ABSTRACT
Hepatitis B virus (HBV) encodes a regulatory protein, termed HBx, that has been intensely studied in the past and shown to play a key role(s) in viral transcription and replication. In addition, a huge body of work exists in the literature related to signal transduction and possible mechanism(s) leading to hepatocarcinogenesis associated with infection. We have previously reported that HBV transcripts are modified by N6-methyladenosine (m6A) at the single consensus DRACH motif at nucleotides (nt) 1905 to 1909 in the epsilon structural element, and this m6A modification affects the HBV life cycle. In this study, we present evidence that additional variants of m6A (DRACH) motifs located within nt 1606 to 1809 correspond to the coding region of HBx mRNA and 3′ untranslated region (UTR) of other viral mRNAs. Using the mutants of additional m6A sites in nt 1606 to 1809 and a depletion strategy of m6A methyltransferases (METTL3/14) and reader proteins (YTHDFs), we show that m6A modification at nt 1616, located in the HBx coding region, regulates HBx protein expression. The HBx RNA and protein expression levels were notably increased by the silencing of m6A reader YTHDF2 and methyltransferases as well as the mutation of m6A sites in the HBx coding region. However, other viral protein expression levels were not affected by the m6A modification at nt 1616. Thus, m6A modifications in the HBx open reading frame (ORF) downregulate HBx protein expression, commonly seen during HBV transfections, transgenic mice, and natural infections of human hepatocytes. These studies identify the functional role of m6A modification in the subtle regulation of HBx protein expression consistent with its possible role in establishing chronic hepatitis.
IMPORTANCE N6-methyladenosien (m6A) modifications recently have been implicated in the HBV life cycle. Previously, we observed that m6A modification occurs in the adenosine at nt 1907 of the HBV genome, and this modification regulates the viral life cycle. Here, we identified an additional m6A site located in nt 1616 of the HBV genome. This modification negatively affects HBx RNA and protein expression. In the absence of m6A methyltransferases (METTL3/14) and reader protein (YTHDF2), the HBx RNA and protein expression were increased. Using HBV mutants that lack m6A in the HBx coding region, we present the unique positional effects of m6A in the regulation of HBx protein expression.
KEYWORDS: hepatitis B virus, N6-methyladenosine, HBx protein, HBV life cycle
INTRODUCTION
Hepatitis B virus (HBV) infection leads to chronic hepatitis and carries the risk of the development of hepatocellular carcinoma (1, 2). Upon its entry into the hepatocytes via Na/taurocholate cotransporting polypeptide (NTCP) receptor, the 3.2-kb partially double-stranded relaxed circular DNA (rcDNA) genome is transported to the nucleus, where it transforms into a covalently closed circular DNA (cccDNA) (1–3). cccDNA serves as a template to produce four viral mRNAs. The HBV DNA genome contains four major promoters and two enhancers that differentially express their transcripts, which include 3.5- to 3.6-kb precore mRNA, 3.5-kb pregenomic RNA (pgRNA), 2.4- and 2.1-kb surface antigen mRNA, and 0.7-kb X mRNA (1, 2). Transcription of these mRNAs is initiated downstream from respective promoters/enhancers in a heterogeneous multitude fashion but ends at a single poly(A) site of the DNA genome. HBV pgRNA becomes encapsidated along with viral polymerase (Pol) into core particles where the reverse transcription (RT) function of Pol converts pgRNA into an rcDNA molecule (1, 2). Enhancer 1 and associated promoter express the X open reading frame (ORF), which encodes a 154-amino-acid protein translated from a 0.7-kb X mRNA. HBx is expressed at almost undetectable levels during infection and in transfected cultured cells, although the HBx antibodies are detected in human sera from HBV-infected patients (1–5). This study addresses the molecular mechanisms behind this layer of RNA regulation.
Interest in the functional properties of HBx protein dates back to the time soon after its molecular cloning from HBV-infected patients’ sera (4, 5). One of its widely agreed-upon functions is in viral replication and transcription that has been studied using myriad approaches and confirmed in vivo (6–8). HBx does not bind DNA but interacts with host transcription factors and coactivators (4, 6–10). These activities place HBx at the episomal HBV cccDNA to regulate the transcription of viral genes from cccDNA (9, 11). Recent interest has focused on the binding of HBx to DNA damage-binding protein 1 (DDB-1) in concert with Cullin 4 and HBx-mediated ubiquitin-proteasomal degradation of the host restriction factor structural maintenance of chromosome 5/6 (SMC5/6) on the cccDNA to initiate transcription of viral genes (12). This has prompted therapeutic interest in HBx due to its association with cccDNA, with functions associated with the initiation of transcription both at the co- and posttranscriptional levels (9). Its elusive role in the development of hepatocellular carcinoma (HCC), which, by all accounts, is indirect, has been studied extensively (1–5).
N6-methyladenosine (m6A) modification of cellular RNA represents one of the most common chemical modifications and is intensely studied and well-characterized (13). This modification regulates various biological processes, including cellular metabolism, stem cell differentiation, stress responses, and cancer development (13). m6A modification occurs cotranscriptionally within the consensus DRACH motifs (where D = A, G, or T, R = A or G, A = methylated adenosine, C = C, and H = A, C, or T) by a complex of methyltransferase-like (METTL) and additional adapter proteins (14). The demethylases (FTO and ALKBH5) remove the m6A modification in cellular RNAs (14). Typically, m6A methylation is enriched near the 3′-untranslated region (UTR) and the stop codons and recognized by m6A reader proteins represented by the YTH domain family (YTHDF) proteins (15, 16). m6A reader proteins regulate m6A-methylated RNA translation and stability by direct interaction with m6A-modified RNAs. m6A methylation has been more frequently identified in the transcripts of DNA viruses and RNA virus genomes than cellular RNAs (17–23). Generally, m6A methyltransferases (METTL3/14) and reader YTHDF proteins regulate the viral replication and translation of both RNA and DNA viruses. In the case of HBV, m6A modification regulates the stability and subcellular localization of viral mRNAs, interferon (IFN)-mediated degradation, and innate immune response (22–25). The newer roles of m6A modification in the viral life cycle are continually being characterized.
Previously, we characterized the major m6A peaks at nucleotides (nt) 1815 to 1950 using methylated RNA immunoprecipitation (MeRIP) sequencing assay and identified a single consensus DRACH motif at nt 1907, located within the lower stem-loop of the epsilon element (Fig. 1A to C) (22). All HBV transcripts acquire the epsilon element in the 3′ end, but pgRNA carries this element at its 5′ end as well due to the terminal redundancy of sequences (Fig. 1B and C). The m6A modification at nt 1907 plays differential roles in the viral life cycle (22). The m6A modification of the 3′ epsilon element reduces viral RNA stability via binding to YTHDF2 reader protein and affects the translation of viral proteins, whereas the m6A methylation of the 5′ epsilon element is required for core-associated viral DNA synthesis (24). An in-depth characterization of this observation is currently being pursued. In addition to major m6A peaks, minor m6A peaks were noted within nt 1606 to 1809, located at all HBV transcripts (Fig. 1A and B), but the functions of these m6A modifications have not been characterized. In this study, we focused on five additional variants of DRACH motifs in the minor m6A peaks (nt 1606 to 1809) reported previously (22). We mutated these sites and observed their effects on HBx protein and mRNA expression. The mutation in the DRACH motif at nt 1616 resulted in a significant increase in the expressions of HBx protein and mRNA. This effect is mediated by the interaction between m6A reader protein YTHDF2 and m6A modification at nt 1616 of the HBx mRNA. These studies provide evidence that the degradation of m6A methylated HBx mRNA catalyzed by YTHDF2 protein leads to modest levels of HBx mRNA (and polypeptide) expression, consistent with HBx expression observed in vitro and in natural infections. This unique regulatory mechanism allows the expression of HBx at levels required to maintain chronic hepatitis and other syndromes associated with the HBV infection.
