Abstract
The degree of DNA banding pattern polymorphism exhibited by vancomycin-resistant Enterococcus faecium (VREM) strains isolated on a renal unit over an 11-month period was investigated. Thirty VREM strains from different patients were analyzed by pulsed-field gel electrophoresis (PFGE; with extended run and optimal pulse times), ribotyping, plasmid profile analysis, biotyping, pyrolysis mass spectrometry, and antibiogram analysis. PFGE resolved 17 banding patterns which formed four distinct clusters at the 82% similarity level. Intercluster band differences ranged from 14 to 31 bands. The strains in one cluster, which contained seven patterns that differed from each other by one to seven bands and from the common pattern by five bands, were confirmed to be a single strain by four of the five other typing methods. The strains in a second cluster with eight patterns, which differed from each other by 1 to 12 bands, contained two subclusters. This subdivision was supported by ribotyping and biotyping. However, it was unclear whether these subclusters represented distinct strains. In one strain, marked polymorphism (patterns that differed from each other by up to four bands) was observed in the ribotype pattern. This study demonstrates the high degree of DNA banding pattern polymorphism found for some strains of VREM and illustrates the complexity involved in defining such strains.
The emergence of enterococci as a major cause of nosocomial infection and their potential for acquiring antibiotic resistance has brought members of this genus, especially Enterococcus faecium, to the forefront of hospital infection control issues. Some regard this organism as the nosocomial pathogen of the 1990s (36).
A large number of phenotypic and genotypic typing methods have been applied to epidemiological investigations of E. faecium (46). Pulsed-field gel electrophoresis (PFGE), especially the contour-clamped homogeneous electric field variety, is viewed by many investigators as the “gold standard” for epidemiological analysis of many bacteria including enterococci (11, 16, 20, 24, 41). However, the lack of standardized running conditions and criteria for interpreting the banding patterns has limited its usefulness, particularly for long-term studies and interlaboratory comparisons.
Recently, general guidelines have been proposed for interpreting PFGE banding patterns and for defining the relatedness of bacteria isolated over periods of up to 3 months. Isolates with patterns that differ from the parental strain pattern, the index strain pattern, or the common pattern by three bands or less are regarded as closely related, those that differ by six bands or less are regarded as possibly related, and those that differ by greater than six bands are regarded as unrelated (41). However, because different species may vary in the degree of polymorphism that they exhibit, the European Study Group on Epidemiological Markers proposed that the degree of relatedness used to indicate working definitions of strains should be adjusted to each species studied and the typing method applied (37).
The present study aimed to establish the degree of DNA banding pattern polymorphism exhibited by strains of vancomycin-resistant E. faecium (VREM) and to determine the level of similarity that should be applied to define “biologically plausible” strains.
MATERIALS AND METHODS
Bacterial isolates.
The 30 VREM isolates studied were isolated from clinical specimens from different patients on the renal facility of a single hospital over a period of 11 months. Seventeen were from urine, four were from intravenous devices, two were from wounds, two were from blood, and one was from peritoneal dialysis fluid; the sources of four isolates were not known. A single colony from a culture of each specimen was selected and stored in 16% glycerol broth at −70°C until tested. The unit involved was extremely busy and was situated in a hospital with the highest bed occupancy rate of any teaching institution in London, United Kingdom. It served acute renal medicine, peritoneal dialysis, hemodialysis, and posttransplantation patients. Patients underwent repeated readmission to any of the four locations within the hospital where renal treatment was provided, including readmission for frequent hemodialysis. The pressure of the workload resulted in patients being subjected to repeated changes of within-ward bed position. Medical records were stored in numerous locations, and pathology reports were not routinely filed. A shortage of infection control personnel coupled with the dispersed nature of record keeping rendered a detailed epidemiological study impossible.
Species identification and biotyping.
The isolates were identified to the species level by a microtiter tray-based method supplemented with motility and pigment production tests (21). The isolates were also tested with the Rapid ID 32 Strep kit according to the manufacturer’s instructions (bioMerieux, Marcy-l’Etoile, France).
PFGE.
PFGE typing of SmaI-digested DNA was performed by a modification of a previously described method (24). Cell suspensions of approximately 2 × 109 cells/ml were mixed with an equal volume of 1.6% low-melting-point agarose at 50°C. The cell-agarose suspension was pipetted into a block mold (6 by 20 by 1 mm) and was allowed to solidify at 4°C. The cells were lysed at 37°C overnight with gentle shaking in lysis buffer (1 mg of lysozyme per ml, 25 μg of RNase per ml, 6 mM Trizma base, 100 mM EDTA, 1 M NaCl, 0.5% Brij 58, 0.2% sodium deoxycholate, 0.5% lauroylsarcosine, and 1 mM MgCl), followed by a further overnight incubation at 50°C in proteolysis buffer (100 μg of proteinase K per ml and 1% lauroylsarcosine in 0.5 M EDTA). The blocks were washed three times for 10 min of each time at 4°C in TE buffer (10 mM Trizma base, 1 mM EDTA) and were stored at 4°C.
