Abstract
Cancer is one of the leading causes of death worldwide, affecting millions of people every year. While chemotherapy remains one of the most common cancer treatments in the world, the severe side effects of chemotherapy drugs impose serious concerns to cancer patients. In many cases, the chemotherapy can be localized to maximize the drug effects; however, the drug systemic circulation induces undesirable side effects. Here, we have developed a highly efficient cellulose-based nanoadsorbent that can capture more than 6000 mg of doxorubicin (DOX), one of the most widely used chemotherapy drugs, per gram of the adsorbent at physiological conditions. Such drug capture capacity is more than 3200% higher than other nanoadsorbents, such as DNA-based platforms. We show how anionic hairy cellulose nanocrystals, also known as electrosterically stabilized nanocrystalline cellulose (ENCC), bind to positively charged drugs in human serum and capture DOX immediately without imposing any cytotoxicity and hemolytic effects. We elucidate how ENCC provides a remarkable platform for biodetoxification at varying pH, ionic strength, ion type, and protein concentration. The outcome of this research may pave the way for developing the next generation in vitro and in vivo drug capture additives and devices.
Keywords: Cellulose nanocrystal, nanocellulose, doxorubicin, drug capture, blood purification, chemotherapy, bioadsorbent
Graphical Abstract
1. Introduction
More than 39% of men and women are diagnosed with cancer at some stage in their lifetime[1] from whom more than 85% will suffer from one or more side effects,[2,3] including anemia, repeated infections, hair loss, jaundice, and fever. Despite the recent advances in drug-free cancer treatment methods, such as immunoengineering,[4,5] chemotherapy drugs remain the standard practice among clinicians.[6–8] As an example, an adult diagnosed with liver cancer may require 60, 75, and 800 mg/m2 of doxorubicin (DOX),[9] cisplatin,[10] and 5-fluorouracil,[11] respectively, administered once a month via intravenous (IV) injection. Such treatment may result in anemia,[10,11] congestive heart failure,[9] cardiomyopathy,[9] seizures,[9,10] myelosuppression,[9–11] and mucositis.[9,11] In some cases, e.g., hepatocellular carcinoma (HCC), localized drug delivery approaches, such as catheter-based minimally invasive injection of chemotherapy drugs and drug eluting materials[12–18] may help increase the treatment efficacy; however, the unused drugs may still reach the circulation system and result in unpreventable damages to healthy tissues.[19,20]
To reduce the off-target effects of cancer drugs during and after localized chemotherapy, eliminating their systemic circulation is necessary. Recently, a few platforms have been proposed to remove unwanted drugs, mainly DOX, from blood, including an endovascular ion exchange device consisting of a porous nylon mesh cylinder filled with the ion exchange resin Dowex,[21] DNA-coated magnetic nanoparticles,[22] DNA-coated polyacrylate,[23] and block copolymers.[24,25] While these strategies have shown some success in removing DOX from blood, their removal efficacy remains exceedingly low, e.g., in the order of a few μg of DOX per mg of adsorbent within several minutes, often necessitating the use of large devices, e.g., ~0.5 m long tubes,[22] to remove physiologically relevant concentrations of DOX.
To overcome the challenges associated with current drug capture materials, specifically the low removal capacity, nanoparticles with a large density of functional groups appear very promising. A large portion of therapeutic drugs, e.g., some chemotherapeutic agents, bactericides, fungicides, virucides, and opioids, contain positively charged moieties,[26] which may bind to oppositely charged substances via electrostatic interactions. However, due to the complex blood composition, charged nanoparticles typically lose their functionality in blood as a result of ionic strength and protein mediated aggregation via several mechanisms, such as electrical double layer screening and bridging.[27,28] Uncharged moieties, such as polyethylene glycol (PEG), have widely been used to protect nanoparticles in blood via steric repulsion and render them stealth;[29–31] however, they may result in compromised drug binding affinity as a result of reduced surface charge.[32] To the best of our knowledge, there is currently no nanoparticle-based super-capacity drug capture system.
We believe that among nanoparticles, nanoscale celluloses, known as nanocellulose, may be an interesting candidate to develop drug capture systems due to their benign chemical structure,[33,34] non-cytotoxic nature,[35–40] and non-biodegradability[41] in the human body. Conventional nanocelluloses are commonly classified as cellulose nanocrystals (CNC) and cellulose nanofibrils (CNF), neither of which may accommodate more than 2 mmol of anionic functional groups, such as carboxylates, per gram due to the inaccessibility of their inner crystalline cellulose chains.[42–48] The emergence of hairy cellulose nanocrystals,[49–51] CNCs decorated with highly functionalized amorphous cellulose chains (hairs) at both ends that are synthesized via the controlled oxidation of cellulose fibers, has opened new opportunities for engineering nanoadsorbents[52] with significantly higher removal capacities than other bio-based materials.
Here, we aim at nanoengineering cellulose to yield anionic hairy cellulose nanocrystals, also known as electrosterically stabilized nanocrystalline cellulose (ENCC), for removing one of the most common chemotherapy drugs, DOX as a model cationic drug with significant side effects,[53–56] in physiological conditions. We will thoroughly investigate the mechanism of drug removal via altering ionic strength, pH, and protein content, and quantify the drug removal efficacy. The ENCC is compared with CNC to shed light on the fundamental advantages of hairy nanocelluloses over conventional ones. In addition, in vitro cytotoxicity and hemocompatibility of ENCC will be investigated.
2. Materials and methods
2.1. Materials
Doxorubicin hydrochloride (DOX) powder was purchased from Oakwood Chemical (USA), cellulose nanocrystal (CNC, freeze-dried solid, including 0.85 wt% sulfur on dry CNC sodium form) was provided by the University of Maine Process Development Center (USA), human serum (sterile filtered male AB plasma), hydrochloric acid (HCl, 37%), bovine serum albumin (BSA, lyophilized powder, 66 kDa), sodium hydroxide (NaOH, ACS reagent, ≥97%), sodium (meta)periodate (NaIO4, ≥99.99%), sodium chlorite (NaClO2, 80%), sodium chloride (NaCl, ACS reagent, ≥99%), ethylene glycol (C2H6O2, 99%), polyethylene glycol (PEG, Mw = 400 Da), calcium chloride (CaCl2, 99.99%), poly-L-lysine (PLL) solution (0.01%, sterile-filtered), Triton X-100, Drabkin’s solution, Alfa Aesar™ paraformaldehyde (4% in PBS), and pure human hemoglobin (lyophilized powder) were supplied by Sigma-Aldrich (USA). Milli-Q water (18.2 MΩ cm at 25 °C) was provided by the Millipore Corporation (USA). Disposable Malvern PANalytical Inc. (DTS1070) folded capillary cells, hydrogen peroxide (H2O2, 30%), and anhydrous ethanol (C2H5OH, 95.27%) were purchased from Fisher Scientific (Canada and USA). Bleached softwood (black spruce) kraft pulp sheets (Q-90) were provided by FPInnovations (Canada). The dialysis membrane (12–14 kDa molecular weight cutoff) was purchased from Spectrum Lab Inc (USA). NIH/3T3 fibroblast cells and human umbilical vein cells (HUVECs) were purchased from the American Type Culture Collection (ATCC, USA). Heat inactivated fetal bovine serum (FBS), Dulbecco’s phosphate-buffered saline (DPBS, 1X), Dulbecco’s modified Eagle medium (DMEM, including 4.5 g of glucose per liter), trypsin-ethylenediaminetetraacetic acid (trypsin-EDTA, 0.5 %, 1X), penicillin/streptomycin (P/S, 100X), and trypan blue solution (0.4 %) were procured from Gibco (USA). PrestoBlue™ cell viability reagent, Live/Dead™ viability/cytotoxicity kit, and Alexa Fluor™ 488 Phalloidin were produced by Invitrogen and provided by ThermoFisher Scientific (USA), and 4′,6-diamidino-2-phenyslindole (DAPI) was purchased from ThermoFisher Scientific. Endothelial cell growth medium-2 (EGM-2) Bulletkit was purchased from Lonza. Tissue culture flasks (75 cm2) were purchased from Corning (USA), and tissue culture-treated polystyrene 96-well plates were obtained from Falcon (USA).
2.2. ENCC synthesis:
ENCC is an anionic hairy cellulose nanocrystal that bears a large amount of dicarboxylates groups on its hairs. We prepared ENCC via the chlorite-mediated oxidation of dialdehyde modified cellulose (DAMC) fibers based on our previous protocols.[50,57] These fibers were prepared by soaking 4 g (25 mmol glucose units) of small (~ 2 cm2) softwood pulp pieces in water overnight, which were disintegrated mechanically using a blender. The pulp slurry was oxidized via a reaction with an aqueous periodate (5.33 g) solution including ~ 266 g water (absorbed water by pulp + added water) in the dark to prevent periodate deactivation. The reaction was stopped after 4 days using 3 mL of ethylene glycol, and the intact DAMC fibers were rinsed with water five times and vacuum filtrated. To synthesize ENCC, 1 g of DAMC fibers was reacted with 1.41 g of sodium chlorite in an aqueous solution containing 1.41 g of hydrogen peroxide and 2.93 g of sodium chloride (total water volume ~ 266 mL). The pH was maintained at ~ 5 via intermittent NaOH (0.5 M) addition for 24 h. After the reaction completion, 0.16 g of ethanol per 1 g of mixture was added, and ENCC was separated via centrifugation (3000×g for 10 min). ENCC was then dialyzed against DI water for at least one day to remove the impurities and salts.
2.3. Conductometric titration:
To measure the carboxylate content of nanocelluloses, we used an established protocol.[58] A dispersion of never-dried ENCC containing 20 mg of solid in 140 mL of Milli-Q water was prepared to which 2 mL of a 20 mM NaCl solution was added, followed by adjusting the pH to 3 using a solution of 0.1 M HCl. The ENCC dispersion was titrated using the Methrom 907 Titrando instrument to add a 10 mM NaOH solution at a rate of 0.1 mL/min until reaching pH ~ 11. The carboxylate group density of ENCC was calculated based on the NaOH volume required to neutralize the weak acid, i.e., the middle region in the titration curve.
