Abstract
In order to determine the suitability of vaccine strains established in the 1960s for a new vaccine, a comprehensive study of strain variation of adenovirus serotype 4 (AV 4) and AV 7 was undertaken. A 1,500-bp region of the hexon gene containing the AV neutralization epitopes from prototype, vaccine, and community-acquired strains and from wild-type strains from military personnel that cause acute respiratory disease (ARD) was sequenced and analyzed. The whole hexon gene from prototype strains, vaccine strains, and selected isolates was sequenced. AV 7 and AV 7a were found to have distinct genotypes, and all vaccine and wild-type strains recovered from 1963 to 1997 had the AV 7a genotype. There was no significant strain variation in the neutralization epitopes of the AV 7a genotype over a 42-year period. The evolution of AV 4 was more complex, with continuous genetic drift punctuated by replacement with a new strain. The current strain of AV 4, which has been in circulation since 1995, is significantly different from the AV 4 prototype and the vaccine strains. Genetic differences were confirmed to be antigenic differences by neutralization tests, which define the new strain as an AV 4 variant. A type-specific PCR for AV 4, AV 7/7a, and AV 21 was developed, and this PCR facilitated the rapid identification of isolates from outbreaks of ARD.
Epidemics of acute respiratory disease (ARD) in military personnel were the most significant cause of morbidity, hospitalizations, and work-time loss in new recruits and trainees through the 1950s (1, 28). The etiology of ARD was discovered to be adenoviruses (AVs), primarily AV serotype 4 (AV 4) and AV 7 (1, 12) and occasionally AV 3, AV 14, and AV 21 (29). In 1956 the first experimental formalin-inactivated adenoviral vaccines, which were made from monkey kidney cell cultures, were tested (13). However, lot-to-lot variation, contamination with simian virus 40, and the possible oncogenicity of AV 3 and AV 7 led to their withdrawal in 1963 (15). A live, enterically coated AV 4 vaccine made from human diploid cells was introduced in that same year (4, 10). Following extensive oncogenicity testing (9), a live enterically coated AV 7 vaccine was introduced in 1971 (26). Both vaccines have been in continuous use since 1971 and have reduced the incidence of ARD caused by AVs in recruits by 80 to 90% (8, 15).
In 1994, vaccine production delays resulted in shortages, leading to outbreaks of ARD in several training centers (19, 27). In 1996, the sole manufacturer of the vaccines, Wyeth-Ayerst, Inc., Marietta, Pa., permanently discontinued production of the vaccines (27). A new manufacturer is being sought by the U.S. Department of Defense, but on-hand vaccine stocks are projected to be exhausted by the winter of 1999, depriving the armed services of effective countermeasures to the excessive morbidity caused by ARD.
Before a new vaccine source is designated, the relationship of the currently circulating strains of AV 4 and AV 7 to the vaccine strains needs to be examined. The vaccine strains were first isolated more than 35 years ago (4, 26). The possibility exists that current strains in circulation may have undergone significant genetic and antigenic drift. Protective neutralizing antibodies to AVs are directed against type-specific epitopes contained in seven hypervariable regions (HVRs) in the viral major coat protein, the hexon (6). Mutation in the HVRs can be extensive and can lead to antigenic shift and drift and the evolution of new serotypes (7).
The intent of this study was to examine the genetic variation among strains of AV 4 and AV 7. In order to map their evolution we sequenced and analyzed a 1,500-bp region that included the seven HVRs from prototype strains established in the 1950s, the vaccine strains from the early 1960s, wild-type strains from patients with community-acquired infections recovered from 1963 to 1996, and strains from recent ARD outbreaks in the military. We sequenced the whole hexon gene from prototype strains, vaccine strains, and two wild-type strains. Genetic variation was confirmed by cross-neutralization tests for antigenic variation. We also developed a type-specific PCR for AV 4, AV 7/7a, and AV 21 to rapidly type isolates from ARD outbreaks.
MATERIALS AND METHODS
Viral strains and isolates.
Viruses from four sources were examined (Table 1). (i) Prototype strains AV 4 (RI-67), AV 7 (Gomen), and AV 7a (S-1058) were from the collection of the Viral and Rickettsial Disease Laboratory (VRDL), California State Department of Health Services, Berkeley. (ii) Vaccine strains AV 4 (CL 68578), lot 4958221, and AV 7 (55142), lot 4958220, were provided by Wyeth Laboratories, Marietta, Pa. (4, 26). (iii) Thirty-eight isolates recovered from military personnel were tested. The isolates from military personnel were identified as AV 4 or AV 7 by the submitting laboratories on the basis of the neutralization test. All isolates from military personnel were tested by a type-specific PCR, and 12 were selected for sequencing. (iv) Seven community-acquired AV 4 isolates and 12 community-acquired AV 7 isolates were obtained from the collection of VRDL and were isolated between 1963 and 1996. Stock virus preparations were made from the vaccine strains and isolates from military personnel by a single passage in A549 cells.
