Abstract
Metabolic processes that fuel the growth of heterotrophic microbial communities are initiated by specialized biopolymer degraders that decompose complex forms of organic matter. It is unclear, however, to what extent degraders structure the downstream assembly of the community that follows polymer breakdown. Investigating a model marine microbial community that degrades chitin, we show that chitinases secreted by different degraders produce oligomers of specific chain lengths that not only select for specialized consumers but also influence the metabolites secreted by these consumers into a shared resource pool. Each species participating in the breakdown cascade exhibits unique hierarchical preferences for substrates, which underlies the sequential colonization of metabolically distinct groups as resource availability changes over time. By identifying the metabolic underpinnings of microbial community assembly, we reveal a hierarchical cross-feeding structure that allows biopolymer degraders to shape the dynamics of community assembly.
Specialized biopolymer degraders structure microbial community assembly through interconnected modes of metabolic cross-feeding.
INTRODUCTION
Microbial communities mediate a staggering number of biological processes that contribute to the health of humans, animals (1), and the planet as a whole (2). The metabolic activity of co-occurring species results in the formation and depletion of nutrients in shared resource pools, where different modes of competition and cooperation affect community composition and form a network of metabolic cross-feeding (3–23). Individual species often provide specific metabolic functions that influence the growth of a broader community, including use of terminal electron acceptors (14, 24, 25), degradation of complex carbon sources (26–29), removal of toxic by-products (30–32), or assimilation of nitrogen and sulfur (18, 33–35). However, we remain with a limited molecular understanding of the extent to which individual species affect the formation and structure of cross-feeding networks and community assembly. Here, we demonstrate multiple metabolic mechanisms by which specialized biopolymer degraders influence the trajectory of community assembly, exemplified with an 18-member community that thrives on chitin, the second most abundant polysaccharide on the planet (36). Similar to other polymer degrading communities (26–28), chitin communities require specialized degraders to supply nutrients to nonspecialized downstream consumers. Our community consists of phylogenetically diverse seawater isolates with distinct metabolic capabilities, which become abundant at different points during assembly of chitin communities. This provides a system that allows us to demonstrate how specialized chitin degraders initiate a hierarchal food web that shapes the community during the assembly process.
RESULTS
Functional classification of bacterial isolates
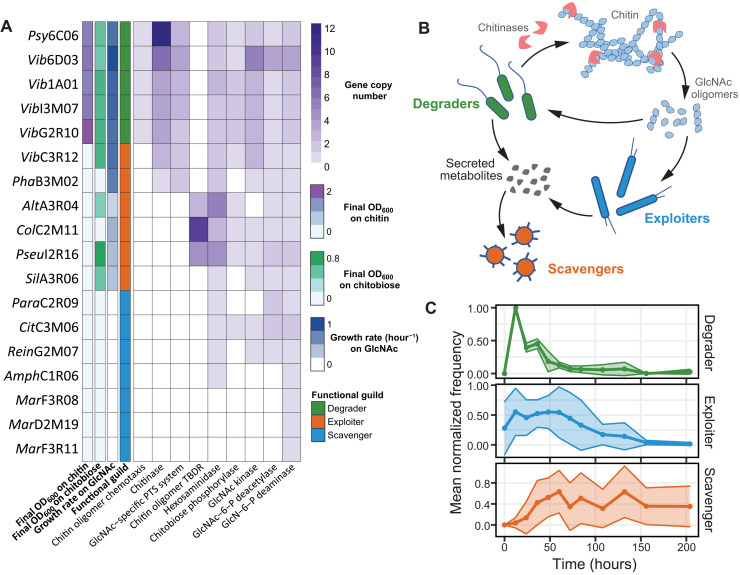
We first classified the 18 isolates, collected from natural polymer degrading communities (table S1) (5, 26), into three functional guilds based on their phenotypic capabilities for chitin utilization (Fig. 1A). These species were tested for the ability to grow on chitin, the chitin monomer N-acetylglucosamine (GlcNAc), and the dimer chitobiose (Fig. 1, A and B, and figs. S1 and S2): Five degraders used all three substrates as a sole carbon and nitrogen source, and six exploiters could not use chitin but grew on GlcNAc, four of which also grew on chitobiose. Seven scavengers could not use any of these