ABSTRACT
Listeria monocytogenes is a major pathogen contributing to foodborne outbreaks with high mortality. Nisin, a natural antimicrobial, has been widely used as a food preservative. However, the mechanisms of L. monocytogenes involved in nisin resistance have not yet to be fully defined. A mariner transposon library was constructed in L. monocytogenes, leading to the identification of 99 genes associated with the innate resistance to nisin via Transposon sequencing (Tn-seq) analysis. To validate the accuracy of the Tn-seq results, we constructed five mutants (ΔyqgS, ΔlafA, ΔvirR, ΔgtcA, and Δlmo1464) in L. monocytogenes. The results revealed that yqgS and lafA, the lipoteichoic acid-related genes, were essential for resistance to nisin, while the gtcA and lmo1464 mutants showed substantially enhanced nisin resistance. Densely wrinkled, collapsed surface and membrane breakdown were shown on ΔyqgS and ΔlafA mutants under nisin treatment. Deletion of yqgS and lafA altered the surface charge, and decreased the resistance to general stress conditions and cell envelope-acting antimicrobials. Furthermore, YqgS and LafA are required for biofilm formation and cell invasion of L. monocytogenes. Collectively, these results reveal novel mechanisms of nisin resistance in L. monocytogenes and may provide unique targets for the development of food-grade inhibitors for nisin-resistant foodborne pathogens.
IMPORTANCE Listeria monocytogenes is an opportunistic Gram-positive pathogen responsible for listeriosis, and is widely present in a variety of foods including ready-to-eat foods, meat, and dairy products. Nisin is the only licensed lantibiotic by the FDA for use as a food-grade inhibitor in over 50 countries. A prior study suggests that L. monocytogenes are more resistant than other Gram-positive pathogens in nisin-mediated bactericidal effects. However, the mechanisms of L. monocytogenes involved in nisin resistance have not yet to be fully defined. Here, we used a mariner transposon library to identify nisin-resistance-related genes on a genome-wide scale via transposon sequencing. We found, for the first time, that YqgS and LafA (Lipoteichoic acid-related proteins) are required for resistance to nisin. Subsequently, we investigated the roles of YqgS and LafA in L. monocytogenes stress resistance, antimicrobial resistance, biofilm formation, and virulence in mammalian cells.
KEYWORDS: Listeria monocytogenes, transposon sequencing (Tn-seq), biofilm, lipoteichoic acid, nisin, pathogenicity
INTRODUCTION
Listeria monocytogenes is an intracellular foodborne pathogen, leading to a severe invasive infection called listeriosis after consumption of contaminated food (1, 2). Listeriosis is a deadly infection resulting in poor survival rates among all foodborne diseases, with up to 20% to 30% mortality (3). L. monocytogenes is ubiquitous in the environment, which is also widely present in a variety of foods including ready-to-eat foods, meat, and dairy products (4–6). Given its ability to adapt to adverse environmental conditions (such as high salt, low temperatures, and pH), the contamination of ready-to-eat foods is particularly concerning (7, 8). Therefore, controlling the survival and reproduction of L. monocytogenes in ready-to-eat foods is a thorny problem, which needs to be urgently addressed.
Bacteriocins are antimicrobial peptides with broad-spectrum antibacterial activities produced by bacteria, which have been used as preservatives to control pathogen propagation in foods for many decades (9, 10). Nisin is the only licensed lantibiotic by the FDA for use as a food grade inhibitor in over 50 countries (11, 12). Generally, nisin, as a cationic antimicrobial peptide, binds to the bacterial cell surface and oligomerizes in cytoplasmic membranes to form pores, leading to the leakage of cellular contents and depolarization of the membrane potential, and eventually cell death (13). In addition, nisin inhibits cell wall synthesis through interacting with lipid II (14). This dual mechanism of action plays a synergistic role in killing bacteria (15).
The intrinsic resistance to nisin of L. monocytogenes was first described in 1991 (16). Remarkably, a prior study suggests that L. monocytogenes is more resistant than other Gram-positive pathogens in nisin-mediated bactericidal effects (17). Despite the mechanisms of nisin have been identified, the association of genes with nisin resistance in L. monocytogenes is not fully understood. Several loci have been identified in L. monocytogenes that can lead to nisin resistance, including the two-component systems LisRK (18), VirRS (19), and LiaR (20), the alternative stress sigma factor SigB (21), the penicillin-binding protein Pbp4 (22), the class three stress gene regulator CtsR (23), the glutamate decarboxylase GadD1 (24), the ABC transporters AnrAB (25) and VirAB (26), lysinylation of phospholipids MprF (27), d-alanyl-teichoic acids biosynthesis DltA (28), and the tellurite resistance protein TelA (29). Although previous studies have reported many genes associated with nisin resistance, a number of nisin-related genes are not yet fully found. With the improvement of sequencing technology, transposon sequencing (Tn-seq), as an efficient microbial functional genomic tool, is widely used to study the association between phenotypes and genotypes in pathogens (30). Compared with conventional mutation approaches, Tn-seq has the advantage of high efficiency, accuracy, and wide coverage (31, 32).
Lipoteichoic acid (LTA), as an important bacterial cell wall component, is a zwitterionic polymer and is typically composed of polyglycerolphosphate backbone chain tethered to the membrane by a glycolipid (Gal-Glc-DAG or Gal-Ptd-6Glc-DAG) (33). In L. monocytogenes, the glycosyltransferases LafA (Lmo2555) and LafB (Lmo2554) are required for the formation of Glc-DAG and Gal-Glc-DAG, respectively(34). YqgS (Lmo0644) and LtaS (Lmo0927) are involved in polyglycerolphosphate backbone synthesis, YqgS transfers the initial glycerolphosphate onto the glycolipid anchor and LtaS extends the glycerolphosphate backbone chain(34). LTA mutants are defective in biofilm formation and reduced virulence in Gram-positive bacteria, such as Staphylococcus aureus, Enterococcus faecalis, and Bacillus subtilis (35–39).
In this study, we used Tn-seq to identify genes involved in nisin resistance via a high-density transposon library of L. monocytogenes EGD-e. In addition to the nisin-resistance-associated genes validated previously, a large amount of novel genes with great significance were identified. YqgS and LafA are involved in the LTA backbone and glycolipid linked to the membrane, respectively (34). Moreover, there has been no systematic study into the relationship between these genes and nisin resistance. Further investigations of ΔyqgS and ΔlafA mutants revealed an associated enhanced surface negative charge and reduced capacity to form biofilm. In addition to revealing the function of YqgS and LafA in various antibiotics and stress resistance, we have also explored the role of LTA in Listeria pathogenicity.
RESULTS
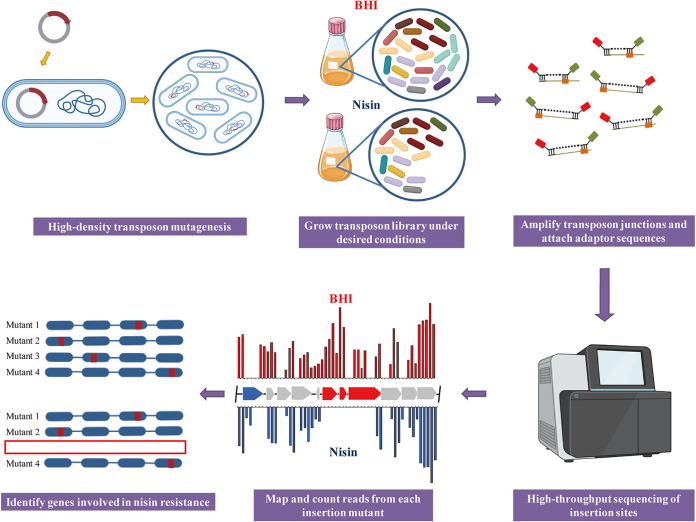
Illustration of the experimental protocols of Tn-seq.
To identify genome-wide genes associated with nisin resistance, a mariner-based transposon insertion library was generated in L. monocytogenes EGDe. The main steps of Tn-seq are illustrated in Fig. 1. In brief, Tn-seq was performed on cultures of the L. monocytogenes transposon mutant library. This library was grown in brain heart infusion (BHI) with or without subinhibitory concentration of nisin (200 μg/mL). Subsequently, DNA was extracted from samples, and we amplified transposon junctions and attached adaptor sequences to construct a DNA library for sequencing. The sequencing results indicated that each sample had at least 10 million Tn-seq readings and no biased insertion sites of the transposon were identified. The distribution of transposon was in an even and high-density pattern, and 43,793 unique insertion sites were identified throughout the genome (Supplementary File 2). Finally, analysis of Tn-seq results was carried out to identify genes associated with nisin resistance.