TABLE 1.
AV 4 and AV 7/7a strains and isolates sequenced for this studya
| Serotype | Strain | Year of isolation | Location (reference) | Age/sex | Syndrome |
|---|---|---|---|---|---|
| AV 4 | RI-67 (p) | 1953 | Missouri (16) | PAP | |
| 55142 (v) | 1963 | Washington, D.C. (5, 30) | 20 yr/M | Febrile respiratory illness | |
| T63-2297 | 1963 | Stockton, Calif. | 40 yr/F | Conjunctivitis | |
| T64-3630 | 1964 | NA | NA | NA | |
| T68-0143 | 1968 | San Jose, Calif. | 10 mo/M | URI | |
| T71-2489 | 1971 | Oakland, Calif. | 2.5 yr/F | PAP | |
| T82-0981 | 1982 | Modesto, Calif. | NA/M | Conjunctivitis | |
| 810223-2 | 1985 | San Francisco, Calif. | >18 yr/F | Seen at VD clinic | |
| 3 isolates | 1988 | EAMC, Ga. | ARD | ||
| Z-G 95-873 | 1995 | Indio, Calif. | 2 mo/F | Meningitis, diarrhea | |
| 1 isolate | 1996 | EAMC, Ga. | ARD | ||
| 3 isolates | 1997 | EAMC, Ga. | ARD | ||
| 1 isolate | 1997 | BAFB, Tex. | ARD | ||
| AV 7/7a | Gomen (p7) | 1954 | Fort Baker, Calif. (2) | Pharyngitis | |
| S-1058 (p7a) | 1955 | Washington, D.C. (3, 28) | Respiratory illness | ||
| CL 68578 (v) | 1963 | Washington, D.C. (5, 30) | 10 mo/F | Respiratory illness | |
| T63-3630 | 1963 | Oakland, Calif. | 9 yr/M | Myocarditis | |
| 66-03-0210 | 1966 | Fort Ord, Calif. | 1 yr/M | Pharyngitis, URI | |
| T69-0705 | 1969 | Oakland, Calif. | 11 yr/M | Myositis | |
| T72-0671 | 1972 | San Mateo, Calif. | 1 yr/M | Rash, fever | |
| T84-0927 | 1984 | Riverside, Calif. | 17 mo/F | Pharyngitis, conjunctivitis | |
| T87-1484 | 1987 | Fresno, Calif. | 12 yr/F | Pneumonia | |
| T92-0229 | 1992 | Sacramento, Calif. | 13 yr/F | Fatal ARD, lupus | |
| T95-0730 | 1995 | San Diego, Calif. | NA/F | Pharyngitis | |
| Kn T96-0620 | 1996 | San Francisco, Calif. | 27 yr/M | Fatal ARD | |
| T96-0731b | 1996 | New Orleans, La. | 2 yr/M | Fatal ARD | |
| T96-0732b | 1996 | New Orleans, La. | 1 yr/M | Severe ARD | |
| T96-0733b | 1996 | New Orleans, La. | 5 yr/M | Severe ARD | |
| 1 isolate | 1996 | EAMC, Ga. | ARD | ||
| 2 isolates | 1996 | BAFB, Tex. | ARD | ||
| 1 isolate | 1997 | BAFB, Tex. | ARD |
Abbreviations: p, prototype strains; v, vaccine strains; PAP, primary atypical pneumonia; NA, not available; URI, upper respiratory infection; VD, venereal disease; EAMC, Eisenhower Army Medical Center; BAFB, Brooks Air Force Base; M, male; F, female.
Isolates from an outbreak at a medical care facility for physically handicapped children. The outbreak resulted in 13 cases of ARD and 7 deaths from severe ARD.
Type-specific PCR.