substrates; hence, they must rely on nutrients secreted by other species to thrive in the community. Previously published genome data of these 18 isolates (5, 26) revealed a wide range of chitin utilization potential (Fig. 1A) that closely matches phenotypes. All five degraders and two exploiters contain chitinases, uptake components specific for GlcNAc, and all genes required for conversion of GlcNAc into glycolytic intermediates: GlcNAc kinase, GlcNAc-6-phosphate deacetylase, and glucosamine-6-phosphate deaminase. The remaining four exploiters contain all metabolic components required to metabolize and transport GlcNAc (Fig. 1A). All seven scavengers lack at least one transporter or catabolic gene required for GlcNAc utilization (Fig. 1A). These varying metabolic capabilities prime guilds to become abundant at different points during community succession due to the different nutrients that become available over time (5). To visualize the trajectory of each functional guild during community assembly, we mapped 16S sequences of all species to 16S ribosomal gene exact sequence variants (ESVs) whose abundance changes were previously determined on chitin particles colonized from a large species pool in seawater (26). The mean trajectories of each functional guild revealed unique dynamics (Fig. 1C), whereby degraders are the first to reach their maximum abundance, followed by exploiters, and lastly scavengers. This succession pattern portrays an order in a trophic cascade, i.e., degraders cause the release of chitin degradation products that are consumed by exploiters, which, in turn, release broader sets of metabolites that support scavengers.
Fig. 1. Successional dynamics of chitin-degrading seawater communities.
(A) Species used in this study. Heatmap depicting (left to right) final optical density at 600 nm (OD600) after growth on colloidal chitin (2 g/liter), final OD600 after growth on 10 mM chitobiose, growth rate on 20 mM GlcNAc, classified functional guild, and copy numbers of genes relevant for chitin degradation. Growth data were obtained from three independent biological replicates. (B) Schematic of functional guilds: Degraders can grow on chitin as a sole carbon source through breakdown of chitin using chitin-degrading enzymes, exploiters can grow on monomeric or oligomeric N-acetylglucosamine (GlcNAc) as a sole carbon source, and scavengers require metabolites secreted by other species to be sustained in the community. (C) 16S sequences of the 18 investigated species are mapped to 16S ribosomal gene ESVs whose abundances were previously determined on chitin particles colonized from a species pool in seawater (26). Mean normalized frequencies of species that comprise each functional guild are plotted over time. TBDR, TolB-dependent receptor.
Downstream metabolic influence of secreted chitinases
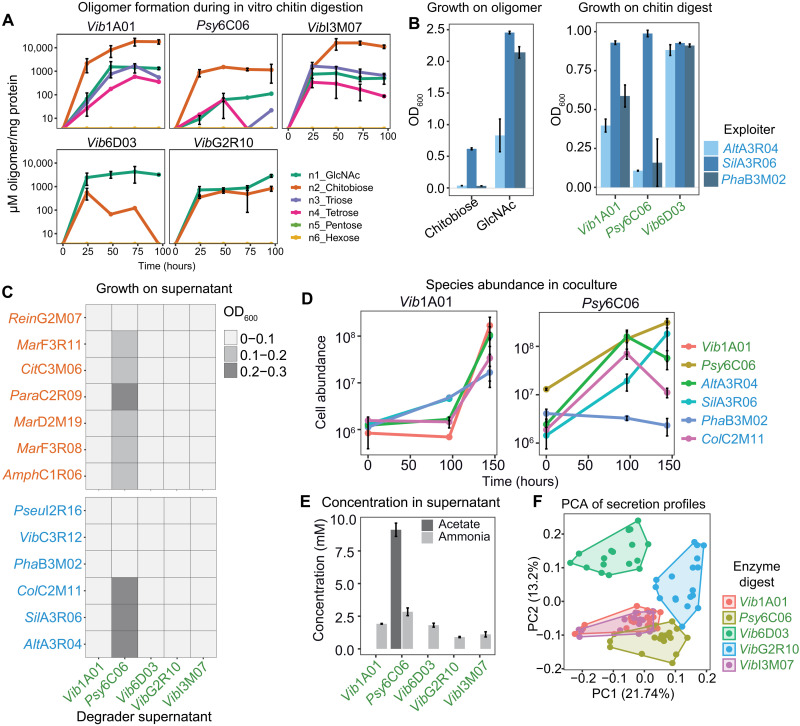
It is likely that degraders form publicly available products that can support the growth of exploiters and possibly select for specific species. To test this, extracellular enzymes were concentrated from culture supernatants of each degrader during growth on chitin and used to digest fresh colloidal chitin. Liquid chromatography–mass spectrometry (LC-MS) quantification of oligosaccharides with six or fewer residues revealed that each digest resulted in a unique oligomer profile, with the disaccharide chitobiose and the monomer GlcNAc as the most abundant products (Fig. 2A). Each of the six exploiters was then grown on the five chitin digests (Fig. 2Band fig. S3). SilA3R06 and three other exploiters grew well