FIG 1.
Schematic illustration of the Tn-seq. First, a high-density transposon mutant library was constructed and cultured in different conditions. Genomic DNA was extracted. Then, sequencing adaptors and barcodes are added to DNA fragments, which could be amplified by using PCR. Subsequently, samples were analyzed by sequencing to map and count reads from each mutant. Finally, genes involved in nisin resistance were identified.
Identification of genes associated with nisin resistance by Tn-seq.
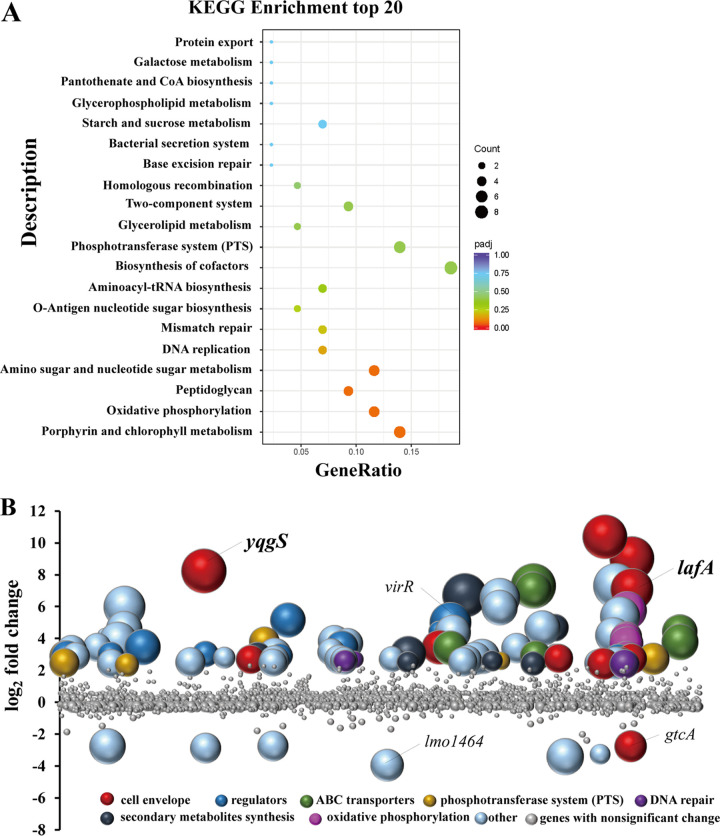
We identified 99 genes, and the number of transposon insertions was significantly different among groups in the presence or absence of nisin (Benjamini-Hochberg < 0.05), which were involved in nisin resistance (Fig. 2A and B). Ninety-two of them were inactivated by transposon insertion, leading to increased nisin sensitivity in the corresponding mutant (Table S2). Namely, the expression products of disrupted genes, some of which were clustered into several particular functional categories (Fig. 2B), were favorable for the resistance of nisin. Eleven genes are related to the cell envelope, which encodes cell-division protein (ftsW), peptidoglycan lytic protein P45 (spl), glycolipid and polyglycerolphosphate lipoteichoic acid synthesis (yqgS and lafABC), d-alanyl-teichoic acids biosynthesis (dltA), lysinylation of phospholipids (mprF), penicillin-binding protein (pbp4), d-alanyl-alanine synthetase (ddl), and UDP-glucose 4-epimerase (galE). Ten regulators genes were identified, including seven putative transcriptional regulators (lacI family, ctsR family, gntR family, marR family, arsR family, lmo0651, and lmo1262), two 2-component response regulators (liaR and virR), and a recombination regulator (recX).
FIG 2.
Identification of genes involved in nisin resistance by Tn-seq analysis. (A) Scatterplot of KEGG enrichment analysis. The enriched biological pathways are shown on the y axis; the ratio of differential genes to the total genes is indicated on the x axis. (B) Tn-seq analysis to identify genes associated with nisin resistance in L. monocytogenes. The bubbles represent genes, the size of the bubble represents the significance of genes with -log10 (BH value) (fold > 5, BH < 0.05). The abscissa represents the position of a gene on the genome and the ordinate indicates the log2 (fold changes) derived from the read-count ratio of libraries grown in BHI to libraries grown in BHI with nisin. Colors represent gene functional categories. All the genes with nonsignificance were marked in gray, which BH were limited to be no more than 0.7 in this figure for a clearer view.
The ABC transporters are known to exert important functions in antimicrobials resistance associated with efflux systems (40–42). Out of the seven newly identified ABC transporter genes, three genes (virA and anrAB) were useful to confer nisin resistance in L. monocytogenes, which were regulated via two-component systems (26). Phosphotransferase system (PTS) regulates carbohydrate metabolism by catalyzing function in sugar transport and phosphorylation (43), mannose PTS permease, and β-glucoside PTS enzyme II were involved in sensitivity to class IIa bacteriocins (44–46). Six putative PTS transports genes (lmo0021, lmo0024, lmo0301, lmo0916, lmo1971, and lmo2649) were identified. The DNA repair genes are essential for DNA damage caused by antimicrobial treatment (47), of which three genes with significant fold decrease in transposon insertions include DNA polymerase (polC), NAD-dependent DNA ligase (ligA), and single-strand DNA-binding protein (lmo2523). Moreover, eight genes involved in the biosynthesis of secondary metabolites were identified, six genes (hemA, hemC, henE, hemH, lmo2113, and lmo0884) of which were clustered in the porphyrin and chlorophyll metabolism pathway. And five susceptibility genes are located in the atpABCDEFG operon (atpA, atpB, atpD, atpG, atpH), which is involved in membrane bioenergetics such as ATP synthase (48), and previous studies suggest that ATP synthesis is related to cell attachment and biofilm stability (49, 50).
Seven genes were identified as contributing to nisin sensitivity (Table S3). That is, the corresponding transposon mutants were significantly enriched in the presence of nisin compared with the untreated group. Of those, two genes (lmo1464 and gtcA) were linked to cell envelope, which encoded diacylglycerol kinase and wall teichoic acid glycosylation protein GtcA, respectively. The other five genes with significant fold decrease are annotated as monovalent cation/H+ antiporter subunit C (lmo2380) and hypothetical proteins (lmo2258, lmo0955, lmo0653, lmo0215). Overall, the transposon library screening has comprehensively identified genes that confer resistance or are important for the mechanisms of action of nisin at the genome-wide scale.
Lipoteichoic acid is essential for L. monocytogenes resistance to nisin but not to bacitracin.
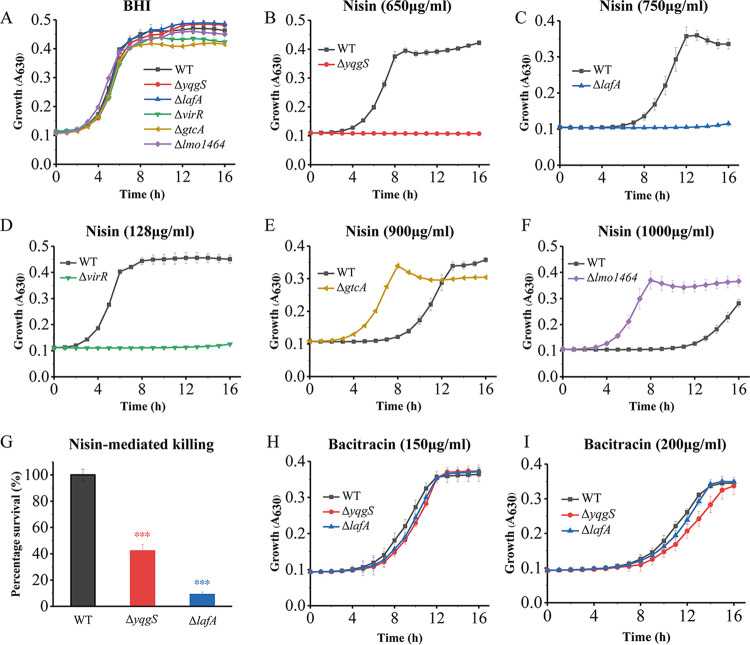
In order to validate the Tn-seq results, we constructed five mutants (ΔyqgS, ΔlafA, ΔvirR, ΔgtcA, and Δlmo1464), which were identified with high fold changes and significant P-values. The VirRS signal transduction system is known to determine the nisin resistance, yet, whether the deficiency in the other four genes changes nisin sensitivity has not been investigated. Under nisin-free conditions, the growth rate or cell density in the stationary phase of mutants was not significantly changed compared with the wild-type strain (Fig. 3A). The growth of yqgS mutant was grossly affected in the presence of nisin (650 μg/mL), and nisin (750 μg/mL) also had a severe effect on the growth of lafA mutant (Fig. 3B and C). In contrast, inactivation of the virR gene has the strongest negative impact on nisin resistance (Fig. 3D). The gtcA and lmo1464 mutants showed significantly enhanced resistance to nisin (Fig. 3E and F). The results of the validation experiment were also consistent with the previous Tn-seq results. In addition, deletion of yqgS and lafA indeed increased L. monocytogenes susceptibility to nisin-mediated killing (Fig. 3G). Next, we determined the growth in mutant and wild-type strains under bacitracin conditions, which also blocks cell wall synthesis by binding to the lipid carrier of peptidoglycan subunits as nisin (51, 52). The growth speed of yqgS and lafA mutants revealed a slight reduction compared with wild-type, although this was not statistically significant (Fig. 3H and I). These genes play a role in glycolipid (lafA) and lipoteichoic acid backbone (yqgS) synthesis, indicating that Lipoteichoic acid is essential for nisin resistance but not to bacitracin in L. monocytogenes.