Isolates from military personnel were typed by a semimultiplex PCR with a single AV generic upstream primer and four downstream primers; the latter four primers comprised three primers that specifically identified AV 4, AV 7/7a, and AV 21 and a downstream generic primer that recognized all serotypes. Generic primers were designed from AV consensus sequences from the hexon major coat protein by using deoxyinosine (I) at positions of ambiguity (6). Generic upstream primer AV1Rm (5′-TICTTTGACATICGIGGIGTICTIGA-3′) and downstream primer AV3L (5′-CTGTCIACIGCCTGITTCCACAT-3′) generate a band of 822 to 870 bp, depending on the serotype. Sequences for type-specific primers were taken from HVRs of the hexon protein. The HVRs contain type-specific residues and generate a unique PCR product band length for AV 4 (510 bp), AV 7 (239 bp) or AV 7a (230 bp), and AV 21 (167 bp). The type-specific primers were AV4HVR5L (5′-CGTAGTTAGCAACAATAITTTTGC-3′), AV7HVR2L (5′-GGCTTGTTGTCTGCAGTAATGTC-3′), and AV21HVR1L (5′-AGATTTTTCTCTTCCTCTTCGTCAGA-3′). The PCR mixture consisted of (i) 2.5 U of Tfl DNA polymerase (Epicenter Technologies, Madison, Wis.), (ii) buffer containing 20 mM (NH4)SO4 and 50 mM Tris-HCl (supplied at a 20× concentration with the enzyme), (iii) 1 mM MgCl2, (iv) 250 μM (each) deoxynucleotide triphosphate, (v) 5 μl of MasterAmp Enhancer containing a single-stranded DNA binding protein (supplied with the enzyme), (vi) 5% glycerol, (vii) 0.1% Triton X-100, and (viii) 25 pmol of each primer (AV1Rm, AV4HVR5L, AV7HVR2L, AV21HVR1L, and AV3L) in a 50-μl reaction volume. The template was 1 to 10 μl of fluid from the cell culture of the original isolate. Two drops of oil was added to each tube. PCR cycling consisted of an initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min.
PCR and direct DNA sequencing.
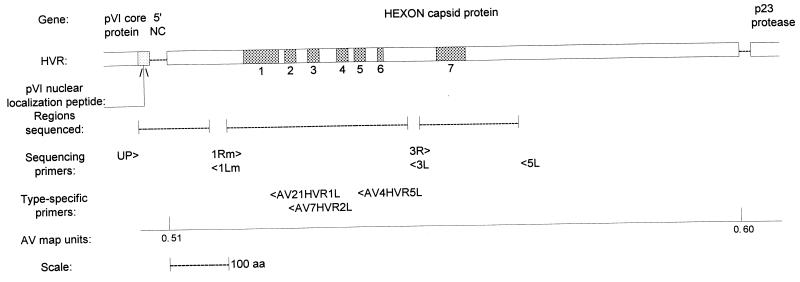
Seventeen strains of AV 4 and 19 strains of AV 7/7a were selected for use in the sequencing of HVRs to include virus strains with broad ranges of temporal distribution and disease manifestation (Table 1). A full-length AV template was prepared by extraction by a method modified from that of Hirt (14). PCR and direct sequencing were performed as described previously (7). Three PCR products were generated from each strain for a total of 1,500 bp, which is slightly more than half of the hexon protein (Fig. 1). The regions sequenced included (i) the conserved pVI core protein nuclear localization peptide, the hexon 5′ noncoding region, and the first 187 highly conserved bases of the hexon protein framed by primers UP (5′-AACAGCATIGTGGGTITGGGIGTG-3′) and AV1Lm (5′-TCIAGIACICCICGIATGTCAAAGIA-3′), (ii) HVRs 1 through 6 framed by primers AV1Rm and AV3L (their sequences are given above), and (iii) HVR 7 framed by primers AV3R (5′-ATGTGGAAICAGGCIGTIGACAG-3′) and AV5L (5′-CGGTGGTGITTIAAIGGITTIACITTGTCCAT-3′). The PCR mixture was as described above but was expanded to a preparative 100-μl reaction volume, with 100 pmol of each primer, 10 μl of MasterAmp Enhancer, and 1 to 5 μl of DNA template but no Triton X-100. PCR cycling was as described above, but the annealing temperature was 45°C. PCR products were cleaned by adsorption to silica (Prep-A-Gene; Bio-Rad Laboratories, Hercules, Calif.). Direct cycle sequencing by incorporation of [33P]dATP was performed with the fmol DNA Sequencing System (Promega, Madison, Wis.) with 10 pmol of primer and 1 to 5 μl of PCR product. The sequencing cycle was as described previously (7). The whole hexon from prototype, vaccine, and two wild-type strains was sequenced with five additional primer sets (6). Gene sequences were aligned with Align Plus (Scientific and Educational Software, State Line, Pa.).
FIG. 1.
Gene map of the AV hexon protein showing HVRs (shaded), the regions sequenced, and generic and type-specific primers. Adapted from reference 7 with permission of the publisher. aa, amino acids.
Neutralization assays.
