 alpha), MAP1LC3B, MAP1LC3C and GABARAPL2 (GABA type A receptor-associated protein like 2) have also been reported to be phosphoregulated by various kinases throughout autophagy progression (Table 2). TBK1 (TANK binding kinase 1) phosphorylates both MAP1LC3C (on Ser 93 and Ser99) and GABARAPL2 (on Ser87 and Ser88), impeding their binding to ATG4 proteases and therefore preventing their deconjugation from PE and their removal from nascent autophagosomes [151]. PKA directly phosphorylates MAP1LC3A and MAP1LC3B on Ser12, which inhibits their recruitment to autophagosomes and results in autophagy inhibition [152]. MAP1LC3B in turn is phosphorylated at Thr50 by STK3 (serine/threonine kinase 3) and STK4 [153,154]. This phosphorylation positively regulates autophagosome-lysosome fusion, thereby promoting autophagy flux. Thr50 in MAP1LC3B is also targeted by the NEK9 (NIMA related kinase 9) [153]. In contrast to the autophagy-enhancing role of STK3 and STK4, NEK9-mediated phosphorylation of Thr50 results in a block of autophagic SQSTM1 (sequestosome 1) degradation. How phosphorylation of Thr50 mediates these seemingly opposing functions in autophagy requires further investigation. Although Atg8 proteins emerge as a phosphorylation hub in autophagy, no phosphatases have been identified that revert these modifications.
Phosphoregulation during autophagosome maturation
The members of the Atg4 cysteine protease family reversibly regulate the association of Atg8 proteins to lipid bilayers. That is, Atg4 proteases post-translationally prime Atg8 proteins by removing a few C-terminal amino acids, exposing a Gly residue that, upon induction of autophagosome biogenesis, will be conjugated to PE [15]. Upon completion, the Atg8–PE present on the autophagosome surface is deconjugated by Atg4 proteases, releasing Atg8 from autophagosomes and promoting their maturation into fusion-competent vesicles [155–157].
The activity of Atg4 proteases on the growing phagophore is tightly regulated by Atg1 and ULK1. Atg1 phosphorylates Atg4 at Ser307 on the surface of autophagosomes to locally inhibit Atg4 proteolytic activity in yeast (Table 1), thereby protecting the Atg8–PE pool required for autophagosome formation [158]. Upon autophagosome completion, Atg1 inactivation or dissociation permits Atg4 to act on Atg8–PE and thus release Atg8 from its PE anchor, allowing the subsequent fusion of the mature autophagosomes with vacuoles (Figure 2) [158]. ULK1-dependent phosphoregulated inhibition of human ATG4B has also been described, however, not on the residue corresponding to yeast Ser307 [159]. ULK1 phosphorylates ATG4B on Ser316, which inhibits its interaction with Atg8 protein family members and their processing. Intriguingly, the PPP2 phosphatase was found to actively dephosphorylate ATG4B at Ser316, thus providing a reversible phospho-switch mechanism to re-activate the protease [159]. Whether PP2A phosphatases reverse Atg4 phosphorylation in yeast remains to be defined. In contrast, ATG4B activity is positively regulated by phosphorylation on Ser34 by AKT1 and AKT2 [160,161] and on Ser383 by STK26 [162]. Also, phosphorylation at Ser383 and Ser392 that enhanced ATG4B activity has been reported, but the responsible kinases have not been identified [163].
Another phosphatase required for autophagosome maturation is Ymr1 (yeast myotubularin related 1). The YMR1 gene encodes Ymr1, the only yeast member of the highly conserved myotubularin family of PtdIns3P phosphatases. Myotubularin phosphatases contain a characteristic CX5R catalytic motif, which is also found in Tyr, dual specificity and SAC phosphatase domain-containing phosphatases [164–166]. This catalytic motif hydrolyzes D-3 or D-4 phosphate groups on different phosphoinositides, but myotubularins show high specificity toward the D-3 position of PtdIns3P and phosphatidylinositol-3,5-biphosphate (PtdIns [3,5]P2, Table 3) [167]. Ymr1 phosphatase activity is specific for PtdIns3P; it is unable to dephosphorylate other phosphoinositides [168]. Ymr1 plays a key role in regulating PtdIns3P levels and their subcellular localization, and therefore in protein sorting pathways that depend on PtdIns3P, such as the endo-lysosomal system [168,169]. Ymr1 is functionally redundant with two synaptojanin-like proteins, Inp52 (inositol polyphosphate 5-phosphatase 52) and Inp53, which can dephosphorylate multiple types of phosphoinositides (Table 3) [168]. Autophagosome formation requires not only the local biosynthesis of PtdIns3P, by PtdIns3K complex I, but also PtdIns3P dissipation. The conversion of PtdIns3P into phosphatidylinositol (PtdIns) is carried out mainly by Ymr1 [168]. Ymr1 is necessary for the normal progression of autophagy [170], and YMR1 deletion leads to a partial block in autophagy, which is further aggravated by INP53 deletion [170]. This phenotype is phenocopied by cells expressing the Ymr1C397S phosphatase-dead variant, showing that Ymr1 phosphatase activity is indeed required for normal autophagy progression [170]. More precisely, the ymr1∆ mutant accumulates complete autophagosomes in the cytoplasm, which are sealed but they are still decorated with Atg proteins (Figure 2) [170]. Altogether, these data indicated that the clearance of PtdIns3P by Ymr1 is necessary for the normal release of the Atg machinery from complete autophagosomes and that this event is a prerequisite for their subsequent fusion with vacuoles. Interestingly, Caenorhabditis elegans MTM-3 (myotubularin myopathy 3) has the same role in autophagy as Ymr1 [171]. Finally, a very recent study suggested that human MTMR8 (myotubularin-related protein 8) and its Drosophila homolog, dMtmr6, may be the functional counterpart of Ymr1 and MTM-3, since cells and tissues lacking these phosphatases accumulate cytoplasmic autophagosomes [172].
