A central pillar of global tuberculosis (TB) control programs is early antimicrobial treatment of paucibacillary infection to prevent its progression to multibacillary disease (1). The rationale is self-evident. This approach is expected to reduce incident disease, both in the infected individual and by reducing risk of onward transmission of infection. It also offers the advantage of shorter treatment with fewer drugs and mitigates against the risk of antimicrobial resistance. The gold standard to identify prevalent disease necessitating antimicrobial treatment has been microbiological diagnosis of Mycobacterium tuberculosis (Mtb) infection by microscopy, culture, PCR, or antigen detection in clinical samples, but the sensitivity of all of these tests is inevitably proportional to the bacillary burden. Our enduring challenge has been how to identify those who will benefit from antimicrobial treatment before we can detect the pathogen. Established tests of T-cell immune memory responses to Mtb antigens have provided reasonable negative predictive value (2), but they do not reflect current infection and consequently offer low positive predictive value (PPV), even in high-transmission settings (3). One innovative approach, at least in low transmission settings, is provided by personalized risk prediction modeling that integrates quantitative measures of T-cell reactivity with other demographic risk factors identified in longitudinal cohorts of at-risk individuals (4). This approach to personalized risk stratification to trigger antimicrobial treatment showed potential to offer greater net benefit than treating everyone with T-cell reactivity but still only identified probability of disease of up to 10% in most and is yet to be tested in clinical trials or evaluated in high-transmission settings.
The accessibility of omics technologies has fueled the search for new biomarkers of TB (5). Among these, quantitation of circulating host mRNA molecules either individually or in combination as a transcriptional signature has gained the most traction—at least one candidate has already been developed into a cartridge-based PCR test (6). The finding that blood transcriptional changes can emerge before conventional diagnosis of TB (7) piqued excitement in the potential for the application of blood transcriptional biomarkers to identify short-term risk of incident disease. Systematic comparison in individual participant meta-analysis showed that the best performing candidate biomarkers were cocorrelated and reflected IFN- or TNF (tumor necrosis factor)–driven host immune responses and may provide up to 10–15% PPV over a 3- to 6-month time horizon (8). One such biomarker comprising a combination of 11 transcripts (RISK11) has been evaluated further using a general population screening approach in South Africa with an a priori defined biomarker threshold to discriminate positive and negative tests. This achieved a PPV for either prevalent or incident disease over 15 months of only 6.6% among HIV-negative subjects in CORTIS (Correlate of Risk Targeted Intervention Study) (9) and 5.7% among HIV-positive subjects in the CORTIS-HR (CORTIS-High Risk) study (10). Low prior probability of prevalent infection in the general population undoubtedly contributed to the low PPV in these studies, but the findings point to two other potential limitations of TB blood transcriptional biomarkers that merit consideration. First, the likelihood that these biomarkers are not specific for immune responses to Mtb infection—after all, IFN and TNF are canonical components of immune responses in many different contexts. Second, the possibility that the presence of an immune response to Mtb infection represented by these biomarkers does not necessarily mean the infection will progress to disease.