FIG 3.
YqgS and LafA are required for nisin resistance but not for bacitracin. (A to F) Growth curves of strain EGDe and different mutants in the absence (A) or presence of nisin with 650 μg/mL (B), 750 μg/mL (C), 128 μg/mL (D), 900 μg/mL (E), and 1,000 μg/mL (F). (G) Survival percentage of L. monocytogenes strains following a nisin challenge (1,000 μg/mL). (H, I) Growth of L. monocytogenes strains in BHI supplemented with 150 μg/mL (H) or 200 μg/mL (I) bacitracin. Data represent mean of three independent experiments. ***, P ≤ 0.001.
Examination of morphology.
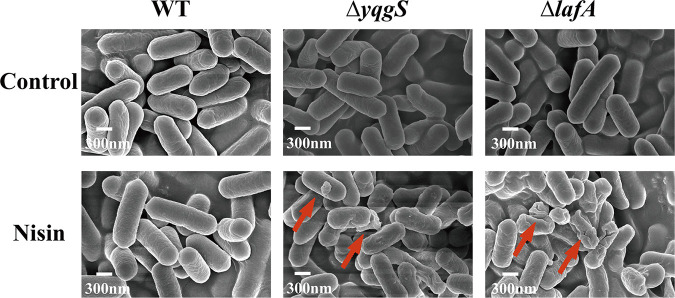
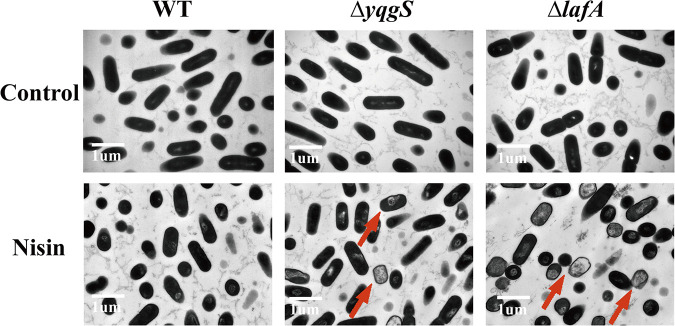
To further explore the effect of nisin on the wild-type and mutant strains, we performed scanning electron microscopy (SEM) and transmission electron microscopy (TEM) scans of the nisin-treated bacterial cultures. No significant difference is observed between the wild-type and mutant strains as displayed in Fig. 4 and 5 (control). After 1-h exposure to nisin (800 μg/mL), wild-type strain showed no visible alteration, but ΔyqgS and ΔlafA mutants exhibited striking changes in cell morphology, including densely wrinkled surface, collapsed surface, membrane breakdown, and cellular content discharge from the cell at a breach point (Fig. 4 and 5). Overall, these results suggested that the ΔyqgS and ΔlafA mutants are more susceptible to nisin-mediated bactericidal action than wild-type.
FIG 4.
The SEM micrograph of L. monocytogenes strains untreated or treated with nisin (800 μg/mL). Arrows indicate the damage of nisin treated L. monocytogenes.
FIG 5.
The TEM micrograph of L. monocytogenes strains untreated or treated with nisin (800 μg/mL). Arrows indicate the damage of nisin treated L. monocytogenes.
YqgS and LafA affect the surface charge of L. monocytogenes.
To determine if the change of surface charge resulted in the reduced amount of nisin adsorbed to the surface of ΔyqgS and ΔlafA mutants, the surface charge of the mutant and wild-type strains was determined by measuring the binding of cytochrome c. As expected, we observed higher levels of cytochrome c binding with the ΔyqgS and ΔlafA mutants than wild-type strain (Fig. 6), indicating that the absence of glycolipid in LTA slightly enhanced the surface negative charge, and the absence of LTA, to a greater extent, increased the negative charge on the bacterial surface. Throughout, the result allowed us to discard electrostatic changes at the bacterial surface as a reason for increased nisin sensitivity on the mutants.
FIG 6.
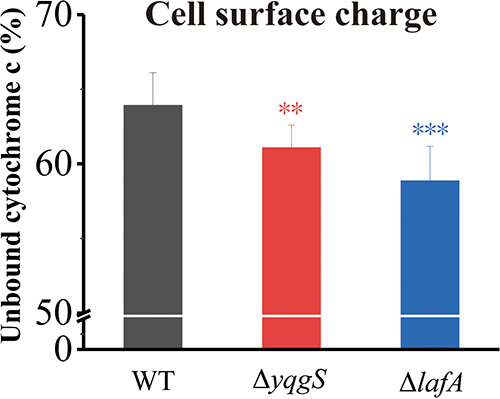
Cell surface charge analysis of L. monocytogenes strains. Data represent the mean ± SD of three independent experiments. **, P ≤ 0.01; ***, P ≤ 0.001.
Influence of YqgS and LafA on biofilm formation in L. monocytogenes.
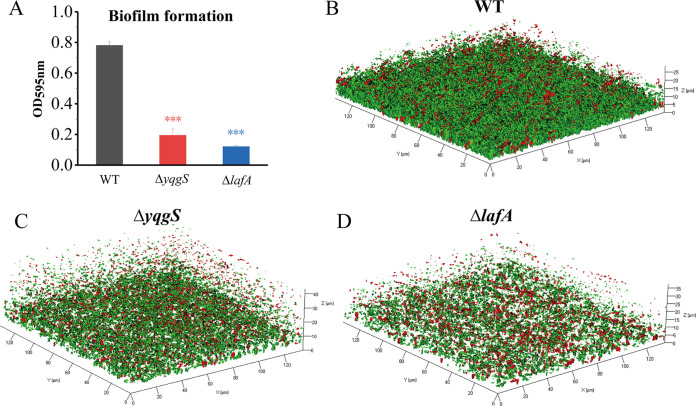
Previous studies showed that the lipoteichoic acid mutation of S. aureus resulted in complete loss of the biofilm-formation capacity on plastic (35). However, the capacity of the L. monocytogenes LTA mutant strain to form biofilm has not been investigated. To inquire whether YqgS and LafA are associated with the formation of biofilms, crystal violet assay was performed (53). We observed that ΔyqgS and ΔlafA mutants displayed a greatly reduced capacity to form biofilm relative to wild-type strain (Fig. 7A). Subsequently, we also investigated the role of YqgS and LafA in biofilm formation by confocal laser scanning microscope (54). Three-dimensional analysis of 2-day-old biofilms confirmed that the yqgS and lafA mutants formed a less dense and relatively thin biofilm compared with wild-type (Fig. 7B to D). Altogether, these results demonstrated that YqgS and LafA are required for biofilm formation.
FIG 7.
YqgS and LafA regulate biofilm formation. (A) Biofilm formation of wild-type, ΔyqgS, and ΔlafA mutant strains, which was measured by the crystal violet assay. Confocal microscopy images of wild-type (B), ΔyqgS (C), and ΔlafA (D) biofilm. Green color cells represent viable cells with intact membranes and red color cells are dead cells. The experiments were performed in triplicate. ***, P ≤ 0.001.
Impact of YqgS and LafA on antibiotic sensitivity and stress resistance.