Half of an agarose block was equilibrated in restriction enzyme buffer for 30 min and was digested with 40 U of SmaI for 2 to 4 h according to the manufacturer’s instructions. The blocks were washed in TE buffer for 60 min at 37°C and were stored at 4°C. The blocks were cut to the well size and were loaded into the wells of a 1.2% agarose gel (gel size, 25 cm [width] by 20 cm [length]). Electrophoresis was performed in 0.5× TBE buffer (44.5 mM Trizma base, 44.5 mM boric acid, 1 mM EDTA) by the contour-clamped homogeneous electric field method with a CHEF-DRII drive module (Bio-Rad Laboratories Ltd., Hemel Hempstead, United Kingdom). To achieve optimal separation of the fragments, SmaI-digested DNA was electrophoresed in two separate gels (run time, 40 h) under two different linear ramped pulse times: 1 to 10 and 10 to 40 s for the separation of fragments below and above 145 kb, respectively. The gels were stained with ethidium bromide (1 μg/ml) for 40 to 60 min and were photographed.
Stability of PFGE banding patterns.
To assess the stability of the PFGE banding patterns, a single colony of one of the isolates was streaked onto a blood agar plate and the plate was incubated at 37°C overnight. A single colony from this plate that had been incubated overnight was then streaked onto a second plate. This was repeated 45 times. On the third subculture a small colonial variant was observed. Subsequently, subcultures of the parent and the small colonial variant were carried out separately. The first and every fifth subsequent subculture were stored in 16% glycerol broth at −70°C until they were tested.
Antibiograms and plasmid profile analysis.
The susceptibilities of the isolates to vancomycin, teicoplanin, chloramphenicol, ciprofloxacin, erythromycin, tetracycline, trimethoprim, and rifampin were determined by an agar incorporation method (44). In addition, high-level resistance to gentamicin (MIC, >2,000 mg/liter), streptomycin (MIC, >2,000 mg/liter), and penicillin (MIC, >100 mg/liter) was tested for by using breakpoint concentrations. Resistance was defined as described by Woodford et al. (48). Plasmid content was determined by an alkaline lysis method as described previously (44).
Ribotyping.
Genomic DNA was extracted by the guanidium thiocyanate method of Pitcher et al. (30) and was digested with BamHI (Gibco BRL, Uxbridge, United Kingdom). The DNA was probed with biotinylated cDNA of the 16S and 23S rRNAs of the type strain (NCTC 775) of Enterococcus faecalis as described previously (47).
PyMS.
Twenty-two isolates, including representative isolates with each PFGE banding pattern, were compared by pyrolysis mass spectrometry (PyMS) and were analyzed as described previously (7, 8, 33).
Analysis of DNA banding patterns.
The DNA banding patterns were analyzed by visual examination, and all loci were scored for the presence or absence of a band. The percent similarity of the banding patterns was estimated with the Dice coefficient (6), and the matrix of similarity coefficients was clustered by the unweighted pair group method with mathematical averages algorithm (34) with multivariate statistical package (Kovach WI. 1993, version 2.1; Kovach Computing Services, Pentraeth, Wales, United Kingdom).
RESULTS
PFGE typing.
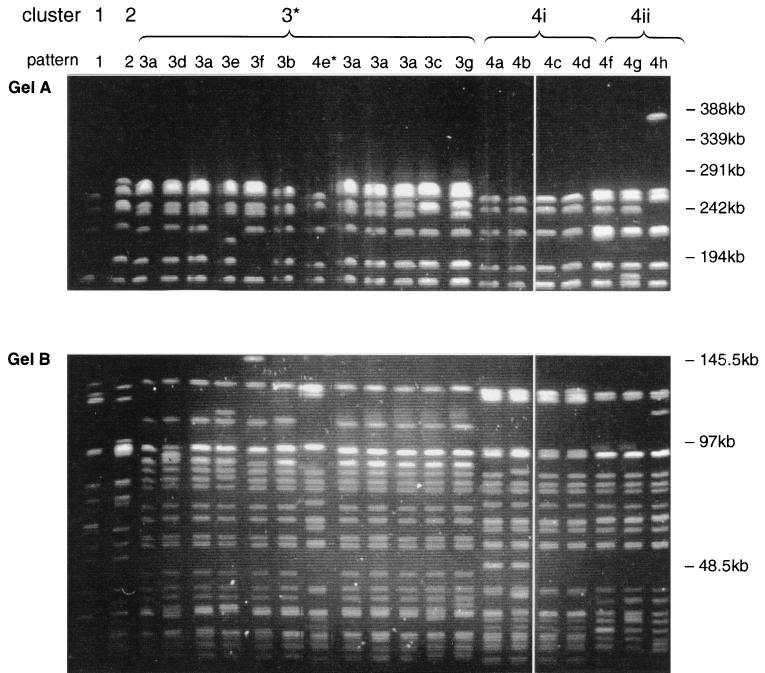
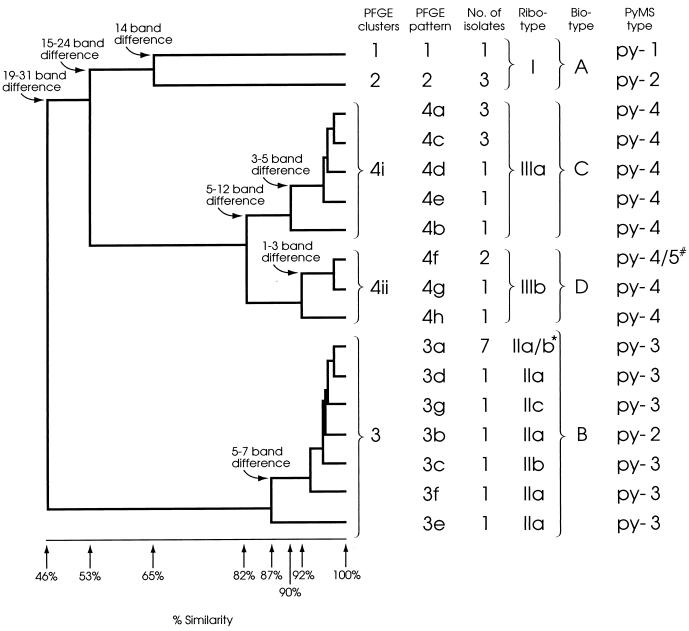