The human myotubularin phosphatase family consists of sixteen genes: MTM1, and MTMR1 to SBF2/MTMR13 and two pseudogenes [169]. Myotubularins specifically bind to and efficiently dephosphorylate PtdIns3P, phosphatidylinositol-3,4-biphosphate and phopshatidylinositol-3,4,5-triphosphate [173]. In higher eukaryotes, PtdIns3P-specific phosphatases such as MTMR3, MTMR6 and MTMR14 have been shown to play a key role during the initial stages of autophagy (Table 3) [174,175]. The overexpression of MTMR3C413S, a dominant-negative mutant, leads to an increase in autophagosome formation in the presence of nutrients but not under starvation conditions. Conversely, overexpression of the wild type variant of this myotubularin reduces autophagic flux under the same conditions, due to formation of smaller autophagosomes [174]. Similarly, depletion of MTMR6 or MTMR14 enhances autophagy flux under starvation conditions. This effect is less prominent in basal growing conditions [174] and is more pronounced in the MTMR14 knockdown cells than the MTMR6 knockdown cells. MTMR14 probably negatively regulates autophagy by inhibiting the association of the PtdIns3P-binding protein WIPI1 with autophagosomal membranes, which in turn affects ATG9A trafficking [175]. Consistently, in vivo knockdown of MTMR14 in zebrafish embryos also increases autophagy flux [176]. Although myotubularin phosphatases seem to also regulate autophagy in mammals, the functional counterpart of Ymr1 has not been identified yet, even if MTMR8 is the presumed candidate. Nonetheless, how PtsIns3P turnover is accomplished upon mammalian autophagosome completion remains to be uncovered.
Phosphoregulation during autophagosome-vacuole/lysosome fusion
The fusion of mature autophagosomes with vacuoles and lysosomes depends soluble N-ethylmaleimide- sensitive factor attachment protein receptor (SNARE) proteins [177,178]. Despite the recent identification of the contributing SNAREs and cofactors, such as tethering factors and RAB GTPases [177,178], little is known about the regulation of this autophagy step. However, recent findings in yeast suggest that this late step of autophagy is also phosphoregulated. Fusion of autophagosomes with the lytic compartment requires the R-SNARE Ykt6 in both yeast and mammals [179–181]. Ykt6 is directly phosphorylated at Ser182 and Ser1832 by Atg1, and another Ytk6 phosphorylation site was identified at Thr158 (Figure 2, Table 1) [53,182,183]. Phosphorylation of these sites inhibits autophagosome-vacuole fusion by preventing Ykt6 bundling with the vacuolar Q-SNAREs Vam3 and Vti1 [182,183]. Therefore, dephosphorylation of Ykt6 on mature autophagosomes is required for their fusion with the vacuole. The responsible phosphatase, however, has not been identified. Interestingly, Ser183 is conserved in mammalian YKT6 (YKT6 v-SNARE homolog), posing the question if YKT6 is also a target of ULK1 in mammals.
Furthermore, PRKCA (protein kinase C alpha) phosphorylates ULK1 on Ser423, which leads to the inhibition of autophagosome-lysosome fusion [184]. PRKCA mediated phosphorylation of ULK1 does not change its kinase activity, but rather it decreases the binding affinity of ULK1 for the autophagosomal SNARE STX17 (syntaxin 17). Unphosphorylated ULK1 recruits STX17 onto autophagosomes, which increases the binding of STX17 to SNAP29 (synaptosome associated protein 29), promoting autophagosome fusion with lysosomes [184]. STX17 also undergoes phosphoregulation, although early during autophagosome formation. Ser202 of STX17 is phosphorylated by TBK1 [185] and this modification promotes the translocation of STX17 from the Golgi to PAS upon starvation. At the PAS, STX17 controls the formation of the ULK1-ATG13-RB1CC1 complex, thereby contributing to autophagy initiation [185].
MTORC1 has been reported to negatively regulate autophagosome-lysosome fusion in mammals by phosphorylating UVRAG (UV radiation resistance associated) product on Ser498 [186]. UVRAG is localized in the ER and endosomes and is known to regulate autophagosome-lysosome fusion by binding to the homotypic fusion and vacuole protein sorting (HOPS) complex. The function of UVRAG in HOPS complex regulation is antagonized by RUBCN (rubicon autophagy regulator), which binds to phosphorylated UVRAG. Therefore, dephosphorylation of UVRAG is required to release this inhibition and allow its binding to the HOPS complex, promoting autophagosome-lysosome fusion [186]. Additionally, MTORC1 negatively regulates autophagosome-lysosome fusion by phosphorylating RUBCNL (RUBCN like autophagy enhancer), a regulator of hepatic autophagy [187]. Phosphorylation of RUBCNL (Ser157) disrupts its association with STX17 and the HOPS complex, thereby preventing autophagosome-lysosome fusion [187].
Another phosphoinositide involved in autophagy is phosphatidylinositol-4-phosphate (PtdIns4P). In yeast, PtdIns4P is synthesized by three kinases, namely the plasma membrane-localized Stt4 (staurosporine and temperature sensitive 4) [188], the Golgi- and nucleus-localized Pik1 (phosphatidylinositol kinase 1) [189] and the vacuole- and plasma membrane-localized Lsb6 (Las seventeen binding protein) [190]. The majority of cellular PtdIns4P is produced by Pik1 and Stt4. Pik1 localized on the Golgi is essential for vesicle-mediated sorting from this organelle, including that of Atg9 since the inactivation of PIK1 causes an impairment of Atg9 vesicle exit from the Golgi, which in turn leads to a block of autophagy [191,192]. Interestingly, Stt4 is also involved in autophagy but the PtsIns4P pool generated by this kinase plays an important role in autophagosome-vacuole fusion [191,192]. This difference in contribution to autophagy has been assigned to Pik1 synthesizing the PtdIns4P ending in the luminal surface of autophagosomes, and Stt4 producing the PtdIns4P decorating the surface of autophagosomes [192].