Stock virus preparations of prototype strains, vaccine strains, and an AV 4 variant were tested in cross-neutralization tests with equivalent virus dosages. The virus dosage was calculated by the method of Reed and Muench (24). Rabbit antisera to prototype strains of AV 7 (Gomen), AV 4 (RI-67), and AV 16 (CH 79) were from the collection of VRDL. National Institutes of Health (NIH) standardized rabbit antiserum to AV 7a (S-1058) was obtained from the American Type Culture Collection (V207A-501-565). Cross-neutralization titers were determined by a semiautomated colorimetric microneutralization assay (5). Antiserum to the prototype AV 16 strain (strain CH 79) was included in the comparison of AV 4 strains because of its high level of cross-reactivity with AV 4 (RI-67) (21, 32).
Nucleotide sequence accession numbers.
The accession numbers for the complete hexon DNA sequences that were entered into the Genbank database are as follows: AV 7 (Gomen), AF065065; AV 7a (S-1058), AF065066; AV 7 vaccine strain (55142), AF065067; AV 7 (Kn T96-0620), AF065068; AV 4 (RI-67), AF065062; AV 4 vaccine strain (CL 68578), AF065063; and AV 4 variant (Z-G 95-873), AF065064.
RESULTS
Type-specific PCR.
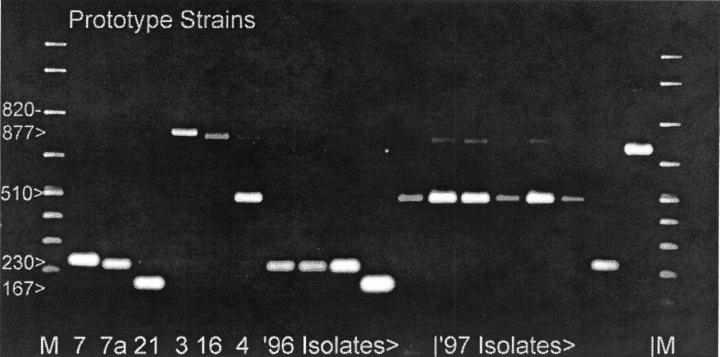
Thirty-eight isolates from military personnel were typed by PCR. The results for 12 of these isolates and their controls are shown in Fig. 2. There was no cross-reactivity between AV 4, AV 7, or AV 21 or with other closely related AV serotypes (AV 3 and AV 16), which generated only generic bands. There was no cross-reactivity with other subgenus B serotypes (AV 11, 14, 34, or 35) or subgenus C serotypes (AV 2 or 5) (data not shown). A few AV 4 and AV 7 strains amplified a faint generic band as well as a strong type-specific band. PCR and neutralization results were concordant for 34 of 38 isolates. Two isolates identified by the submitting laboratories as AV 7 were AV 4 and AV 21 strains, respectively, by PCR. Two strains previously identified as AV 4 were AV 7 and non-AV4/7/21, respectively, by PCR. Determination of the serotypes of the isolates with discordant results by DNA sequencing confirmed the PCR results for all four isolates. The serotypes segregated primarily with year of isolation. The three isolates recovered in 1988 were AV 4. Regardless of the geographic source, 10 isolates recovered in 1996 and early 1997 were AV 7, with 1 AV 4 isolate and 1 AV 21 isolate. Twenty-two isolates recovered in 1997 were AV 4, with 1 AV 2 isolate and 1 AV 7 isolate. The AV 2 isolate was identified as an isolate other than an AV 4, AV 7, or AV 21 isolate by PCR and typed and confirmed by neutralization assay and sequencing.
FIG. 2.
Type-specific PCR of military isolates and prototype strains. The wells are loaded with 5 μl (10%) of each PCR product. Lane M, molecular mass ladder. Numbers on the left are in base pairs.
Molecular analysis of AV 7/7a strain variability.