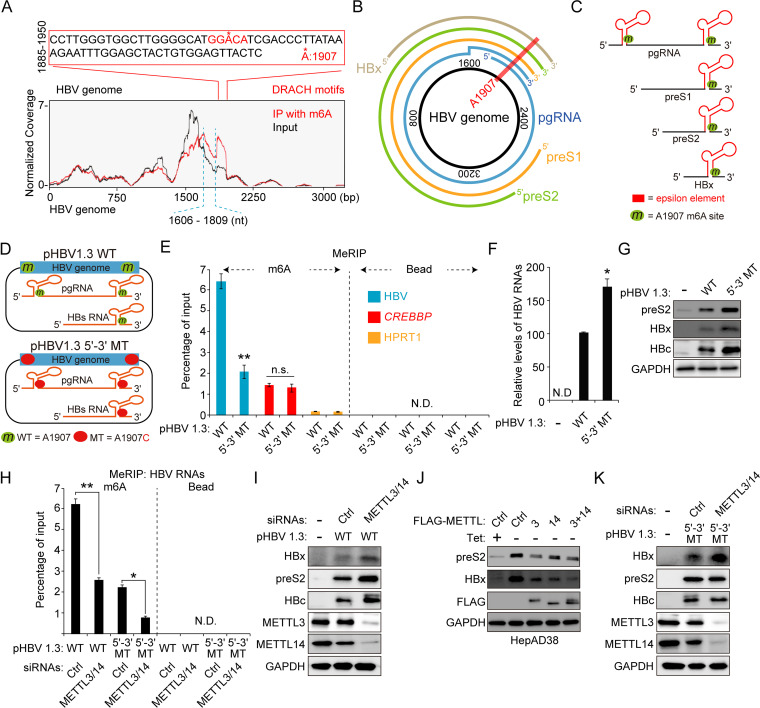
FIG 1.
Regulation of HBx protein expression by m6A modification sites other than the nt 1907 m6A site. (A) The m6A peaks are identified in nt 1606 to 1809 and nt 1885 to 1950 of the HBV genome using methylated RNA immunoprecipitation (IP) sequencing. The major peak in 1885 to 1950 includes a single m6A consensus motif at nt 1907, located on the lower stem-loop of the epsilon element. The minor peak corresponds to nt 1606 to 1809. (B) The location of the 1907 m6A site in the pictorial representation of transcripts within the HBV genome is indicated by a red line across the HBV DNA/transcripts. (C) Schematic showing the position of the 1907 m6A site, indicated by the green circle in all of the HBV RNAs. (D) Schematics indicate the 5′ and 3′ m6A sites in the epsilon elements of HBV pgRNA. Green circles indicate the m6A site, and red circles indicate the A1907C mutation in HBV pgRNA. pHBV 1.3 5′–3′ MT contains A1907C mutation at the 5′ and 3′ ends. (E to G) The indicated plasmids were transfected in Huh7 cells for 72 h. m6A methylated RNAs were immunoprecipitated from total RNA using an anti-m6A antibody. (E) m6A-methylated HBV RNAs, CREBBP, and HPRT1 RNAs were analyzed by input RNA levels by RT-qPCR. CREBBP and HPRT1 serve as positive and negative controls (Ctrl), respectively. (F) The HBV RNA levels were analyzed by RT-qPCR. (G) The indicated proteins were analyzed by immunoblotting. (H) Huh7 cells expressing the HBV WT or 5′–3′ MT were transfected with METTL3/14 siRNAs. Total RNA was immunoprecipitated using an anti-m6A antibody. m6A-methylated RNAs were analyzed by RT-qPCR. (I) Huh7 cells expressing the HBV WT were transfected with METTL3/14 siRNAs. After 48 h, cellular lysates were isolated and the indicated proteins were analyzed by Western blotting. (J) The indicated plasmids were transfected into HepAD38 cells grown in the absence or presence of tetracycline for 48 h. Cellular lysates were prepared from these cells and analyzed by Western blotting. (K) Huh7 cells expressing the HBV 5′–3′ MT were transfected with METTL3/14 siRNAs. After 48 h, cellular lysates were isolated and the indicated proteins were analyzed by Western blotting. In panels E, F, and H, the error bars represent the SDs from three independent experiments. The P values were calculated via an unpaired Student's t test. *, P < 0.05; **, P < 0.01. WT, wild type; MT, mutant; ND, nondetect.
RESULTS
m6A modifications occur at nt 1606 to 1809 of HBV genome.
We conducted an MeRIP reverse transcription-quantitative PCR (RT-qPCR) assay using the m6A-specific antibody in HBV-transfected cells to determine whether m6A modifications occur at DRACH motifs in a minor m6A peak (nt 1606 to 1809) of the HBV genome in addition to the m6A site at nt 1907. For MeRIP–RT-qPCR assay, we transfected pHBV 1.3 5′–3′ mutant (MT) plasmid, containing A1907C mutation in both 5′ and 3′ epsilon elements, into Huh7 cells to exclude the effect of m6A modification at nt 1907 (Fig. 1D). HBV RNAs from pHBV 1.3 wild-type (WT) or 5′–3′ MT-transfected Huh7 cells were immunoprecipitated using the m6A-specific antibodies, and m6A-methylated HBV RNAs from immunoprecipitates were amplified by RT-qPCR (Fig. 1E). Interestingly, we observed that m6A modification still occurred in HBV RNAs transcribed from pHBV 1.3 5′–3′ MT. In the MeRIP RT-qPCR assay, CREBBP and HPRT1 were used as positive and negative controls, respectively. The mutation of nt 1907 increased viral RNA and protein levels (Fig. 1F and G), as previously reported. These results suggest that HBV transcripts are m6A methylated at other variant DRACH motifs in nt 1609 to 1809 in addition to the m6A site at nt 1907.
METTL3/14 affect HBx protein expression through m6A modifications at nt 1606 to 1809 of the HBV genome.
We next investigated the effect of m6A methyltransferases (METTL3/14) on m6A modifications in the minor m6A peaks (nt 1606 to 1809). Short interfering RNA (siRNA)-mediated depletion of METTL3/14 substantially reduced the levels of m6A methylated HBV mRNAs in both WT and 5′–3′ MT-transfected cells (Fig. 1H), indicating that METTL3 and -14 catalyze the m6A modifications in the minor m6A peaks. Because it was reported that the m6A modification at nt 1907 reduces viral RNA stability and protein expression (22), depletion of METTL3/14 increased viral RNA and protein levels in HBV WT-transfected cells (Fig. 1I). On the contrary, overexpression of METTL3/14 reduced HBV protein expression (Fig. 1J). Surprisingly, the silencing of METTL3/14 increased only HBx protein expression in HBV 5′–3′ MT transfected cells but did not affect preS2 and core protein levels (Fig. 1K). Taken together, these results demonstrate that m6A modifications occur at additional variant DRACH motifs in nt 1606 to 1809 other than the 1907 m6A site, and m6A methylations in additional DRACH motifs regulate specifically HBx protein expression but do not affect other viral protein expression levels.
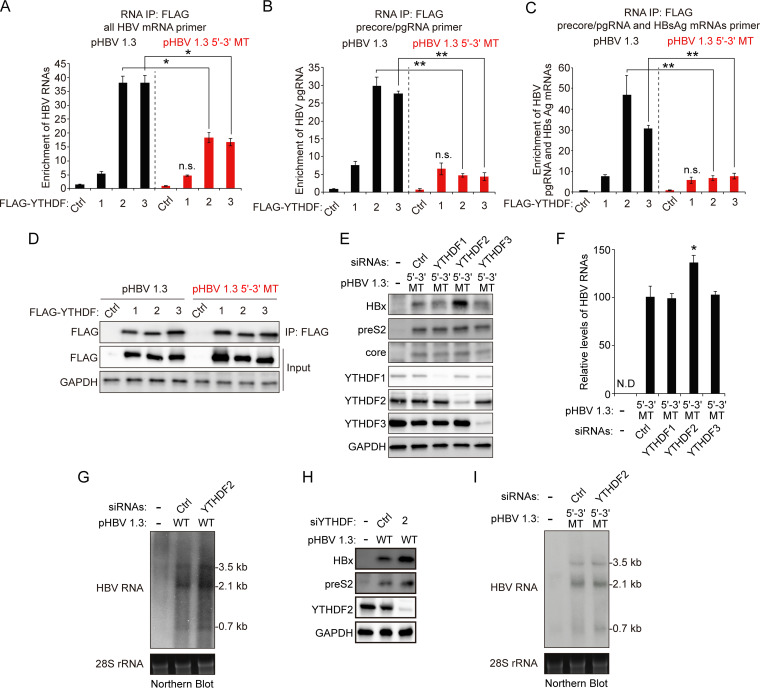
YTHDF2 m6A reader protein regulates HBx mRNA and protein expression via interaction with m6A sites in HBx coding region.
