Abstract
Identifying the potential for SARS-CoV-2 reinfection is crucial for understanding possible long-term epidemic dynamics. We analysed longitudinal PCR and serological testing data from a prospective cohort of 4,411 United States employees in 4 states between April 2020 and February 2021. We conducted a multivariable logistic regression investigating the association between baseline serological status and subsequent PCR test result in order to calculate an odds ratio for reinfection. We estimated an odds ratio for reinfection ranging from 0.14 (95% CI: 0.019 to 0.63) to 0.28 (95% CI: 0.05 to 1.1), implying that the presence of SARS-CoV-2 antibodies at baseline is associated with around 72% to 86% reduced odds of a subsequent PCR positive test based on our point estimates. This suggests that primary infection with SARS-CoV-2 provides protection against reinfection in the majority of individuals, at least over a 6-month time period. We also highlight 2 major sources of bias and uncertainty to be considered when estimating the relative risk of reinfection, confounders and the choice of baseline time point, and show how to account for both in reinfection analysis.
Identifying the potential for SARS-CoV-2 reinfection is crucial for understanding possible long-term epidemic dynamics. Analysis of a seroepidemiological cohort suggests that primary infection with SARS-CoV-2 protects against reinfection in the majority of individuals, at least over a six month period.
Introduction
The rapid global spread of COVID-19 throughout 2020 occurred as a result of the introduction of a highly transmissible virus, SARS-CoV-2, into populations with little preexisting immunity [1]. Identifying the extent and duration of protective immunity afforded by natural infection is therefore of crucial importance for understanding possible long-term epidemic dynamics of SARS-CoV-2 [2].
Studies have estimated that over 95% of symptomatic COVID-19 cases develop antibodies against SARS-CoV-2, with most individuals developing antibodies within 3 weeks of symptom onset [3,4]. Several serological studies have also characterised individual-level immune dynamics, with some finding evidence for antibody waning and others for sustained antibody responses over several months [5–10]. Antibody kinetics are thought to vary between individuals and are possibly associated with severity of illness, where asymptomatic or mildly symptomatic individuals may develop lower levels of antibodies that wane more rapidly [3,7,11]. While neutralising antibodies are thought to be associated with protection from reinfection, there are still limited studies on the impact of postinfection seropositivity on future reinfection risk [12]. Confirmed cases of reinfection with SARS-CoV-2 have been reported since August 2020 [13]. However, existing large studies examining the relative risk of reinfection in antibody positive individuals have typically involved specific cohorts who may not be representative of the wider community, such as closed communities or healthcare worker cohorts [14–17]. To evaluate the relative risk of SARS-CoV-2 infection and reinfection over time, we analysed PCR and serological testing data from a prospective cohort of SpaceX employees in the US between April 2020 and February 2021 [18,19].
Results
Of 4,411 individuals enrolled, 309 individuals tested seropositive during the study period (Fig 1). This resulted in an overall adjusted percentage ever seropositive of 8.2% (95% CI: 7.3% to 9.1%) by the end of August 2020, after the final round of serological testing (Fig 2B). Here, imperfect test sensitivity and specificity were adjusted for using the Rogan–Gladen correction [20]. We defined a possible reinfection as a new positive PCR test more than 30 days after initial seropositive result. This identified 14 possible reinfections with a median time of 66.5 days between initial seropositive test and PCR positive test (Fig 2C).
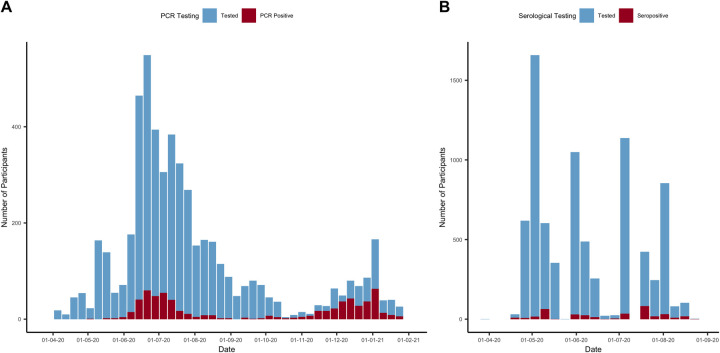
Fig 1.