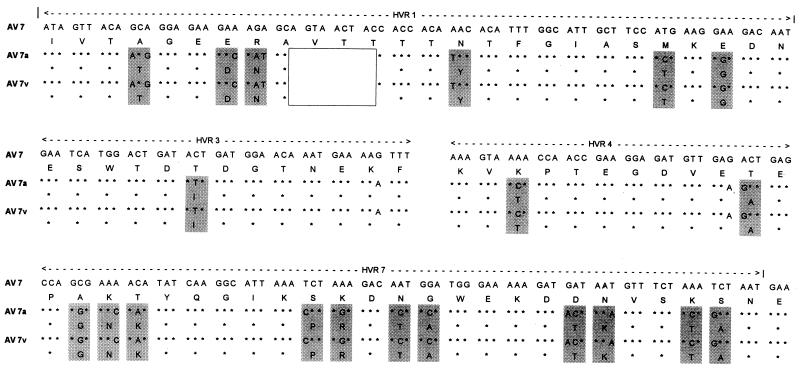
Comparison of the sequences of the whole hexons of AV 7 (Gomen) and AV 7a (S-1058) revealed that they were two genetically distinct strains (or had distinct genotypes), with 38 coding and 68 noncoding differences across the hexon gene, including a 9-bp deletion from HVR1 in AV 7a (Fig. 3). The level of nucleic acid homology between AV 7 and AV 7a was 96%, and the level of protein homology was 97%. From the sequence analysis of the whole hexons of the vaccine strain (55142) and isolate Kn T96-0620 and 1,500 bp of 15 wild-type isolates, it was clear that all of the vaccine, community-acquired, and military personnel strains recovered from 1963 to 1997 were of the AV 7a genotype. Not a single isolate resembled the prototype AV 7 (Gomen) strain. The mutation rate of the AV 7a genotype strains was extremely low. Over a 42-year period (1955 to 1997) there were six single-base differences among the 18 strains with the AV 7a genotype. For the prototype AV 7a strain (strain S-1058), 3 of the 1,500 sequenced bases were unique to that strain. The vaccine strain (strain 55142) had one coding change that was unique to that strain. Among the 16 wild-type isolates there were two base changes: one HVR 7 coding change that was shared by three strains recovered from 1995 to 1996 (strains T95-0730 and T96-0732 and one isolate recovered from a member of the military in 1996) and one unique noncoding change in an isolate from a patient with fatal case of ARD in 1996 (Kn T96-0620).
FIG. 3.
Major coding differences in the HVRs between AV 7 (Gomen), AV 7a (S-1958), and the AV 7 vaccine strain (55142) indicated by shading. The deletion characteristic of AV 7a strains is indicated by the unfilled box.
AV 4 genetic variability.
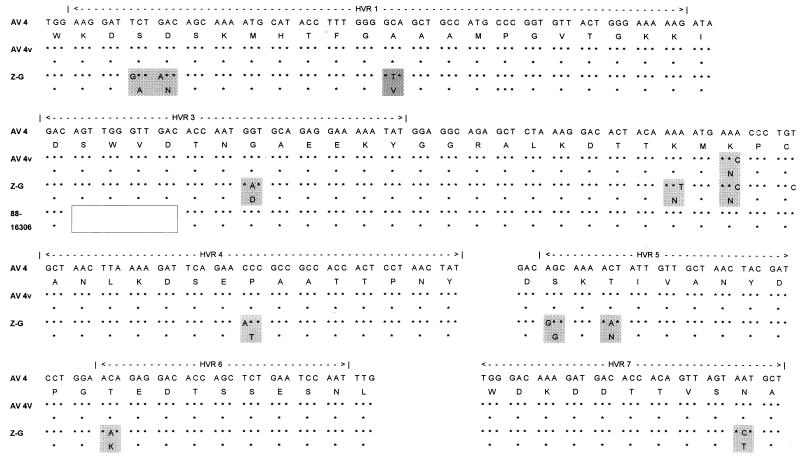
Sequence analysis of the prototype AV 4 strain (strain RI-67), vaccine strain 55142, and 15 archival and military personnel isolates demonstrated a more complex evolution. The prototype strain, all isolates recovered from 1963 to 1971, and a single isolate recovered in 1996 were identical. The vaccine strain had one coding change that it shared with one isolate recovered in 1985 and all isolates recovered after 1995. An isolate recovered in 1982 had a second coding change that appeared in all isolates recovered later. The isolate recovered in 1985 contained a unique coding change. In 1988 a unique strain was isolated from three specimens at a single location (Eisenhower Army Medical Center). That strain had incorporated the 1982 coding change and contained a characteristic deletion of 12 bp (four codons) from HVR 3. In 1995 another unique strain (strain Z-G 95-873) appeared. That strain incorporated the two earlier coding changes and, in addition, contained 9 coding and 13 noncoding changes. Nine of the coding changes occurred in HVRs 1, 3, 4, 5, 6, and 7 and two occurred in the matrix residues between HVRs 3 and 4 (Fig. 4). All AV 4 isolates recovered since 1995 except one isolate recovered a member of the military in 1996 were this new strain. The isolate recovered from a member of the military resembled earlier strains.
FIG. 4.
Major coding changes in the HVRs among AV 4 strains indicated by shading: prototype (RI-67), vaccine strain (CL 68578), and the AV 4 variant (Z-G 95-0620). The unique deletion from the strains recovered in 1988, represented by strain 88-16306, is indicated by unfilled box.
Neutralization assays.