Similar to yeast, mammals require PtdIns4P for normal progression of autophagy. In mammals, PtdIns4P is synthesized by four kinases, two of which, PI4K2A (phosphatidylinositol 4-kinase type 2 alpha) and PI4KB (phosphatidylinositol 4-kinase beta), localize on the Golgi apparatus (Table 3) [193]. PI4K2A is also recruited onto autophagosomes by associating with GABARAP (GABA type A receptor-associated protein) proteins to produce PtdIns4P locally, which is required to promote autophagosome-lysosome fusion [194,195]. PI4KB, in contrast, is present on the ATG9A-positive compartments that are transported from the Golgi to the PAS under amino acid starvation conditions [196]. At the PAS, PI4KB interacts with ATG13 suggesting a role in the initial stages of autophagosome formation, possibly participating in the assembly of ATG machinery via the synthesis of PtsIns4P [196].
The PtdIns4P phosphatase, Sac1 (suppressor of actin), has very recently been shown to play a role in autophagosome-vacuole fusion [197]. Sac1 is a type-II transmembrane protein that predominantly localizes to the ER and Golgi [197]. Sac1 was identified in a high-throughput screen for autophagy defective mutants in yeast [198]. In the absence of SAC1, an aberrant increase in PtdIns4P levels in the Golgi leads to Atg9 vesicles carrying PtdIns4P and, consequently, to an abnormal incorporation of this lipid into autophagosomes (Table 3) [198]. This abrogates autophagosome-vacuole fusion by blocking the recruitment of SNAREs, such as Vam7 (vacuolar morphogenesis 7) and Ykt6, causing an accumulation of complete autophagosomes in the cytoplasm [198]. A similar autophagy defect was observed upon knockdown of its homolog in C. elegans embryos or in HeLa human cervical cancer cells [198]. Another report, however, reached a different conclusion about the role of human SACM1L (SAC1 like phosphatidylinositide phosphatase) in autophagy [199]. In particular, knockdown of SAC1 phosphatase enhanced the autophagic flux in African green monkey kidney fibroblast-like COS-7 cells [199]. TMEM39A (transmembrane protein 39A) is an ER transmembrane protein that binds SACM1L and COPII-coated vesicles, acting as an adapter that allows SAC1 trafficking to early Golgi compartments, where it dephosphorylates PtdIns4P into PtdIns. Knockdown of TMEM39A leads to an arrest of SACM1L in the ER and a concomitant increase in PtdIns4P levels in both late endosomes and lysosomes. This enhances the recruitment of the HOPS complex, which in turn facilitates the assembly of the SNARE complex that is involved in autophagosome fusion with lysosomes [199]. Additional investigations are needed to understand the exact role of SACM1L phosphatases in autophagy.
Conclusions
Although this review mainly focused on the kinases and phosphatases that directly phosphorylate and dephosphorylate the core Atg machinery, there are several other kinases and phosphatases that act upstream of the Atg1/ULK kinase complex and regulate autophagy, also at the transcriptional level.
Over the years, research has revealed that autophagy is central for many pathophysiological functions. The proper functioning of kinases and the correct phosphorylation of their targets have emerged as integral for the whole autophagy process. For this reason, kinase inhibitors that act on autophagy kinases have gained interest, and several potential kinase inhibitors to target autophagy have been reported. These reports were recently reviewed and discussed elsewhere [200]. Despite a central role in regulating a multitude of signaling pathways, the promiscuity of phosphatases has made them an unattractive target for drug discovery [201,202]. However, the recent discovery that the pharmacological modulation of their regulatory subunits allows the specific regulation of phosphatases has changed this perception [201,202]. Similarly, the role of phosphatases in autophagy has been only marginally investigated due to the notion that they are highly redundant. Recent studies, however, have revealed that different phosphatases play specific roles in the regulation of the diverse steps of autophagy. Specific inhibition of these phosphatases by targeting regulatory subunits or motifs present on the substrate would be desirable.
The studies conducted so far have highlighted the regulatory significance of several phosphosites on the core Atg machinery. Numerous kinases and a few counteracting phosphatases have been shown to act in concert to fine-tune autophagy in response to the nutritional or environmental cues. In particular, ULK1 and BECN1 have emerged as signaling hubs undergoing extensive phosphoregulation (Figure 3). All these studies have likely revealed only the tip of the iceberg, as a recent systematic and comprehensive analysis in yeast has revealed that most components of the core Atg machinery are phosphorylated by Atg1 and TORC1, but also additional other kinases [53]. Thus, the phosphoregulation of autophagy remains to be comprehensively decoded. Advances in this direction will be important to develop better molecules for the optimal and appropriate modulation of autophagy in specific pathological situations.
Acknowledgements
All the authors are part of the DRIVE consortium, which is supported by a Marie Skłodowska-Curie ETN grant under the European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement No 765912). The Kraft laboratory has also received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), SFB 1381 (Project-ID 403222702), SFB 1177 (Project-ID 259130777), Germany’s Excellence Strategy (CIBSS - EXC-2189- Project ID 390939984) and the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement No 769065. F.R. is also supported by ALW Open Programme (ALWOP.310), ENW KLEIN-1 (OCENW.KLEIN.118) and ZonMW TOP (91217002) grants. This work reflects only the authors’ view and the European Union’s Horizon 2020 research and innovation programme is not responsible for any use that may be made of the information it contains. The authors declare no competing financial interests.