2.4. Atomic force microscopy (AFM) imaging:
To image CNC and ENCC, we adopted the sample preparation and imaging parameters from literature.[59] In brief, a PLL solution (0.1% w/v) was placed on a freshly cleaved mica disc for 5 min, rinsed with Milli-Q water, and the disc was mounted on a magnetic stainless steel disc. This is a necessary treatment to ensure the deposition of negatively charged nanocelluloses on a positively charged polymer. To deposit the nanocelluloses, a droplet of dispersion (concentration = 0.1 mg mL−1) was incubated on the PLL-treated mica for 5 min, followed by the removal of excess liquid via gentle Milli-Q water rinse and air-drying overnight. The samples were imaged under the tapping mode using an AFM (Bruker Dimension Icon) equipped with a silicon nitride probe (Scanasyst-Air). For image processing, the NanoScope Analysis software (version 2.0) was used, and the nanocellulose dimensions were measured using Gwyddion (version 2.56).[60]
2.5. Hydrodynamic size and ζ-potential measurements:
A Zetasizer Nano ZS from Malvern Panalytical Ltd was used to conduct dynamic light scattering (DLS) and electrokinetic measurements in folded capillary cells (DTS1070). Samples were prepared with an ENCC concentration of 0.1 mg mL−1 in 1 mg mL−1 of NaCl solution. A final DOX concentration of 0.03 mg mL−1 was used to have an ~ 1:10 molar ratio of DOX to the carboxylate of ENCC. For ζ-potential measurements on DOX, the concentration was lowered to 0.003 mg mL−1. At least three measurements were taken for each DLS experiment, with 11 runs per measurement. For the ζ-potential tests, the number of measurements was at least three, with up to 100 runs per measurement.
2.6. DOX capture:
DOX capturing experiments were carried out in varying media. Each sample had a total volume of 500 μL, except the removal tests in human serum for which 250 μL was used. The concentration of ENCC was fixed at 195 μg mL−1 for all samples. The media included DPBS, Milli-Q water with adjusted pH ~ 4 using HCl, Milli-Q water with adjusted monovalent ion concentration (171 mM) using an NaCl solution, Milli-Q water with adjusted divalent ion concentration (0.9 or 90 mM) using a CaCl2 solution, DPBS including BSA (35 mg mL−1), and human serum. Samples were prepared by adding the nanocellulose dispersion to a DOX solution at DOX : COO− of nanocellulose ratios between ~ 0.46 – 2.07 mol mol−1, covering ratios well below and above the stoichiometric (1:1 mol mol−1) value. The concentration of DOX in the supernatant of each sample was measured after centrifuging the samples for 2 min at 2000 rpm and 20 °C using 2 μL aliquots tested by a the NanoDrop One microvolume spectrophotometer (ThermoFisher Scientific, USA) at 480 nm. Standard curves were obtained for the freshly prepared DOX stock solutions in the same media as the removal tests, which were used to quantify the free drug in the supernatant after adding the nanocelluloses. For the time dependent measurements, the samples were centrifuged at pre-defined time points, aliquots were withdrawn and analyzed under the same conditions as equilibrium experiments. For control experiments, nanocellulose dispersions were replaced with an equivalent volume of MilliQ water.
2.7. In vitro cytotoxicity tests:
The cytotoxicity of ENCC against HUVECs was assessed by incubating the nanoparticles with two-dimensionally (2D) cultured cells. HUVECs were cultured in EGM-2 Bulletkit cell culture medium at 37 °C and 5% CO2, followed by trypsinization using 0.5 % trypsin-EDTA and seeding into 96-well plates at a density of 2.5×103 cells per well. After cell adhesion ~ 2 h post seeding, the culture media were replaced with 50 μL of complete media containing varying ENCC concentrations. ENCC-containing media and ENCC-free media (control) were refreshed every other day. Cell viability was assessed after 1 day of exposure to ENCC via incubating the cells in a solution of DPBS including 0.05% v/v calcein AM (green staining of live cells) and 0.2% v/v ethidium homodimer (red staining of dead cells) (Live/Dead viability kit, Invitrogen) for 10 min. Following the incubation, the cells were washed twice with DPBS and maintained in 50 μL of DPBS to be examined under a Zeiss fluorescence microscope (Axio Observer 5, Zeiss, Germany). To examine the morphology, cells were fixed at day 3 with 4% v/v paraformaldehyde for 15 min and washed three times with DPBS. A solution of 0.3% v/v Triton X-100 in DPBS was used to permeabilize the fixed cells while a solution of BSA (1% w/v) in DPBS was used for 30 min at room temperature to block non-specific binding. The cells were then maintained at 4 °C until the final staining procedure was completed. The cytoskeleton was stained with a 1:40 dilution of Alexa Fluor 488 in a BSA solution (1% w/v) and incubated for 40 min at room temperature. Subsequently, the nuclei were counterstained with a 1:1000 dilution of DAPI in DPBS and incubated for 15 min at room temperature in dark. Images were acquired using a Zeiss fluorescence microscope at the excitation/emission wavelengths of ~ 494/515 nm (calcein), ~ 528/617 nm (ethidium homodimer-1), ~ 495/518 nm (Alexa Fluor™ 488 Phalloidin), and ~ 358/461 nm (DAPI). The metabolic activity of HUVECs was measured after 3 days of exposure to ENCC. Cells were incubated with 10% PrestoBlue™ cell viability reagent for 90 min in the culture media at 37 °C and 5% CO2. Subsequently, the supernatant was collected from each sample and transferred to 96-wells plates. The fluorescence (excitation/emission wavelengths of ~ 530/590 nm) of supernatant was determined using the BioTek Synergy 2 microplate reader (USA). Measurements were performed in triplicate and the background signal (PrestoBlue™-containing cell-free media) was subtracted from the fluorescence values.
2.8. Hemolysis assessment:
The hemolytic activity of ENCC was measured via exposing human whole blood, withdrawn from 3 healthy volunteer donors by another lab, to varying concentrations of ENCC and measuring the concentration of released hemoglobin in plasma according to the American Society for Testing and Materials (ASTM) E2524–08 (2013) standard analysis of hemolytic activity of nanoparticles.[61] A colorimetric technique was used to measure the oxidation of methemoglobin by cyanide (Drabkin’s reagent) at an absorbance of ~ 540 nm. A calibration curve was obtained using 0.025–1 mg/L of pure human hemoglobin to quantify the concentration of released hemoglobin in heparinized human whole blood exposed to ENCC. The blood samples were diluted in DPBS to have 10 ± 2 mg of hemoglobin per mL. ENCC was added to a microcentrifuge tube, followed by the addition of DPBS (800 μL) and the diluted blood (100 μL). After gently mixing the samples, they were incubated for 3 h ± 15 min at 37 °C and centrifuged for 15 min at 14000 rpm and ambient temperature. A portion of supernatant (~ 100 μL) was pipetted into a 96-well plate and mixed with 100 μL of the Drabkin’s reagent. The reaction was continued for 15 min at room temperature in dark. A microplate reader (540 nm, BioTek UV/vis Synnergy 2, USA) was then used to measure the relative absorbance of each well against the reagent blank to correct the background (blood-free samples) absorbance. The hemolysis percentage was measured based on the hemoglobin concentration in the samples divided by the total blood hemoglobin (~ 10 mg mL−1). Positive and negative controls were Triton X-100 (1 % v/v) and PEG (4.4 % v/v), respectively.
2.9. Biodetoxification tests:
To determine the biodetoxification effect of ENCC, 5×103 NIH/3T3 cells or HUVECs were seeded in 96-well plates with 200 μL of DMEM supplemented with FBS (10% v/v) and 1% v/v of Pen-Strep or EGM bullet kit HUVEC media, respectively. The cells were allowed to attach to the bottom of wells for 1 h, the media were aspirated, and the cells were rinsed with 200 μL of DPBS, followed by the addition of 200 μL of corresponding media including DOX (final concentration = 400 μg mL−1), which was previously detoxified by adding ENCC (final concentration = 1 mg mL−1), centrifuged at 1000 rpm for 3 min, and separated from the precipitate. The control experiments were conducted in corresponding DOX-free cell culture media or DOX-containing media without ENCC treatment. Each group consisted of 5 wells. After 8 h, the media was aspirated, the cells were rinsed twice with 200 μL of DPBS, followed by the addition of 100 μL of PrestoBlue™ reagent to each well. The cells were incubated for 1 h, and then the PrestoBlue™ absorbance was recorded using the microplate reader. Immediately afterwards, the cells were again rinsed twice with 200 μL of DPBS, followed by the addition of 150 μL of the Live/Dead reagents. The cells were incubated for 90 min and imaged using the fluorescence microscope. The fluorescence images were analyzed using the ImageJ Software.[62]
2.10. Statistical analysis:
The data of at least three measurements are presented as mean values ± standard deviation. We performed the two-way ANOVA (analysis of variance) and identified the statistically significant data when p ≤ 0.05. Statistically significant values are presented as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and **** p ≤ 0.0001.