The 30 E. faecium isolates had 17 banding patterns by PFGE, and 19 to 23 (average 21) bands were distinguished per isolate (Fig. 1). Only one isolate gave bands above 291 kb. Cluster analysis revealed four distinct clusters (clusters 1 to 4) at the 82% similarity level, and intercluster band variation ranged from 14 to 31 bands (Fig. 2). The seven patterns in cluster 3 differed from each other by up to seven bands and differed from the common pattern in this cluster (pulsed-field type 3a [PF3a]) by five bands. Cluster 4 contained eight patterns which differed from each other by up to 12 bands. No single common pattern was observed. Interestingly, the patterns could be subdivided into two closely related subclusters (subclusters 4i and 4ii) which formed at similarity levels of 90 and 92%, respectively (Fig. 2). The patterns within each subcluster differed from each other by one to five and one to three bands, respectively. These subclusters were also observed when the banding pattern data were analyzed with several other similarity coefficients (Jacaard, Yule, and Simple Matching [35]) and clustering algorithms (nearest neighbor, furthest neighbor, and weighted pair group [34]) and when the data were analyzed in different sequences.
FIG. 1.
PFGE of SmaI-digested DNA from VREM showing representative pattern of all 17 patterns that were resolved. (Gel A) Linear ramped pulse time of 10 to 40 s with a run time of 40 h. Only well-separated bands above 145 kb are shown. (Gel B) Linear ramped pulse time of 1 to 10 s with a run time of 40 h. Only well-separated bands below 145 kb are shown. *, pattern 4e is included among the cluster 3 patterns on this gel.
FIG. 2.
Dendrogram produced following Dice and UPGMA analysis of the PFGE patterns of SmaI-digested DNA. The percent similarities and band differences between and within clusters are shown. The corresponding ribotype, biotype, and PyMS type of each isolate are also included. *, two of the PFGE pattern 3a isolates were ribotype IIa and five were ribotype IIb; #, one of the PFGE pattern 4f isolates was PyMS type py-4 and the other was PyMS type py-5.
Stability of PFGE banding patterns.
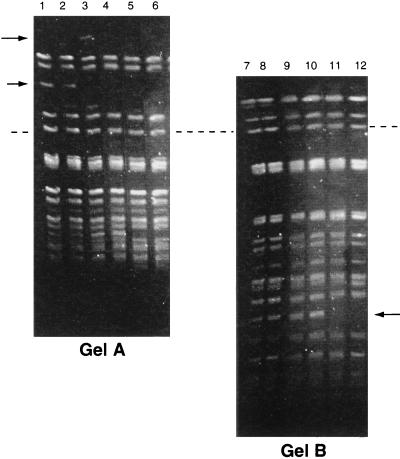
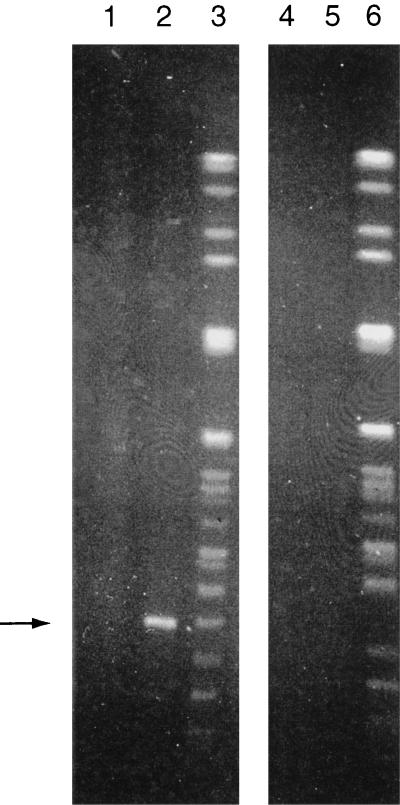
No differences were observed between the banding patterns of different subcultures of the parent colony, even though after the 40th subculture it became susceptible to vancomycin (data not shown). Single colonies of the small colonial variant always produced banding patterns that were a mixture of those of both parent and small colonies on subculture. On the fifth subculture, the banding pattern varied at two loci; one with a missing band (194 to 242.5kb) and one with a new band (242.5 to 291 kb) (Fig. 3, arrows). The latter band was not observed on subsequent subculture. On the 15th subculture, another band (approximately 48.5 kb) disappeared (Fig. 3, arrows). PFGE of a SmaI-digested plasmid preparation showed that the latter band was a plasmid (Fig. 4, arrow). No further changes were observed.
FIG. 3.
PFGE banding patterns of the small colonial variant of an isolate of VREM following 20 serial single colony subcultures. (Gel A) Linear ramped pulse time of 10 to 40 s with a run time of 40 h. (Gel B) Linear ramped pulse time of 1 to 10 s with a run time of 40 h. Lanes 1 and 7, original culture; lanes 2 and 8, 1st subculture; lanes 3 and 9, 5th subculture; lanes 4 and 10, 10th subculture; lanes 5 and 11, 15th subculture; lanes 6 and 12, 20th subculture. Arrows, bands which appeared and/or disappeared during subculture; dashed line, the positions of the same band on both gels.