The results of cross-neutralization tests with AV 7 (Gomen) and AV 7a (S-1058) strains were as follows: NIH antiserum to AV 7a gave a homotypic titer of 5,120 and a titer of 640 against AV7 (Gomen); this is an eightfold difference between strains. The vaccine strain was neutralized at a titer of 5,120 as well. The antiserum to AV 7 (Gomen) did not have the same specificity, giving titers of 1,280 for both AV 7 (Gomen) and the AV 7a strain (S-1058). The titer for the vaccine strain was not significantly different from that for the prototype AV 7a strain. The genetic differences between AV 4 strains were also reflected in their neutralization titers. The AV 4 (RI-67) antiserum gave a homotypic titer of 10,240, and a titer of 2,560 was obtained with the Z-G variant; this was a fourfold reduction. The titer for the vaccine strain was not significantly different from that for the prototype strain. The titer with AV 16 antiserum was 2,560 against AV 4 and 80 against the Z-G variant; this was a 32-fold reduction. The titer for the vaccine strain was not significantly different from that for the prototype strain.
DISCUSSION
The AV serotypes of isolates from military personnel were associated with year of isolation, regardless of location, with 1996 being a predominantly AV 7 year and 1997 being a predominantly AV 4 year. The wave of AV 4 infections in 1997 has been confirmed by others (27). The cycling of AV 4 and AV 7 in training centers has been reported previously (16). The type-specific PCR, as formulated, is a timely and cost-effective alternative to serologic identification of isolates when a small number of serotypes is suspected, as in outbreaks of ARD among military personnel. It has the potential to be a timely and cost-effective diagnostic tool, as well, when used with specimens submitted for virus isolation. AVs lend themselves to a multiplex PCR approach because the seven HVRs contain sequences unique to each serotype, and the present PCR configuration could be expanded to include additional serotypes. The AV 21-specific primer was included with AV 4 and AV 7/7a because AV 21 was earlier shown to be a significant cause of ARD in military recruits (29).
The implications of this information for AV 7/7a vaccine design and delivery are clear. There is essentially no variability among strains with the AV 7a genotype. The AV 7a vaccine strain is identical to all of the isolates collected from military personnel and most wild-type strains isolated from the civilian population. We have recently examined the mutation rate in this region of the hexon in strains of a subgenus D AV that were isolated over a 6-year period and estimated a mutation rate of one mutation per 2,500 bases per year (6). The mutation rate for the AV 7a genotype appears to be much lower, with a total of six mutations in 42 years. Given such a low mutation rate, the profound differences between AV 7 and AV 7a indicate that many, many years have passed since their divergence.
The existence of a distinct subtype of AV 7 (subtype AV 7-H) was first proposed in 1957 on the basis of cross-neutralization tests with AV 7 (20). Designated AV 7a in 1958 (25), its validity has been debated almost since it was first reported. Some reports have found support for these differences (2, 11), and others have not (23, 31). In cross-neutralization tests, the critical determinants of differentiation were the equivalence of virus dosage (2, 25) and the specificity of antiserum (20, 26). In our neutralization tests, with virus dosage controlled, the specificity of the antiserum determined significant differences. The AV 7 (Gomen) hyperimmune antiserum used here was prepared by using a multiple series of immunizations in order to produce the highest titer, but specificity was sacrificed.
More recent characterization of AV 7/7a has been based on restriction enzyme analysis (REA) of the whole 36-kbp genome, and AV 7b-h have been added to the original AV 7 and 7a (17). REA has proven to be a useful tool for epidemiological investigation. However, hexon gene sequences for two AV 7b strains and three AV 7d strains culled from DNA sequence data banks (18) showed the AV 7a hexon sequence in the HVRs with the characteristic deletion in HVR 1. Differences in restriction endonuclease cleavage sites did not indicate differences in neutralization epitopes for AV 7a. Our data establish that AV 7 and AV 7a are genetically distinct subtypes. The protein homology between disparate serotypes AV 3 and AV 7 is only slightly less at 94.3% (22). The AV 7a genotype has predominated since the 1960s, with neutralization epitopes virtually unchanged. The AV 7a vaccine strain can offer protection against current wild-type viruses in circulation now and in the foreseeable future.