To study the role of YqgS and LafA as LTA-related proteins in antibiotic resistance, we assessed the antimicrobial sensitivity of ΔyqgS and ΔlafA mutants by antibiotic disk agar-based assays. The assay results demonstrated that genetic inactivation of YqgS and LafA results in markedly increased antibiotics susceptibility, including oxacillin, polymixinB, cefuroxime, ceftriaxone, and carbenicillin (Table 1). An inactivating mutation at the yqgS gene can also increase penicillin sensitivity but no significant changes in resistance to ampicillin, cefotaxime, ciprofloxacin, amikacin, erythromycin, and chloramphenicol. In addition, significant decreases in cefotaxime resistance were detected in ΔlafA mutant. Meanwhile, mutations in the LTA system were linked to increased resistance to vancomycin and gentamicin. It is thus apparent that YqgS and LafA greatly contribute to the resistance of L. monocytogenes to various important cell envelope-targeting antimicrobials by affecting bacterial cell envelope integrity.
TABLE 1.
Relative sensitivities of mutants and wild-type strains to a variety of antibiotics
| Antibiotic | Mean size of the zone of inhibition (mm) ± SDa |
||
|---|---|---|---|
| EGD-e | ΔyqgS mutant | ΔlafA mutant | |
| Penicillin | 34.00 ± 0.72 | 37.50 ± 0.50** | 34.50 ± 0.50 |
| Ampicillin | 30.93 ± 0.12 | 31.03 ± 1.00 | 31.67 ± 0.58 |
| Vancomycin | 20.13 ± 0.15 | 18.83 ± 0.29** | 18.10 ± 0.10*** |
| Oxacillin | 9.83 ± 0.47 | 12.13 ± 0.35** | 11.90 ± 0.26** |
| PolymixinB | 7.00 ± 0.00 | 9.80 ± 0.70** | 10.83 ± 0.35*** |
| Cefuroxime | 21.00 ± 0.20 | 25.50 ± 0.50*** | 30.67 ± 0.58*** |
| Ceftriaxone | 20.56 ± 0.25 | 21.27 ± 0.25* | 26.03 ± 0.06*** |
| Carbenicillin | 33.93 ± 0.12 | 36.50 ± 0.50*** | 35.40 ± 0.35** |
| Cefotaxime | 20.63 ± 0.15 | 21.10 ± 0.36 | 24.73 ± 0.64*** |
| Gentamycin | 24.83 ± 0.47 | 23.10 ± 0.36** | 22.83 ± 0.35** |
| Ciprofloxacin | 23.17 ± 0.29 | 23.03 ± 0.06 | 23.43 ± 0.40 |
| Amikacin | 21.93 ± 0.12 | 21.80 ± 0.82 | 21.80 ± 0.26 |
| Erythromycin | 28.27 ± 0.25 | 27.80 ± 0.44 | 28.03 ± 0.25 |
| Clindamycin | 19.77 ± 0.68 | 17.93 ± 0.12** | 18.57 ± 0.51 |
| Chloramphenicol | 26.777 ± 0.40 | 26.60 ± 0.36 | 24.83 ± 0.15** |
Results are mean ± SD of three independent experiments.*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To evaluate the function of YqgS and LafA on stress resistance, we further assessed the effects of environmental pressure from food processing (acid, alkaline, salts, and oxidant) on bacterial growth in different strains. The resistance of ΔyqgS and ΔlafA mutants to acid (Fig. 8A) and salts (Fig. 8C) was slightly decreased compared with wild-type strain. The growth of mutants was severely affected under alkaline conditions (Fig. 8B). These results above suggest that the destruction of cell wall-related gene functions may lead to a drastic effect on coping pH change for L. monocytogenes, and indicate that pH is a critical factor for enhanced bacteria killing in particular circumstances. In addition, the antioxidant capacity of ΔlafA mutant was considerably attenuated (Fig. 8D). Overall, YqgS and LafA are dispensable for general stress resistance in L. monocytogenes.
FIG 8.
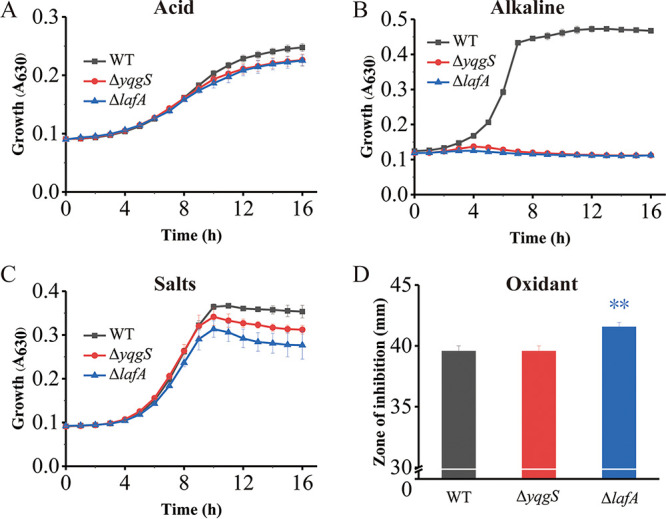
Impact of yqgS and lafA deletion on growth under unfavorable conditions. The growth curves of L. monocytogenes wild-type, ΔyqgS, and ΔlafA mutants in unfavorable conditions, including pH 6.0 (A), pH 9.5 (B), 750 mM NaCl (C). (D) The size of the inhibition zone of the WT and mutants with 30% H2O2 (15 μL).
YqgS and LafA are required for intracellular multiplication.
Given the fact that YqgS and LafA increase the surface negative charge and play an important role in biofilm formation, we hypothesized that the LTA-related genes are essential for L. monocytogenes pathogenicity. To test this assumption, we infected the RAW264.7 murine macrophage-like cells with different strains (wild-type, ΔyqgS, and ΔlafA mutant) and determined the number of intracellular bacteria as a function of time. No obvious differences were detected in adherence from all the strains (Fig. 9A), but the multiplication rates of the mutants exhibited partial defects upon internalization in the RAW (Fig. 9B). In sum, these results showed that deletion of yqgS and lafA had no effect on adhesion, but these are crucial in efficient survival and replication of L. monocytogenes in macrophages.
FIG 9.
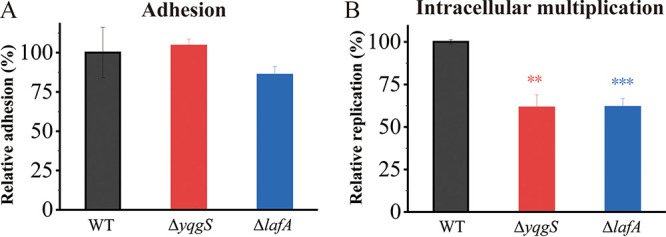
RAW264.7 macrophages were infected with wild-type, ΔyqgS, and ΔlafA mutant strains. (A) Adhesion of RAW264.7 by different strains as CFU counts relative to wild-type (fixed at 100%). (B) Intracellular multiplication behavior of mutants and wild-type strains in RAW264.7. Results are mean ± SD of three independent experiments. **, P ≤ 0.01; ***, P ≤ 0.001.
DISCUSSION
In this study, 99 genes associated with nisin resistance have been identified in L. monocytogenes at a genome-wide scale via Tn-seq. Of these, 92 genes contribute to nisin resistance, and only seven genes exert negative roles on nisin resistance. Previous studies have demonstrated that the cell envelope-related genes of dltA, mprF, and lmo2229 contributed to the resistance of L. monocytogenes to nisin (22, 27, 28, 55). The DltA mediates the d-alanylation of teichoic acids and the MprF is required for lysinylation of phospholipids, which reduce the surface negative charge of bacterial cells, decrease the binding of CAMPs, and consequently enhance resistance to nisin (27, 56–59). The pbp4 mutant showed increased nisin sensitivity likely via regulation of peptidoglycan peptide cross-links (22, 55). The changes of the cytoplasmic membrane, such as the presence of less diphosphatidylglycerol and more phosphatidylethanolamine, were generally related to the impaired nisin effectiveness (60, 61). Therefore, in this study we focused on two cell envelope-related genes, which were involved in glycolipid (lafA) and lipoteichoic acid backbone (yqgS) synthesis.
Resistance to nisin is attributed to stress response mechanisms, and it has been reported previously that two-component systems (lisRK, liaRS, and virRS) perform important functions in innate lantibiotic resistance (18, 26, 62–64). Strikingly, out of the 27 genes regulated by liaRS (65), seven genes (lmo0954, virA, lmo1966, telA, pbp4, lmo2484, and lmo2485) were identified as nisin-resistance genes, the three resistance genes of which were previously described including virA (26, 64), telA (29), and pbp4 (22), encoding ABC transporter permease, toxic ion resistance protein, and penicillin-binding protein, respectively. In addition, five VirR-regulated genes (anrAB, dltA, mprF, and lmo2156) were correlated with resistance against nisin, which also were verified experimentally in previous studies (27, 28, 64) except for the gene of lmo2156.