The YTHDF proteins are cellular m6A RNA-binding proteins (readers) that regulate the translation and stability of m6A-methylated RNA (16). Hence, we determined whether YTHDF proteins bind to additional m6A sites of HBV RNAs to affect HBx protein expression. Huh7 cells expressing HBV WT or 5′–3′ MT genome were transfected with plasmids encoding FLAG-YTHDF1, YTHDF2, and YTHDF3, and these cells were irradiated by UV to produce cross-linked RNA-protein complexes (Fig. 2A to D) (26). The cellular lysates were extracted from these cells and subjected to immunoprecipitation assay using an anti-FLAG antibody. The results show that HBV WT mRNAs were enriched in both YTHDF2 and YTHDF3 immunoprecipitates relative to the control and YTHDF1 (Fig. 2A). Interestingly, the HBV mRNAs transcribed from the HBV 5′–3′ MT genome were also recognized by YTHDF2 and YTHDF3, indicating that YTHDF2/3 proteins interact with HBV RNAs by m6A modifications in the minor m6A peaks of nt 1610 to 1809. We further performed RT-qPCR assay using specific primers for precore/pgRNA or precore/pgRNA and HBsAg mRNAs and observed that the A1907C mutation specifically disrupts the interaction between YTHDF2/3 and pgRNA/HBsAg mRNAs (Fig. 2B and C). These results suggest that YTHDF2/3 proteins bind to HBx mRNA via additional m6A sites in nt 1606 to 1809. To determine whether YTHDF proteins affect HBx protein expression via these m6A sites, we silenced the expression of YTHDF proteins using specific siRNAs in HBV 5′–3′ MT-expressing cells. Importantly, we found that the silencing of YTHDF2 significantly increased the HBx protein expression, while the expression of the preS2 and core proteins remained unaffected (Fig. 2E). However, depletion of YTHDF2 slightly increased HBV mRNAs levels compared to YTHDF1 and -3 (Fig. 2F). We further confirmed these results using the Northern blot assay (Fig. 2G and H). Depletion of YTHDF2 increased all HBV mRNA and protein levels in HBV WT-transfected cells (Fig. 2G and H). Importantly, in the case of HBV 5′–3′ MT-transfected cells, HBx mRNAs levels were increased by the silencing of YTHDF2, but pgRNA and HBsAg mRNA levels were not altered (Fig. 2I). Taken together, these results suggest that additional m6A modification occurs at nt 1606 to 1809 of HBx mRNA by m6A methyltransferases other than the nt 1907 DRACH motif and that YTHDF2 protein affects HBx mRNA and protein expression via the interaction with additional m6A modifications.
FIG 2.
Silencing of m6A reader YTHDF2 increases HBx mRNA and protein expression via interaction with m6A sites of the HBx coding region. (A to D) Huh7 cells expressing the HBV WT or 5′–3′ MT were transfected with the FLAG-YTHDF expression plasmids. After 48 h, total RNA and cell lysates were isolated from these cells. Total RNA was immunoprecipitated using an anti-FLAG antibody. (A to C) The immunoprecipitated HBV RNA levels were analyzed by the indicated primers. (D) The indicated proteins were analyzed by Western blotting. (E and F) Huh7 cells were transfected with pHBV 1.3 5′–3′ MT plasmid and then treated with YTHDF2 siRNAs for 48 h. Cell lysates and total RNA were isolated and analyzed by immunoblotting (E) and RT-qPCR (F). (G and H) Huh7 cells expressing HBV WT genome were transfected with the YTHDF2 siRNAs. After 48 h, total RNA and cellular lysates were extracted from these cells for Northern blotting (G) and Western blotting (H). (I) Huh7 cells expressing HBV 5′–3′ MT genome were transfected with the YTHDF2 siRNAs. After 48 h, total RNA and cellular lysates were extracted from these cells for Northern blotting. In panels A to C and F, the error bars represent the SDs from three independent experiments. The P values are calculated via an unpaired Student's t test. *, P < 0.05; **, P < 0.01. WT, wild type; MT, mutant; IP, immunoprecipitation; n.s., nonsignificant; ND, not detected.
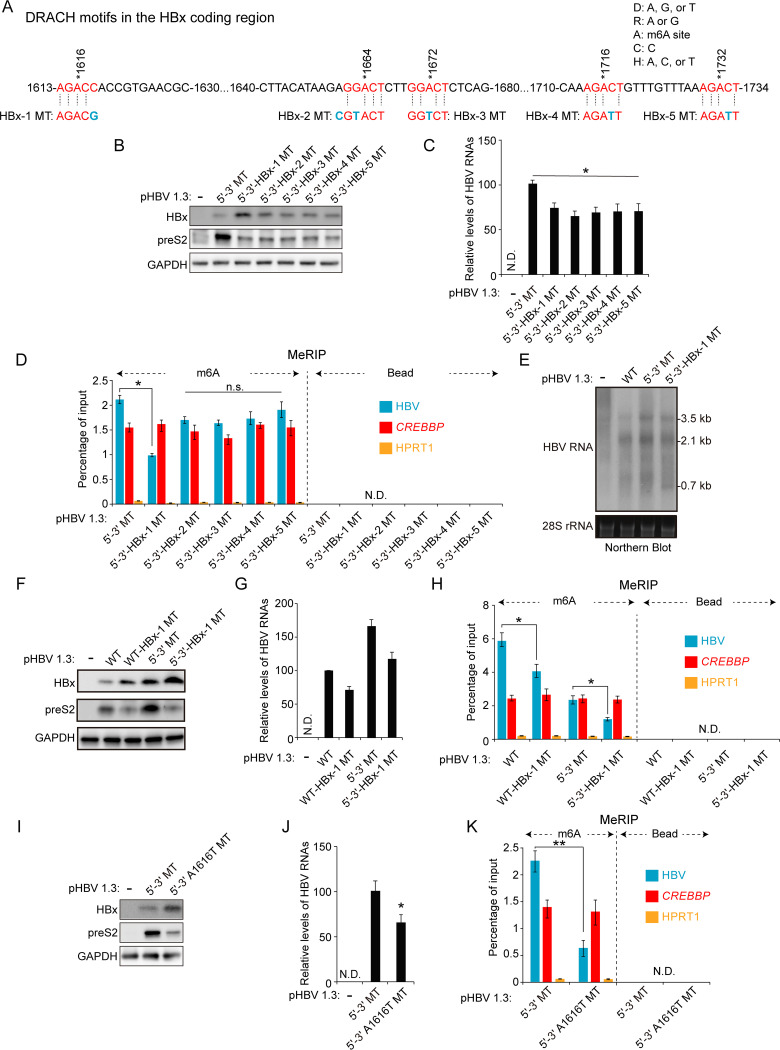
m6A modification at nt 1616 affects HBx mRNA and protein expression.