3. Results and discussion
3.1. Engineering hairy cellulose nanocrystals
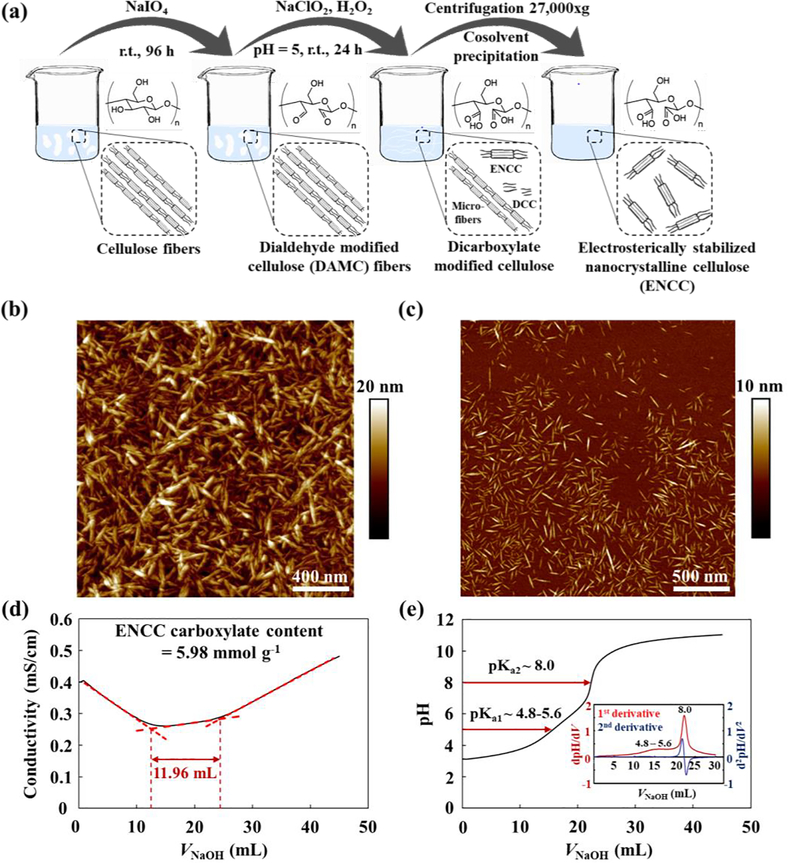
Fig. 1a shows the schematic of ENCC production from cellulose fibers via a two-step periodate-chlorite oxidation. The first reaction results in the production of intact DAMC fibers, which are then chemically disintegrated via chlorite-mediated oxidation of aldehyde groups to carboxylate, yielding a mixture of ENCC, dicarboxylated cellulose (DCC), and unfibrillated cellulose (microfibers). ENCC is separated from the system via ethanol-mediated precipitation. AFM images of conventional CNC and ENCC are shown in Figs. 1b and 1c, respectively, attesting to a similar crystalline body. The length and diameter (height) of CNC are 153.9±43.7 nm and 6.2±2.1 nm, respectively, which are comparable with ENCC length, 104.5±29.7 nm, and diameter, 3.5±1.3 nm. The conductometric titration of ENCC, presented in Fig. 1d, attests to about 5.98 mmol of carboxylate groups per gram of ENCC. This is about 600% higher than the conventional 2,2,6,6-tetramethylpiperidine 1-oxyl, 2,2,6,6-tetramethyl-1-piperidinyloxy (TEMPO)-mediated oxidized CNC.[63] We did not register any weak acid in the titration of CNC. During the conductometric titration, we recorded the solution pH as NaOH was added. The dispersion pH versus NaOH volume is presented in Fig. 1e with insets showing the first and second derivates of pH with respect to NaOH volume. According to this figure, two pKa values are registered for ENCC, associated with the adjacent carboxylate groups on the C2 and C3 of glucose rings, i.e., dicarboxylate groups. ENCC is an anionic hairy cellulose nanocrystal, which benefits from electrostatic and steric colloidal stability due to the highly negatively charged carboxylate groups and excluded volume of hairs, respectively, thus called electrosterically stabilized.
Fig. 1. ENCC synthesis and characterization.
(a) ENCC synthesis involving two-step oxidation using periodate to initially open the glucose ring from C2-C3 bond, converting them to dialdehyde groups, followed by chlorite-mediated oxidation to carboxylate groups. AFM images of (b) CNC and (c) ENCC. Titration of ENCC (20 mg) based on (d) conductivity and (e) pH versus added base (NaOH, 10 mM) volume. The inset in panel (e) shows the first and second derivatives of pH versus NaOH volume.
3.2. Chemotherapy drug capture in DPBS
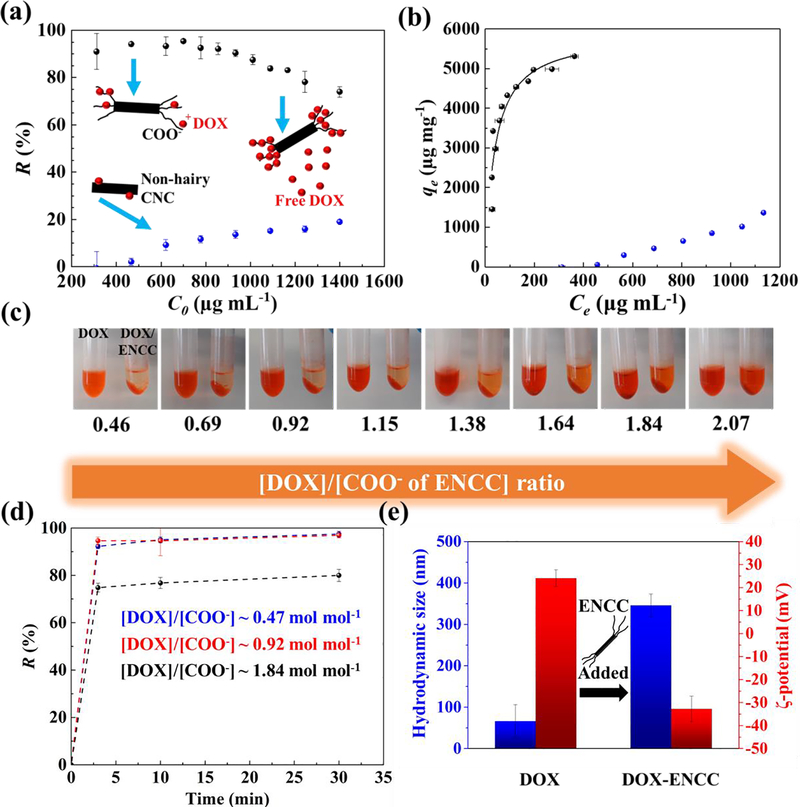
To study the interactions between the nanoadsorbent ENCC and chemotherapy drug DOX, ENCC was added to DOX solutions in DPBS with varying initial DOX concentrations (C0). The concentration of DOX was tuned in a way that the molar ratio of DOX to the negatively charged groups (COO− ~ 5.98 mmol g−1) of ENCC varied from ~ 0.46 to ~ 2.07. Fig. 2a shows the removal percentage of DOX, R (%) = 100 × (C0 − Ce)/C0, versus the initial DOX concentration (C0) when the nanoadsorbent concentration is ~ 195 μg mL−1 (COO− concentration ~ 1.166 mM). The equilibrium DOX concentration (Ce) was measured via sampling the supernatant after centrifuging ENCC-DOX samples. When the drug concentration was ≤ 800 μg mL−1 (~1.4 mM), more than 90% of the drug was removed by the ENCC. Increasing the drug concentration beyond 800 μg mL−1 saturated the nanoadsorbent, decreasing the removal percentage to ~ 70% at a DOX concentration of 1400 μg mL−1. The inset of Fig. 2a schematically shows the non-saturated and saturated states of ENCC. When the carboxylate groups of ENCC were not fully saturated by the positively-charged drug, most of the drug molecules were captured. Increasing the drug concentration eventually saturated the charged functional groups of ENCC, decreasing the drug removal percentage. As a control, the drug capture capability of conventional, non-hairy CNC was evaluated. CNC is commonly prepared by the strong acid (sulfuric acid)-mediated hydrolysis of cellulose fibers.[64] As can be seen in Fig. 2a, the removal percentage of CNC is below 20%, and it is unable to effectively capture DOX as a result of the low density of charged groups (~ 0.3 mmol of sulfate half ester groups, according to the provider).
Fig. 2. Drug capture in aqueous media.
(a) DOX removal R = (C0 − Ce)/C0 by ENCC or CNC in DPBS versus initial DOX concentration C0, obtained from measuring the equilibrium DOX concentration Ce using a microvolume UV-vis spectrophotometer. (b) DOX capture capacity of ENCC (black symbols) or CNC (blue symbols) defined as qe = (C0 − Ce) V/mENCC versus equilibrium DOX concentration Ce and the corresponding Langmuir fit (curve). Sample volume V ~ 500 μL. The DOX removal capacity of ENCC in DPBS is >5200 μg mg−1, and CNC is unable to effectively capture DOX due to the lack of high-density anionic groups. The background error is corrected. (c) Optical images of DOX solutions with concentrations ranging from ~ 300 μg mL−1 to ~1400 μg mL−1 before and after the addition of ENCC with a final concentration ~ 195 μg mL−1. This wide range of concentrations corresponds to a broad ratio of DOX to the carboxylate content of ENCC (~ 0.46–2.07). (d) Time-dependent DOX capture using ENCC, showing an extremely fast drug removal taking place almost immediately after ENCC-DOX contact. Here, DOX concentration [DOX] ~ 311, 622, and 1244 μg mL−1, and ENCC concentration [ENCC] ~ 195 μg mL−1, and the ratio of DOX to the COO− of ENCC ~ 0.47, 0.92, 1.82 mol mol−1. (e) Colloidal properties of DOX significantly change upon adsorption onto ENCC. Hydrodynamic size of DOX aggregates ~ 66 nm increases to ~ 350 nm, an indication of ENCC-mediated aggregation. The ζ-potential of colloidal DOX changes from ~ +24 mV to ~ −32 mV as a result of adsorption to the negatively-charged ENCC. Note that [DOX]/[COO−] ~ 0.1, therefore ENCC (concentration ~ 100 μg mL−1) is not saturated, forming stable colloidal aggregates.