(A) Number of PCR tests and PCR positive tests in the cohort between April 5, 2020 and January 31, 2021 from 3,296 participants. (B) Number of serological tests and seropositive tests between March 29, 2020 and August 23, 2020 from 4,411 participants. Data underlying this figure can be found in https://github.com/EmilieFinch/covid-reinfection.
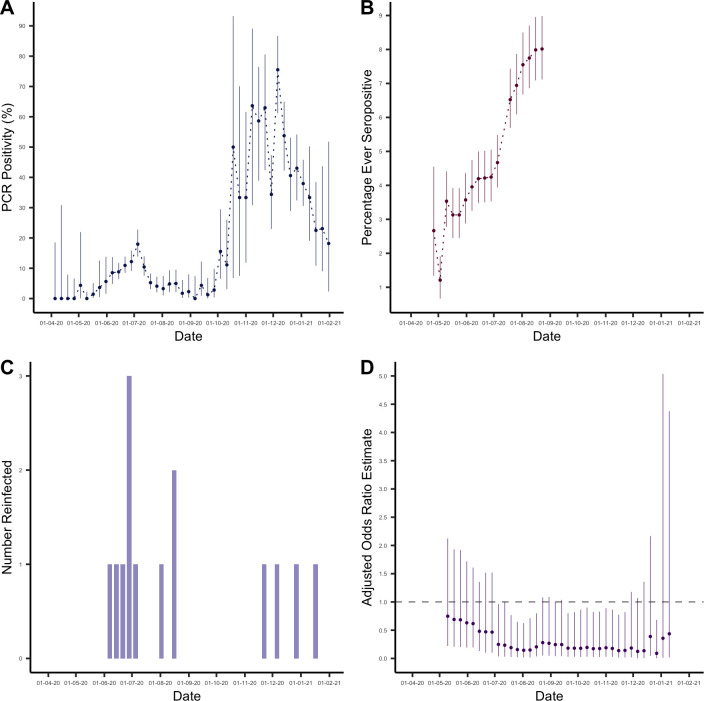
Fig 2.
(A) PCR positivity (%) in the cohort between April 5, 2020 and January 31, 2021. (B) Percentage ever seropositive in the cohort (number ever seropositive/cumulative number enrolled) between March 29, 2020 and August 23, 2020. Note that the percentage ever positive decreases initially as participants continue to be enrolled in the study. (C) Number of possible reinfections in cohort over time (defined as a new positive PCR test more than 30 days after initial seropositive result). (D) Odds ratio estimates comparing odds of reinfection in the seropositive group with odds of primary infection in the seronegative group, estimated using logistic regression and adjusted for potential confounders. The estimates are presented with their associated 95% CIs and with the cutoff week used to define baseline seroprevalence on the x-axis. Data underlying this figure can be found in https://github.com/EmilieFinch/covid-reinfection.
SARS-CoV-2 infection and reinfection
We estimated the odds ratio for SARS-CoV-2 reinfection using multivariable logistic regression, to adjust for any background individual-level variation in the risk of infection (see Methods). This required us to choose a cutoff week in order to define baseline seroprevalence and the subsequent observation period for PCR testing. To examine how our estimate for the odds ratio for reinfection varied depending on the cutoff week chosen, we repeated the analysis using every possible cutoff week.
We considered that the most robust estimation of the odds ratio for reinfection would occur midepidemic when using cutoff weeks in between 2 “waves” of the epidemic seen in the study cohort. We validated this methodological assumption by conducting a simulation study (see Fig A in S2 Text).
We defined a midepidemic period in between 2 epidemic waves where PCR positivity in the study cohort was below the WHO specified threshold of 5%, which occurred between July 26, 2020 and September 27, 2020 (Fig 2A). During these cutoff weeks, estimates of the odds ratio for reinfection (Fig 2D) ranged from 0.14 (95% CI: 0.019 to 0.63) to 0.28 (95% CI: 0.05 to 1.1). Our point estimates suggest that the presence of SARS-CoV-2 antibodies confers around 72% to 86% protection against reinfection with SARS-CoV-2, at least over a 6-month period. As a sensitivity analysis, we conducted the same analysis but excluding records where individuals had recorded a specific trigger reason for testing such as symptom onset or potential exposure (and so reflecting individuals tested at random). Considering the weeks between July 26, 2020 and September 27, 2020, we found estimates of the odds ratio for reinfection ranged from 0.18 (95% CI: 0.024 to 0.80) to 0.36 (95% CI: 0.06 to 1.5).