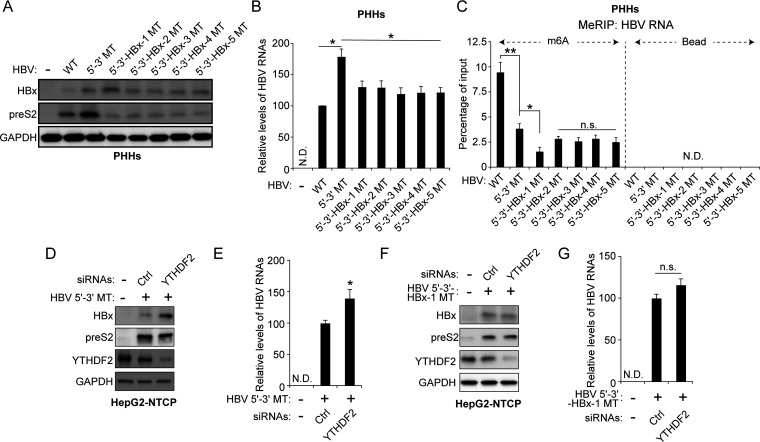
m6A modification occurs at a distinct consensus DRACH motif, but not all DRACH motifs are m6A methylated (15). Thus, we searched for consensus DRACH motifs or their variant sequences within the minor m6A peak bordering nt 1609 and 1809 in the HBV genomes. The sequence-based m6A modification site predictor (SRAMP) revealed that minor m6A peaks included five DRACH motifs at A1616, A1664, A1672, A1716, and A1732 (Fig. 3A) (27). These variants of the DRACH motif were mutated without changing the amino acids (silent mutations) to determine which m6A sites are important in HBx protein expression. These mutants were generated in the pHBV 1.3 5′–3′ MT plasmid to exclude the effect of m6A modification at nt 1907 on HBx protein expression. We transfected the HBx mutant plasmids (pHBV 1.3 5′–3′ HBx-1, HBx-2, HBx-3, HBx-4, and HBx-5 MT) into Huh7 cells and analyzed HBx protein expression levels (Fig. 3B). Of the 5 mutants, only the HBx-1 MT (nt 1616) showed an increase in HBx expression, whereas preS2 expression levels were reduced by all HBx mutations compared with HBV 5′–3′ MT. Similarly, HBV RNA levels analyzed by RT-qPCR showed a slight decrease in all cells transfected with each HBx mutant (Fig. 3C). MeRIP analysis showed that HBx-1 mutation reduced m6A methylation of HBV RNAs while other mutations (HBx-2, -3, -4, and -5 MT) did not affect m6A methylated HBV RNA levels (Fig. 3D). We then conducted the Northern blot assay to determine whether HBx-1 mutation affects HBx mRNA expression levels (Fig. 3E). The result shows that pgRNA and HBsAg mRNAs were slightly decreased in HBx-1 MT-transfected cells, but HBx-1 mutation clearly increased HBx mRNA levels. In addition, we generated HBx-1 mutation based on pHBV 1.3 WT plasmids and observed similar results (Fig. 3F to H). HBx-1 mutation in pHBV 1.3 WT also increased HBx protein expression but reduced m6A-methylated HBV RNA levels. When we designed the HBx-1 MT plasmid, the G (guanosine) at nt 1618 was changed to C (cytosine) to produce a silent mutation (Fig. 3A). We also generated the A1616T mutation in which A (adenosine) at nt 1616 was mutated to T (thymine). This mutation contains the change of amino acid from Thr (threonine) to Ser (serine). Similarly, the A1616T mutation decreased m6A-methylated HBV RNA levels while it increased HBx protein expression (Fig. 3I to K). Moreover, we observed that preS2 protein expression was reduced by A1616T mutation. The reduced preS2 expression levels by mutations in the HBx coding region seem not to be dependent on m6A modification, because HBx-2, -3, -4, and -5 mutations reduced preS2 levels regardless of no alteration of m6A-methylated HBV RNA levels. Next, we carried out these analyses in the HBV infection system using primary human hepatocytes (PHHs). PHHs were infected with HBV WT, 5′–3′ MT, or HBx MT series infectious particles, and we analyzed HBV protein and m6A-methylated HBV RNA levels (Fig. 4A to C). Similarly, HBx-1 mutation increased HBx protein expression but decreased preS2 levels in HBV-infected PHHs. As depicted in Fig. 4C, the m6A-methylated HBV RNA level was reduced in HBx-1 MT-infected PHHs. However, other HBx mutations did not affect HBx protein expression but decreased preS2 protein expression without the alteration of m6A levels. Therefore, these results suggest that m6A modification occurs at nt 1616, and this modification specifically reduces HBx mRNA and protein expression.
FIG 3.
Of the additional variant m6A sites in the HBx coding region, m6A modification at nt 1616 of the HBV genome regulates HBx protein expression. (A) The location of m6A consensus DRACH motifs within the minor m6A peak (1613 to 1734 nt) of the HBV genome is shown in red. Blue characters indicate mutations to disrupt the DRACH motifs in the HBx coding region. HBx-1, -2, -3, -4, or -5 MT contains the mutation of the DRACH motif at the position indicated in blue. (B to D) The indicated HBV expression plasmids were transfected into Huh7 cells for 72 h. Cell lysates and total RNA were extracted from these cells. (B) The indicated proteins were analyzed by immunoblotting. (C) HBV RNAs were analyzed by RT-qPCR. (D) m6A-methylated RNAs were immunoprecipitated from total RNA using an anti-m6A antibody, and m6A-methylated HBV RNA levels were normalized to input RNA levels by RT-qPCR. (E) Huh7 cells were transfected with pHBV 1.3 WT, 5′–3′ MT, or 5′–3′-HBx-1 MT plasmid for 72 h. Cellular HBV RNAs were analyzed by Northern blotting. (F to H) Huh7 cells were transfected with the indicated plasmids for 72 h. Cell lysates and total RNA were extracted from these cells. (F) The indicated protein expression levels were analyzed by Western blotting. (G) HBV RNA levels were analyzed by RT-qPCR. (H) m6A-methylated RNAs were immunoprecipitated from total RNA using an anti-m6A antibody and m6A-methylated HBV RNAs were normalized by input RNA levels by RT-qPCR. (I to K) Huh7 cells were transfected with pHBV 1.3 5′–3′ MT or pHBV 1.3 5′–3′-A1616T MT plasmid for 72 h. (I) The indicated proteins were analyzed by Western blotting. (J) HBV RNA levels were analyzed by RT-qPCR. (K) m6A methylated HBV RNAs were immunoprecipitated from total RNA using an anti-m6A antibody and normalized by input RNA levels by RT-qPCR. In panels C, D, G, H, J, and K, the error bars represent the SDs from three independent experiments. The P values were calculated via an unpaired Student's t test. *, P < 0.05; **, P < 0.01. n.s., nonsignificant; ND, not detected.
FIG 4.
Regulation of HBx protein by m6A modification in the HBV-infected PHHs and HepG2-NTCP cells. (A to C) HBV particles prepared from the indicated HBV genome-transfected cells were used to infect primary human hepatocytes (PHHs) for 10 days. Total RNA and cellular lysates were extracted from these cells. (A) The indicated proteins were analyzed by Western blotting. (B) HBV RNA levels were analyzed by RT-qPCR. (C) Total RNAs extracted from the cells shown in panel A were immunoprecipitated using an anti-m6A antibody to identify m6A methylated HBV RNAs. These were normalized by input RNA levels by RT-qPCR. (D to G) HepG2-NTCP cells were infected with infectious virus particles prepared from HBV 5′–3′ MT- or 5′–3′-HBx-1 MT-expressing cells for 8 days. HepG2-NTCP cells were also transfected with control and/or YTHDF2 siRNAs for 48 h. Total RNA and cellular lysates were isolated from these cells. (D and F) The indicated proteins were analyzed by Western blotting. (E and G) HBV RNA levels were assayed by RT-qPCR. In panels B, C, E, and G, the error bars represent the SDs from three independent experiments. The P values were calculated via an unpaired Student's t test. *, P < 0.05; **, P < 0.01; n.s., nonsignificant; ND, not detected.
Because YTHDF2 affected HBx protein expression by the interaction with additional m6A modifications in nt 1606 to 1809 (Fig. 2), we determined whether YTHDF2 protein affects HBx protein expression by m6A modification at nt 1616. We infected HepG2-NTCP cells with virus particles from the HBV 5′–3′ MT or HBx-1 MT. These cells were then transfected with YTHDF2 siRNAs. As expected, the silencing of YTHDF2 expression increased HBx protein and HBV RNA levels in HBV 5′–3′ MT-infected cells (Fig. 4D and E). In contrast, HBx expression was not affected by depletion of YTHDF2 in HBx-1 MT-infected cells (Fig. 4F and G). Altogether, these results demonstrate that YTHDF2 plays a key role in the maintenance of regulated levels of HBx RNA and protein by recognizing m6A methylation at nt 1616 of HBx mRNA and causing its degradation.