Funding Statement
This work was supported by the H2020 European Research Council [769065]; NWO [OCENW.KLEIN.118]; NWO [ALWOP.310]; DFG Germany’s Excellence Strategy [390939984]; H2020 Marie Skłodowska-Curie Actions [765912]; DFG SBF 1177 [259130777]; DFG SFB 1381 [403222702]; H2020 Marie Skłodowska-Curie Actions [765912]; ZonMw [91217002].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Xu H, Wang Y, Lin S, et al. PTMD: a database of human disease-associated post-translational modifications. Genomics, Proteomics & Bioinformatics. 2018;16(4):244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Deribe YL, Pawson T, Dikic I.. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17(6):666–672. [DOI] [PubMed] [Google Scholar]
- [3].Wang YC, Peterson SE, Loring JF.. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014;24(2):143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2012;367(1602):2513–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Karthikeyan S, Zhou Q, Osterman AL, et al. Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism. Biochemistry. 2003;42(43):12532–12538. [DOI] [PubMed] [Google Scholar]
- [6].Topolcan O, Holubec L Jr.. The role of thymidine kinase in cancer diseases. Expert Opin Med Diagn. 2008;2(2):129–141. [DOI] [PubMed] [Google Scholar]
- [7].Heath CM, Stahl PD, Barbieri MA. Lipid kinases play crucial and multiple roles in membrane trafficking and signaling. Histol Histopathol. 2003;18(3):989–998. [DOI] [PubMed] [Google Scholar]
- [8].Fabbro D, Cowan-Jacob SW, Moebitz H. Ten things you should know about protein kinases: IUPHAR review 14. Br J Pharmacol. 2015;172:2675–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li X, Wilmanns M, Thornton J, et al. Elucidating human phosphatase-substrate networks. Sci Signal. 2013;6(275):rs10. [DOI] [PubMed] [Google Scholar]
- [10].Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58(1):453–508. [DOI] [PubMed] [Google Scholar]
- [11].Roy J, Cyert MS. Cracking the phosphatase code: docking interactions determine substrate specificity. Sci Signal. 2009;2(100):re9. [DOI] [PubMed] [Google Scholar]
- [12].Sacco F, Perfetto L, Castagnoli L, et al. The human phosphatase interactome: an intricate family portrait. FEBS Lett. 2012;586(17):2732–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139(3):468–484. [DOI] [PubMed] [Google Scholar]
- [14].Hardman G, Perkins S, Brownridge PJ, et al. Strong anion exchange-mediated phosphoproteomics reveals extensive human non-canonical phosphorylation. Embo J. 2019;38(21):e100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol. 2020;21:439–458. [DOI] [PubMed] [Google Scholar]
- [16].Jing K, Lim K. Why is autophagy important in human diseases? Exp Mol Med. 2012;44(2):69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Van Beek N, Klionsky DJ, Reggiori F. Genetic aberrations in macroautophagy genes leading to diseases. Biochim Biophys Acta. 2018;1865(5):803–816. [DOI] [PubMed] [Google Scholar]
- [20].Kirkin V, Rogov VV. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76(2):268–285. [DOI] [PubMed] [Google Scholar]
- [21].Gonzalez A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. Embo J. 2017;36(4):397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1(1):11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Klionsky DJ, Cregg JM, Dunn WA Jr., et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–545. [DOI] [PubMed] [Google Scholar]
- [24].Kim J, Huang W-P, Stromhaug PE, et al. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277(1):763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Suzuki K, Akioka M, Kondo-Kakuta C, et al. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126(11):2534–2544. [DOI] [PubMed] [Google Scholar]
- [26].Suzuki K, Kirisako T, Kamada Y, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. Embo J. 2001;20(21):5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Graef M, Friedman JR, Graham C, et al. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24(18):2918–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yla-Anttila P, Vihinen H, Jokitalo E, et al. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5(8):1180–1185. [DOI] [PubMed] [Google Scholar]
- [30].Hayashi-Nishino M, Fujita N, Noda T, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11(12):1433–1437. [DOI] [PubMed] [Google Scholar]
- [31].Hollenstein DM, Gomez-Sanchez R, Ciftci A, et al. Vac8 spatially confines autophagosome formation at the vacuole in S. cerevisiae. J Cell Sci. 2019;132(22):jcs235002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. [DOI] [PubMed] [Google Scholar]
- [33].Matsuura A, Tsukada M, Wada Y, et al. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192(2):245–250. [DOI] [PubMed] [Google Scholar]
- [34].Torggler R, Papinski D, Brach T, et al. Two Independent pathways within selective autophagy converge to activate Atg1 kinase at the vacuole. Mol Cell. 2016;64(2):221–235. [DOI] [PubMed] [Google Scholar]
- [35].Kawamata T, Kamada Y, Kabeya Y, et al. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19(5):2039–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yamamoto H, Fujioka Y, Suzuki SW, et al. The Intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev Cell. 2016;38(1):86–99. [DOI] [PubMed] [Google Scholar]
- [38].Kabeya Y, Kamada Y, Baba M, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16(5):2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kijanska M, Dohnal I, Reiter W, et al. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010;6(8):1168–1178. [DOI] [PubMed] [Google Scholar]
- [40].Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in saccharomyces cerevisiae. Genetics. 2010;185(3):871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fujioka Y, Alam JM, Noshiro D, et al. Phase separation organizes the site of autophagosome formation. Nature. 2020;578(7794):301–305. [DOI] [PubMed] [Google Scholar]
- [42].Young AR, Chan E, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119(18):3888–3900. [DOI] [PubMed] [Google Scholar]
- [43].Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy [research support, non-U.S. Gov’t]. Mol Biol Cell. 2009;20(7):1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ganley IG, Lam Du H, Wang J, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ravenhill BJ, Boyle KB, Von Muhlinen N, et al. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol Cell. 2019;74(2):320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lazarus MB, Novotny CJ, Shokat KM. Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem Biol. 2015;10(1):257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dorsey FC, Rose KL, Coenen S, et al. Mapping the phosphorylation sites of Ulk1. J Proteome Res. 2009;8(11):5253–5263. [DOI] [PubMed] [Google Scholar]
- [50].Liu CC, Lin YC, Chen YH, et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell. 2016;61(1):84–97. [DOI] [PubMed] [Google Scholar]