The DOX removal capacity of ENCC or CNC, qe, defined as the amount of removed drug per unit mass of the nanoadsorbent is presented versus the drug equilibrium concentration in Fig. 2b. By increasing the DOX concentration, initially, most of the drug was captured independent of its concentration, resulting in a low equilibrium drug concentration. As the drug concentration increased, ENCC was saturated, and the equilibrium drug concentration significantly increased. The plateau in Fig. 2b shows the maximum removal capacity of ENCC, i.e., >5200 μg (~ 9 μmol) drug per 1 mg of ENCC, which has ~ 5.98 μmol of carboxylate. The DOX removal capacity of ENCC is higher than the stochiometric value, possibly because DOX molecules in solution tend to aggregate.[65] Fitting qe-Ce data to the Langmuir isotherm[66] according to the following equation (Eqn. 1):[67]
| Eqn. 1 |
yields an equilibrium constant K = τads / τads ~ 0.0066 and a maximum removal capacity qm ~ 5899 μg mg−1, with the coefficient of determination R2 ~ 0.878. Note that τads and τdes are the adsorption and desorption characteristic times, respectively. The drug removal capacity of CNC, shown in Figure 2b remains below ~ 1000 μg mg−1, which is significantly lower than ENCC.
Note that the increase in the removal capacity of CNC at high DOX concentration is possibly due to the aggregation of DOX,[65] and not the CNC effect, as reflected in the near-zero removal capacity at DOX concentrations below 500 mg mL−1. Such a striking difference attests to the superiority that the anionic hairs impart to ENCC, enabling it to perform as a potential nanosponge for biodetoxification applications.
Fig. 2c shows the images of DOX solutions after adding ENCC (right) compared with the ENCC-free DOX solutions (left). Regardless of the DOX/ENCC ratio within the molar ratio of DOX : COO− of ENCC ~ 0.47–2.07, ENCC effectively precipitates the drug, decreasing the drug concentration in the supernatant. When the ratio of DOX concentration to the COO− of ENCC is less than 1 mol mol−1, the supernatant is almost clear, turning red at higher drug concentrations due to the free, uncaptured drug. The kinetics of drug capture was studied by measuring the DOX concentration in the supernatant over time. Fig. 2d shows the DOX removal percentage versus time. In less than 3 min, the drug removal equilibrates, which shows an extremely fast (almost instantaneous) drug capture. In fact, the first few minutes post nanoadsorbent-drug contact were associated with manually processing the sample and measuring the drug concentration. In the absence of such operator-related time limit, the interaction between ENCC and DOX is immediate (within seconds) as visually seen in Fig. 2c.
The adsorption of DOX to ENCC was examined by studying the colloidal properties of drug aggregates upon ENCC addition. Fig. 2e shows the hydrodynamic size and ζ-potential of DOX aggregates. Before ENCC addition, the drug aggregates have a hydrodynamic size of ~ 66 nm and ζ-potential ~ +24 mV, which are consistent with literature.[65] When ENCC was added, the size significantly increased to >350 nm due to the DOX-mediated ENCC aggregation. The ratio of DOX to the COO− of ENCC was maintained low (~ 0.1) in a way that ENCC remained excess, i.e., it was not saturated, and no precipitation was formed. The ζ-potential of ENCC-DOX complexes dropped to ~ −32 mV, which was due to the negatively charged, stable DOX-ENCC aggregates.
3.3. Effects of pH and ionic strength on chemotherapy drug capture
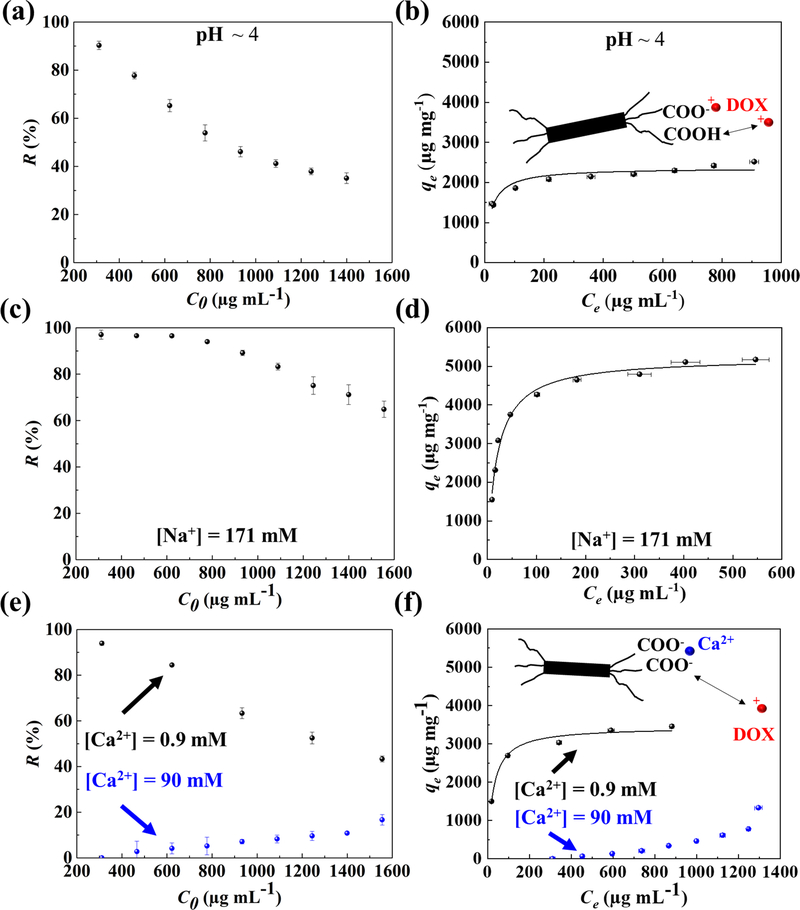
To shed light on the mechanism of ENCC-mediated drug capture, the effects of pH and ionic strength were investigated. Fig. 3 presents the drug removal capability of ENCC at a low pH as well as in the presence of mono- or divalent cations. Fig. 3a shows the drug removal percentage at pH ~ 4. The removal immediately starts to decrease as the concentration of drug increases. At this pH, some of the carboxylate groups of ENCC are protonated and therefore cannot interact with the positively charged drug. The adjacent carboxylates of ENCC located on the C2 and C3 of opened glucose ring have pKa ~ 4.8–5.6 and 8, which is also reported in our previous work.[67] According to Eqn. 2,
| (2) |
almost all of the COOH groups with pKa = 8 are protonated, and only ~25% of the carboxylate groups with pKa ~ 4.8 are deprotonated. Therefore, at pH ~ 4, only 12.5 % of the acidic groups (~0.75 mmol COO− per gram of ENCC) are deprotonated. Fig. 3b presents the removal capacity of ENCC at pH ~ 4. At this pH, the removal capacity of ENCC is reduced to < 2400 μg mg−1, less than half of the removal capacity at pH = 7.4, which is higher than the stoichiometric ratio. The Langmuir fit yields K ~ 0.0080 and qm ~ 2365 μg mg−1, with R2 ~ 0.907. This K is higher than the K in DPBS, indicating that the adsorption occurs slower (larger τads/τdes). Besides the aggregation of DOX, the excess DOX may partially shift the equilibrium of carboxylic acid according to the Le Chatelier’s principle, increasing the removal capacity beyond the stoichiometric ratio. Such an effect has previously been reported for the interaction between copper ions with ENCC.[52]
Fig. 3. Effects of pH and ionic strength on ENCC-mediated DOX capture.
(a) DOX removal percentage versus initial DOX concentration, and (b) DOX capture capacity of ENCC versus equilibrium DOX concentration at pH ~ 4. Decreasing pH to 4 resulted in the protonation of carboxylic acid groups, which decreased the available DOX binding sites on ENCC, decreasing the removal percentage and capture capacity. The capture capacity at pH ~ 4 is less than half of that at pH ~ 7.4 (Fig. 2b). The effect of monovalent ion, Na+, on the (c) removal percentage and (d) capture capacity of ENCC, showing no significant impact because the equilibrium of carboxylate groups is not affected by monovalent ions. Effect of divalent ion, Ca2+, on the (e) removal percentage and (f) capture capacity of ENCC. The presence of 0.9 and 90 mM of Ca2+ decreased the DOX capture capacity of ENCC by a factor of 35% and 600%, respectively. This proves that the main drug capture mechanism is DOX-ENCC electrostatic attraction, disturbed by the divalent ion-mediated bridging of ENCC. In panels b, d, and f, the curve shows the Langmuir fit.
The concentration of sodium (Na+) in blood is ~ 135–145 mM. [31,68] The effect of monovalent ions, such as Na+ on the drug removal was investigated in Fig. 3c and 3d. At a Na+ concentration above the physiological level (171 mM), the DOX removal percentage (Fig. 3c) and capture capacity (Fig. 3d) were similar to the Na+-free medium (Figs. 2a and 2b). The presence of monovalent ions in the media does not block the ENCC charge and only shrinks the electrical double layer around the protruding dicarboxylate-modified cellulose chains. Therefore, sodium and likely other monovalent ions in the blood do not hinder the drug capture capacity of ENCC. The Langmuir fit yields K ~ 0.0036 and qm ~ 5225 μg mg−1, with R2 ~ 0.986. Fig. 3e and 3f present the drug removal percentage and capacity of ENCC in the presence of 0.9 or 90 mM of a divalent cation, in this case calcium (Ca2+). The removal percentage immediately decreased when the divalent ion was added to the medium due to the calcium-mediated charge neutralization of ENCC.[69] At 0.9 mM of Ca2+, the Langmuir fit yielded K ~ 0.0075 and qm ~ 3440 μg mg−1, with R2 ~ 0.981, and when the calcium ions reached 90 mM, the removal capacity was < 1000 μg mg−1. In contrast to Na+, Ca2+ competes with the positively charged drug molecules in binding to the COO− of ENCC, decreasing the drug capture capacity of ENCC.