FIG. 4.
PFGE of total DNA and plasmid preparation by using ramped pulse times of 1 to 10 s for 30 h followed by 10 to 30 s for 15 h. Lanes 1 to 3, isolate with PFGE pattern 4a; lanes 4 to 6, isolate with PFGE pattern 4c; lanes 1 and 4, undigested plasmid preparation; lanes 2 and 5, plasmid preparation digested with SmaI; lanes 3 and 6, total DNA digested with SmaI. Arrow, plasmid band.
Ribotyping.
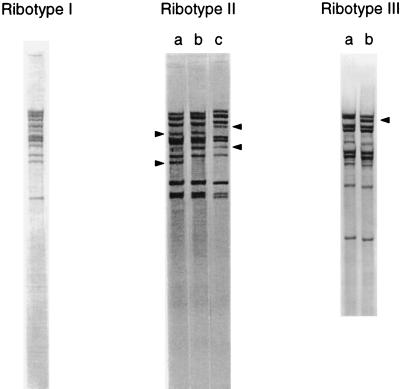
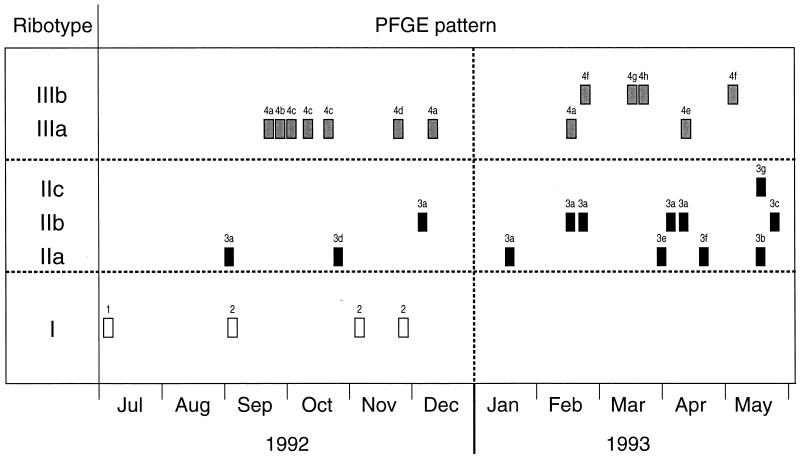
Ribotyping resolved 10 to 11 fragments per isolate and divided the 30 E. faecium isolates into six groups with distinct banding patterns which formed three clusters; ribotypes I, II and III (Fig. 5). Intercluster banding patterns varied by 4 to 13 bands. Ribotype I corresponded to PFGE clusters 1 and 2. The ribotype II cluster contained three similar patterns (subtypes IIa, IIb, and IIc) and corresponded to PFGE cluster 3. Ribotypes IIa and IIb and ribotypes IIb and IIc each differed by two bands, and ribotypes IIa and IIc differed by four bands (Fig. 5, and arrows). Ribotypes IIb and IIc appeared 3 and 8 months, respectively, after the first isolation of ribotype IIa (Fig. 6). Several isolates of ribotypes IIa and IIb had identical PFGE banding patterns, and the PFGE pattern for the single ribotype IIc isolate differed from this PFGE pattern by only one band. These isolates were also identical by biotyping. It seemed appropriate, therefore, to regard these three patterns as subtypes of ribotype II, despite the relatively large band differences between them. The two patterns of ribotype III (subtypes IIIa and IIIb) differed from each other by a single band (Fig. 5, arrows) and corresponded to PFGE cluster 4. Interestingly, ribotypes IIIa and IIIb correlated exactly with PFGE subclusters 4i and 4ii, respectively (Fig. 2). The polymorphism observed within both ribotypes II and III was progressive with time (Fig. 6).
FIG. 5.
Ribotyping patterns of the VREM strains. Arrowheads, alteration in bands responsible for the subtypes (a, b, and c) of ribotype II and the subtypes (a and b) of ribotype III.
FIG. 6.
Time chart showing the date when each of the 30 VREM isolates was isolated on the renal unit and the corresponding ribotype and PFGE pattern for each strain.
Biotyping.
All isolates gave identical results in the majority of biochemical reactions tested. However, variation was observed in five tests (mannitol, raffinose, cyclodextrine, β-mannosidase, and melibiose), and four biotypes that differed from each other by the results of at least two of the tests were distinguished (Table 1). The clusters identified by biotyping correlated well with the PFGE clusters but failed to distinguish isolates within clusters 1 and 2. Biotyping did, however, clearly distinguish subclusters 4i and 4ii, with differences observed by all five tests (Fig. 2).
TABLE 1.
Biotypes and corresponding PFGE clusters of VREM
| PFGE cluster | Biotype | Result of tests with the following biochemicals:
|
||||
|---|---|---|---|---|---|---|
| Mannitol | Raffinose | Cyclodextrine | β-Mannosidase | Melibiosea | ||
| 1 | A | + | − | + | + | − |
| 2 | A | + | − | + | + | − |
| 3 | B | + | − | + | − | + |
| 4i | C | − | − | − | − | − |
| 4ii | D | + | + | + | + | + |
One isolate each in clusters 3 and 4i gave a result opposite of the indicated result.