The reidentification of locus previously shown to be related to nisin resistance confirms the reliability of the screening, but two known nisin-resistance genes (lisK and gadD1) have not been identified. The signal transduction system LisK and the glutamate decarboxylase GadD1 determined the sensitivity of L. monocytogenes LO28 to nisin (18), but the corresponding genes were not identified in L. monocytogenes EGDe, presumably due to the genetic difference of strains or variation in experimental conditions (66). In addition, the newly identified genes (yqgS, lafA, gtcA, and lmo1464) were deleted to validate the accuracy of Tn-seq analysis. We have demonstrated the role of GtcA and Lmo1464 in nisin sensitivity, and these mutants showed significantly enhanced resistance to nisin. However, the detailed mechanism still needs to be investigated.
LTAs are important cell wall polymers usually composed of polyglycerolphosphate backbone chain that is embedded in the membrane via a glycolipid, which plays critical roles in the maintenance of antimicrobial resistance and pathogenicity of Gram-positive pathogens (39, 67). LafABC, involved in glycolipid synthesis, are required for LTA synthesis in L. monocytogenes. The LTA primase YqgS (also called LtaP) and synthase LtaS (Lmo0927) are responsible for LTA backbone synthesis (34). We identified all the LTA-related genes in our Tn-seq analysis except for ltaS, which is potentially attributable to the severe growth defect of ltaS mutant (34). LTA-related genes (yqgS and lafA) are essential for L. monocytogenes resistance to nisin but not to bacitracin. LTA was previously reported to be involved in nisin resistance of Streptococcus bovis and S. aureus (68, 69).
Generally, nisin binds to the bacterial cell surface and oligomerizes in cytoplasmic membranes to form pores, leading to the leakage of cellular contents and cell death (13). The binding affinity of nisin toward the surface of the bacteria is driven by electrostatic forces between cationic peptides and negatively charged cell envelope (70), and modification of LTAs is known to accumulate positive charge on the bacterial surface to decrease the effectiveness of nisin (28, 71). The increased negative charge on the surface of the yqgS and lafA mutants may be part of the reason for the increased nisin sensitivity. YqgS and LafA significantly contribute to the resistance of cell envelope-acting antimicrobials and unfavorable conditions. Notably, several studies have linked nisin and cephalosporin resistance in L. monocytogenes (18, 20, 25, 29). The identification of the genes related to cephalosporin resistance may promote the development of strategies to cope with the high resistance to these antibacterial agents (72).
L. monocytogenes colonizes food processing environments to form biofilms, which lead to biofilm-mediated antimicrobial resistance and stress tolerance, with negative implications for food safety (73–75). Biofilms could be responsible for persistent food contamination of bacteria, which in turn leads to the outbreak of Listeria infections (76, 77).The ΔyqgS and ΔlafA mutants displayed a greatly reduced capacity to form biofilm relative to wild-type strain. Bacterial adhesion, as a process indispensable to biofilm formation, is dictated by several factors, such as electrostatic and hydrophobic interactions, temperature, steric hindrance, and so on (78, 79). Given the negative charge of most bacteria and inert surfaces, electrostatic interactions are detrimental to the process of bacterial adhesion. The decline in biofilm forming capability of the ΔyqgS and ΔlafA mutants might be in part explained by the increased electrostatic repulsion.
LTA expression on group B streptococci plays an important role in human brain microvascular endothelial cells’ invasiveness (80), and LTA deficient mutant was less pathogenic than S. aureus wild-type (36). YqgS and LafA are crucial in efficient survival and replication of L. monocytogenes in macrophages. The mechanistic basis of the decreased pathogenic nature of the LTA mutation has not been resolved but might be connected to the alterations in envelope charge and envelope integrity, which increased sensitivity to antibacterial effectors produced by macrophages.
In conclusion, this study provided comprehensive identification of genes required for nisin resistance, which revealed novel mechanisms of nisin resistance in L. monocytogenes. LTA-related genes (yqgS and lafA) are required for biofilm formation, surface charge maintenance, and intracellular multiplication, which also significantly contribute to the resistance of cell envelope-acting antimicrobials and unfavorable conditions. These findings may provide novel targets for the development of food-grade inhibitors for nisin-resistant foodborne pathogens.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The L. monocytogenes strain EGDe (American Tissue Culture Collection) and the Escherichia coli strains EC1000 were used in this study. Unless otherwise noted, the bacteria were cultured in BHI (Hopebiol) at 37°C with shaking. Under necessary conditions, the concentrations of antibiotics used are as follows: gentamicin 25 μg/mL for L. monocytogenes and E. coli, chloramphenicol 5 μg/mL for L. monocytogenes, spectinomycin 100 μg/mL for L. monocytogenes and E. coli. All strains and plasmids used in this study are listed in Table S1.
Construction of mariner transposon mutant library in L. monocytogenes.
To create a high-density transposon mutant library, the temperature-sensitive plasmid pGPA2 (81) was electroporated into L. monocytogenes. The mutant library was generated according to previously described methods (82). In brief, the plasmid-containing strains were incubated overnight in BHI with chloramphenicol at 30°C, and 200 μL of this culture was added into the BHI containing gentamicin (25 μg/mL) and nisin (25 ng/mL) for overnight culture. Then two successive passages were passed in BHI without antibiotics at 37°C. Subsequently, cultures were stored at −80°C with 50% (vol/vol) glycerol as mutant library stocks.
Tn-seq analysis of genes involved in nisin resistance.
To identify genes that are involved in nisin resistance, 40 μL aliquots of the mutant library in L. monocytogenes were inoculated in BHI containing gentamicin (25 μg/mL) for overnight culture. Then three experimental replicates of approximately 2 × 107 CFU were inoculated into 100 mL BHI or 100 mL BHI with 200 μg/mL nisin (Aladdin), which were incubated at 37°C for 16 h and then further processed for Tn-seq as described previously (83). Tn-seq sequencing was performed on Illumina Hiseq-PE150 (Personalbio), generating an average of 1G high clean data per sample.
Tn-seq data analysis was performed as previously described. In short, barcodes were split using the Galaxy platform and 16-nucleotide fragments of each read were mapped to the L. monocytogenes EGDe genome using Bowtie 2. The genome was subsequently divided in 25-bp windows and each alignment was sorted and indexed by IGV. Insertions were counted per window and then summed over the genes. Read counts per gene were adjusted to cover only the first 90% of the gene, because the final 10% of a gene were discarded as these insertions may not inactivate gene function. Subsequently, read counts were normalized to the total number of reads which mapped to the genome in each replicate, by calculating the normalized read-count reads per kilobase per million input reads (RKPM); RPTAM = (number of reads mapped to a gene × 106)/(total mapped input reads in the sample × number of TA sites in this gene). The statistical analysis on the RPTAM values was performed by Cyber-T. Genes that significantly contributed to either nisin susceptibility or nisin resistance were determined when the Benjamini-Hochberg (BH) corrected P-value was < 0.05 and the difference in abundance of the transposon mutant during growth in BHI with or without nisin was > 5. Tn-seq data was provided in supplementary file 2.
Construction of mutant strains.
Plasmid pWS3 was used to generate mutations through a process of double homologous recombination in L. monocytogenes, with a method based on the Cre-lox recombination system (82). The upstream and downstream homology arms of yqgS were PCR amplified from chromosomal DNA with yqgS_up_F/R and yqgS_down_F/R, respectively. The gentamicin maker was PCR amplified from pAT392 with pAT392_lox66_genta_F/R. These three DNA fragments were cloned in tandem (up-gm-down) on SmaI site of the pWS3 vector with NovoRec plus One step PCR Cloning Kit (novoprotein), resulting in plasmid pWS3- yqgS which used EC1000 as the cloning host. The recombinant plasmid with homologous arm and gentamicin maker was introduced in L. monocytogenes EGDe by electroporation. Positive clones were inoculated in prewarmed BHI medium without antibiotics and grown overnight at 37°C to get marked deletion mutants. To obtain the markerless mutants, the shuttle plasmid pWS3-erm-cre was electrotransformed into the marked mutants. Then cell culture was plated on BHI agar plates and incubated at 37°C to get the markerless mutations, verified by PCR and sequencing.