YTHDF2 interacts with the m6A site of HBx mRNA to affect their protein expression.
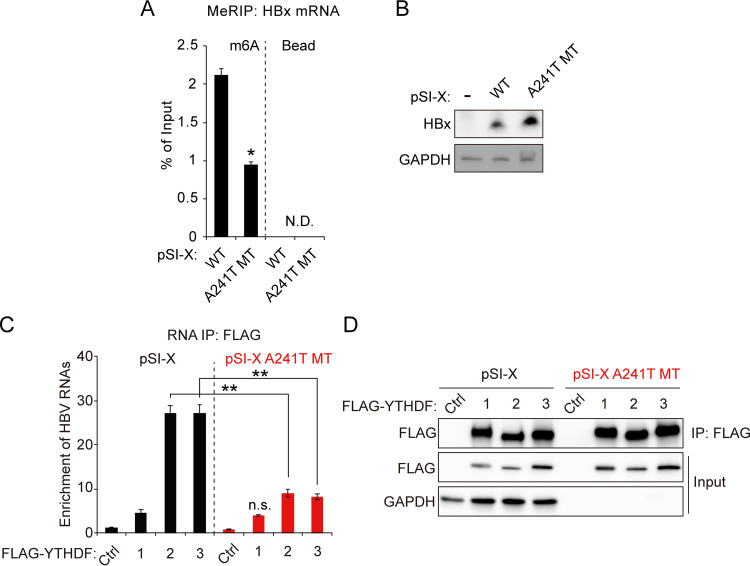
We also used the HBx expression plasmid, pSI-X, to investigate its regulation by m6A modification at nt 1661. To determine whether YTHDF2 interacts with the m6A methylation at nt 1616 of HBx mRNA, we generated the A241T mutation in the HBx expression plasmid (pSI-X) containing only the coding region of HBx (28). The m6A site at nt 241 of the pSI-X plasmid corresponds to the m6A site at nt 1616 of the HBV expression vector. We conducted the MeRIP-RT-qPCR assay using total RNA from pSI-X or pSI-X A241T MT-transfected cells (Fig. 5A). The results show that the A241T mutation reduced the m6A modification levels of HBx mRNA. On the other hand, HBx protein expression was significantly increased by the A241T mutation (Fig. 5B). We further investigated whether the A241T mutation affects the interaction between HBx mRNA and YTHDF2/3 proteins (Fig. 5C and D). We performed immunoprecipitation experiments using cell lysates from the Huh7 cells in which FLAG-YTHDF plasmids and pSI-X or pSI-X A241T MT were cotransfected. RT-qPCR analysis shows that the A241T mutation decreased the enrichment of HBx mRNA by YTHDF2/3 proteins. Of interest is the m6A modification of HBx RNA produced from the HBx expression plasmid (pSI-X)-transfected cells, indicating that DRACH motifs in the HBx ORF can affect their m6A modification in the absence of whole HBV genome expression. These data clearly demonstrate that the m6A modification at nt 1616 of HBV RNAs recruits YTHDF2, resulting in the decrease of HBx mRNA and protein levels.
FIG 5.
m6A modification in HBx coding region affects HBx expression by interaction with YTHDF2. (A and B) Huh7 cells were transfected with pSI-X WT or A241T MT plasmid for 48 h. (A) Total RNA and cell lysates were extracted. m6A methylated RNAs were enriched by an anti-m6A antibody and normalized by input RNA levels using RT-qPCR. (B) The indicated proteins were assayed by immunoblotting. (C and D) Huh7 cells expressing pSI-X WT or pSI-X A241T MT were transfected with FLAG-YTHDFs expression plasmids for 48 h. The RNA-protein complexes were immunoprecipitated using an anti-FLAG antibody. (C) Immunoprecipitated HBx mRNA levels were normalized by input HBx mRNA using RT-qPCR. (D) The indicated proteins were analyzed by Western blotting. In panels A and C the error bars represent the SDs from three independent experiments. The P values were calculated via an unpaired Student's t test. *, P < 0.05; **, P < 0.01. IP, immunoprecipitation; n.s., nonsignificant; N.D., not detected.
DISCUSSION
The m6A RNA modification regulates the functions of both cellular and viral RNAs and affects a wide variety of functions (15–23). Previously, we identified the functional roles of the m6A site at nt 1907, located in the lower stem-loop of the epsilon element (22). Our current and revised view is that m6A methylation of the 5′ epsilon structure plays a positive role in the events of core particle assembly, while the m6A modification of the 3′ epsilon structure decreases viral RNA stability and translation via its interaction with YTHDF2 (22, 24). Since an additional m6A peak was identified in the HBx coding region, we investigated whether these variant consensus DRACH motifs specific to N6-methyladenosine modification in the HBx coding region are chemically modified. We used MeRIP sequencing to align the m6A site on the HBV genome and identified several m6A consensus motifs located within the HBx coding region that are the sites for m6A modification (Fig. 1 and 3). We found that m6A modification occurs at these additional variants of DRACH motifs, located in the HBx coding region by cellular m6A methyltransferases, and that this modification negatively affected HBx protein expression (Fig. 1). By generating HBV mutants that lack m6A in the HBx coding region, we were able to define the unique positional effects of m6A on HBx protein expression. Thus far, of the five possible m6A sites, only the m6A site at nt 1616 showed any functional relevance affecting levels of HBx mRNA and protein expression (Fig. 3 and 4). Depletions of the m6A methyltransferases (METTL3/14) and m6A reader (YTHDF2) also resulted in a similar increase of HBx protein levels (Fig. 1 and 2). The mutation at nt 1907 (5′–3′ HBV MT) resulted in the increased HBV mRNAs as well as viral protein levels, including HBx, but more importantly, the mutation at nt 1616 (HBx-1 MT) increased only HBx mRNA and protein expression (Fig. 3). HBx mRNA is methylated at the motifs within its coding sequences as well as at its 3′ UTR at nt 1907, making it more unstable and subject to increased degradation via YTHDF proteins. This is consistent with its reduced expression seen in HBV-infected liver tissues, transgenic mice, and HBV transfection studies (1, 2, 29).
Our previous studies shed light on a plethora of viral functions that are associated with m6A modification of HBV RNA at nt 1907 (22). m6A modification affects the innate immune response and IFN-mediated degradation of HBV RNAs, promotes the synthesis of viral DNA, and affects host mRNA methylation profile and cytoplasmic transport of HBV RNAs (18, 23–25, 30–32). The current studies add a new functional role of m6A modification in the regulation of HBx mRNA and protein expression by the m6A site located within its coding sequences.
HBx plays a critical role in viral transcription and replication as a regulatory protein (6, 7, 9, 11, 12). HBx directly binds to host transcription factors and coactivators associated with cccDNA to promote viral transcription. It also affects the host transcription profile by the same mechanism. Another notable observation we previously described is that during HBV expression or infection, HBx binds and recruits METTL3/14 complex onto the cccDNA to promote cotranscriptional m6A methylation of viral transcripts (32). HBx-defective HBV expression fails to accumulate m6A-modified viral transcripts. Here, we showed that m6A at the HBx coding region, in addition to nt 1907, renders HBx mRNA less stable, leading to a decrease in HBx protein expression. Thus, HBx regulates itself (autoregulates) via a feedback loop associated with m6A modification, and this self-regulation may affect viral transcription and replication. This additional layer of regulation may dictate the role of HBx in the maintenance of chronic infection via regulating m6A modification of viral transcripts.