PyMS.
With the exception of two isolates, PyMS distinguished, at the 95% confidence limits, four groups that corresponded to PFGE clusters 1 to 4 (Fig. 2). The exceptions were a PFGE cluster 3 isolate that appeared to have greater similarity to isolates of PFGE cluster 2 and a PFGE subcluster 4ii isolate which was distinct from all the other isolates.
Plasmid profiles.
Twenty-one distinct plasmid profiles were found among the 30 isolates, with each isolate containing between four and nine plasmids. Four clusters corresponding to PFGE clusters 1 to 4 were distinguished (data not shown). Two clusters corresponding to PFGE clusters 3 and 4 grouped at low similarity levels (59 and 61% respectively) as a result of the extensive variation in the profiles within these clusters. For example, the 13 isolates of PFGE cluster 3 and the 7 isolates of PFGE subcluster 4i were each divided into seven unique plasmid profiles that differed from each other by up to five and six plasmids, respectively.
Antibiograms.
All isolates were resistant to 7 of the 12 antibiotics tested (vancomycin, teicoplanin, ampicillin, ciprofloxacin, erythromycin, rifampin, and trimethoprim), and all except 1 of the isolates were resistant to high levels of penicillin. The majority of the isolates were also resistant to chloramphenicol, tetracycline, and high levels of gentamicin and streptomycin. Although nine unique antibiograms were observed, there was little correlation between these antibiograms and the groupings obtained by the other typing methods; the same antibiogram was observed for isolates in more than one PFGE cluster, and isolates of a single PFGE subtype (subtype 3a) were subdivided into four groups according to their antibiograms.
DISCUSSION
PFGE remains the method of choice for epidemiological typing of E. faecium. It is more discriminatory than other methods, all isolates are typeable, and good reproducibility is obtained (16, 20, 22). However, the level of polymorphism acceptable in isolates of the same strain is still an open question, as is also the case for many other species. Pennington (28) compared this question with the equally controversial issue of the definition of a species and concluded that the definition of strains involves considerable uncertainty and requires a sizable judgmental element. This subjective element is also acknowledged in the recently proposed guidelines for the interpretation of PFGE patterns (41). A philosophical principle, the consilience of induction, is suggested as the criterion for evaluation of the naturalness of strain groupings; natural groups must be identical or very similar when tested by methods that measure independent markers (28). This was the approach adopted in this study.
PFGE resolved the 30 VREM isolates examined into four distinct clusters. PyMS and plasmid typing supported the grouping of PFGE clusters 1 and 2. The ribotypes and biotypes of the isolates in these clusters are the most frequently observed ribotypes and biotypes among the E. faecium strains referred to our laboratory (20a) and hence represent a lack of discrimination by these methods rather than the identities of isolates in clusters 1 and 2. Isolates of PFGE cluster 3, although clearly distinct from those of the other clusters, differed from each other by a relatively large number of bands (one to seven bands). The guidelines for the interpretation of PFGE patterns presuppose that band differences are calculated with references to the commonest strain, the index strain, or the parental strain pattern (41), which has also been referred to as the “modal” band difference (43). PFGE cluster 3 patterns differed from the common pattern (PF3a) by up to five bands, which falls outside the criteria (three band differences) proposed for the identification of closely related isolates. These isolates were, however, grouped together by ribotyping, biotyping, PyMS (except for two isolates), and plasmid typing, which, together with the geographical and temporal relatedness of these isolates, indicated that they represented a single strain. Such large intrastrain band differences have been reported for other species. Differences of six bands were found for a β-lactamase-producing strain of E. faecalis isolated over a 7-year period (32) and a strain of Pseudomonas aeruginosa isolated over a 3-year period (12).
The strain-defining criteria proposed in the guidelines were specifically devised to cover studies that span periods of less than 3 months. This study, which covered 11 months, suggests that in outbreaks that cover extended periods a three-band-difference rule for the definition of strains of E. faecium should not be followed rigidly. The guidelines themselves allow differences of up to six bands (two genetic events) if there is good epidemiological evidence to suggest the relatedness of the isolates (41).
The criteria used to define strains by ribotyping and other DNA typing methods, such as plasmid typing and randomly amplified polymorphic DNA analysis with PCR, are much more problematic, and guidelines have not yet been proposed (40). In the majority of ribotyping and other probe-based typing studies, it is arbitrarily assumed that strains with patterns that differ by even a single band represent genetically and epidemiologically unrelated strains (11, 23, 47). In some ribotyping studies one (1) or two (18, 26, 27, 29) band differences have been allowed for isolates considered to be the same strain. In this study the data indicated that E. faecium isolates which differed in their ribotype patterns by up to four bands represented a single strain.
The conclusion regarding the strain boundaries of the PFGE cluster 4 isolates is not clear and illustrates the judgmental element alluded to by Pennington (28). Patterns within this cluster differed by a large number of bands; 12 bands if all patterns are considered and 9 bands if only a common pattern is considered. However, the cluster formed at a similarity level (82%) which falls within the strain boundary levels reported for other species (3, 23, 38, 39). If isolates of the same strain may differ by two genetic events (6 band differences) within a 3-month period (41), then within an 11-month period, two further genetic events could have occurred, resulting in up to 12 band differences, as observed in this cluster. There is, however, evidence which suggests that the PFGE cluster 4 isolates may represent two distinct strains. Dendrograms generated with a variety of similarity coefficients and clustering algorithms all divided this cluster into two subclusters. Furthermore, the biotyping data and the ribotype subtypes correlated exactly with the PFGE subclusters. It is difficult to judge the weight which should be given to the latter typing data results since the ribotype subtypes differed by only one band and large changes in biotype may occur following a single genetic event, such as the introduction of a transposon (5). The strain designations of these isolates, therefore, remained unresolved. For practical infection control purposes, it may be argued that it is preferable to consider isolates which may be unrelated as the outbreak strain rather than risk missing a source of cross-infection (9).