The same strategy was used to remove the lafA, gtcA, and lmo1464 genes in L. monocytogenes to get the corresponding markerless mutations. Primers used in this study are listed in Table S1. All plasmid constructs and gene deletions were confirmed by DNA sequencing.
Growth curves and nisin-mediated killing.
To draw growth curves of L. monocytogenes strains (wild-type and mutants), overnight cultures were diluted 100-fold in fresh BHI and BHI containing appropriate nisin or bacitracin. The absorbance of 630 nm (A630 nm) was measured every 1 h for 16 h. The same strategy was used to draw growth curves of L. monocytogenes wild-type, ΔyqgS, and ΔlafA mutants in unfavorable conditions.
Overnight cultures of L. monocytogenes strains were inoculated into the fresh BHI medium at a ratio of 1:100 and grew to exponential phase (OD600nm = 0.6 to 0.8), and then spun down and resuspended in phosphate buffer saline (PBS) to OD600nm = 0.65, next nisin (1,000 μg/mL) was added. Viable bacterial CFU counts were conducted after 2 h.
Transmission electron microscopy and scanning electron microscopy.
The L. monocytogenes strains were grown to exponential phase (OD600nm = 0.8), followed by nisin (800 μg/mL) treatment in experimental groups for 1 h, after which the bacteria cultures were spun down and fixed with 2.5% glutaraldehyde in PBS for more than 4 h. The samples were washed three times in PBS, postfixed with 1% OsO4 for 1 h, and washed three times. Then the dehydration of samples used a graded series of ethanol (30%, 50%, 70%, 80%, 90%, 95%, 100%). The samples were dehydrated by alcohol for 20 min, and dehydrated in Hitachi Model HCP-2 critical point dryer. Later we can coat with gold-palladium in Hitachi Model E-1010 ion sputter for 4 to 5 min and observe in Hitachi Model SU-8010 SEM. The samples were transferred to absolute acetone for dehydration, placed in 1:1 mixture of absolute acetone and the final Spurr resin mixture, then transferred to 1:3 mixture of absolute acetone and the final resin mixture and to the final Spurr resin mixture for overnight. Specimens were heated at 70°C, then sectioned in LEICA EM UC7 ultratome, and sections were stained by uranyl acetate and alkaline lead citrate for 5 to 10 min respectively and observed in Hitachi Model H-7650 TEM.
Determination of bacterial surface charge.
Bacterial surface charge was performed as described (84). Overnight cultures of L. monocytogenes strains were inoculated into the fresh BHI medium and grew to exponential phase (OD600nm = 0.6 to 0.8), then washed twice in 20 mM MOPS buffer (pH 7.0) and adjusted to OD600nm = 0.7. Bacterial aliquots were concentrated in half volume of 0.5 mg/mL equine cytochrome c dissolved in 20 mM MOPS buffer. After 10 min of incubation in the dark, the bacteria were centrifuged (14,000 rpm, 3 min) and the supernatant liquid was transferred to a 96-well plate. The absorbance was measured at 530 nm. The absorbance of samples only containing MOPS buffer was recorded as 100% binding, and the value of samples containing cytochrome c but lacking bacteria was recorded as 100% binding.
Quantification of biofilm formation.
The overnight cultures were inoculated into the fresh BHI, adjusting the bacterial liquid concentration to 2 × 106 CFU/mL. Then 200 μL of this bacterial liquid was absorbed into 96-well plate and incubated at 37°C for 48 h. The biofilm was quantified with the crystal violet (CV) assay (85). Briefly, culture was removed and washed with PBS three times to remove unattached cells. The wells were air-dried for 30 min, then 200 μL of methanol were added and set for 30 min. The wells were stained with 200 μL of 1% (wt/vol) crystal violet. After 30 min, rinse the well with 200 μL PBS for three times to remove excess staining and air dry the well. Finally, 200 μL glacial acetic acid (33%) was added to dissolve the crystalline violet bound to the biofilm. The absorbance of 595 nm was measured with the FLx800 Absorbance Reader (Biotech, USA). Experiments are performed at least in triplicate.
Fluorescence staining was used to observe the formation of biofilm under confocal microscope. 4 × 106 CFU were added to 2 mL BHI with confocal dish and incubated at 37°C for 48 h. Cultures were discarded and dishes were washed thrice with PBS. Next, 200 μL SYTO9/PI dye solution was added to the culture dishes and treated for 30 min in the dark. After washing the dishes with PBS once, they were observed under a Confocal Laser Scanning Microscope LSM700.
Antibiotic disk assays.
The antibiotic disk diffusion assays were performed as previously studied (18, 86). Briefly, the overnight cultures were diluted to OD600nm = 0.1 and swabbed onto BHI plates. Six-millimeter antibiotic disks were placed onto the corresponding BHI plates and incubated at 37°C for 24 h. The zones of inhibition were measured.
The size of inhibition zone of H2O2 was performed as mentioned above. It should be noted that 15 μL H2O2 (33%) was added to a 6-mL blank disk and left to dry for an hour. Subsequently, the H2O2 disks were placed onto the corresponding BHI plates and incubated at 37°C for 24 h to measure the size of inhibition zone.
RAW264.7 macrophage infection.
Invasion and intracellular multiplication assays in RAW were performed as described (87). The L. monocytogenes strains were grown to exponential phase (OD600nm = 0.6 to 0.8), washed three times in dulbecco's modified eagle medium (DMEM), and diluted to MOI ≈25. Bacterial suspensions were transferred to macrophages and incubated for 1 h at 37°C, then washed three times and added fresh DMEM with gentamicin (25 μg/mL) for 1.5 h. Cells were lysed in 0.1% Triton X-100 and serially diluted for counting in BHI at different time points (0, 1, and 8 h).
Statistical analyses.
To ensure the significance of the results, the data were analyzed using Student’s t test. The differences were considered statistically significant at the 95% level of confidence (P < 0.05).
Data availability.
Illumina sequencing reads of L. monocytogenes in Tn-seq have been submitted in the NCBI server (Accession No. SRR17303189).
ACKNOWLEDGMENTS
This study was supported by the Project of Shandong Province Higher Educational Outstanding Youth Innovation Team (2019KJF011), the Natural Science Foundation of Shandong Province, China (ZR2019ZD21), the Taishan Scholars Program of Shandong Province, China (ts20190955), and the National Key Research and Development Program of China (2019YFE0103900; 2018YFC1200100).
We declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplemental material is available online only.
Contributor Information
Xinglin Zhang, Email: zhangxinglin@lyu.edu.cn.
Pablo Power, Universidad de Buenos Aires, Facultad de Farmacia y Bioquimica.