- [51].Budovskaya YV, Stephan JS, Deminoff SJ, et al. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 2005;102(39):13933–13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yeh YY, Shah KH, Chou CC, et al. The identification and analysis of phosphorylation sites on the Atg1 protein kinase. Autophagy. 2011;7(7):716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hu Z, Raucci S, Jaquenoud M, et al. Multilayered control of protein turnover by TORC1 and Atg1. Cell Rep. 2019;28(13):3486–3496. [DOI] [PubMed] [Google Scholar]
- [54].Shang L, Chen S, Du F, et al. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108(12):4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wong PM, Feng Y, Wang J, et al. Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nat Commun. 2015;6(1):8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Torii S, Yoshida T, Arakawa S, et al. Identification of PPM1D as an essential Ulk1 phosphatase for genotoxic stress-induced autophagy. EMBO Rep. 2016;17(11):1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Torii S, Yamaguchi H, Nakanishi A, et al. Identification of a phosphorylation site on Ulk1 required for genotoxic stress-induced alternative autophagy. Nat Commun. 2020;11(1):1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1 [research support, N.I.H., Extramural]. Nat Cell Biol. 2011;13(2):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mack HI, Zheng B, Asara J, et al. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8(8):1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Odle RI, Walker SA, Oxley D, et al. An mTORC1-to-CDK1 switch maintains autophagy suppression during mitosis. Mol Cell. 2020;77(2):228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li Z, Tian X, Ji X, et al. ULK1-ATG13 and their mitotic phospho-regulation by CDK1 connect autophagy to cell cycle. PLoS Biol. 2020;18(6):e3000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lu H, Xiao J, Ke C, et al. TOPK inhibits autophagy by phosphorylating ULK1 and promotes glioma resistance to TMZ. Cell Death Dis. 2019;10(8):583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bach M, Larance M, James DE, et al. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011;440(2):283–291. [DOI] [PubMed] [Google Scholar]
- [65].Kamada Y, Yoshino K, Kondo C, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fujioka Y, Suzuki SW, Yamamoto H, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21(6):513–521. [DOI] [PubMed] [Google Scholar]
- [67].Rao Y, Perna MG, Hofmann B, et al. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat Commun. 2016;7(1):10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Stephan JS, Yeh YY, Ramachandran V, et al. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106(40):17049–17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5(5):649–662. [DOI] [PubMed] [Google Scholar]
- [70].Puente C, Hendrickson RC, Jiang X. Nutrient-regulated phosphorylation of ATG13 inhibits starvation-induced autophagy. J Biol Chem. 2016;291(11):6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Joo JH, Dorsey FC, Joshi A, et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell. 2011;43(4):572–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Egan DF, Chun MG, Vamos M, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59(2):285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mao K, Chew LH, Inoue-Aono Y, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A. 2013;110(31):E2875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yao W, Li Y, Wu L, et al. Atg11 is required for initiation of glucose starvation-induced autophagy. Autophagy. 2020;16(12):2206–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Feng W, Wu T, Dan X, et al. Phosphorylation of Atg31 is required for autophagy. Protein Cell. 2015;6(4):288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chen S, Wang C, Yeo S, et al. Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev. 2016;30(7):856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5(1):85–87. [DOI] [PubMed] [Google Scholar]
- [78].Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in arabidopsis. Plant Cell. 2014;26(2):788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Steffan JS. Does Huntingtin play a role in selective macroautophagy? Cell Cycle. 2010;9(17):3401–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Nishimura T, Tamura N, Kono N, et al. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. Embo J. 2017;36(12):1719–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Turco E, Witt M, Abert C, et al. FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol Cell. 2019;74(2):330–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Vargas JNS, Wang C, Bunker E, et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell. 2019;74(2):347–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Smith MD, Harley ME, Kemp AJ, et al. CCPG1 Is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell. 2018;44(2):217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Suzuki H, Kaizuka T, Mizushima N, et al. Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat Struct Mol Biol. 2015;22(7):572–580. [DOI] [PubMed] [Google Scholar]
- [85].Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353(3):417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ronne H, Carlberg M, Hu GZ, et al. Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis Mol Cell Biol. 1991;11(10):4876–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lin FC, Arndt KT. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis Embo J. 1995;14(12):2745–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wei H, Ashby DG, Moreno CS, et al. Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J Biol Chem. 2001;276(2):1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zabrocki P, Van Hoof C, Goris J, et al. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol Microbiol. 2002;43(4):835–842. [DOI] [PubMed] [Google Scholar]
- [90].Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10(15):1904–1916. [DOI] [PubMed] [Google Scholar]
- [91].Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. Embo J. 1999;18(10):2782–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Yeasmin AM, Waliullah TM, Kondo A, et al. Orchestrated action of PP2A antagonizes Atg13 phosphorylation and promotes autophagy after the inactivation of TORC1. PLoS One. 2016;11(12):e0166636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yorimitsu T, He C, Wang K, et al. Tap42-associated protein phosphatase type 2A negatively regulates induction of autophagy. Autophagy. 2009;5(5):616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Arino J, Velazquez D, Casamayor A. Ser/Thr protein phosphatases in fungi: structure, regulation and function. Microb Cell. 2019;6(5):217–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Schuhmacher D, Sontag JM, Sontag E. Protein phosphatase 2A: more than a passenger in the regulation of epithelial cell-cell junctions. Front Cell Dev Biol. 2019;7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Taylor GS, Liu Y, Baskerville C, et al. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem. 1997;272(38):24054–24063. [DOI] [PubMed] [Google Scholar]