In the adjusted analyses, odds ratio estimates for reinfection converged to similar values for cutoffs spanning a period after the first peak of infection in early July. By this point, sufficient numbers of participants had been both recruited and tested seropositive (see Fig 2B) that we had enough data to distinguish infection dynamics in seropositive and seronegative groups. Adjusted odds ratio estimates for reinfection then lost precision when using late cutoff weeks from mid-December onwards due to increasingly small numbers of participants experiencing PCR infection after the cutoff point, consistent with our simulation study (see S2 Text).
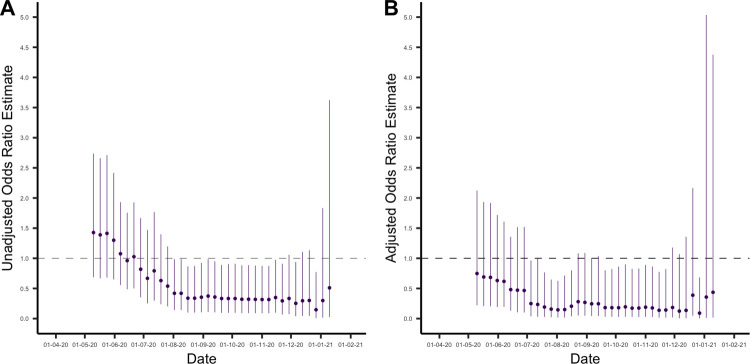
Unadjusted odds ratio estimates tended to overestimate the odds ratio for reinfection compared with primary infection, particularly when using early cutoff weeks (Fig 3). Notably, with early cutoff weeks the unadjusted analysis estimated a higher odds of reinfection compared to primary infection, albeit with wide CIs. This is the result of a subset of individuals who are at higher risk of initial seroconversion (who would be included in analyses at earlier time thresholds) and also at higher risk of later reinfection, giving a biased estimate of the association between antibodies and subsequent infection when using earlier cutoff weeks.
Fig 3.
(A) Unadjusted odds ratio estimates comparing odds of reinfection in the seropositive group with odds of primary infection in the seronegative group. The estimates are presented with their associated 95% CIs and with the cutoff week used to define baseline seroprevalence on the x-axis. (B) Odds ratio estimates comparing odds of reinfection in the seropositive group with odds of primary infection in the seronegative group, estimated using logistic regression and adjusted for potential confounders. The estimates are presented with their associated 95% CIs and with the cutoff week used to define baseline seroprevalence on the x-axis. Data underlying this figure can be found in https://github.com/EmilieFinch/covid-reinfection.
Discussion
We identified 14 possible reinfections out of 309 seropositive individuals in the prospective seroepidemiological cohort between April 2020 and February 2021, estimating an odds ratio for reinfection ranging from 0.14 (95% CI: 0.019 to 0.63) to 0.28 (95% CI: 0.05 to 1.1). This provides evidence that primary infection with SARS-CoV-2 results in protection against reinfection in the majority of individuals, at least over a sixth month time period. Our findings are broadly consistent with estimates of 0.17 (95% CI 0.13 to 0.24) odds ratio [14] and 0.11 (0.03 to 0.44) incidence rate ratio [15] for healthcare workers, 0.18 (0.11 to 0.28) incidence rate ratio for military recruits [16], and 0.195 (95% CI 0.155 to 0.246) incidence rate ratio from a Danish population-level study [21].
Our analysis addressed 2 key sources of bias and uncertainty in estimating the relative risk of reinfection. First, confounders may inflate estimates; if a specific subset of the cohort is at higher risk of infection (e.g., due to underlying health conditions or increased risk of exposure), these participants will be more likely to be both initially seropositive and to have a subsequent reinfection. Second, the time period considered could increase uncertainty; defining the baseline seroprevalence at an early time point means few will be seropositive, whereas defining it at a later point means there is less time to observe possible reinfections. We accounted for these 2 factors by adjusting for key confounders to calculate an adjusted odds ratio for reinfection. We then investigated how changing the cutoff date to define baseline seroprevalence impacted the accuracy of the adjusted odds ratio calculated. We assumed that for a 2-wave epidemic scenario, a cutoff week in the period in between the 2 waves of infection risk would result in the most robust estimates of the odds ratio for reinfection, which we validated using a simulation study (see S2 Text). This suggests that the robustness of estimates of the relative risk of reinfection will be sensitive to the study period chosen, relative to population-level epidemic dynamics.