Furthermore, HBx is also known to be involved in the development of liver disease and HCC (1, 5). We reported that HBx guides m6A methyltransferases onto the chromosomal locus of phosphatase tensin homolog (PTEN), a tumor suppressor, and induces m6A methylation in PTEN mRNA, promoting the degradation of its RNA (31). Recently, the roles of m6A methylation have been reported in liver disease, including HCC (33). m6A demethylase (FTO) may be associated with nonalcoholic fatty liver disease (NAFLD), a risk factor predisposing patients to HCC formation (34). METTL3 and -14 also regulate gene expression related to the development of HCC (35). In this respect, HBx may affect host m6A patterns and regulate host gene expression associated with hepatocellular carcinoma, contributing to the development of HCC. It remains to be investigated whether the functions of host gene expression are regulated by alteration in their m6A modification dependent on HBx expression, as we observed for PTEN expression (31). Several reports highlighted the role of YTHDF proteins in HCC and other cancers (36–39). It was shown that YTHDF2 is mutated in HCC and other cancers (35, 38). Such YTHDF mutations will lead to losing their ability to affect RNA stability and translation (38). If such mechanisms exist to inactivate RNA decay in cancer, then this mechanism could increase HBx RNA and protein expression levels in virus-associated HCCs due to the absence of YTHDF2-mediated degradation. Increased levels of HBx protein may contribute to increased signal transduction pathways, among others, and accelerate the course of the normal infectious process toward progression to hepatocarcinogenesis. Thus, the increased HBx expression resulting from the loss of YTHDF2 function may contribute to HBV-associated HCC. Such a scenario is supported by the vast body of literature in which ectopic HBx expression was associated with playing an indirect role in hepatocarcinogenesis (5–9, 12).
Interestingly, mutations of the DRACH motifs in the HBx coding region reduced preS2 protein expression (Fig. 3). These effects may not be due to the deficiency of m6A modification in HBV transcripts, because depletion of YTHDF2 did not affect preS2 levels in HBV 5′–3′ MT-transfected cells (Fig. 2). The m6A sites in the HBx coding region are located in the 3′-UTR of HBsAg mRNAs. The 3′-UTR structure of viral mRNAs and cellular mRNAs is involved in their stability and translation activity (40, 41). Thus, the mutations of DRACH motifs in the HBx coding region may affect the RNA secondary structure of the viral mRNA 3′ UTR, leading to a decrease in HBV transcripts and preS2 expression levels (Fig. 3 and 4). This hypothesis is supported by the finding that naturally occurring single-nucleotide mutations at nt 1753, 1762, 1764, and 1766 affect HBV replication by downregulating HBs protein expression (42). Another possibility is that other m6A reader proteins such as YTHDC2 can recognize the 3′-UTR of preS2 mRNA by m6A modification at nt 1606 to 1809, which inhibits the recognition by YTHDF2/3. It is known that YTHDC2 positively regulates the translation of m6A-methylated RNA (43). If so, then the reduced preS2 expression by HBx-1 mutation may have resulted from the inability of YTHDC2 to interact with preS2 mRNA. In addition, a recent report described in-frame deletions in the preS region that often happen in the later stage of chronic HBV infection (44). These deletions inhibit the transcription of HBsAg mRNA by promoter occlusion, which is accompanied by increases in precore and HBx mRNA expression (45). In this respect, the maintenance of the m6A sites in the HBV genome aids in the progression of infection to chronic hepatitis by regulating viral protein expression.
MeRIP analysis showed that m6A-methylated HBV RNA is still present in cells expressing both nt 1907 and 1616 mutant genomes (HBV 5′–3′ HBx-1 MT; Fig. 3D and 4C). These results imply that HBV RNA could be m6A methylated in other DRACH motifs in addition to nt 1907 and 1616. Another possibility is the specificity or cross-reactivity of base-specific antibodies. In this study, we used commercially available m6A-specific polyclonal antibodies from Synaptic Systems that showed moderate cross-reactivity with m5C modification (46). Thus, the faint m6A levels detected from HBV 5′–3′ HBx-1 MT-transfected cells may be from m5C-methylated HBV RNA. In this regard, it has been reported that the human immunodeficiency virus (HIV) genome is m5C methylated, and this modification regulates viral gene expression (47). m5C modification of HBV transcripts has not been studied.
Targeting HBx in the treatment of HBV infection is emerging as a viable therapeutic strategy because HBx plays a central role in viral replication. The work presented here highlights an additional role of m6A in maintaining and regulating HBx expression. Thus, m6A methylation can be the target for disruption of HBx protein for HBV therapeutics. Furthermore, targeted m6A methyltransferases (METT3/14) and demethylase (FTO and ALKBH5) are being considered anticancer reagents (48, 49). This study offers new avenues for possible therapeutic intervention of the function of HBx aimed at curing chronic HBV infection.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents.
The pHBV 1.3-mer plasmid was a kind gift from Wang-Shick Ryu (Yonsei University, South Korea) and obtained from Addgene (65459). The pHBV 1.3-mer 5′–3′ MT plasmid was previously constructed. The pHBV 1.3-mer 5′–3′-HBx-1 MT, 5′–3′-HBx-2 MT, 5′–3′-HBx-3 MT, 5′–3′-HBx-4 MT, 5′–3′-HBx-5 MT, and pSI-A241T MT plasmids were generated by site-directed mutagenesis. The pSI-X plasmid was provided by Betty L. Slagle (Baylor College of Medicine, Houston, TX, USA). FLAG-YTHDF (1, 2, and 3) plasmids were a kind gift from Stacy M. Horner (Duke University Medical Center, Durham, NC, USA). Antibodies were anti-preS2 (number SC-23944) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; number SC-47724) antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA), anti-METTL3 (number 15073-1-AP) antibody from Proteintech Group (Rosemont, IL, USA), anti-METTL14 (number HPA038002) antibody from Sigma-Aldrich (San Jose, CA, USA), anti-FLAG (number 14793) antibody from Cell Signaling Technology (Danvers, MA, USA), anti-m6A antibody from Synaptic Systems (Göttingen, Germany), and anti-core and anti-precore antibodies from Jing-Hsiung James Ou (University of Southern California, CA, USA). Anti-HBx antibody was obtained from Gilead Sciences, Inc. (Foster City, CA, USA). Anti-preS2 and HBx antibodies were diluted at a 1:200 ratio in 5% bovine serum albumin (BSA) buffer for immunoblotting. The other antibodies were used at a 1:1,000 ratio in 5% BSA buffer for immunoblotting. The ON-TARGET plus siRNAs of METTL3 (number L-005170-02-0005), METTL14 (number L-014169-02-0005), YTHDF1 (number L-018095-02-0005), YTHDF2 (number L-021009-02-0005), and YTHDF3 (number L-017080-01-0005) were obtained from Dharmacon (Lafayette, CO, USA).
Cell culture and transfection.
Huh7 and HepG2-NTCP cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The HepG2-NTCP cells were provided by Wenhui Li (National Institute of Biological Sciences, Beijing, China). PHHs were obtained from Thermo Fisher Scientific. PHHs were maintained according to the manufacturer’s protocol. The medium was supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.1 mM nonessential amino acids under standard culture conditions (5% CO2, 37°C). Plasmids were transfected into cells using Mirus TransIT-LT1 reagent (Mirus, Madison, WI, USA) according to the manufacturer’s protocol. Huh7 cells and HepG2-NTCP cells were transfected with 10 nM either the targeting or control siRNAs using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA, USA). The siRNA transfection was performed according to the manufacturer’s protocol.
Virus production and cell infection.
HBV particles were harvested from the supernatants of pHBV 1.3-mer, pHBV 1.3-mer 5′–3′ MT, or HBx MT series plasmids expressing Huh7 cells. The culture medium was centrifuged at 4°C, 10,000 × g for 15 min. The clarified supernatants were incubated with 5% polyethylene glycol (PEG) 8000 overnight at 4°C and then centrifuged at 4,000 rpm for 30 min at 4°C. Pellet was redissolved in a serum-free culture medium in a 1% volume of the original supernatant. For infection, the PHHs and HepG2-NTCP cells were split in collagen-coated plates and incubated for 24 h with HBV particles, which are diluted in a serum-free culture medium with 4% PEG 8000 and 2% dimethyl sulfoxide (DMSO). After being incubated with HBV particles, the cells were washed with a culture medium. Cells were incubated for 10 days in medium containing 2% DMSO, changed every 2 days.