- [97].Kondo A, Mostofa MG, Miyake K, et al. Cdc14 phosphatase promotes TORC1-regulated autophagy in yeast. J Mol Biol. 2018;430(11):1671–1684. [DOI] [PubMed] [Google Scholar]
- [98].Gray CH, Good V, Tonks N, et al. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. Embo J. 2003;22(14):3524–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Manzano-Lopez J, Monje-Casas F. The multiple roles of the Cdc14 phosphatase in cell cycle control. Int J Mol Sci. 2020;21(3):709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Li L, Ernsting BR, Wishart MJ, et al. A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J Biol Chem. 1997;272(47):29403–29406. [DOI] [PubMed] [Google Scholar]
- [101].Ruan H, Yan Z, Sun H, et al. The YCR079w gene confers a rapamycin-resistant function and encodes the sixth type 2C protein phosphatase in Saccharomyces cerevisiae. FEMS Yeast Res. 2007;7(2):209–215. [DOI] [PubMed] [Google Scholar]
- [102].Jiang L, Whiteway M, Ramos C, et al. The YHR076w gene encodes a type 2C protein phosphatase and represents the seventh PP2C gene in budding yeast. FEBS Lett. 2002;527(1–3):323–325. [DOI] [PubMed] [Google Scholar]
- [103].Cheng A, Ross KE, Kaldis P, et al. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 1999;13(22):2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Arino J, Casamayor A, Gonzalez A. Type 2C protein phosphatases in fungi. Eukaryot Cell. 2011;10(1):21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Young C, Mapes J, Hanneman J, et al. Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaryot Cell. 2002;1(6):1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Warmka J, Hanneman J, Lee J, et al. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol Cell Biol. 2001;21(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369(6477):242–245. [DOI] [PubMed] [Google Scholar]
- [108].Welihinda AA, Tirasophon W, Green SR, et al. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol. 1998;18(4):1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Leroy C, Lee SE, Vaze MB, et al. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol Cell. 2003;11(3):827–835. [DOI] [PubMed] [Google Scholar]
- [110].Memisoglu G, Eapen VV, Yang Y, et al. PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Proc Natl Acad Sci U S A. 2019;116(5):1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Lammers T, Lavi S. Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit Rev Biochem Mol Biol. 2007;42(6):437–461. [DOI] [PubMed] [Google Scholar]
- [112].Papinski D, Schuschnig M, Reiter W, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53(3):471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Mari M, Griffith J, Rieter E, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190(6):1005–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Reggiori F, Tucker KA, Stromhaug PE, et al. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6(1):79–90. [DOI] [PubMed] [Google Scholar]
- [116].Feng Y, Backues SK, Baba M, et al. Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy. 2016;12(4):648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhou C, Ma K, Gao R, et al. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017;27(2):184–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Weerasekara VK, Panek DJ, Broadbent DG, et al. Metabolic-stress-induced rearrangement of the 14-3-3zeta interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3zeta interaction with phosphorylated Atg9. Mol Cell Biol. 2014;34(24):4379–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. [DOI] [PubMed] [Google Scholar]
- [120].Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6(6):764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Vicinanza M, Korolchuk VI, Ashkenazi A, et al. PI(5)P regulates autophagosome biogenesis. Mol Cell. 2015;57(2):219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell. 2015;59(3):372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Park JM, Jung CH, Seo M, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12(3):547–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Wold MS, Lim J, Lachance V, et al. ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington’s disease models. Mol Neurodegener. 2016;11(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9(12):1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy [research support, non-U.S. Gov’t]. J Cell Biol. 2010;191(1):155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Furuya T, Kim M, Lipinski M, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 2010;38(4):500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012;19(5):788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Fujiwara N, Usui T, Ohama T, et al. Regulation of Beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3 (DAPK3). J Biol Chem. 2016;291(20):10858–10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Menon MB, Dhamija S. Beclin 1 phosphorylation - at the center of autophagy regulation. Front Cell Dev Biol. 2018;6:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Hill SM, Wrobel L, Rubinsztein DC. Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation. Cell Death Differ. 2019;26(4):617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Lu J, He L, Behrends C, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun. 2014;5(1):3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Zhong Y, Morris DH, Jin L, et al. Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J Biol Chem. 2014;289(38):26021–26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Araki Y, Ku WC, Akioka M, et al. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203(2):299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Cao Y, Wang Y, Abi Saab WF, et al. NRBF2 regulates macroautophagy as a component of Vps34 complex I. Biochem J. 2014;461(2):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ma X, Zhang S, He L, et al. MTORC1-mediated NRBF2 phosphorylation functions as a switch for the class III PtdIns3K and autophagy. Autophagy. 2017;13(3):592–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Proikas-Cezanne T, Waddell S, Gaugel A, et al. WIPI-1a (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23(58):9314–9325. [DOI] [PubMed] [Google Scholar]
- [138].Polson HE, De Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6(4):506–522. [DOI] [PubMed] [Google Scholar]
- [139].Krick R, Tolstrup J, Appelles A, et al. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006;580(19):4632–4638. [DOI] [PubMed] [Google Scholar]
- [140].Wan W, You Z, Zhou L, et al. mTORC1-regulated and HUWE1-mediated WIPI2 degradation controls autophagy flux. Mol Cell. 2018;72(2):303–315. [DOI] [PubMed] [Google Scholar]
- [141].Obara K, Sekito T, Niimi K, et al. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283(35):23972–23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Papinski D, Kraft C. Atg1 kinase organizes autophagosome formation by phosphorylating Atg9. Autophagy. 2014;10(7):1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2020;63:1–10. [DOI] [PubMed] [Google Scholar]