There are several limitations to the underlying data that should be considered when interpreting these findings. This prospective cohort was recruited opportunistically from employees at one US company and is unlikely to be representative of the general population. However, as we did not identify any workplace outbreaks, infections in this cohort are likely to be more reflective of community transmission than in healthcare worker cohorts or other specialised populations. Additionally, we only considered possible reinfections (as opposed to probable or true reinfections). As possible reinfections did not meet a stringent case definition, such as confirmation through genomic sequencing, they may include cases of prolonged viral shedding following an initial infection. This would result in an overestimation of the odds ratio for reinfection and so our analysis reflects the minimum possible effect of antibodies on future SARS-CoV-2 infection risk. Finally, the date of infection among seropositive participants is unknown, limiting inference on exact duration of protection.
As well as quantifying the relative risk of reinfection over a 6-month period among a prospectively followed workplace population, our study highlights the importance of accounting for both individual-level heterogeneity in infection risk and population-level variation in epidemic dynamics when assessing the potential for reinfections.
Methods
Seroepidemiological cohort description
We used data from a seroepidemiological study of US employees at SpaceX, as described previously [19]. In brief, this study involved employees from 7 work locations in California, Florida, Texas, and Washington State, with ages ranging from 18 to 71. A total of 4,411 employees volunteered to participate in the study and were enrolled from approximately 8,400 total employees. All employees were invited to participate by email, and there were no exclusion criteria. Study participants were offered SARS-CoV-2 IgG receptor binding domain (RBD) antibody testing with an in-house ELISA assay with 82.4% sensitivity and 99.6% specificity [22]. Serological samples were taken during 4 rounds of testing between April and September 2020. A questionnaire including demographic, symptom, and exposure information was conducted at enrolment and with each round of serological testing. Individuals continued to be enrolled throughout the study period, and around half of the total participants (48%) were tested at more than 1 time point. Participants occupied a range of job positions within SpaceX including office-based and factory-based jobs. Additionally, symptomatic and asymptomatic PCR testing were widely available for employees using the Infinity BiologiX (IBX) TaqPath rRT-PCR assay, with data available from April 2020 to January 2021. Employees could request a test for any reason, and testing was also specifically performed for symptomatic individuals, individuals with potential exposure, and mission critical employees. Both serology and PCR testing data were available for 1,800 individuals.
Ethics statement
The study protocol was approved by the Western Institutional Review Board (ref 20200991). The use of deidentified data and biological samples was approved by the Mass General Brigham Healthcare Institutional Review Board (ref 2020P001166). Secondary data analysis was approved by the LSHTM Observational Research Ethics Committee (ref 22466). All participants provided written informed consent.
Statistical analysis
To estimate the odds ratio for SARS-CoV-2 reinfection, we conducted multivariable logistic regression analysis investigating the association between baseline serological status and subsequent PCR test result, given a test was sought.
The choice of cutoff week used to define participants’ baseline seroprevalence and the subsequent observation period for PCR testing have important implications in the estimation of the odds ratio for reinfection. For instance, a cutoff week early in the study period will result in few seropositive individuals, while a cutoff week later in the study period leaves less time to observe subsequent PCR testing and detect possible reinfections, impacting the accuracy of estimates. To assess how the choice of cutoff week affected estimates of the odds ratio for reinfection, we repeated the multivariable logistic regression for every possible cutoff week. We assumed the most robust estimation of the relative risk of reinfection would occur in the between the 2 “waves” of infection risk seen in the study cohort. We validated this assumption by conducting a simulation analysis using a known underlying probability distribution of infection and reinfection (see S2 Text).
Potential confounding variables included age, sex, race, ethnicity, BMI, state, work location, job category, household size, history of chronic disease, history of smoking, and test frequency. We used a backwards selection procedure to select which variables to adjust for in our analyses, minimising root mean square error (RMSE) at each step [23]. Age and sex were considered “forced” variables, which we decided to control for a priori and were adjusted for in all analyses [24,25]. We conducted variable selection separately for each cutoff week and the variable sets adjusted for in each regression analysis are listed in Table A in S1 Text. For most cutoff weeks (specifically those between May 19, 2020 and November 22, 2020), all potential confounders were adjusted for, while early weeks (between April 26, 2020 and May 3, 2020) and late weeks (between December 20, 2020 and January 17, 2021) adjusted for a subset of potential confounders.