3.4. Effect of plasma proteins on chemotherapy drug capture
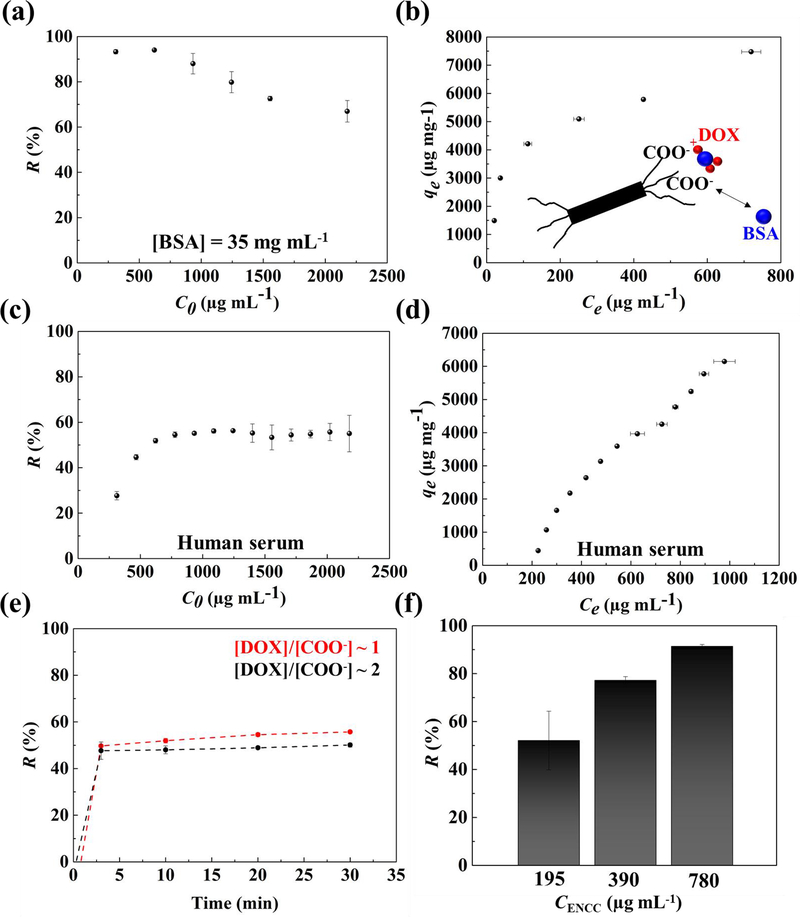
An important constituent of blood is plasma proteins. We evaluated the drug capture capacity of ENCC in the presence of physiologically relevant concentrations of BSA (35 mg mL−1)[70] as a model system for serum albumin. The drug removal percentage and capacity in the presence of BSA are shown in Figs. 4a and 4b, respectively. The removal percentage remained >90% up to a drug concentration <1000 μg mL−1, which is higher than the removal percentage in the absence of BSA (Fig. 1a). Compared to the BSA-free system (Fig. 1b), the removal capacity (Fig. 4b) was also enhanced in the presence of BSA and reached >7500 μg mg−1 of ENCC. The underlying mechanism for BSA-mediated enhanced drug removal can be explained via the attractive interactions between the positively charged drug and negatively charged BSA:[71] the drug is adsorbed by BSA, which is then picked up by ENCC. Accordingly, ENCC picks up the drug-BSA complex, which may include more drugs than the stoichiometric capacity of ENCC in a BSA-free system. This may help overcome the Ca2+-mediated reduction of DOX capture capacity of ENCC.
Fig. 4. ENCC-mediated DOX capture in physiological media.
Effect of BSA on the DOX (a) removal percentage and (b) capture capacity of ENCC, showing that the serum albumin not only does not impair the drug capture capability of ENCC, but also enhances the removal capacity as a result of DOX-protein binding. The DOX (c) removal percentage and (d) capture capacity of ENCC in human serum. The DOX capture capacity of ENCC in human serum is remarkably high, possibly as a synergistic effect of drug-protein complex formation. (e) The drug capture time scale in human serum is extremely short, allowing for almost immediate removal of the chemotherapy drug. (f) Increasing the ENCC concentration increases the DOX removal capacity, allowing for almost complete elimination of the drug.
We investigated the drug capture capacity of ENCC in a physiologically relevant medium, i.e., human serum. Fig. 4c and 4d show the drug removal percentage and capacity of ENCC in human serum, respectively. The drug removal percentage remained at ~ 60% when the initial drug concentration was increased beyond 500 μg mL−1. As more drug was added to the media, more drug was removed, possibly as a result of drug-protein complex formation, followed by the ENCC-mediated capture. Interestingly, at low drug concentrations (< 500 μg mL−1), the removal percentage was noticeably low, which may be due to the stable, negatively charged drug-protein complexes, remaining in the solution. The drug removal capacity of ENCC in human serum shown in Fig. 4d increased by increasing the drug concentration, which may be attributed to the effect of serum proteins. Increasing the drug concentration, facilitates the drug-protein complex formation, decorating the proteins with more positively charged DOX molecules, which are then adsorbed to ENCC. Human serum is typically comprised of proteins (e.g. albumin, globulins, and fibrinogen), hormones and enzymes, lipids and carbohydrates.[68,72] Accordingly, while the divalent ions may have negative effects on the drug capture capacity of ENCC, the presence of a range of proteins in the serum enhance the drug removal.
The kinetics of drug capture in human serum is shown in Fig. 4e, suggesting that the capture took place almost immediately, similar to the non-physiological medium (Fig. 2d). To increase the removal percentage of drug, the amount of ENCC was increased. Fig. 4f shows the effect of ENCC content on the drug removal percentage in human serum. The drug removal percentage can readily be increased from ~ 50% to >90% by increasing the ENCC concentration from 195 to 780 μg mL−1. Accordingly, ENCC can successfully remove the majority of chemotherapy drug from the serum.
3.5. Cytotoxicity assessment of hairy cellulose nanocrystals
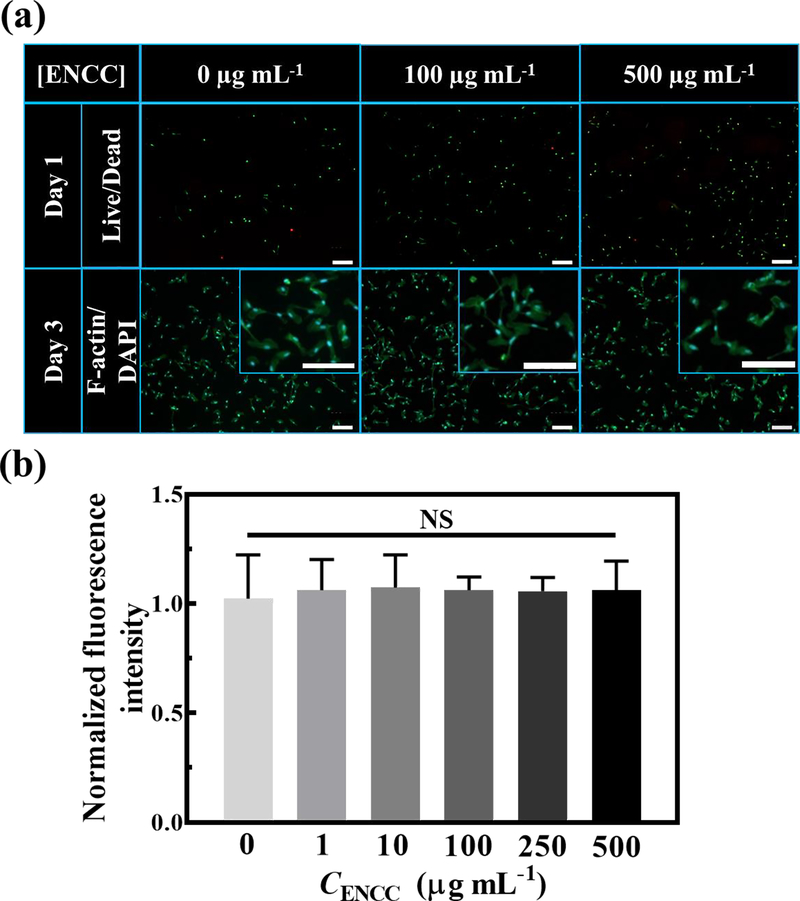
The cytotoxicity of ENCC against human umbilical vein endothelial cells (HUVECs) was examined in vitro. Fig. 5 presents the live/dead and F-actin/DAPI staining and metabolic activity of HUVECs post exposure to varying concentrations of ENCC compared with the ENCC-free control samples. As can be seen in Fig. 5a, the incubation of up to 500 μg of ENCC per mL of cell culture medium yielded similar cell viability as the ENCC-free system after 24 h, none of which resulted in significant cell death. Staining the cytoskeleton and nuclei of cells with F-actin and DAPI, respectively, showed that the ENCC did not have any adverse effects on the cell cytoskeleton and nuclei, e.g., cells spread successfully, and the nuclei remained intact. Fig. 5b shows that after 3 days of exposure to up to 500 μg of ENCC per mL of cell culture medium, the normalized fluorescent intensity, a representation of cell metabolic activity according to the PrestoBlue™ cell viability reagent, remained comparable to that of ENCC-free cells, which underwent no significant loss. Accordingly, ENCC does not impose any toxicity risk to HUVECs, enabling it to effectively interact with the drug.
Fig. 5. ENCC cytotoxicity.
(a) Effect of ENCC concentration on the HUVECs, shown with live (green)/dead (red) and F-actin (green)/DAPI (blue) staining after 24 h and 72 h post nanoparticle exposure, respectively. Magnified F-actin/DAPI images are shown in the insets. Almost all the cells are viable and have undergone spreading and elongation with no significant damage to their nuclei. The scale bars are 200 μm. (b) Metabolic activity of HUVECs after 72 h exposure to various concentrations of ENCC normalized with the metabolic activity in the absence of ENCC, obtained using the PrestoBlue™ assay. The unchanged metabolic activity of HUVECs show that ENCC is not toxic against endothelial cells.