- [144].Dooley HC, Razi M, Polson HE, et al. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55(2):238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Diamanti MA, Gupta J, Bennecke M, et al. IKKalpha controls ATG16L1 degradation to prevent ER stress during inflammation. J Exp Med. 2017;214(2):423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Alsaadi RM, Losier TT, Tian W, et al. ULK1-mediated phosphorylation of ATG16L1 promotes xenophagy, but destabilizes the ATG16L1 Crohn’s mutant. EMBO Rep. 2019;20(7):e46885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Zhao X, Nedvetsky P, Stanchi F, et al. Endothelial PKA activity regulates angiogenesis by limiting autophagy through phosphorylation of ATG16L1. Elife. 2019;8:e46380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Song H, Pu J, Wang L, et al. ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation. Autophagy. 2015;11(8):1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Feng X, Zhang H, Meng L, et al. Hypoxia-induced acetylation of PAK1 enhances autophagy and promotes brain tumorigenesis via phosphorylating ATG5. Autophagy. 2021;17(3):723–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Keil E, Hocker R, Schuster M, et al. Phosphorylation of Atg5 by the Gadd45beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ. 2013;20(2):321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Herhaus L, Bhaskara RM, Lystad AH, et al. TBK1-mediated phosphorylation of LC3C and GABARAP-L2 controls autophagosome shedding by ATG4 protease. EMBO Rep. 2020;21(1):e48317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Cherra SJ 3rd, Kulich SM, Uechi G, et al. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190(4):533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Shrestha BK, Skytte Rasmussen M, Abudu YP, et al. NIMA-related kinase 9-mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1. J Biol Chem. 2020;295(5):1240–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wilkinson DS, Jariwala JS, Anderson E, et al. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol Cell. 2015;57(1):55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Nakatogawa H, Ishii J, Asai E, et al. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis [Research Support, Non-U.S. Gov’t]. Autophagy. 2012;8(2):177–186. [DOI] [PubMed] [Google Scholar]
- [156].Abreu S, Kriegenburg F, Gómez‐Sánchez R, et al. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO reports. 2017;18(5):765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Nair U, Yen WL, Mari M, et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8(5):780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Sanchez-Wandelmer J, Kriegenburg F, Rohringer S, et al. Atg4 proteolytic activity can be inhibited by Atg1 phosphorylation. Nat Commun. 2017;8(1):295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Pengo N, Agrotis A, Prak K, et al. A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat Commun. 2017;8(1):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Ni Z, He J, Wu Y, et al. AKT-mediated phosphorylation of ATG4B impairs mitochondrial activity and enhances the Warburg effect in hepatocellular carcinoma cells. Autophagy. 2018;14(4):685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Pengo N, Prak K, Costa JR, et al. Identification of kinases and phosphatases that regulate ATG4B activity by siRNA and small molecule screening in cells. Front Cell Dev Biol. 2018;6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Huang T, Kim CK, Alvarez AA, et al. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell. 2017;32(6):840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Yang Z, Wilkie-Grantham RP, Yanagi T, et al. ATG4B (Autophagin-1) phosphorylation modulates autophagy. J Biol Chem. 2015;290(44):26549–26561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Kerk D, Moorhead GB. A phylogenetic survey of myotubularin genes of eukaryotes: distribution, protein structure, evolution, and gene expression. BMC Evol Biol. 2010;10(1):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Hughes WE, Cooke FT, Parker PJ. Sac phosphatase domain proteins. Biochem J. 2000;350(2):337–352. [PMC free article] [PubMed] [Google Scholar]
- [166].Nandurkar HH, Huysmans R. The myotubularin family: novel phosphoinositide regulators. IUBMB Life. 2002;53(1):37–43. [DOI] [PubMed] [Google Scholar]
- [167].Schaletzky J, Dove SK, Short B, et al. Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr Biol. 2003;13(6):504–509. [DOI] [PubMed] [Google Scholar]
- [168].Parrish WR, Stefan CJ, Emr SD. Essential role for the myotubularin-related phosphatase Ymr1p and the synaptojanin-like phosphatases Sjl2p and Sjl3p in regulation of phosphatidylinositol 3-phosphate in yeast. Mol Biol Cell. 2004;15(8):3567–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Laporte J, Bedez F, Bolino A, et al. Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet. 2003;12(2):R285–92. [DOI] [PubMed] [Google Scholar]
- [170].Cebollero E, Van Der Vaart A, Zhao M, et al. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012;22(17):1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Wu Y, Cheng S, Zhao H, et al. PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep. 2014;15(9):973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Allen EA, Amato C, Fortier TM, et al. A conserved myotubularin-related phosphatase regulates autophagy by maintaining autophagic flux. J Cell Biol. 2020;219(11):e201909073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Itoh T, Takenawa T. Phosphoinositide-binding domains: functional units for temporal and spatial regulation of intracellular signalling. Cell Signal. 2002;14(9):733–743. [DOI] [PubMed] [Google Scholar]
- [174].Taguchi-Atarashi N, Hamasaki M, Matsunaga K, et al. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11(4):468–478. [DOI] [PubMed] [Google Scholar]
- [175].Vergne I, Roberts E, Elmaoued RA, et al. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. Embo J. 2009;28(15):2244–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Gibbs EM, Feldman EL, Dowling JJ. The role of MTMR14 in autophagy and in muscle disease. Autophagy. 2010;6(6):819–820. [DOI] [PubMed] [Google Scholar]
- [177].Reggiori F, Ungermann C. Autophagosome maturation and fusion. J Mol Biol. 2017;429(4):486–496. [DOI] [PubMed] [Google Scholar]