The pattern of evolution in AV 4 is more complex than that in AV 7a and appears to resemble that of influenza virus. A small but constant genetic drift is punctuated by the periodic appearance of a new strain that replaces former strains. Genomic variation of AV 4, determined by REA, has been reported by others (3). The current strain in circulation is significantly different from the prototype and vaccine strains. Nine of the coding changes occur in the HVRs which contain the neutralization epitopes and are reflected in the decrease in neutralization titer. The fourfold reduction in neutralization titer compared to that for the prototype strain indicates that this strain is an AV 4 variant. As reported by others (21, 32), AV 16 antiserum exhibited a high level of cross-reactivity with the AV 4 prototype strain. The reduction in cross-reactivity of AV 16 with the AV 4 variant (Z-G) was dramatic, indicating that one or more of the coding changes in the variant occurred in neutralization epitopes shared by AV 4 and AV 16. The degree of protection provided by the vaccine strain against the currently circulating variant cannot be determined by molecular methods. Certainly, AV 4 has drifted genetically and antigenically and will continue to do so. It is probable that AV 4 may drift sufficiently that the vaccine will not offer adequate protection in the foreseeable future. Molecular surveillance of circulating AV 4 strains is indicated to determine whether a major antigenic change has taken place so that a new AV 4 vaccine strain can be implemented in a timely fashion.
ACKNOWLEDGMENTS
We thank the following for providing viruses included in this study: K. Mills McNeil and Loise Dunn, Eisenhower Army Medical Center, Fort Gordon, Ga.; Linda Canas, Armstrong Laboratory, Brooks Air Force Base, Tex.; and Robert Gohd, Children’s Hospital, New Orleans, La. We thank Mamta Tahiliani and Lynn Suer for technical assistance.
This work was supported by the Henry M. Jackson Foundation for the Advancement of Military Medicine, Rockville, Md.
REFERENCES
- 1.Berge T O, England B, Mauris C, Shuey H E, Lennette E H. Etiology of acute respiratory disease among service personnel at Fort Ord, California. Am J Hyg. 1955;62:283–294. doi: 10.1093/oxfordjournals.aje.a119779. [DOI] [PubMed] [Google Scholar]
- 2.Binn L N, Hilleman M R, Rodriquez J E, Glabere R R. Antigenic relationships among adenoviruses with appraisal of reliability of complement-fixation test for typing isolates. J Immunol. 1958;80:501–508. [PubMed] [Google Scholar]
- 3.Cooper R J, Bailey A S, Killough R, Richmond S J. Genome analysis of adenovirus 4 isolated over a six year period. J Med Virol. 1993;39:62–66. doi: 10.1002/jmv.1890390112. [DOI] [PubMed] [Google Scholar]
- 4.Couch R B, Chanock R M, Cates T B, Lang D J, Knight V, Huebner R J. Immunization with types 4 and 7 adenovirus by selective infection of the intestinal tract. Annu Rev Respir Dis. 1963;88(Suppl.):394–403. doi: 10.1164/arrd.1963.88.3P2.394. [DOI] [PubMed] [Google Scholar]
- 5.Crawford-Miksza L K, Schnurr D P. A quantitative spectrophotometric microneutralization assay for the characterization of adenoviruses. J Clin Microbiol. 1994;32:2231–2234. doi: 10.1128/jcm.32.9.2331-2334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford-Miksza L K, Schnurr D P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol. 1996;70:1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford-Miksza L K, Schnurr D P. Adenovirus serotype evolution is driven by illegitimate recombination in the hypervariable regions of the hexon protein. Virology. 1996;224:357–367. doi: 10.1006/viro.1996.0543. [DOI] [PubMed] [Google Scholar]
- 8.Gaydos C A, Gaydos J C. Adenovirus vaccines in the military. Milit Med. 1995;160:300–303. [PubMed] [Google Scholar]
- 9.Girardi A J, Hilleman M R, Zwickey R E. Tests in hamsters for oncogenic quality of ordinary viruses including adenovirus type 7. Proc Soc Exp Biol Med. 1964;115:1141–1150. doi: 10.3181/00379727-115-29138. [DOI] [PubMed] [Google Scholar]
- 10.Gutekunst R R, White R J, Edmondson W P, Chanock R M. Immunization with live type 4 adenovirus: determination of infectious virus dose and protective effect of enteric infection. Am J Epidemiol. 1967;86:341–349. doi: 10.1093/oxfordjournals.aje.a120744. [DOI] [PubMed] [Google Scholar]