REFERENCES
- 1.Radoshevich L, Cossart P. 2018. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol 16:32–46. doi: 10.1038/nrmicro.2017.126. [DOI] [PubMed] [Google Scholar]
- 2.Schlech WF, Lavigne PM, Bortolussi RA, Allen AC, Haldane EV, Wort AJ, Hightower AW, Johnson SE, King SH, Nicholls ES, Broome CV. 1983. Epidemic listeriosis - evidence for transmission by food. N Engl J Med 308:203–206. doi: 10.1056/NEJM198301273080407. [DOI] [PubMed] [Google Scholar]
- 3.de Noordhout CM, Devleesschauwer B, Angulo FJ, Verbeke G, Haagsma J, Kirk M, Havelaar A, Speybroeck N. 2014. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infectious Diseases 14:1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goulet V, Jacquet C, Vaillant V, Rebiere I, Mouret E, Lorente C, Maillot E, Stainer F, Rocourt J. 1995. Listeriosis from consumption of raw-milk cheese. Lancet 345:1581–1582. doi: 10.1016/s0140-6736(95)91135-9. [DOI] [PubMed] [Google Scholar]
- 5.Lachmann R, Halbedel S, Adler M, Becker N, Allerberger F, Holzer A, Boone I, Falkenhorst G, Kleta S, Al Dahouk S, Stark K, Luber P, Flieger A, Wilking H. 2021. Nationwide outbreak of invasive listeriosis associated with consumption of meat products in health care facilities, Germany, 2014-2019. Clin Microbiol Infect 27:1035.e1–1035.e5. doi: 10.1016/j.cmi.2020.09.020. [DOI] [PubMed] [Google Scholar]
- 6.McLauchlin J, Aird H, Amar C, Barker C, Dallman T, Lai S, Painset A, Willis C. 2021. An outbreak of human listeriosis associated with frozen sweet corn consumption: Investigations in the UK. Int J Food Microbiol 338:108994. doi: 10.1016/j.ijfoodmicro.2020.108994. [DOI] [PubMed] [Google Scholar]
- 7.Hyden P, Pietzka A, Lennkh A, Murer A, Springer B, Blaschitz M, Indra A, Huhulescu S, Allerberger F, Ruppitsch W, Sensen CW. 2016. Whole genome sequence-based serogrouping of Listeria monocytogenes isolates. J Biotechnol 235:181–186. doi: 10.1016/j.jbiotec.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Pradhan AK, Ivanek R, Grohn YT, Geornaras I, Sofos JN, Wiedmann M. 2009. Quantitative risk assessment for Listeria monocytogenes in selected categories of deli meats: impact of lactate and diacetate on listeriosis cases and deaths. J Food Prot 72:978–989. doi: 10.4315/0362-028x-72.5.978. [DOI] [PubMed] [Google Scholar]
- 9.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 10.Yang SC, Lin CH, Sung CT, Fang JY. 2014. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol 5:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumariya R, Garsa AK, Rajput YS, Sood SK, Akhtar N, Patel S. 2019. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb Pathog 128:171–177. doi: 10.1016/j.micpath.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Cleveland J, Montville TJ, Nes IF, Chikindas ML. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71:1–20. doi: 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 13.Barbosa AAT, de Melo MR, da Silva CMR, Jain S, Dolabella SS. 2021. Nisin resistance in Gram-positive bacteria and approaches to circumvent resistance for successful therapeutic use. Crit Rev Microbiol 47:376–385. doi: 10.1080/1040841X.2021.1893264. [DOI] [PubMed] [Google Scholar]
- 14.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 15.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris LJ, Fleming HP, Klaenhammer TR. 1991. Sensitivity and resistance of listeria-monocytogenes Atcc-19115, Scott-a, and Ual500 to Nisin. J Food Prot 54:836–840. doi: 10.4315/0362-028X-54.11.836. [DOI] [PubMed] [Google Scholar]
- 17.Field D, Quigley L, O'Connor PM, Rea MC, Daly K, Cotter PD, Hill C, Ross RP. 2010. Studies with bioengineered Nisin peptides highlight the broad-spectrum potency of Nisin V. Microb Biotechnol 3:473–486. doi: 10.1111/j.1751-7915.2010.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotter PD, Guinane CM, Hill C. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob Agents Chemother 46:2784–2790. doi: 10.1128/AAC.46.9.2784-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang JH, Wiedmann M, Boor KJ, Bergholz TM. 2015. VirR-mediated resistance of Listeria monocytogenes against food antimicrobials and cross-protection induced by exposure to organic acid salts. Appl Environ Microbiol 81:4553–4562. doi: 10.1128/AEM.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gravesen A, Sorensen K, Aarestrup FM, Knochel S. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb Drug Resist 7:127–135. doi: 10.1089/10766290152045002. [DOI] [PubMed] [Google Scholar]
- 21.Begley M, Hill C, Ross RP. 2006. Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on SigB. Appl Environ Microbiol 72:2231–2234. doi: 10.1128/AEM.72.3.2231-2234.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gravesen A, Kallipolitis B, Holmstrom K, Hoiby PE, Ramnath M, Knochel S. 2004. pbp2229-mediated nisin resistance mechanism in Listeria monocytogenes confers cross-protection to class IIa bacteriocins and affects virulence gene expression. Appl Environ Microbiol 70:1669–1679. doi: 10.1128/AEM.70.3.1669-1679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joerger RD, Chen HQ, Kniel KE. 2006. Characterization of a spontaneous, pressure-tolerant Listeria monocytogenes Scott A ctsR deletion mutant. Foodborne Pathog Dis 3:196–202. doi: 10.1089/fpd.2006.3.196. [DOI] [PubMed] [Google Scholar]
- 24.Begley M, Cotter PD, Hill C, Ross RP. 2010. Glutamate decarboxylase-mediated nisin resistance in Listeria monocytogenes. Appl Environ Microbiol 76:6541–6546. doi: 10.1128/AEM.00203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins B, Curtis N, Cotter PD, Hill C, Ross RP. 2010. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob Agents Chemother 54:4416–4423. doi: 10.1128/AAC.00503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grubaugh D, Regeimbal JM, Ghosh P, Zhou Y, Lauer P, Dubensky TW, Higgins DE. 2018. The VirAB ABC transporter is required for VirR regulation of Listeria monocytogenes virulence and resistance to nisin. Infect Immun 86. doi: 10.1128/IAI.00901-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. 2006. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol 62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 28.Henderson LO, Erazo Flores BJ, Skeens J, Kent D, Murphy SI, Wiedmann M, Guariglia-Oropeza V. 2020. Nevertheless, she resisted - role of the environment on Listeria monocytogenes sensitivity to Nisin treatment in a laboratory cheese model Front Microbiol 11:635. doi: 10.3389/fmicb.2020.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins B, Joyce S, Hill C, Cotter PD, Ross RP. 2010. TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob Agents Chemother 54:4658–4663. doi: 10.1128/AAC.00290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon YM, Ricke SC, Mandal RK. 2016. Transposon sequencing: methods and expanding applications. Appl Microbiol Biotechnol 100:31–43. doi: 10.1007/s00253-015-7037-8. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Liu G, Wu Y, Pang X, Wu Y, Qinshu, Niu J, Chen Q, Zhang X. 2021. Transposon sequencing: A powerful tool for the functional genomic study of food-borne pathogens. Trends in Food Science & Technology doi: 10.1016/j.tifs.2021.06.032. [DOI] [Google Scholar]
- 33.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 34.Webb AJ, Karatsa-Dodgson M, Grundling A. 2009. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol Microbiol 74:299–314. doi: 10.1111/j.1365-2958.2009.06829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fedtke I, Mader D, Kohler T, Moll H, Nicholson G, Biswas R, Henseler K, Gotz F, Zahringer U, Peschel A. 2007. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol Microbiol 65:1078–1091. doi: 10.1111/j.1365-2958.2007.05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheen TR, Ebrahimi CM, Hiemstra IH, Barlow SB, Peschel A, Doran KS. 2010. Penetration of the blood-brain barrier by Staphylococcus aureus: contribution of membrane-anchored lipoteichoic acid. J Mol Med (Berl) 88:633–639. doi: 10.1007/s00109-010-0630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theilacker C, Sanchez-Carballo P, Toma I, Fabretti F, Sava I, Kropec A, Holst O, Huebner J. 2009. Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis. Mol Microbiol 71:1055–1069. doi: 10.1111/j.1365-2958.2008.06587.x. [DOI] [PubMed] [Google Scholar]
- 38.Salzberg LI, Helmann JD. 2008. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J Bacteriol 190:7797–7807. doi: 10.1128/JB.00720-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reichmann NT, Grundling A. 2011. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett 319:97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Downes MT, Mehla J, Ananthaswamy N, Wakschlag A, Lamonde M, Dine E, Ambudkar SV, Golin J. 2013. The transmission interface of the Saccharomyces cerevisiae multidrug transporter Pdr5: Val-656 located in intracellular loop 2 plays a major role in drug resistance. Antimicrob Agents Chemother 57:1025–1034. doi: 10.1128/AAC.02133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filipic B, Golic N, Jovcic B, Tolinacki M, Bay DC, Turner RJ, Antic-Stankovic J, Kojic M, Topisirovic L. 2013. The cmbT gene encodes a novel major facilitator multidrug resistance transporter in Lactococcus lactis. Res Microbiol 164:46–54. doi: 10.1016/j.resmic.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Baucheron S, Imberechts H, Chaslus-Dancla E, Cloeckaert A. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar typhimurium phage type DT204. Microb Drug Resist 8:281–289. doi: 10.1089/10766290260469543. [DOI] [PubMed] [Google Scholar]