3.6. Hemolytic effects of hairy cellulose nanocrystals
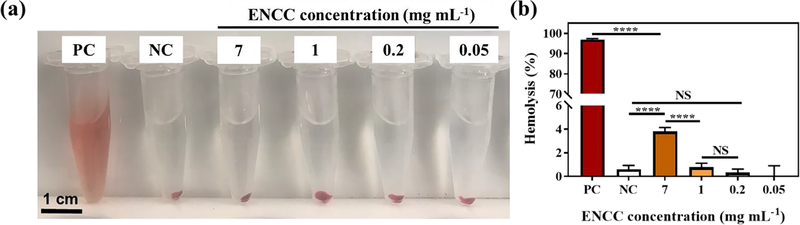
The interaction between ENCC and red blood cells (RBCs) was studied via a standard hemolysis assay (see Materials and Methods). Figs. 6a shows the optical images of blood exposed to ENCC, which was compared to the negative control (NC, PEG) and positive control (PC, Triton X-100). PEG does not induce significant hemolysis, whereas Triton X-100 results in severe RBC damage and hemoglobin leakage, which can be seen in the stable red color of supernatant post-centrifugation. After centrifuging the blood exposed to up to 7 mg mL−1 of ENCC, the supernatant looks as clear as the negative control, showing no noticeable RBC damage. Fig. 6b presents the hemolysis percentage for varying ENCC concentrations, which are compared with the NC and PC. The PC almost fully destroys the RBCs, releasing the hemoglobin, resulting in >95% hemolysis. The ENCC-mediated hemolysis remained less than 4% up to 7 mg mL−1 of ENCC, which is below the maximum percentage allowed, i.e., 5%. This concentration translates into capturing > 42 mg of DOX by using only 1 mL of a 0.7% ENCC dispersion without any adverse effects on the cell viability and RBC integrity.
Fig. 6. Assessment of the hemolytic properties of ENCC.
(a) Optical images of blood exposed to varying concentrations of ENCC compared with the negative control (NC, PEG) and positive control (PC, Triton X-100). The disruption of RBC is seen as stable red color in the supernatant post-centrifugation. Up to 7 mg mL−1 of ENCC does not have any significant effect on the color of supernatant attesting to the integrity of RBCs. (b) Quantification of hemolysis percentage via measuring the absorbance of supernatant. While the positive control vigorously disrupts the RBCs resulting in >95% hemolysis, up to 7 mg mL−1 of ENCC yields less than 4% hemolysis, which is below the accepted threshold (5%).
3.7. Hairy cellulose nanocrystal-enabled biodetoxification
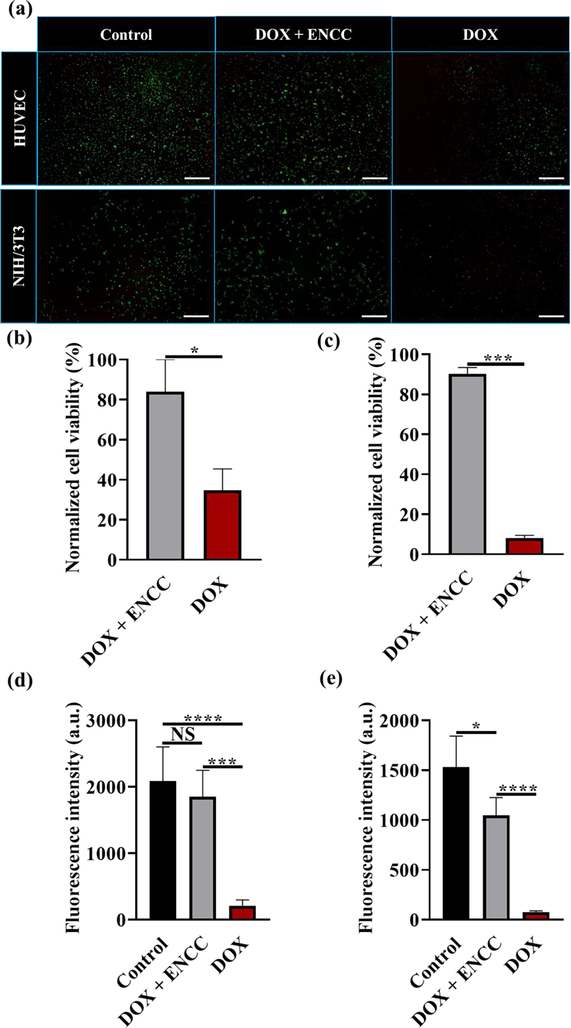
The biodetoxification effect of ENCC was evaluated in vitro using two cell lines. HUVECs and NIH/3T3 cells were cultured to attach to the well plates (2D), followed by adding DOX-containing media pre- and post ENCC treatment and culturing for 8 h. Fig. 7 presents the Live/Dead staining, quantification of cell viability, and the metabolic activity of cells post exposure to DOX with and without ENCC-mediated detoxification. When the cells were exposed to 400 μg of DOX per mL of cell culture media, their viability was compromised due to the DOX-mediated DNA damage.[73] This is reflected in the red staining of HUVECs and NIH/3T3 fibroblasts in Fig. 7a. When the drug containing media was treated with ENCC, followed by removing the precipitated drug via centrifugation, the majority of cells maintained their viability and the Live staining (green color) showed a similar result as the non-toxified (DOX-free) control. The image analysis enabled us to quantify the cell viability via counting the live and total cell numbers. Figs. 7b and 7c present the cell viability percentage of HUVECs and NIH/3T3 fibroblasts normalized with the viability of control samples, respectively. Adding the drug to the media decreases the cell viability to ~ 35% and <10% for HUVECs and NIH/3T3 cells, respectively, whereas the ENCC-mediated detoxification of media maintains more than 80% of both cells viable. The difference between HUVEC and NIH/3T3 fibroblast viability after DOX treatment can be explained by the higher resistance of HUVECs to DOX compared to 3T3 fibroblast cells. HUVECs can develop a resistance to DOX due to increased P-glycoprotein expression.[74] Furthermore, DOX concentrations above 2.5×10−5 M inhibit fibroblast proliferation and DOX concentrations above 5×10−5 M induce fibroblast cellular toxicity.[75] As we used DOX solutions with a concentration of 400 μg mL−1 (7×10−4 M), the dose caused high cell toxicity in NIH/3T3 cells, resulting in the lower cell viability compared to HUVECs, which were protected due to their increased resistance. The metabolic activities of HUVECs and NIH/3T3 cells, presented in Figs. 7d and 7e, respectively, showed that the drug decreased the metabolic activity of these cells by more than 2000%, whereas treating the media with ENCC resulted in no significant decrease in the metabolic activity of HUVECs or only 30% decrease in the metabolic activity of fibroblasts. Together, these results show that the ENCC can protect the cells from drug-associated damages.
Fig. 7. In vitro biodetoxification effect of ENCC.
(a) Live/Dead staining of HUVECs and NIH/3T3 fibroblasts after 8 h culturing in cell culture media (control), ENCC (1 mg mL−1)-treated DOX (400 μg mL−1)-containing media, and untreated DOX (400 μg mL−1)-containing media. The scale bars are 200 μm. Normalized viability of (b) HUVECs and (c) NIH/3T3 fibroblasts calculated from analyzing the fluorescence images in panel (a). Metabolic activity of (d) HUVECs and (e) NIH/3T3 cultured in similar conditions as (a), measured using the PrestoBlue™ cell viability reagent.
4. Conclusions
Preventing the systemic circulation of localized drugs, e.g., in localized cancer chemotherapy, can help reduce the drug side effects while allowing frequent, high-dosage treatments. Blood as the main carrier of drugs is particularly challenging to treat due to the existence of a broad range of biomolecules and cells. Developing materials that can specifically capture clinically-relevant concentrations of chemotherapy drugs at physiological conditions without imposing adverse effects on cells is of particular challenge. We have nanoengineered cellulose fibers to produce biocompatible anionic hairy cellulose nanocrystals, known as electrosterically stabilized nanocrystalline cellulose (ENCC), that remove more than 6000 μg of DOX, one of the most common chemotherapy drugs, per one mg of nanoparticles. This is more than 3200% improvement compared to some of the most recent nanoadsorbents, such as DNA-based materials. The ENCC immediately separates the drug from the physiological media, enabling the fast, high-efficiency, and biocompatible biodetoxification of blood in vitro/ex vivo. This technology can set the stage for next generation low-cost drug capture devices.
Highlights.
Nanoengineered cellulose provides a super-capacity drug nanoadsorbent.
Dicarboxylated hairy cellulose nanocrystals capture > 6000 mg of DOX per gram.
The drug capture capacity is 3 orders of magnitude higher than DNA.
Hairy nanocelluloses provide a remarkable platform for biodetoxification.
Hairy nanocelluloses may enable next generation drug capture devices.
5. Acknowledgments
A.K. acknowledges funding from the National Institutes of Health (1R01EB024403-01). A.S. would like to acknowledge the financial support from the Canadian Institutes of Health Research (CIHR) post-doctoral fellowship and the startup fund from The Pennsylvania State University.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].Cancer Statistics, Natl. Cancer Inst. (2020). https://www.cancer.gov/about-cancer/understanding/statistics.