- [178].Zhao YG, Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. J Cell Biol. 2019;218(3):757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Matsui T, Jiang P, Nakano S, et al. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol. 2018;217(8):2633–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Bas L, Papinski D, Licheva M, et al. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J Cell Biol. 2018;217(10):3656–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Gao J, Reggiori F, Ungermann C. A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J Cell Biol. 2018;217(10):3670–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Gao J, Kurre R, Rose J, et al. Function of the SNARE Ykt6 on autophagosomes requires the Dsl1 complex and the Atg1 kinase complex. EMBO Rep. 2020;21(12):e50733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Barz S, Kriegenburg F, Henning A, et al. Atg1 kinase regulates autophagosome-vacuole fusion by controlling SNARE bundling. EMBO Rep. 2020;21(12):e51869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Wang C, Wang H, Zhang D, et al. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat Commun. 2018;9(1):3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Kumar S, Gu Y, Abudu YP, et al. Phosphorylation of syntaxin 17 by TBK1 controls autophagy initiation. Dev Cell. 2019;49(1):130–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Kim YM, Jung CH, Seo M, et al. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell. 2015;57(2):207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Cheng X, Ma X, Ding X, et al. Pacer mediates the function of class III PI3K and HOPS complexes in autophagosome maturation by engaging Stx17. Mol Cell. 2017;65(6):1029–1043. [DOI] [PubMed] [Google Scholar]
- [188].Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2(5):593–605. [DOI] [PubMed] [Google Scholar]
- [189].Strahl T, Hama H, DeWald DB, et al. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171(6):967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Han GS, Audhya A, Markley DJ, et al. The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. J Biol Chem. 2002;277(49):47709–47718. [DOI] [PubMed] [Google Scholar]
- [191].Wang K, Yang Z, Liu X, et al. Phosphatidylinositol 4-kinases are required for autophagic membrane trafficking. J Biol Chem. 2012;287(45):37964–37972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Kurokawa Y, Konishi R, Yoshida A, et al. Essential and distinct roles of phosphatidylinositol 4-kinases, Pik1p and Stt4p, in yeast autophagy. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(9):1214–1225. [DOI] [PubMed] [Google Scholar]
- [193].Boura E, Nencka R. Phosphatidylinositol 4-kinases: function, structure, and inhibition. Exp Cell Res. 2015;337(2):136–145. [DOI] [PubMed] [Google Scholar]
- [194].Wang H, Sun HQ, Zhu X, et al. GABARAPs regulate PI4P-dependent autophagosome: lysosomefusion. Proc Natl Acad Sci U S A. 2015;112(22):7015–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Kurokawa Y, Yoshida A, Fujii E, et al. Phosphatidylinositol 4-phosphate on Rab7-positive autophagosomes revealed by the freeze-fracture replica labeling. Traffic. 2019;20(1):82–95. [DOI] [PubMed] [Google Scholar]
- [196].Judith D, Jefferies HBJ, Boeing S, et al. ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase IIIbeta. J Cell Biol. 2019;218(5):1634–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Del Bel LM, Brill JA. Sac1, a lipid phosphatase at the interface of vesicular and nonvesicular transport. Traffic. 2018;19(5):301–318. [DOI] [PubMed] [Google Scholar]
- [198].Zhang H, Zhou J, Xiao P, et al. PtdIns4P restriction by hydrolase SAC1 decides specific fusion of autophagosomes with lysosomes. Autophagy. in press 2020;1–11. DOI: 10.1080/15548627.2020.1796321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Miao G, Zhang Y, Chen D, et al. The ER-localized transmembrane protein TMEM39A/SUSR2 regulates autophagy by controlling the trafficking of the PtdIns(4)P phosphatase SAC1. Mol Cell. 2020;77(3):618–632. [DOI] [PubMed] [Google Scholar]
- [200].Xiang H, Zhang J, Lin C, et al. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm Sin B. 2020;10(4):569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Kohn M. Turn and Face the Strange: a New View on Phosphatases. ACS Cent Sci. 2020;6(4):467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Bertolotti A. The split protein phosphatase system. Biochem J. 2018;475(23):3707–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282(17):13123–13132. [DOI] [PubMed] [Google Scholar]
- [204].Furuya N, Yu J, Byfield M, et al. The evolutionarily conserved domain of beclin 1 is required for Vps34 binding, autophagy, and tumor suppressor function. Autophagy. 2005;1(1):46–52. [DOI] [PubMed] [Google Scholar]
- [205].Li X, He L, Che KH, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3(1):662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Birgisdottir AB, Mouilleron S, Bhujabal Z, et al. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy. 2019;15(8):1333–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Park JM, Seo M, Jung CH, et al. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy. 2018;14(4):584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Kumar A, Shaha C. SESN2 facilitates mitophagy by helping Parkin translocation through ULK1 mediated Beclin1 phosphorylation. Sci Rep. 2018;8(1):615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Qian X, Li X, Cai Q, et al. Phosphoglycerate kinase 1 phosphorylates Beclin1 to Induce autophagy. Mol Cell. 2017;65(5):917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Wei Y, An Z, Zou Z, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife. 2015;4:e05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Li X, Wu XQ, Deng R, et al. CaMKII-mediated Beclin 1 phosphorylation regulates autophagy that promotes degradation of Id and neuroblastoma cell differentiation. Nat Commun. 2017;8(1):1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Kim J, Kim YC, Fang C, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1–2):290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Maejima Y, Kyoi S, Zhai P, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19(11):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Shiloh R, Gilad Y, Ber Y, et al. Non-canonical activation of DAPK2 by AMPK constitutes a new pathway linking metabolic stress to autophagy. Nat Commun. 2018;9(1):1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Zalckvar E, Berissi H, Mizrachy L, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10(3):285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Gurkar AU, Chu K, Raj L, et al. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun. 2013;4(1):2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Wei Y, Zou Z, Becker N, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154(6):1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Cheng Z, Zhu Q, Dee R, et al. Focal adhesion kinase-mediated phosphorylation of beclin1 protein suppresses cardiomyocyte autophagy and initiates hypertrophic growth. J Biol Chem. 2017;292(6):2065–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Yu C, Gorantla SP, Muller-Rudorf A, et al. Phosphorylation of BECLIN-1 by BCR-ABL suppresses autophagy in chronic myeloid leukemia. Haematologica. 2020;105(5):1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Wang RC, Wei Y, An Z, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338(6109):956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Zhang D, Wang W, Sun X, et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy. 2016;12(9):1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Sun T, Li X, Zhang P, et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6(1):7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-Kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17(4):1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]