Real-time RT-qPCR.
Total RNA was isolated using an RNeasy minikit (Qiagen, Valencia, CA, USA). The cDNAs were synthesized from extracted total RNA using iScript reverse transcription supermix (Bio-Rad, Hercules, CA, USA). The quantitative PCR was assessed with Ssoadvanced universal SYBR green supermix (Bio-Rad). Each viral RNA and mRNA expression level, normalized to GAPDH, was analyzed using the ΔΔCT method. The primers used for RT-qPCR were HBV RNA (forward primer, 5′-CTCCCCGTCTGTGCCTTCT-3′; reverse primer, 5′-GCCCCAAAGCCACCCAAG-3′), HBV pgRNA (forward primer, 5′-CTCAATCTCGGGAATCTCAATGT-3′; reverse primer, 5′-TGGATAAAACCTAGGAGGCATAAT-3′), HBV pgRNA and HBsAg mRNAs (forward primer, 5′-ATGTTGCCCGTTTGTCCTCT-3′; reverse primer, 5′-GCCCTACGAACCACTGAACA-3′), and GAPDH (forward primer, 5′-CCTGCACCACCAACTGCTTA-3′; reverse primer, 5′-CATGAGTCCTTCCACGATACCA-3′).
MeRIP sequencing and RT-qPCR assay.
Total RNA was extracted from three independent samples for MeRIP sequencing. Poly(A) RNA was enriched from total RNA using a poly(A) spin mRNA isolation kit (New England Biolabs, Ipswich, MA, USA) and fragmented using RNA fragmentation reagent (Ambion. Waltham, MA, USA). The fragmented RNA was heated to 75°C for 5 min and placed on ice for 3 min. Anti-m6A antibody (Synaptic Systems) conjugated to protein G Dynabeads (Thermo Fisher Scientific) was incubated with the prepared RNA samples in MeRIP buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 0.1% NP-40) overnight at 4°C. The immunoprecipitated RNA-bead complexes were washed with MeRIP buffer 5 times, and bound RNA was eluted in MeRIP buffer containing 6.7 mM m6A 5′-monophosphate sodium salt (Sigma-Aldrich). Eluted RNA was cleaned by TRIzol (Thermo Fisher Scientific). The cDNA libraries were synthesized from this RNA for Illumina sequencing using a TruSeq RNA sequencing kit (Illumina, San Diego, CA, USA). The method of deep-sequencing analysis was previously described. MeRIP RT-qPCR followed the same protocol, except that total RNA was not fragmented. Eluted RNA was reverse transcribed into cDNA and subjected to RT-qPCR.
Western blotting and immunoprecipitation.
Cells were incubated with an NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl) supplemented with a protease inhibitor (Thermo Fisher Scientific) for 15 min at 4°C. After centrifugation for 20 min at 14,000 rpm at 4°C, cell lysates were transferred to a new microcentrifuge tube. HBV core protein was immunoprecipitated using anti-core antibody conjugated to protein G Dynabeads (Thermo Fisher Scientific) from cell lysates following incubation for 2 h on a rotator at 4°C. Immunoprecipitated protein-bead complexes were washed with lysis buffer 3 times. Lysates or immunoprecipitates were resolved by SDS-PAGE gel and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% BSA for 90 min, following by overnight incubation with primary antibodies at 4°C. After washing, the membranes were incubated with secondary antibodies conjugated to horseradish peroxidase (Cell Signaling Technology) for 1 h. Antibody complexes were detected using a chemiluminescence substrate (Millipore, Burlington, MA, USA). Chemiluminescence signals were detected using the ChemiDoc MP imaging system (Bio-Rad).
RNA-protein UV cross-linking assay.
Cells were exposed to 245-nm UV at 250 mJ/cm and then harvested. Cell pellets were resuspended with SDS lysis buffer (0.5% SDS, 50 mM Tris-HCl, pH 6.8, 1 mM EDTA, 1 mM dithiothreitol, 150 mM NaCl) supplemented with a protease inhibitor and RNase inhibitor (Thermo Fisher Scientific). Lysates were immunoprecipitated with anti-core antibody conjugated to protein G Dynabeads (Thermo Fisher Scientific) for 2 h on the rotator at 4°C. The bead complex was washed with NP-40 lysis buffer 5 times. After the wash step, beads were resuspended with NP-40 lysis buffer. Half of the bead was incubated with proteinase K for 90 min at 37°C, and then immunoprecipitated RNA was extracted using the RNeasy minikit (Qiagen). The other half of the bead was added to protein sample buffer to be analyzed with a Western blot assay.
Northern blot assay for HBV RNA.
The 20 μg total RNA was resolved by electrophoresis on 1.2% formaldehyde agarose gel and transferred to a positively charged nylon membrane (Thermo Fisher Scientific). The membrane was fixed by UV cross-linking. The fixed membrane was prehybridized for 30 min, incubated by ULTRAhyb hybridization buffer (Invitrogen) for 30 min, and then hybridized with an internally radiolabeled double-stranded DNA probe in ULTRAhyb hybridization buffer (Thermo Fisher Scientific) overnight. The probe was generated using [α-32P]dCTP and a DECAprimeII kit (Thermo Fisher Scientific). After washing, the membrane was exposed to BIOMAX XAR film (Sigma-Aldrich). The probe template was amplified by PCR using forward (5′-ATGGCTGCTAGGCTGTGCTGCC-3′) and reverse (5′-ATAAGGGTCGATGTCCATGCCCCAAAG-3′) primers from the pHBV1.3-mer plasmid.
Statistical analysis.
All results are representative of three independent experiments. For each result, error bars represent the standard deviations (SD) from at least three independent experiments. The P value was calculated using a one-tailed unpaired Student's t test.
Data availability.
All study data are included in the article.
ACKNOWLEDGMENTS
We thank Jing-Hsiung James Ou (University of Southern California, Los Angeles, CA) for providing anticore antibodies. We thank Betty L. Slagle (Baylor College of Medicine, Huston, TX) for the generous gift of pSI-X plasmid. We thank Gilead Sciences, Inc., for the generous gift of anti-HBx antibody.
This study was supported by NIH grants AI125350 and AI139234 to A.S.
We declare that we have no conflicts of interest.
Contributor Information
Aleem Siddiqui, Email: asiddiqui@health.ucsd.edu.