- [2].Pearce A, Haas M, Viney R, Pearson S-A, Haywood P, Brown C, Ward R, Incidence and severity of self-reported chemotherapy side effects in routine care: A prospective cohort study, PLoS One. 12 (2017) e0184360. doi: 10.1371/journal.pone.0184360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Statistics, 2021, CA. Cancer J. Clin. 71 (2021) 7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- [4].Waldman AD, Fritz JM, Lenardo MJ, A guide to cancer immunotherapy: from T cell basic science to clinical practice, Nat. Rev. Immunol. 20 (2020) 651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shao K, Singha S, Clemente-casares X, Tsai S, Yang Y, Nanoparticle-based immunotherapy for cancer, ACS Nano. 9 (2015) 16–30. doi: 10.1021/nn5062029. [DOI] [PubMed] [Google Scholar]
- [6].Shen J, Lu Z, Wang J, Zhang T, Yang J, Li Y, Liu G, Zhang X, Advances of Nanoparticles for Leukemia Treatment, ACS Biomater. Sci. Eng. 6 (2020) 6478–6489. doi: 10.1021/acsbiomaterials.0c01040. [DOI] [PubMed] [Google Scholar]
- [7].Rodríguez-Nogales C, González-Fernández Y, Aldaz A, Couvreur P, Blanco-Prieto MJ, Nanomedicines for pediatric cancers, ACS Nano. 12 (2018) 7482–7496. doi: 10.1021/acsnano.8b03684. [DOI] [PubMed] [Google Scholar]
- [8].Ediriwickrema A, Saltzman WM, Nanotherapy for Cancer: Targeting and Multifunctionality in the Future of Cancer Therapies, ACS Biomater. Sci. Eng. 1 (2015) 64–78. doi: 10.1021/ab500084g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].FDA, Daptomycin Highlights of Prescribing Information, 2020.
- [10].FDA, Cisplatin Package Insert, 2009.
- [11].FDA, Fluorouracil Highlights of Prescribing Information, 2016.
- [12].De Souza R, Zahedi P, Allen CJ, Piquette-Miller M, Polymeric drug delivery systems for localized cancer chemotherapy, Drug Deliv. 17 (2010) 365–375. doi: 10.3109/10717541003762854. [DOI] [PubMed] [Google Scholar]
- [13].Llovet JM, Bruix J, Systematic review of randomized trials for unresectable hepatocellular carcinoma: chemoembolization improves survival, Hepatology. 37 (2003) 429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- [14].Vogl TJ, Naguib NNN, Nour-Eldin N-EA, Rao P, Emami AH, Zangos S, Nabil M, Abdelkader A, Review on transarterial chemoembolization in hepatocellular carcinoma: palliative, combined, neoadjuvant, bridging, and symptomatic indications, Eur. J. Radiol. 72 (2009) 505–516. doi: 10.1016/j.ejrad.2008.08.007. [DOI] [PubMed] [Google Scholar]
- [15].Imai N, Ishigami M, Ishizu Y, Kuzuya T, Honda T, Hayashi K, Hirooka Y, Goto H, Transarterial chemoembolization for hepatocellular carcinoma: A review of techniques, World J. Hepatol. 6 (2014) 844. doi: 10.4254/wjh.v6.i12.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Massmann A, Rodt T, Marquardt S, Seidel R, Thomas K, Wacker F, Richter GM, Kauczor HU, Bücker A, Pereira PL, Transarterial chemoembolization (TACE) for colorectal liver metastases—current status and critical review, Langenbeck’s Arch. Surg. 400 (2015) 641–659. doi: 10.1007/s00423-015-1308-9. [DOI] [PubMed] [Google Scholar]
- [17].Sacco R, Tapete G, Simonetti N, Sellitri R, Natali V, Melissari S, Cabibbo G, Biscaglia L, Bresci G, Giacomelli L, Transarterial chemoembolization for the treatment of hepatocellular carcinoma: a review, J. Hepatocell. Carcinoma. 4 (2017) 105. doi: 10.2147/JHC.S103661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tam KY, Leung KCF, Wang YXJ, Chemoembolization agents for cancer treatment, Eur. J. Pharm. Sci. 44 (2011) 1–10. doi: 10.1016/j.ejps.2011.06.013. [DOI] [PubMed] [Google Scholar]
- [19].Ichikawa Y, Ghanefar M, Bayeva M, Wu R, Khechaduri A, Prasad SVN, Mutharasan RK, Naik TJ, Ardehali H, Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation, J. Clin. Invest. 124 (2014) 617–630. doi: 10.1172/JCI72931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jain D, Cardiotoxicity of doxorubicin and other anthracycline derivatives, J. Nucl. Cardiol. 7 (2000) 53–62. doi: 10.1067/mnc.2000.103324. [DOI] [PubMed] [Google Scholar]
- [21].Yee C, McCoy D, Yu J, Losey A, Jordan C, Moore T, Stillson C, Oh HJ, Kilbride B, Roy S, Patel A, Wilson MW, Hetts SW, Endovascular Ion Exchange Chemofiltration Device Reduces Off-Target Doxorubicin Exposure in a Hepatic Intra-arterial Chemotherapy Model, Radiol. Imaging Cancer. 1 (2019) e190009. doi: 10.1148/rycan.2019190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Blumenfeld CM, Schulz MD, Aboian MS, Wilson MW, Moore T, Hetts SW, Grubbs RH, Drug capture materials based on genomic DNA-functionalized magnetic nanoparticles, Nat. Commun. 9 (2018) 1–7. doi: 10.1038/s41467-018-05305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yee DW, Hetts SW, Greer JR, 3D-Printed Drug Capture Materials Based on Genomic DNA Coatings, ACS Appl. Mater. Interfaces. (2021). doi: 10.1021/acsami.1c05209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen XC, Oh HJ, Yu JF, Yang JK, Petzetakis N, Patel AS, Hetts SW, Balsara NP, Block Copolymer Membranes for Efficient Capture of a Chemotherapy Drug, ACS Macro Lett. 5 (2016) 936–941. doi: 10.1021/acsmacrolett.6b00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Oh HJ, Aboian MS, Yi MYJ, Maslyn JA, Loo WS, Jiang X, Parkinson DY, Wilson MW, Moore T, Yee CR, Robbins GR, Barth FM, Desimone JM, Hetts SW, Balsara NP, 3D Printed Absorber for Capturing Chemotherapy Drugs before They Spread through the Body, ACS Cent. Sci. 5 (2019) 419–427. doi: 10.1021/acscentsci.8b00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Karpiński TM, Selected medicines used in iontophoresis, Pharmaceutics. 10 (2018). doi: 10.3390/pharmaceutics10040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rausch K, Reuter A, Fischer K, Schmidt M, Evaluation of nanoparticle aggregation in human blood serum, Biomacromolecules. 11 (2010) 2836–2839. doi: 10.1021/bm100971q. [DOI] [PubMed] [Google Scholar]
- [28].Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC, Brown D, Alkilany AM, Farokhzad OC, Mahmoudi M, Cellular uptake of nanoparticles: journey inside the cell, Chem. Soc. Rev. 46 (2017) 4218–4244. doi: 10.1039/c6cs00636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Buchakjian MR, Ginader T, Tasche KK, Pagedar NA, Smith BJ, Sperry SM, PEGylation as a strategy for improving nanoparticle-based drug and gene delivery, Adv. Drug Deliv. Rev. 99 (2018) 28–51. doi: 10.1177/0194599818773070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu Y, Shipton MK, Ryan J, Kaufman ED, Franzen S, Feldheim DL, Synthesis, stability, and cellular internalization of gold nanoparticles containing mixed peptide-poly(ethylene glycol) monolayers, Anal. Chem. 79 (2007) 2221–2229. doi: 10.1021/ac061578f. [DOI] [PubMed] [Google Scholar]
- [31].Moore TL, Rodriguez-Lorenzo L, Hirsch V, Balog S, Urban D, Jud C, Rothen-Rutishauser B, Lattuada M, Petri-Fink A, Nanoparticle colloidal stability in cell culture media and impact on cellular interactions, Chem. Soc. Rev. 44 (2015) 6287–6305. doi: 10.1039/c4cs00487f. [DOI] [PubMed] [Google Scholar]
- [32].Doane TL, Chuang C-H, Hill RJ, Burda C, Nanoparticle ζ-potentials, Acc. Chem. Res. 45 (2012) 317–326. doi: 10.1021/ar200113c. [DOI] [PubMed] [Google Scholar]
- [33].Kim J-H, Shim BS, Kim HS, Lee Y-J, Min S-K, Jang D, Abas Z, Kim J, Review of nanocellulose for sustainable future materials, Int. J. Precis. Eng. Manuf. Technol. 2 (2015) 197–213. doi: 10.1007/s40684-015-0024-9. [DOI] [Google Scholar]
- [34].Endes C, Camarero-Espinosa S, Mueller S, Foster EJ, Petri-Fink A, Rothen-Rutishauser B, Weder C, Clift MJD, A critical review of the current knowledge regarding the biological impact of nanocellulose, J. Nanobiotechnology. 14 (2016) 78. doi: 10.1186/s12951-016-0230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Roman M, Toxicity of Cellulose Nanocrystals: A Review, Ind. Biotechnol. 11 (2015) 25–33. doi: 10.1089/ind.2014.0024. [DOI] [Google Scholar]
- [36].Clift MJD, Foster EJ, Vanhecke D, Studer D, Wick P, Gehr P, Rothen-Rutishauser B, Weder C, Investigating the interaction of cellulose nanofibers derived from cotton with a sophisticated 3D human lung cell coculture, Biomacromolecules. 12 (2011) 3666–3673. doi: 10.1021/bm200865j. [DOI] [PubMed] [Google Scholar]