In contrast to findings of the present study, Bonten et al. (2) reported that the differences in the PFGE patterns of related and unrelated strains of E. faecium was clear-cut. Related isolates differed by 4 bands or fewer while unrelated isolates differed by 10 or more bands. This has also been reported for P. aeruginosa by Hla et al. (15), who found that related and unrelated isolates were clearly distinguished by both band differences and percent similarities of banding patterns. The small intrastrain variation in PFGE patterns observed by Bonten et al. (2) for E. faecium and by Hla et al. (15) for P. aeruginosa compared with the large variation reported in this study may reflect differences in the fluidities of the genomes of the strains studied. However, since Bonten et al. (2) and Hla et al. (15) observed this in a number of different strains, another possible explanation is that the electrophoretic conditions adopted in those studies did not allow resolution of all the differences that were present. The electrophoresis run times in those studies were half those used in the present study.
Although the frequencies of occurrence of the genetic events that modify banding patterns are unknown, we have observed two independent genetic events after 45 in vitro serial subcultures, one of which resulted in the loss of a plasmid. The changes observed in vitro suggest that we may expect greater polymorphism in the genomes of isolates recovered from their natural habitats, in which interaction and competition with other organisms of the same or different species and genera may occur. Banding pattern polymorphism is commonly attributed to either mutations affecting a restriction enzyme target site (three-band difference) or DNA rearrangements involving deletions or insertions (two-band difference) (14, 41). To this should be added the loss or gain of plasmids (one-band difference), as demonstrated for E. faecium in this study and has also been demonstrated for E. faecalis (32). DNA banding pattern polymorphisms due to insertion sequences (25), transposons (4, 42), and phages (19, 31) have been reported. In contrast, it is also apparent that epidemiologically relevant changes, such as the loss of resistance to vancomycin observed during serial subculture in this study, may not be reflected in the DNA banding pattern. In the course of an outbreak, Woodford et al. (45) found VREM strains with identical PFGE patterns but different plasmid profiles and van genotypes.
Prior to the publication of the guidelines for PFGE typing (41), there had been little consensus regarding strain definition, even for strains within the same species. For instance, Talon et al. (39) proposed a nine-band-difference rule for P. aeruginosa, Grothues et al. (12) and Struelens et al. (38) proposed a six-band-difference rule, and Grundmann et al. (13) proposed a three-band-difference rule. This may, in part, be due to a lack of standardization in the description of band pattern differences. Some workers (38, 41) count the total number of band differences and take into account differences at every band locus (as adopted in this study), while others count band shifts (9) and, hence, will report smaller numbers of band differences. Inconsistency also arises when the band difference obtained by the comparison of all subtypes (42) rather than the modal band difference (43) is reported. The latter approach, as proposed in the guidelines (41) and reemphasized recently (10), gives a better representation of the relatedness of isolates. Isolates derived from the modal pattern by a single but independent genetic event (two- to three-band difference) will differ from each other by two genetic events (four- to six-band difference). In this study, the modal band difference within PFGE clusters 3 and 4 was 5 and 8 or 9 bands, respectively, compared with absolute differences of 7 and 12 bands, respectively.
The usefulness of plasmid typing and antibiogram analysis for long-term epidemiological studies appears to be limited. In this study the groupings obtained according to the different antibiograms did not correlate with any of the groupings obtained by the other typing methods that were used, and although plasmid typing supported the clusters obtained by the other typing methods, there was a high degree of variation within each cluster. There is no consensus in the literature on the usefulness of these two methods for the epidemiological typing of enterococci (17).
In conclusion, we have demonstrated a large degree of DNA banding pattern polymorphism within strains of VREM and have confirmed that isolates belonging to a single strain may differ from each other by four bands by ribotyping and by seven bands by PFGE typing.