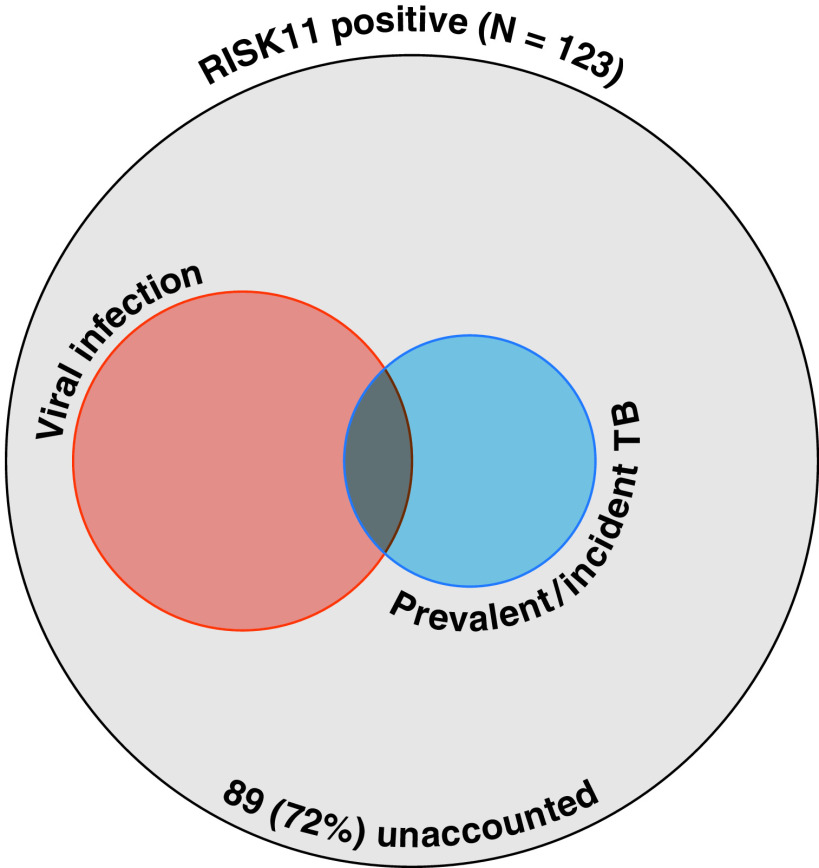
In this issue of the Journal, Mulenga and colleagues (pp. 1463–1472) have extended their analyses of the CORTIS studies to provide valuable new insights into these hypotheses (11). First, they show that 73–85% of HIV-negative individuals who are RISK11 positive at recruitment revert to become RISK11 negative by 3–12 months’ follow up, irrespective of whether they received TB preventative treatment. This suggests that the majority of RISK11-positive results in this context may reflect a self-limiting immune response. If this represented spontaneous resolution of the Mtb infection, we might expect immunocompromised HIV-positive individuals to be more likely to reveal a differential effect of antimicrobial treatment on reversion of RISK11 score. In contrast to this hypothesis, among HIV-infected RISK11-positive individuals, isoniazid preventive treatment also had no effect on reversion to becoming RISK11 negative over 3 months’ follow-up. Interestingly, the 3-month reversion rate of ∼42% in all HIV-infected participants was significantly lower than that of HIV-negative individuals. Among HIV-infected subjects at enrollment, lack of antiretroviral therapy was associated with higher rates of RISK11 positivity. In follow-up, RISK11-positive, antiretroviral therapy–naive individuals showed lower rates of reversion to negative, and RISK11-negative individuals showed higher rates of conversion to positive. Untreated HIV is characterized by IFN-associated changes in the blood transcriptome (12) and these findings all point to uncontrolled HIV as a nonspecific driver of increased RISK11 scores. So, can high RISK11 scores in HIV-negative individuals be attributed to other coincidental infections? Multiplex PCR testing of upper respiratory tract samples among HIV-negative individuals coenrolled to the CORTIS study (who were therefore also investigated for TB) revealed that viral, but not bacterial, infections were associated with elevated RISK11 scores. In fact, RISK11 scores failed to discriminate viral infection from prevalent TB. Importantly, 24/123 (20%) coenrolled RISK11-positive participants had evidence of viral infection, whereas 13/123 (11%) had prevalent or incident TB. The majority (72%) of positive RISK11 scores were therefore unexplained (Figure 1).
Figure 1.
Venn diagram showing proportions of RISK11-positive participants in the HIV-negative cohort coenrolled in the CORTIS trial who had evidence of viral infection or prevalent or incident TB. Data are derived from Mulenga and colleagues (11). CORTIS = Correlate of Risk Targeted Intervention Study; TB = tuberculosis.
It remains to be seen if any of the existing TB blood transcriptional biomarkers offer better specificity than RISK11, but this seems unlikely given the degree of cocorrelation in head-to-head comparisons and the underlying biology they represent. Several signatures did show promising accuracy for prevalent TB in a systematic head-to-head analysis among an observational cohort of symptomatic patients attending a community-based TB clinic in South Africa (13). It may be possible to improve on their specificity for TB, for example, by combining with biomarkers of respiratory viral infection (14) for multiclass classification, or by deriving new signatures to discriminate TB from viral infections directly in larger, more generalizable training data sets, though this may be challenging in view of the common IFN-driven responses in both infections. Until then, the application of the current biomarkers for TB will need to overcome the limitations of specificity, for example, by targeted application in high-risk groups such as recent household contacts (in whom the prior probability of TB may be expected to be higher than that of incidental viral infection) and by integration into multivariable risk prediction tools (4).