As a sensitivity analysis, we performed the same analysis but excluding records where individuals had recorded a specific reason for test such as onset of symptoms or potential exposure to a COVID-19 case. As such, this sensitivity analysis included only individuals tested at random.
We investigated the propensity to be tested among seronegative and seropositive individuals for each cutoff week by examining the percentage of those enrolled in the study by each cutoff week who had at least 1 test in the subsequent observation period and found that for cutoff weeks from mid-July onwards they were broadly similar between the 2 groups (Fig 4). However, the average distribution of test frequency differed between the seropositive and seronegative groups, with higher frequency of testing more common in the seronegative group. To account for this, we included PCR test frequency as a potential confounder in our analysis, defined as the number of PCR tests each individual took during the observation period (1 to 2, 3 to 5, or 6+). Protection against infection with SARS-CoV-2 conferred by the presence of antibodies was estimated such that
Fig 4. Propensity to seek a PCR test between the seronegative and seropositive groups, for each cutoff week considered in the main analysis.
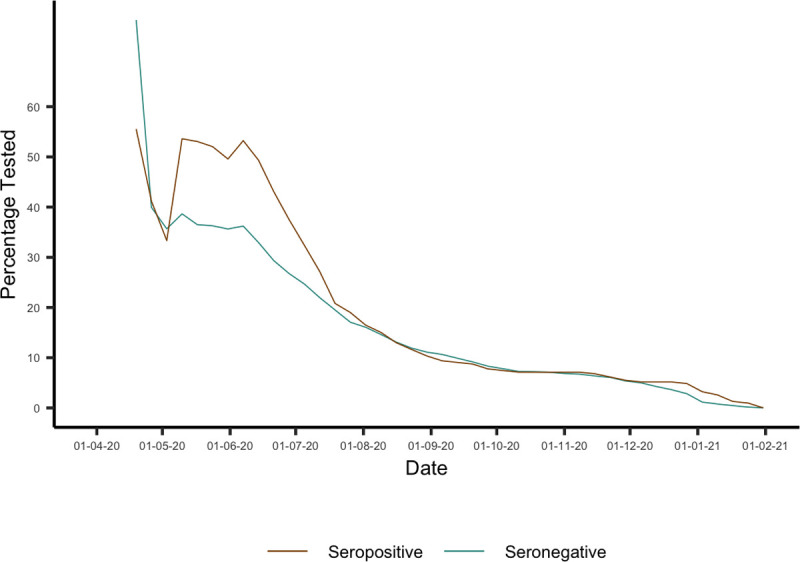
This was calculated as the percentage of those enrolled by the cutoff week shown on the x-axis who received at least 1 PCR test in the subsequent observation period. Data underlying this figure can be found in https://github.com/EmilieFinch/covid-reinfection.
Analysis was conducted in R version 4.0.3. Code to reproduce the figures and simulation analysis presented here can be found at https://github.com/EmilieFinch/covid-reinfection.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
We thank SpaceX employees Lindsay Chapman, Jordan Steinhart, Suzanne Siebert, and Kyle Meade for their valuable support, in addition to the dedication and commitment of the many SpaceX employees that volunteered to participate in this study.
CMMID COVID-19 working group information
The following authors were part of the Centre for Mathematical Modelling of Infectious Disease COVID-19 Working Group. Each contributed in processing, cleaning, and interpretation of data, interpreted findings, contributed to the manuscript, and approved the work for publication: Jiayao Lei, Sebastian Funk, Fiona Yueqian Sun, Amy Gimma, Emily S Nightingale, Graham Medley, Sam Abbott, Fabienne Krauer, Nicholas G. Davies, Mark Jit, Akira Endo, Oliver Brady, Anna M Foss, Yung-Wai Desmond Chan, Thibaut Jombart, Kevin van Zandvoort, Rosalind M Eggo, Yang Liu, Gwenan M Knight, Carl A B Pearson, Kaja Abbas, Katherine E. Atkins, Samuel Clifford, Mihaly Koltai, Yalda Jafari, Damien C Tully, Christopher I Jarvis, Kathleen O’Reilly, Nikos I Bosse, Kiesha Prem, Billy J Quilty, Simon R Procter, Rosanna C Barnard, William Waites, Ciara McCarthy, James D Munday, David Hodgson, W John Edmunds, Alicia Rosello, C Julian Villabona-Arenas, Hamish P Gibbs, Stefan Flasche, Timothy W Russell, Sophie R Meakin, Joel Hellewell, Naomi R Waterlow, Matthew Quaife, and Frank G Sandmann.