- 43.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hechard Y, Pelletier C, Cenatiempo Y, Frere J. 2001. Analysis of sigma(54)-dependent genes in Enterococcus faecalis: a mannose PTS permease (EII(Man)) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology (Reading) 147:1575–1580. doi: 10.1099/00221287-147-6-1575. [DOI] [PubMed] [Google Scholar]
- 45.Gravesen A, Ramnath M, Rechinger KB, Andersen N, Jansch L, Hechard Y, Hastings JW, Knochel S. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology (Reading) 148:2361–2369. doi: 10.1099/00221287-148-8-2361. [DOI] [PubMed] [Google Scholar]
- 46.Opsata M, Nes IF, Holo H. 2010. Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol 10:224. doi: 10.1186/1471-2180-10-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 48.D’Alessandro M, Turina P, Melandri BA. 2008. Intrinsic uncoupling in the ATP synthase of Escherichia coli. Biochim Biophys Acta 1777:1518–1527. doi: 10.1016/j.bbabio.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Xu H, Teo K, Neo H, Liu Y. 2012. Chemically inhibited ATP synthesis promoted detachment of different-age biofilms from membrane surface. Appl Microbiol Biotechnol 95:1073–1082. doi: 10.1007/s00253-011-3770-9. [DOI] [PubMed] [Google Scholar]
- 50.Saville RM, Rakshe S, Haagensen JA, Shukla S, Spormann AM. 2011. Energy-dependent stability of Shewanella oneidensis MR-1 biofilms. J Bacteriol 193:3257–3264. doi: 10.1128/JB.00251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C 55 -isoprenyl pyrophosphate. Proc Natl Acad Sci USA 68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 53.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinheiro J, Lisboa J, Pombinho R, Carvalho F, Carreaux A, Brito C, Pontinen A, Korkeala H, Dos Santos NMS, Morais-Cabral JH, Sousa S, Cabanes D. 2018. MouR controls the expression of the Listeria monocytogenes Agr system and mediates virulence. Nucleic Acids Res 46:9338–9352. doi: 10.1093/nar/gky624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guinane CM, Cotter PD, Ross RP, Hill C. 2006. Contribution of penicillin-binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob Agents Chemother 50:2824–2828. doi: 10.1128/AAC.00167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. 1995. Incorporation of D-Alanine into lipoteichoic acid and wall teichoic-acid in Bacillus-Subtilis - identification of genes and regulation. J Biol Chem 270:15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- 57.Nilsson M, Rybtke M, Givskov M, Hoiby N, Twetman S, Tolker-Nielsen T. 2016. The dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int J Antimicrob Agents 48:298–304. doi: 10.1016/j.ijantimicag.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 58.Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. 2006. A functional dlt operon, encoding proteins required for incorporation of D-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol 188:5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF Consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verheul A, Russell NJ, Van THR, Rombouts FM, Abee T. 1997. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl Environ Microbiol 63:3451–3457. doi: 10.1128/aem.63.9.3451-3457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crandall AD, Montville TJ. 1998. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl Environ Microbiol 64:231–237. doi: 10.1128/AEM.64.1.231-237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mascher T, Zimmer SL, Smith TA, Helmann JD. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergholz TM, Tang S, Wiedmann M, Boor KJ. 2013. Nisin resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl Environ Microbiol 79:5682–5688. doi: 10.1128/AEM.01797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang X, Geng Y, Ren S, Yu T, Li Y, Liu G, Wang H, Meng H, Shi L. 2019. The VirAB-VirSR-AnrAB multicomponent system is involved in resistance of Listeria monocytogenes EGD-e to cephalosporins, bacitracin, nisin, benzalkonium chloride, and ethidium bromide. Appl Environ Microbiol 85. doi: 10.1128/AEM.01470-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fritsch F, Mauder N, Williams T, Weiser J, Oberle M, Beier D. 2011. The cell envelope stress response mediated by the LiaFSRLm three-component system of Listeria monocytogenes is controlled via the phosphatase activity of the bifunctional histidine kinase LiaSLm. Microbiology (Reading) 157:373–386. doi: 10.1099/mic.0.044776-0. [DOI] [PubMed] [Google Scholar]
- 66.Becavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu ZF, Kuhbacher A, Brisse S, Pucciarelli MG, Garcia-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerda J, Moszer I, Bierne H, Cossart P. 2014. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic differences underlying variations in pathogenicity. mBio 5. doi: 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Percy MG, Grundling A. 2014. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 68.Mantovani HC, Russell JB. 2001. Nisin resistance of Streptococcus bovis. Appl Environ Microbiol 67:808–813. doi: 10.1128/AEM.67.2.808-813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 70.Koprivnjak T, Peschel A, Gelb MH, Liang NS, Weiss JP. 2002. Role of charge properties of bacterial envelope in bactericidal action of human group IIA phospholipase A2 against Staphylococcus aureus. J Biol Chem 277:47636–47644. doi: 10.1074/jbc.M205104200. [DOI] [PubMed] [Google Scholar]
- 71.Martinez B, Obeso JM, Rodriguez A, Garcia P. 2008. Nisin-bacteriophage crossresistance in Staphylococcus aureus. Int J Food Microbiol 122:253–258. doi: 10.1016/j.ijfoodmicro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 72.Nielsen PK, Andersen AZ, Mols M, van der Veen S, Abee T, Kallipolitis BH. 2012. Genome-wide transcriptional profiling of the cell envelope stress response and the role of LisRK and CesRK in Listeria monocytogenes. Microbiology (Reading) 158:963–974. doi: 10.1099/mic.0.055467-0. [DOI] [PubMed] [Google Scholar]
- 73.Pilchova T, Hernould M, Prevost H, Demnerova K, Pazlarova J, Tresse O. 2014. Influence of food processing environments on structure initiation of static biofilm of Listeria monocytogenes. Food Control 35:366–372. doi: 10.1016/j.foodcont.2013.07.021. [DOI] [Google Scholar]
- 74.Jahid IK, Ha SD. 2012. A review of microbial biofilms of produce: Future challenge to food safety. Food Sci Biotechnol 21:299–316. doi: 10.1007/s10068-012-0041-1. [DOI] [Google Scholar]
- 75.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 76.Beresford MR, Andrew PW, Shama G. 2001. Listeria monocytogenes adheres to many materials found in food-processing environments. J Appl Microbiol 90:1000–1005. doi: 10.1046/j.1365-2672.2001.01330.x. [DOI] [PubMed] [Google Scholar]
- 77.Renier S, Hebraud M, Desvaux M. 2011. Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram-positive foodborne pathogen. Environ Microbiol 13:835–850. doi: 10.1111/j.1462-2920.2010.02378.x. [DOI] [PubMed] [Google Scholar]
- 78.Carpentier B, Cerf O. 1993. Biofilms and their consequences, with particular reference to hygiene in the food-industry. J Appl Bacteriol 75:499–511. doi: 10.1111/j.1365-2672.1993.tb01587.x. [DOI] [PubMed] [Google Scholar]
- 79.Dunne WM. 2002. Bacterial adhesion: Seen any good biofilms lately? Clin Microbiol Rev 15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doran KS, Engelson EJ, Khosravi A, Maisey HC, Fedtke I, Equils O, Michelsen KS, Arditi M, Peschel A, Nizet V. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J Clin Invest 115:2499–2507. doi: 10.1172/JCI23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang X, de Maat V, Guzman Prieto AM, Prajsnar TK, Bayjanov JR, de Been M, Rogers MRC, Bonten MJM, Mesnage S, Willems RJL, van Schaik W. 2017. RNA-seq and Tn-seq reveal fitness determinants of vancomycin-resistant Enterococcus faecium during growth in human serum. BMC Genomics 18:893. doi: 10.1186/s12864-017-4299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Paganelli FL, Bierschenk D, Kuipers A, Bonten MJ, Willems RJ, van Schaik W. 2012. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet 8:e1002804. doi: 10.1371/journal.pgen.1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burghout P, Zomer A, van der Gaast-de Jongh CE, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Hermans PW. 2013. Streptococcus pneumoniae folate biosynthesis responds to environmental CO2 levels. J Bacteriol 195:1573–1582. doi: 10.1128/JB.01942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carvalho F, Atilano ML, Pombinho R, Covas G, Gallo RL, Filipe SR, Sousa S, Cabanes D. 2015. L-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog 11:e1004919. doi: 10.1371/journal.ppat.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Djordjevic D, Wiedmann M, McLandsborough LA. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol 68:2950–2958. doi: 10.1128/AEM.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kallipolitis BH, Ingmer H, Gahan CG, Hill C, Sogaard-Andersen L. 2003. CesRK, a two-component signal transduction system in Listeria monocytogenes, responds to the presence of cell wall-acting antibiotics and affects beta-lactam resistance. Antimicrob Agents Chemother 47:3421–3429. doi: 10.1128/AAC.47.11.3421-3429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prevost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA 104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02095-21_Supp_2_seq14.xlsx, XLSX file, 0.8 MB (877KB, xlsx)
Supplemental material. Download SPECTRUM02095-21_Supp_1_seq4.pdf, PDF file, 0.3 MB (334.8KB, pdf)
Data Availability Statement
Illumina sequencing reads of L. monocytogenes in Tn-seq have been submitted in the NCBI server (Accession No. SRR17303189).