REFERENCES
- 1.Arpin C, Coze C, Rogues A M, Gachie J P, Bebear C, Quentin C. Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit. J Clin Microbiol. 1996;34:2163–2169. doi: 10.1128/jcm.34.9.2163-2169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonten M J M, Hayden M K, Nathan C, Rice T W, Weinstein R A. Stability of vancomycin-resistant enterococcal genotypes isolated from long-term-colonized patients. J Infect Dis. 1998;177:378–382. doi: 10.1086/514196. [DOI] [PubMed] [Google Scholar]
- 3.Chetoui H, Melin P, Struelens M J, Delhalle E, Mutro Nigro M, de Ryck R, de Mol P. Comparison of biotyping, ribotyping, and pulsed-field gel electrophoresis for investigation of a common-source outbreak of Burkholderia pickettii bacteremia. J Clin Microbiol. 1997;35:1398–1403. doi: 10.1128/jcm.35.6.1398-1403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow J W, Perri M B, Thal L A, Zervos M J. Mobilization of the penicillinase gene in Enterococcus faecalis. Antimicrob Agents Chemother. 1993;37:1187–1189. doi: 10.1128/aac.37.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clermont D, Delbos F, de Cespedes G, Horaud T. Old and new (Tn3708) mobile chromosomal elements in streptococci and enterococci. In: Ferretti J J, Gilmore M S, Klaenhammer T R, Brown F, editors. Genetics of streptococci, enterococci and lactococci. Basel, Switzerland: Krager; 1995. pp. 55–61. [PubMed] [Google Scholar]
- 6.Dice L R. Measures of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 7.Freeman R, Goodfellow M, Gould F K, Hudson S J, Lightfoot N E. Pyrolysis mass spectrometry (Py-MS) for the rapid epidemiological typing of clinically significant bacterial pathogens. J Med Microbiol. 1990;32:283–286. doi: 10.1099/00222615-32-4-283. [DOI] [PubMed] [Google Scholar]
- 8.Freeman R, Gould F K, Ryan D W, Chamberlain J, Sisson P R. Nosocomial infection due to enterococci attributed to a fluidised microsphere bed. The value of pyrolysis mass spectrometry. J Hosp Infect. 1994;27:187–193. doi: 10.1016/0195-6701(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 9.Goering R V. Molecular epidemiology of nosocomial infection: analysis of chromosomal restriction fragment patterns by pulsed-field gel electrophoresis. Infect Control Hosp Epidemiol. 1993;14:595–600. doi: 10.1086/646645. [DOI] [PubMed] [Google Scholar]
- 10.Goering R V, Tenover F C. Epidemiological interpretation of chromosomal macrorestriction fragment patterns analyzed by pulsed-field gel electrophoresis. J Clin Microbiol. 1997;35:2432–2433. doi: 10.1128/jcm.35.9.2432-2433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordillo M E, Singh K V, Murray B E. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J Clin Microbiol. 1993;31:1570–1574. doi: 10.1128/jcm.31.6.1570-1574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grothues D, Koopman V, von der Hardt H, Tummler B. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol. 1988;26:1973–1977. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundmann H, Schneider C, Hartung D, Daschner F D, Pitt T L. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:528–534. doi: 10.1128/jcm.33.3.528-534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall L M. Are point mutations or DNA rearrangements responsible for the restriction fragment length polymorphisms that are used to type bacteria? Microbiology. 1994;140:197–204. doi: 10.1099/13500872-140-1-197. [DOI] [PubMed] [Google Scholar]
- 15.Hla S W, Hui K P, Tan W C, Ho B. Genome macrorestriction analysis of sequential Pseudomonas aeruginosa isolates from bronchiectasis patients without cystic fibrosis. J Clin Microbiol. 1996;34:575–578. doi: 10.1128/jcm.34.3.575-578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn I, Burman L G, Haeggman S, Tullus K, Murray B E. Biochemical fingerprinting compared with ribotyping and pulsed-field gel electrophoresis of DNA for epidemiological typing of enterococci. J Clin Microbiol. 1995;33:2812–2817. doi: 10.1128/jcm.33.11.2812-2817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavery A, Rossney A S, Morrison D, Power A, Keane C T. Incidence and detection of multidrug-resistant enterococci. J Med Microbiol. 1997;46:1–7. doi: 10.1099/00222615-46-2-150. [DOI] [PubMed] [Google Scholar]
- 18.Lew A E, Desmarchelier P M. Molecular typing of Pseudomonas pseudomallei: restriction fragment length polymorphisms of rRNA genes. J Clin Microbiol. 1997;31:533–539. doi: 10.1128/jcm.31.3.533-539.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lina B, Bes M, Vandenesch F, Greenland T, Etienne J, Fleurette J. Role of bacteriophages in genomic variability of related coagulase-negative staphylococci. FEMS Microbiol Lett. 1993;109:273–278. doi: 10.1016/0378-1097(93)90032-w. [DOI] [PubMed] [Google Scholar]
- 20.Miranda A G, Singh K V, Murray B E. DNA fingerprinting of Enterococcus faecium by pulsed-field gel electrophoresis may be a useful epidemiologic tool. J Clin Microbiol. 1991;29:2752–2757. doi: 10.1128/jcm.29.12.2752-2757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Morrison, D. Unpublished observations.