Funding statements for the CMMID COVID-19 working group are as follows. KvZ: KvZ is supported by the UK Foreign, Commonwealth and Development Office (FCDO)/Wellcome Trust Epidemic Preparedness Coronavirus research programme (ref. 221303/Z/20/Z), and Elrha’s Research for Health in Humanitarian Crises (R2HC) Programme, which aims to improve health outcomes by strengthening the evidence base for public health interventions in humanitarian crises. The R2HC programme is funded by the UK Government (FCDO), the Wellcome Trust, and the UK National Institute for Health Research (NIHR). SC: Wellcome Trust (grant: 208812/Z/17/Z). FYS: NIHR EPIC grant (16/137/109). Sfunk: Wellcome Trust (grant: 210758/Z/18/Z), NIHR (NIHR200908). GFM: NTD Modelling Consortium by the Bill and Melinda Gates Foundation (OPP1184344). YJ: LSHTM, DHSC/UKRI COVID-19 Rapid Response Initiative. SRM: Wellcome Trust (grant: 210758/Z/18/Z). WJE: European Commission (EpiPose 101003688), NIHR (NIHR200908). MQ: European Research Council Starting Grant (Action Number #757699); Bill and Melinda Gates Foundation (INV-001754). NRW: Medical Research Council (grant number MR/N013638/1). RME: HDR UK (grant: MR/S003975/1), MRC (grant: MC_PC 19065), NIHR (grant: NIHR200908). NGD: UKRI Research England; NIHR Health Protection Research Unit in Immunisation (NIHR200929); UK MRC (MC_PC_19065). JYL: Bill & Melinda Gates Foundation (INV-003174). MK: Foreign, Commonwealth and Development Office / Wellcome Trust. FK: Innovation Fund of the Joint Federal Committee (Grant number 01VSF18015), Wellcome Trust (UNS110424). DCT: No funding declared. JDM: Wellcome Trust (grant: 210758/Z/18/Z). AS: No funding declared. AMF: No funding declared. KP: Gates (INV-003174), European Commission (101003688). Sflasche: Wellcome Trust (grant: 208812/Z/17/Z). SA: Wellcome Trust (grant: 210758/Z/18/Z). BJQ: This research was partly funded by the National Institute for Health Research (NIHR) (16/137/109 & 16/136/46) using UK aid from the UK Government to support global health research. BJQ is supported in part by a grant from the Bill and Melinda Gates Foundation (OPP1139859). TJ: RCUK/ESRC (grant: ES/P010873/1); UK PH RST; NIHR HPRU Modelling & Health Economics (NIHR200908). AR: NIHR (grant: PR-OD- 1017–20002). GMK: UK Medical Research Council (grant: MR/P014658/1). MJ: Gates (INV- 003174, INV-016832), NIHR (16/137/109, NIHR200929, NIHR200908), European Commission (EpiPose 101003688). YL: Gates (INV-003174), NIHR (16/137/109), European Commission (101003688). JW: NIHR Health Protection Research Unit and NIHR HTA. JH: Wellcome Trust (grant: 210758/Z/18/Z). KO’R: Bill and Melinda Gates Foundation (OPP1191821). YWDC: No funding declared. TWR: Wellcome Trust (grant: 206250/Z/17/Z). CIJ: Global Challenges Research Fund (GCRF) project “RECAP” managed through RCUK and ESRC (ES/P010873/1). SRP: Bill and Melinda Gates Foundation (INV-016832). AE: The Nakajima Foundation. ESN: Gates (OPP1183986). NIB: Health Protection Research Unit (grant code NIHR200908). CJVA: European Research Council Starting Grant (Action number 757688). FGS: NIHR Health Protection Research Unit in Modelling & Health Economics, and in Immunisation. AG: European Commission (EpiPose 101003688). KA: Bill & Melinda Gates Foundation (OPP1157270, INV-016832). WW: MRC (grant MR/V027956/1). KEA: European Research Council Starting Grant (Action number 757688). RCB: European Commission (EpiPose 101003688). PK: This research was partly funded by the Royal Society under award RP\EA\180004, European Commission (101003688), Bill & Melinda Gates Foundation (INV-003174). HPG: This research was produced by CSIGN which is part of the EDCTP2 programme supported by the European Union (grant number RIA2020EF-2983-CSIGN).