Footnotes
Originally Published in Press as DOI: 10.1164/rccm.202109-2146ED on October 27, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.World Health Organization. https://www.who.int/teams/global-tuberculosis-programme/the-end-tb-strategy
- 2. Abubakar I, Drobniewski F, Southern J, Sitch AJ, Jackson C, Lipman M, et al. PREDICT Study Team. Prognostic value of interferon-γ release assays and tuberculin skin test in predicting the development of active tuberculosis (UK PREDICT TB): a prospective cohort study. Lancet Infect Dis . 2018;18:1077–1087. doi: 10.1016/S1473-3099(18)30355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mahomed H, Hawkridge T, Verver S, Abrahams D, Geiter L, Hatherill M, et al. The tuberculin skin test versus QuantiFERON TB Gold® in predicting tuberculosis disease in an adolescent cohort study in South Africa. PLoS One . 2011;6:e17984. doi: 10.1371/journal.pone.0017984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gupta RK, Calderwood CJ, Yavlinsky A, Krutikov M, Quartagno M, Aichelburg MC, et al. Discovery and validation of a personalized risk predictor for incident tuberculosis in low transmission settings. Nat Med . 2020;26:1941–1949. doi: 10.1038/s41591-020-1076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haas CT, Roe JK, Pollara G, Mehta M, Noursadeghi M. Diagnostic ‘omics’ for active tuberculosis. BMC Med . 2016;14:37. doi: 10.1186/s12916-016-0583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Södersten E, Ongarello S, Mantsoki A, Wyss R, Persing DH, Banderby S, et al. Diagnostic accuracy study of a novel blood-based assay for identification of tuberculosis in people living with HIV. J Clin Microbiol . 2021;59:e01643–e20. doi: 10.1128/JCM.01643-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, et al. ACS and GC6-74 cohort study groups. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet . 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta RK, Turner CT, Venturini C, Esmail H, Rangaka MX, Copas A, et al. Concise whole blood transcriptional signatures for incipient tuberculosis: a systematic review and patient-level pooled meta-analysis. Lancet Respir Med . 2020;8:395–406. doi: 10.1016/S2213-2600(19)30282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scriba TJ, Fiore-Gartland A, Penn-Nicholson A, Mulenga H, Kimbung Mbandi S, Borate B, et al. CORTIS-01 Study Team. Biomarker-guided tuberculosis preventive therapy (CORTIS): a randomised controlled trial. Lancet Infect Dis . 2021;21:354–365. doi: 10.1016/S1473-3099(20)30914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mendelsohn SC, Fiore-Gartland A, Penn-Nicholson A, Mulenga H, Mbandi SK, Borate B, et al. CORTIS-HR Study Team. Validation of a host blood transcriptomic biomarker for pulmonary tuberculosis in people living with HIV: a prospective diagnostic and prognostic accuracy study. Lancet Glob Health . 2021;9:e841–e853. doi: 10.1016/S2214-109X(21)00045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mulenga H, Musvosvi M, Mendelsohn SC, Penn-Nicholson A, Kimbung Mbandi S, Fiore-Gartland A, et al. CORTIS Study Team. Longitudinal dynamics of a blood transcriptomic signature of tuberculosis. Am J Respir Crit Care Med . 2021;204 doi: 10.1164/rccm.202103-0548OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turner CT, Brown J, Shaw E, Uddin I, Tsaliki E, Roe JK, et al. Persistent T cell repertoire perturbation and T cell activation in HIV after long term treatment. Front Immunol . 2021;12:634489. doi: 10.3389/fimmu.2021.634489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turner CT, Gupta RK, Tsaliki E, Roe JK, Mondal P, Nwayo G, et al. Blood transcriptional biomarkers for active pulmonary tuberculosis in a high-burden setting: a prospective, observational, diagnostic accuracy study. Lancet Respir Med . 2020;8:407–419. doi: 10.1016/S2213-2600(19)30469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta RK, Rosenheim J, Bell LC, Chandran A, Guerra-Assuncao JA, Pollara G, et al. COVIDsortium Investigators. Blood transcriptional biomarkers of acute viral infection for detection of pre-symptomatic SARS-CoV-2 infection: a nested, case-control diagnostic accuracy study. Lancet Microbe . 2021;2:e508–e517. doi: 10.1016/S2666-5247(21)00146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]