- 21.Morrison D, Woodford N, Cookson B D. Enterococci as emerging pathogens of humans. J Appl Microbiol. 1997;83:89S–99S. doi: 10.1046/j.1365-2672.83.s1.10.x. [DOI] [PubMed] [Google Scholar]
- 22.Morrison D, Woodford N, Solaun M, Riley U, Bignardi G, Goldberg L, Cookson B D, Johnson A P. Abstracts of the Third International Meeting on Bacterial Epidemiological Markers, Cambridge, United Kingdom. 1994. The use of pulsed-field gel electrophoresis and ribotyping to study the epidemiology of a cluster of multi-resistant Enterococcus faecium causing infection on a renal unit, abstr. PA31. [Google Scholar]
- 23.Morvan A, Aubert S, Godard C, El Solh N. Contribution of a typing method based on IS256 probing of SmaI-digested cellular DNA to discrimination of European phage type 77 methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1997;35:1415–1423. doi: 10.1128/jcm.35.6.1415-1423.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. . (Erratum, 29:418, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naas T, Blot M, Fitch W M, Arber W. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol Biol Evol. 1995;212:198–207. doi: 10.1093/oxfordjournals.molbev.a040198. [DOI] [PubMed] [Google Scholar]
- 26.Owen R J, Bell G D, Desai M, Moreno M, Gant P W, Jones P H, Linton D. Biotype and molecular fingerprints of metronidazole-resistant strains of Helicobacter pylori from antral gastric mucosa. J Med Microbiol. 1993;38:6–12. doi: 10.1099/00222615-38-1-6. [DOI] [PubMed] [Google Scholar]
- 27.Owen R J, Hunton C, Bickley J, Moreno M, Linton D. Ribosomal RNA gene restriction patterns of Helicobacter pylori: analysis and appraisal of HaeIII digests as a molecular typing system. Epidemiol Infect. 1992;109:35–47. [PMC free article] [PubMed] [Google Scholar]
- 28.Pennington T H. Molecular systematics and traditional medical microbiologists—problems and solutions. J Med Microbiol. 1994;41:371–373. doi: 10.1099/00222615-41-6-371. [DOI] [PubMed] [Google Scholar]
- 29.Pitcher D G, Johnson A P, Allerberger F, Woodford N, George R C. An investigation of nosocomial infection with Corynebacterium jeikeium in surgical patients using a ribosomal ribonucleic acid gene probe. Eur J Clin Microbiol Infect Dis. 1990;9:643–648. doi: 10.1007/BF01964264. [DOI] [PubMed] [Google Scholar]
- 30.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 31.Reith S, Marples R R, Cookson B. Abstracts of the Third International Meeting on Bacterial Epidemiological Markers, Cambridge, United Kingdom. 1994. The epidemiology and evolution in typing characteristics of a new epidemic methicillin-resistant Staphylococcus aureus (EMRSA-16), abstr. PA32. [Google Scholar]
- 32.Seetulsingh P S, Tomayko J F, Coudron P E, Markowitz S M, Skinner C, Singh K V, Murray B E. Chromosomal DNA restriction endonuclease digestion patterns of β-lactamase-producing Enterococcus faecalis isolates collected from a single hospital over a 7-year period. J Clin Microbiol. 1996;34:1892–1896. doi: 10.1128/jcm.34.8.1892-1896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sisson P R, Freeman R, Lightfoot N E. Pyrolysis mass spectrometry of microorganisms. PHLS Microbiol Digest. 1992;9:65–68. [Google Scholar]
- 34.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman & Co.; 1973. The estimation of taxonomic resemblance; pp. 114–187. [Google Scholar]
- 35.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman & Co.; 1973. Taxonomic structure; pp. 188–308. [Google Scholar]
- 36.Spera R V, Jr, Farber B F. Multiply-resistant Enterococcus faecium. The nosocomial pathogen of the 1990s. JAMA. 1992;268:2563–2564. [PubMed] [Google Scholar]
- 37.Struelens M J Members of the European Study Group on Epidemiological Markers (ESGEM) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2:2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 38.Struelens M J, Schwam V, Deplano A, Baran D. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J Clin Microbiol. 1993;31:2320–2326. doi: 10.1128/jcm.31.9.2320-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talon D, Cailleaux M, Thouverez M, Michel-Briand Y. Discriminatory power and usefulness of pulsed-field gel electrophoresis in epidemiological studies of Pseudomonas aeruginosa. J Hosp Infect. 1996;32:135–145. doi: 10.1016/s0195-6701(96)90055-9. [DOI] [PubMed] [Google Scholar]
- 40.Tenover F C, Arbeit R D, Goering R V. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect Control Hosp Epidemiol. 1997;18:426–439. doi: 10.1086/647644. [DOI] [PubMed] [Google Scholar]
- 41.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D P, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thal L A, Silverman J, Donabedian S, Zervos M J. The effect of Tn916 insertions on contour-clamped homogeneous electrophoresis patterns of Enterococcus faecalis. J Clin Microbiol. 1997;35:969–972. doi: 10.1128/jcm.35.4.969-972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomayko J F, Murray B E. Analysis of Enterococcus faecalis isolates from intercontinental sources by multilocus enzyme electrophoresis and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:2903–2907. doi: 10.1128/jcm.33.11.2903-2907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uttley A H, George R C, Naidoo J, Woodford N, Johnson A P, Collins C H, Morrison D, Gilfillan A J, Fitch L E, Heptonstall J. High-level vancomycin-resistant enterococci causing hospital infections. Epidemiol Infect. 1989;103:173–181. doi: 10.1017/s0950268800030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodford N, Chadwick P R, Morrison D, Cookson B D. Strains of glycopeptide-resistant Enterococcus faecium can alter their van genotypes during an outbreak. J Clin Microbiol. 1997;35:2966–2968. doi: 10.1128/jcm.35.11.2966-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodford N, Johnson A P, Morrison D, Speller D C E. Current perspectives of glycopeptide resistance. Clin Microbiol Rev. 1995;8:585–615. doi: 10.1128/cmr.8.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodford N, Morrison D, Johnson A P, Briant V, George R C, Cookson B. Application of DNA probes for rRNA and vanA genes to investigation of a nosocomial cluster of vancomycin-resistant enterococci. J Clin Microbiol. 1993;31:653–658. doi: 10.1128/jcm.31.3.653-658.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodford N, Morrison D, Johnson A P, George R C. Antimicrobial resistance amongst enterococci isolated in the United Kingdom: a reference laboratory perspective. J Antimicrob Chemother. 1993;32:344–346. doi: 10.1093/jac/32.2.344. [DOI] [PubMed] [Google Scholar]