- [37].Yanamala N, Farcas MT, Hatfield MK, Kisin ER, Kagan VE, Geraci CL, Shvedova AA, In vivo evaluation of the pulmonary toxicity of cellulose nanocrystals: A renewable and sustainable nanomaterial of the future, ACS Sustain. Chem. Eng. 2 (2014) 1691–1698. doi: 10.1021/sc500153k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Domingues RMA, Gomes ME, Reis RL, The potential of cellulose nanocrystals in tissue engineering strategies, Biomacromolecules. 15 (2014) 2327–2346. doi: 10.1021/bm500524s. [DOI] [PubMed] [Google Scholar]
- [39].Cullen RT, Miller BG, Jones AD, Davis JMG, Toxicity of cellulose fibres, Ann. Occup. Hyg. 46 (2002) 81–84. doi: 10.1093/annhyg/46.suppl-1.81. [DOI] [Google Scholar]
- [40].Sheikhi A, Hayashi J, Eichenbaum J, Gutin M, Kuntjoro N, Recent advances in nanoengineering cellulose for cargo delivery, J. Control. Release. 294 (2019) 53–76. doi: 10.1016/j.jconrel.2018.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cummings JH, Cellulose and the human gut., Gut. 25 (1984) 805–810. doi: 10.1136/gut.25.8.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Moon RJ, Martini A, Nairn J, Simonsen J, Youngblood J, Cellulose nanomaterials review: structure, properties and nanocomposites, Chem. Soc. Rev. 40 (2011) 3941–3994. doi: 10.1039/c0cs00108b. [DOI] [PubMed] [Google Scholar]
- [43].Roy D, Semsarilar M, Guthrie JT, Perrier S, Cellulose modification by polymer grafting: a review, Chem. Soc. Rev. 38 (2009) 2046–2064. doi: 10.1039/B808639G. [DOI] [PubMed] [Google Scholar]
- [44].Foster EJ, Moon RJ, Agarwal UP, Bortner MJ, Bras J, Camarero-Espinosa S, Chan KJ, Clift MJD, Cranston ED, Eichhorn SJ, Current characterization methods for cellulose nanomaterials, Chem. Soc. Rev. (2018). doi: 10.1039/c6cs00895j. [DOI] [PubMed] [Google Scholar]
- [45].Tao H, Lavoine N, Jiang F, Tang J, Lin N, Reducing end modification on cellulose nanocrystals: Strategy, characterization, applications and challenges, Nanoscale Horizons. 5 (2020) 607–627. doi: 10.1039/d0nh00016g. [DOI] [PubMed] [Google Scholar]
- [46].Zhu H, Luo W, Ciesielski PN, Fang Z, Zhu JY, Henriksson G, Himmel ME, Hu L, Wood-Derived Materials for Green Electronics, Biological Devices, and Energy Applications, Chem. Rev. 116 (2016) 9305–9374. doi: 10.1021/acs.chemrev.6b00225. [DOI] [PubMed] [Google Scholar]
- [47].Klemm D, Kramer F, Moritz S, Lindström T, Ankerfors M, Gray D, Dorris A, Nanocelluloses: A new family of nature-based materials, Angew. Chem. Int. Ed. 50 (2011) 5438–5466. doi: 10.1002/anie.201001273. [DOI] [PubMed] [Google Scholar]
- [48].Sheikhi A, Emerging cellulose-based nanomaterials and nanocomposites, in: Nanomater. Polym. Nanocomposites, Elsevier, 2019: pp. 307–351. doi: 10.1016/B978-0-12-814615-6.00009-6. [DOI] [Google Scholar]
- [49].van de Ven TGM, Sheikhi A, Hairy cellulose nanocrystalloids: A novel class of nanocellulose, Nanoscale. 8 (2016) 15101–15114. doi: 10.1039/c6nr01570k. [DOI] [PubMed] [Google Scholar]
- [50].Sheikhi A, Yang H, Alam MN, van de Ven TGM, Highly stable, functional hairy nanoparticles and biopolymers from wood fibers: Towards sustainable nanotechnology, J. Vis. Exp. 113 (2016) e54133. doi: 10.3791/54133. [DOI] [PubMed] [Google Scholar]
- [51].Sheikhi A, van de Ven TGM, Colloidal aspects of Janus-like hairy cellulose nanocrystalloids, Curr. Opin. Colloid Interface Sci. 29 (2017) 21–31. doi: 10.1016/j.cocis.2017.02.001. [DOI] [Google Scholar]
- [52].Sheikhi A, Safari S, Yang H, van de Ven TGM, Copper removal using electrosterically stabilized nanocrystalline cellulose, ACS Appl. Mater. Interfaces. 7 (2015) 11301–11308. doi: 10.1021/acsami.5b01619. [DOI] [PubMed] [Google Scholar]
- [53].van der Zanden SY, Qiao X, Neefjes J, New insights into the activities and toxicities of the old anticancer drug doxorubicin, FEBS J. (2020). doi: 10.1111/febs.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pugazhendhi A, Edison TNJI, Velmurugan BK, Jacob JA, Karuppusamy I, Toxicity of Doxorubicin (Dox) to different experimental organ systems, Life Sci. 200 (2018) 26–30. doi: 10.1016/j.lfs.2018.03.023. [DOI] [PubMed] [Google Scholar]
- [55].Keizer HG, Pinedo HM, Schuurhuis GJ, Joenje H, Doxorubicin (adriamycin): a critical review of free radical-dependent mechanisms of cytotoxicity, Pharmacol. Ther. 47 (1990) 219–231. doi: 10.1016/0163-7258(90)90088-J. [DOI] [PubMed] [Google Scholar]
- [56].Songbo M, Lang H, Xinyong C, Bin X, Ping Z, Liang S, Oxidative stress injury in doxorubicin-induced cardiotoxicity, Toxicol. Lett. 307 (2019) 41–48. doi: 10.1016/j.toxlet.2019.02.013. [DOI] [PubMed] [Google Scholar]
- [57].Yang H, Tejado A, Alam N, Antal M, van de Ven TGM, Films prepared from electrosterically stabilized nanocrystalline cellulose, Langmuir. 28 (2012) 7834–7842. doi: 10.1021/la2049663. [DOI] [PubMed] [Google Scholar]
- [58].Araki J, Wada M, Kuga S, Steric stabilization of a cellulose microcrystal suspension by poly (ethylene glycol) grafting, Cellulose. 17 (2001) 21–27. doi: 10.1021/la001070m. [DOI] [Google Scholar]
- [59].Tavakolian M, Wiebe H, Sadeghi MA, Van De Ven TGM, Dye Removal Using Hairy Nanocellulose: Experimental and Theoretical Investigations, ACS Appl. Mater. Interfaces. 12 (2020) 5040–5049. doi: 10.1021/acsami.9b18679. [DOI] [PubMed] [Google Scholar]
- [60].Nečas D, Klapetek P, Gwyddion: an open-source software for SPM data analysis, Open Phys. 10 (2012) 181–188. doi: 10.2478/s11534-011-0096-2. [DOI] [Google Scholar]
- [61].ASTM E2524 – 08 Standard Test Method for Analysis of Hemolytic Properties of Nanoparticles, ASTM Int. (2008). [Google Scholar]
- [62].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat. Methods. 9 (2012) 671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fraschini C, Chauve G, Bouchard J, TEMPO-mediated surface oxidation of cellulose nanocrystals (CNCs), Cellulose. 24 (2017) 2775–2790. doi: 10.1007/s10570-017-1319-5. [DOI] [Google Scholar]
- [64].Lu P, Lo Hsieh Y, Preparation and properties of cellulose nanocrystals: Rods, spheres, and network, Carbohydr. Polym. 82 (2010) 329–336. doi: 10.1016/j.carbpol.2010.04.073. [DOI] [Google Scholar]
- [65].Fülöp Z, Gref R, Loftsson T, A permeation method for detection of self-aggregation of doxorubicin in aqueous environment, Int. J. Pharm. 454 (2013) 559–561. doi: 10.1016/j.ijpharm.2013.06.058. [DOI] [PubMed] [Google Scholar]
- [66].Langmuir I, The adsorption of gases on plane surfaces of glass, mica and platinum., J. Am. Chem. Soc. 40 (1918) 1361–1403. [Google Scholar]
- [67].Yang H, Sheikhi A, van de Ven TGM, Reusable Green Aerogels from Cross-Linked Hairy Nanocrystalline Cellulose and Modified Chitosan for Dye Removal, Langmuir. 32 (2016) 11771–11779. doi: 10.1021/acs.langmuir.6b03084. [DOI] [PubMed] [Google Scholar]
- [68].Biga LM, Dawson S, Harwell A, Hopkins R, Kaufmann J, LeMaster M, Matern P, Morrison-Graham K, Quick D, Runyeon J, Anatomy & Physiology, in: Anat. Physiol, 2017: pp. 1054–1063, 1727–1737. doi: 10.5399/osu/1116. [DOI] [Google Scholar]
- [69].Sheikhi A, Kakkar A, van de Ven TGM, A leaf out of Nature’s book: hairy nanocelluloses for bioinspired mineralization, Cryst. Growth Des. 16 (2016) 4627–4634. doi: 10.1021/acs.cgd.6b00713. [DOI] [Google Scholar]
- [70].Peters T, All about albumin: biochemistry, Genet. Med. Appl. (Academic Press. 1996). (1996). [Google Scholar]
- [71].Fologea D, Ledden B, McNabb DS, Li J, Electrical characterization of protein molecules by a solid-state nanopore, Appl. Phys. Lett. 91 (2007) 1–9. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS, The human serum metabolome, PLoS One. 6 (2011). doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kim H-S, Lee Y-S, Kim D-K, Doxorubicin exerts cytotoxic effects through cell cycle arrest and Fas-mediated cell death, Pharmacology. 84 (2009) 300–309. doi: 10.1159/000245937. [DOI] [PubMed] [Google Scholar]
- [74].Huang L, Perrault C, Coelho-Martins J, Hu C, Dulong C, Varna M, Liu J, Jin J, Soria C, Cazin L, Induction of acquired drug resistance in endothelial cells and its involvement in anticancer therapy, J. Hematol. Oncol. 6 (2013) 49. doi: 10.1186/1756-8722-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Izaguirre L, Pinilla I, Gonzalvo F, Pérez S, Honrubia FM, Effect of doxorubicin on fibroblast migration and proliferation, Ann. Ophthalmol. 35 (2003) 48–52. doi: 10.1385/AO:35:1:48. [DOI] [Google Scholar]