J.-H. James Ou, University of Southern California
REFERENCES
- 1.Hu J, Protzer U, Siddiqui A. 2019. Revisiting hepatitis B virus: challenges of curative therapies. J Virol 93:e01032-19. 10.1128/JVI.01032-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479-480:672–686. 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan H, Zhong GC, Xu GW, He WH, Jing ZY, Gao ZC, Huang Y, Qi YH, Peng B, Wang HM, Fu LR, Song M, Chen P, Gao WQ, Ren BJ, Sun YY, Cai T, Feng XF, Sui JH, Li WH. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard MJ, Schneider RJ. 2004. The enigmatic X gene of hepatitis B virus. J Virol 78:12725–12734. 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slagle BL, Andrisani OM, Bouchard MJ, Lee CG, Ou JH, Siddiqui A. 2015. Technical standards for hepatitis B virus X protein (HBx) research. Hepatology 61:1416–1424. 10.1002/hep.27360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slagle BL, Bouchard MJ. 2016. Hepatitis B virus X and regulation of viral gene expression. Cold Spring Harb Perspect Med 6:a021402. 10.1101/cshperspect.a021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U. 2011. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol 55:996–1003. 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Qadri I, Conaway JW, Conaway RC, Schaack J, Siddiqui A. 1996. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci USA 93:10578–10583. 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. 2009. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA 106:19975–19979. 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balsano C, Billet O, Bennoun M, Cavard C, Zider A, Grimber G, Natoli G, Briand P, Levrero M. 1994. Hepatitis-B virus-X gene-product acts as a transactivator in-vivo. J Hepatol 21:103–109. 10.1016/s0168-8278(94)80144-4. [DOI] [PubMed] [Google Scholar]
- 11.Maguire HF, Hoeffler JP, Siddiqui A. 1991. Hbv X-protein alters the DNA-binding specificity of Creb and Atf-2 by protein-protein interactions. Science 252:842–844. 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 12.Murphy CM, Xu YP, Li F, Nio K, Reszka-Blanco N, Li XD, Wu YX, Yu YB, Xiong Y, Su LS. 2016. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep 16:2846–2854. 10.1016/j.celrep.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200. 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue Y, Liu J, He C. 2015. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 29:1343–1355. 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, Wei J, He C. 2019. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74:640–650. 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer KD, Jaffrey SR. 2017. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol 33:319–342. 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzales-van Horn SR, Sarnow P. 2017. Making the mark: the role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe 21:661–669. 10.1016/j.chom.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim GW, Siddiqui A. 2021. The role of N6-methyladenosine modification in the life cycle and disease pathogenesis of hepatitis B and C viruses. Exp Mol Med 53:339–345. 10.1038/s12276-021-00581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, Ilkayeva OR, Law BA, Holley CL, Garcia-Blanco MA, Evans MJ, Suthar MS, Bradrick SS, Mason CE, Horner SM. 2016. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20:654–665. 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1:16011. 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim GW, Siddiqui A. 2021. N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc Natl Acad Sci USA 118:e2022024118. 10.1073/pnas.2022024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, Kim SJ, Mason CE, Horner SM, Siddiqui A. 2018. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci USA 115:8829–8834. 10.1073/pnas.1808319115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imam H, Kim GW, Siddiqui A. 2020. Epitranscriptomic(N6-methyladenosine) modification of viral RNA and virus-host interactions. Front Cell Infect Microbiol 10:584283. 10.3389/fcimb.2020.584283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim GW, Imam H, Siddiqui A. 2021. The RNA binding proteins YTHDC1 and FMRP regulate the nuclear export of N(6)-methyladenosine-modified hepatitis B virus transcripts and affect the viral life cycle. J Virol 95:e0009721. 10.1128/JVI.00097-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim GW, Imam H, Khan M, Siddiqui A. 2020. N-6-methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem 295:13123–13133. 10.1074/jbc.RA120.014260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poria DK, Ray PS. 2017. RNA-protein UV-crosslinking assay. Bio Protoc 7:e2193. 10.21769/BioProtoc.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. 2016. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res 44:e91. 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keasler VV, Hodgson AJ, Madden CR, Slagle BL. 2009. Hepatitis B virus HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically-injected mice. Virology 390:122–129. 10.1016/j.virol.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Chen WL, Louie SG, Yen TS, Ou JH. 2007. Hepatitis B virus promotes hepatocarcinogenesis in transgenic mice. Hepatology 45:16–21. 10.1002/hep.21445. [DOI] [PubMed] [Google Scholar]
- 30.Imam H, Kim GW, Mir SA, Khan M, Siddiqui A. 2020. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified hepatitis B virus transcripts. PLoS Pathog 16:e1008338. 10.1371/journal.ppat.1008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim GW, Imam H, Khan M, Mir SA, Kim SJ, Yoon SK, Hur W, Siddiqui A. 2021. HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology 73:533–547. 10.1002/hep.31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim GW, Siddiqui A. 2021. Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs. Proc Natl Acad Sci USA 118:e2019455118. 10.1073/pnas.2019455118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Z, Meng J, Su R, Zhang J, Chen J, Ma X, Xia Q. 2020. Epitranscriptomics in liver disease: basic concepts and therapeutic potential. J Hepatol 73:664–679. 10.1016/j.jhep.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Mittenbuhler MJ, Saedler K, Nolte H, Kern L, Zhou J, Qian SB, Meder L, Ullrich RT, Bruning JC, Wunderlich FT. 2020. Hepatic FTO is dispensable for the regulation of metabolism but counteracts HCC development in vivo. Mol Metab 42:101085. 10.1016/j.molmet.2020.101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen MN, Wei L, Law CT, Tsang FHC, Shen JL, Cheng CLH, Tsang LH, Ho DWH, Chiu DKC, Lee JMF, Wong CCL, Ng IOL, Wong CM. 2018. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67:2254–2270. 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 36.Shi RK, Ying SL, Li YD, Zhu LY, Wang X, Jin HC. 2021. Linking the YTH domain to cancer: the importance of YTH family proteins in epigenetics. Cell Death Dis 12:346. 10.1038/s41419-021-03625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang CZ, Huang SZ, Zhuang HK, Ruan SY, Zhou ZX, Huang KJ, Ji F, Ma ZY, Hou BH, He XS. 2020. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 39:4507–4518. 10.1038/s41388-020-1303-7. [DOI] [PubMed] [Google Scholar]
- 38.Hou JJ, Zhang H, Liu J, Zhao ZJ, Wang JY, Lu ZK, Hu B, Zhou JK, Zhao ZC, Feng MX, Zhang HY, Shen B, Huang XX, Sun BC, He C, Xia Q. 2019. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer 18:163. 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Sheng H, Ma F, Wu Q, Huang J, Chen Q, Sheng L, Zhu X, Zhu X, Xu M. 2021. RNA m(6)A reader YTHDF2 facilitates lung adenocarcinoma cell proliferation and metastasis by targeting the AXIN1/Wnt/beta-catenin signaling. Cell Death Dis 12:479. 10.1038/s41419-021-03763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu XB, Bartel DP. 2017. Widespread influence of 3 '-end structures on mammalian mRNA processing and stability. Cell 169:905–917. 10.1016/j.cell.2017.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasekhian M, Roohvand F, Habtemariam S, Marzbany M, Kazemimanesh M. 2021. The role of 3'UTR of RNA viruses on mRNA stability and translation enhancement. Mini Rev Med Chem 21:2389–2398. 10.2174/1389557521666210217092305. [DOI] [PubMed] [Google Scholar]
- 42.Parekh S, Zoulim F, Ahn SH, Tsai A, Li J, Kawai S, Khan N, Trepo C, Wands J, Tong S. 2003. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol 77:6601–6612. 10.1128/jvi.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B, Qian SB. 2019. m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun 10:5332. 10.1038/s41467-019-13317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollicino T, Cacciola I, Saffioti F, Raimondo G. 2014. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol 61:408–417. 10.1016/j.jhep.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 45.Fu S, Zhang J, Yuan Q, Wang Q, Deng Q, Li J, Xia N, Wang Y, Wen Y, Tong S. 2021. Lost small envelope protein expression from naturally occurring preS1 deletion mutants of hepatitis B virus is often accompanied by increased HBx and core protein expression as well as genome replication. J Virol 95:e0066021. 10.1128/JVI.00660-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weichmann F, Hett R, Schepers A, Ito-Kureha T, Flatley A, Slama K, Hastert FD, Angstman NB, Cardoso MC, Konig J, Huttelmaier S, Dieterich C, Canzar S, Helm M, Heissmeyer V, Feederle R, Meister G. 2020. Validation strategies for antibodies targeting modified ribonucleotides. RNA 26:1489–1506. 10.1261/rna.076026.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26:217–227. 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D, Hendrick AG, Webster NA, Andrews B, Fosbeary R, Guest P, Irigoyen N, Eleftheriou M, Gozdecka M, Dias JML, Bannister AJ, Vick B, Jeremias I, Vassiliou GS, Rausch O, Tzelepis K, Kouzarides T. 2021. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593:597–601. 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su R, Dong L, Li YC, Gao M, Han L, Wunderlich M, Deng XL, Li HZ, Huang Y, Gao L, Li CY, Zhao ZC, Robinson S, Tan B, Qing Y, Qin X, Prince E, Xie J, Qin HJ, Li W, Shen C, Sun J, Kulkarni P, Weng HY, Huang HL, Chen ZH, Zhang B, Wu XW, Olsen MJ, Muschen M, Marcucci G, Salgia R, Li L, Fathi AT, Li ZJ, Mulloy JC, Wei MJ, Horne D, Chen JJ. 2020. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell 38:79–96. 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article.