This research is funded by the Department of Health and Social Care using UK Aid funding and is managed by the NIHR. CABP: CABP is supported by the Bill & Melinda Gates Foundation (OPP1184344) and the UK Foreign, Commonwealth and Development Office (FCDO)/Wellcome Trust Epidemic Preparedness Coronavirus research programme (ref. 221303/Z/20/Z). OJB: Wellcome Trust (grant: 206471/Z/17/Z). WJE: European Commission (EpiPose 101003688), NIHR (NIHR200908, MRC: MC_PC_19065). CIJ: Global Challenges Research Fund (GCRF) project “RECAP” managed through RCUK and ESRC (ES/P010873/1). NGD: UKRI Research England; NIHR Health Protection Research Unit in Immunisation (NIHR200929); UK MRC (MC_PC_19065).
Disclaimers
The views expressed in this publication are those of the author(s) and not necessarily those of the NIHR or the UK Department of Health and Social Care. The views and opinions of authors expressed herein do not necessarily state or reflect those of EDCTP. The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health and Social Care (PR-OD-1017-20001).
Abbreviations
- COVID-19
Coronavirus Disease 2019
- IBX
Infinity BiologiX
- RBD
receptor binding domain
- RMSE
root mean square error
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
Data Availability
The data that support the findings of this study are available on request by email to the Harvard Humanitarian Initiative at hhi@harvard.edu. The data are not publicly available due to their containing information that could compromise the privacy of research participants. Code and aggregated data to reproduce the analysis and figures presented can be found at https://github.com/EmilieFinch/covid-reinfection.
Funding Statement
The authors received funding from the following sources: EF was funded by the Medical Research Council (MR/N013638/1); AJK was supported by Wellcome Trust (206250/Z/17/Z) and National Institute for Health Research (NIHR200908); RL was funded by a Royal Society Dorothy Hodgkin Fellowship (https://royalsociety.org). EN was supported by the US Centers for Disease Control and Prevention (U01 U01GH002238). AM was supported by the Translational Research Institute for Space Health through NASA Cooperative Agreement (https://www.nasa.gov/hrp/tri; NNX16AO69A). GA was supported by the Massachusetts Consortium on Pathogen Readiness (https://masscpr.hms.harvard.edu/; MassCPR), the National Institutes of Health (3R37AI080289-11S1, R01AI146785, U19AI42790-01, U19AI135995-02, 1U01CA260476-01) and the Musk Foundation (http://www.muskfoundation.org/). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baker RE, Yang W, Vecchi GA, Metcalf CJE, Grenfell BT. Susceptible supply limits the role of climate in the early SARS-CoV-2 pandemic. Science. 2020;369(6501):315–9. doi: 10.1126/science.abc2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabino EC, Buss LF, Carvalho MPS, Prete CA, Crispim MAE, Fraiji NA, et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397(10273):452–5. doi: 10.1016/S0140-6736(21)00183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5(12):1598–607. doi: 10.1038/s41564-020-00813-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–8. doi: 10.1038/s41591-020-0897-1 [DOI] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383(18):1724–34. doi: 10.1056/NEJMoa2026116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, Elliott J, Hofmann C, Hausner MA, et al. Rapid Decay of Anti–SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N Engl J Med [Internet]. 2020. Jul 21 [cited 2021 Apr 28]. Available from: https://www.nejm.org/doi/10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chia WN, Zhu F, Ong SWX, Young BE, Fong S-W, Bert NL, et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe [Internet]. 2021. [cited 2021 Apr 28]. Available from: https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(21)00025-2/abstract. doi: 10.1016/S2666-5247(21)00025-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duysburgh E, Mortgat L, Barbezange C, Dierick K, Fischer N, Heyndrickx L, et al. Persistence of IgG response to SARS-CoV-2. Lancet Infect Dis. 2021;21(2):163–4. doi: 10.1016/S1473-3099(20)30943-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science [Internet]. 2021. [cited 2021 Apr 28];371(6529). Available from: https://science.sciencemag.org/content/371/6529/eabf4063. doi: 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward H, Cooke G, Atchison C, Whitaker M, Elliott J, Moshe M, et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv. 2020;2020.10.26.20219725. [Google Scholar]