- 11.Harris D J, Wulff H, Ray C G, Poland J D, Chin T D Y, Wenner H A. Viruses and disease. III. An outbreak of adenovirus type 7a in a children’s home. Am J Epidemiol. 1971;93:399–402. doi: 10.1093/oxfordjournals.aje.a121273. [DOI] [PubMed] [Google Scholar]
- 12.Hilleman M R, Werner J H. Recovery of a new agent from patients with acute respiratory illness. Proc Soc Exp Biol Med. 1954;85:183–188. doi: 10.3181/00379727-85-20825. [DOI] [PubMed] [Google Scholar]
- 13.Hilleman M R, Stallones R A, Gauld R L, Warfield M S, Anderson S A. Prevention of acute respiratory illness in recruits by adenovirus (RI-APC-ARD) vaccine. Proc Soc Exp Biol Med. 1956;92:377–383. doi: 10.3181/00379727-92-22484. [DOI] [PubMed] [Google Scholar]
- 14.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 15.Lee S-G, Hung P P. Vaccines for control of respiratory disease caused by adenoviruses. Rev Med Virol. 1993;3:209–216. [Google Scholar]
- 16.Lennette E H, Stallones R A, Holquin A H. Pattern of respiratory virus infections in army recruits. Am J Hyg. 1961;74:225–233. doi: 10.1093/oxfordjournals.aje.a120215. [DOI] [PubMed] [Google Scholar]
- 17.Li Q-G, Wadell G. Analysis of 15 different genome types of adenovirus type 7 isolated on five continents. J Virol. 1986;60:331–335. doi: 10.1128/jvi.60.1.331-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q-G, Wadell G. GenBank accession numbers HAU77390 to HAU77394. 1996. [Google Scholar]
- 19.Ludwig S L, Brundage J F, Kelley P W, Nang R, Towle C, Schnurr D P, Crawford-Miksza L K, Gaydos J C. Prevalence of antibodies to adenovirus serotypes 4 and 7 among unimmunized US Army trainees: results of a retrospective nationwide seroprevalence survey. J Infect Dis. 1998;178:1776–1778. doi: 10.1086/314498. [DOI] [PubMed] [Google Scholar]
- 20.Pereira H G, Kelly B. Studies on natural and experimental infections by adenoviruses. Proc R Soc Med. 1957;50:755–757. [Google Scholar]
- 21.Pring-Akerblom P, Trijssenaar F E J, Adrian T. Sequence characterization and comparison of adenovirus subgenus B and E hexons. Virology. 1995;212:232–236. doi: 10.1006/viro.1995.1474. [DOI] [PubMed] [Google Scholar]
- 22.Pring-Akerblom P, Trijssenaar F E J, Adrian T. Hexon sequence of adenovirus type 7 and comparison with other serotypes of subgenus B. Res Virol. 1995;146:383–388. doi: 10.1016/0923-2516(96)80897-1. [DOI] [PubMed] [Google Scholar]
- 23.Rafajko R R. Studies on serological relationships between strains of adenovirus types 3 and 7. Proc Soc Exp Biol Med. 1967;124:580–585. doi: 10.3181/00379727-124-31797. [DOI] [PubMed] [Google Scholar]
- 24.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 25.Rowe W P, Hartley J W, Huebner R J. Serotype composition of the adenovirus group. Proc Soc Exp Biol Med. 1958;97:465–470. doi: 10.3181/00379727-97-23776. [DOI] [PubMed] [Google Scholar]
- 26.Top F H, Grossman R A, Bartelloni P J, Segal H E, Dudding B A, Russell P K, Buescher E L. Immunization with live types 7 and 4 adenovirus vaccines. I. Safety, infectivity, antigenicity, and potency of adenovirus type 7 vaccine in humans. J Infect Dis. 1971;124:148–154. doi: 10.1093/infdis/124.2.148. [DOI] [PubMed] [Google Scholar]
- 27.U.S. Army Center for Health Promotion and Preventive Medicine. Adenovirus, type 4, among military trainees, Fort Jackson, SC, Fort Gordon, GA. Med Surveill Monthly Rep. 1997;3:12–13. [Google Scholar]
- 28.U.S. Army Commission on Acute Respiratory Diseases. Acute respiratory disease among new recruits. Am J Pub Health. 1946;36:439–450. [Google Scholar]
- 29.Van der Veen J, Dijkman J H. Association of type 21 adenovirus with acute respiratory illness in military recruits. Am J Hyg. 1962;76:149–159. doi: 10.1093/oxfordjournals.aje.a120270. [DOI] [PubMed] [Google Scholar]
- 30.Van der Veen J. The role of adenoviruses in respiratory disease. Am Rev Respir Dis. 1963;88:167–181. doi: 10.1164/arrd.1963.88.3P2.167. [DOI] [PubMed] [Google Scholar]
- 31.Wigand R. Does adenovirus subtype 7a exist? Arch Virol. 1976;50:335–337. doi: 10.1007/BF01317958. [DOI] [PubMed] [Google Scholar]
- 32.Wigand R. Pitfalls in the identification of adenoviruses. J Virol Methods. 1987;16:161–169. doi: 10.1016/0166-0934(87)90001-2. [DOI] [PubMed] [Google Scholar]