- 11.Long Q-X, Tang X-J, Shi Q-L, Li Q, Deng H-J, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–4. doi: 10.1038/s41591-020-0965-6 [DOI] [PubMed] [Google Scholar]
- 12.Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–11. doi: 10.1038/s41591-021-01377-8 [DOI] [PubMed] [Google Scholar]
- 13.To KK-W, Hung IF-N, Ip JD, Chu AW-H, Chan W-M, Tam AR, et al. Coronavirus Disease 2019 (COVID-19) Re-infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin Infect Dis [Internet]. 2020. [cited 2021 Apr 28];(ciaa1275). Available from: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall VJ, Foulkes S, Charlett A, Atti A, Monk EJ, Simmons R, et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet [Internet]. 2021. [cited 2021 Apr 14];0(0). Available from: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)00675-9/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lumley SF, O’Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, et al. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N Engl J Med. 2021. Feb 11;384(6):533–40. doi: 10.1056/NEJMoa2034545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letizia AG, Ge Y, Vangeti S, Goforth C, Weir DL, Kuzmina NA, et al. SARS-CoV-2 seropositivity and subsequent infection risk in healthy young adults: a prospective cohort study. medRxiv. 2021;2021.01.26.21250535. doi: 10.1016/S2213-2600(21)00158-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang M-L, et al. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J Clin Microbiol [Internet]. 2020. [cited 2021 Mar 3];58(11). Available from: https://jcm.asm.org/content/58/11/e02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartsch YC, Fischinger S, Siddiqui SM, Chen Z, Yu J, Gebre M, et al. Discrete SARS-CoV-2 antibody titers track with functional humoral stability. Nat Commun. 2021;12(1):1018. doi: 10.1038/s41467-021-21336-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilles EJ, Siddiqui SM, Fischinger S, Bartsch YC, de St. Aubin M, Zhou G, et al. Epidemiological and Immunological Features of Obesity and SARS-CoV-2. Viruses. 2021;13 (11):2235. doi: 10.3390/v13112235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogan WJ, Gladen B. Estimating prevalence from the results of a screening test. Am J Epidemiol. 1978;107(1):71–6. doi: 10.1093/oxfordjournals.aje.a112510 [DOI] [PubMed] [Google Scholar]
- 21.Hansen CH, Michlmayr D, Gubbels SM, Mølbak K, Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet [Internet]. 2021. [cited 2021 Mar 24];0(0). Available from: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)00575-4/abstract. doi: 10.1016/S0140-6736(21)00575-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy V, Fischinger S, Atyeo C, Slein M, Loos C, Balazs A, et al. SARS-CoV-2-specific ELISA development. J Immunol Methods. 2020. 1;484–485:112832. doi: 10.1016/j.jim.2020.112832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenland S, Daniel R, Pearce N. Outcome modelling strategies in epidemiology: traditional methods and basic alternatives. Int J Epidemiol 2016;45(2):565–75. doi: 10.1093/ije/dyw040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies NG, Klepac P, Liu Y, Prem K, Jit M, CMMID COVID-19 working group, et al. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat Med. 2020;26(8):1205–11. doi: 10.1038/s41591-020-0962-9 [DOI] [PubMed] [Google Scholar]
- 25.Pijls BG, Jolani S, Atherley A, Derckx RT, Dijkstra JIR, Franssen GHL, et al. Demographic risk factors for COVID-19 infection, severity, ICU admission and death: a meta-analysis of 59 studies. BMJ Open. 2021;11(1):e044640. doi: 10.1136/bmjopen-2020-044640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
The data that support the findings of this study are available on request by email to the Harvard Humanitarian Initiative at hhi@harvard.edu. The data are not publicly available due to their containing information that could compromise the privacy of research participants. Code and aggregated data to reproduce the analysis and figures presented can be found at https://github.com/EmilieFinch/covid-reinfection.