on all digests. While PhaB3M02 and AltA3R04 hardly grew on the Psy6C06 digest that contained chitobiose as the major oligomer (Fig. 2A), these grew on Vib1A01 and Psy6C06 digests containing larger quantities of GlcNAc and other oligomers (Fig. 2A). Consistently, PhaB3M02 and AltA3R04 did not grow on chitobiose as the sole carbon source (Fig. 2B and fig. S3). Inspired by this observation, we tested whether the differences in hydrolysis product profiles among degraders could control which exploiters grow in coculture. For this purpose, we grew degraders Vib1A01 and Psy6C06 in coculture with the four exploiters. In accord with the enzyme digest data (Fig. 2A), Vib1A01, which produces a range of oligomers, supported the growth of all species in coculture (Fig. 2D). The chitobiose producer Psy6C06 was able to support all exploiters except PhaB3M02, showing that the composition of degradation products can influence the abundance of specific exploiters. Unexpectedly, the exploiter AltA3R04 also grew in coculture with Psy6C06, although it grew poorly on the enzyme digest of Psy6C06 and showed no growth on chitobiose (Fig. 2B). This suggests that additional metabolites were secreted by Psy6C06 or another exploiter to support growth of AltA3R04 but not of PhaB3M02 and, more generally, that multiple resource pools are formed during chitin degradation that allow downstream growth of less specialized species.
Fig. 2. Chitin degradation products influence exploiter population and their secreted resources.
(A) Quantification of GlcNAc oligonucleotide during in vitro digestion of chitin (5 g/liter) by secreted enzymes from each degrader. (B) OD600 of three exploiters after 36-hour growth on 20 mM GlcNAc, 10 mM chitobiose, and three enzyme digests. (C) Growth of scavengers and exploiters after 36 hours on the cell-free supernatant produced by each of the five degraders upon growth on colloidal chitin. (D) Absolute abundance of species in cocultures of one degrader (Vib1A01 or Psy6C06) and four exploiters during grown on colloidal chitin, as determined by quantitative polymerase chain reaction (qPCR). (E) Enzymatic quantification of ammonia and acetate in degrader supernatants upon growth on colloidal chitin. (F) Principal components analysis (PCA) exploiter metabolite secretion profiles after growth on each colloidal chitin digest. Colors represent the digest used as a growth substrate. All error bars represent SD from three biological replicates.
To understand the mechanism that facilitated the growth of AltA3R06 in coculture, we investigated whether degraders secrete additional metabolites during growth on chitin to provide a carbon or nitrogen source independent of GlcNAc oligomers. We grew the five degraders on colloidal chitin until early stationary phase and inoculated the cell-free culture supernatants individually with six exploiters and seven scavengers (Fig. 2C). While four of the supernatants did not support growth of any species to detectable optical densities, presumably because degraders consumed available limiting nutrients required for growth, the Psy6C06 supernatant supported the growth of six scavengers and three exploiters. LC-MS–targeted metabolomics revealed that GlcNAc and chitobiose concentrations were below the limit of detection in all five supernatants. However, nine metabolites were present in at least one supernatant, including succinate, glutamate, aspartate, and propionic acid (fig. S4 and dataset S1). Because catabolism of GlcNAc into the glycolytic intermediate glucose 6-phosphate entails deacetylation and deamination, acetate or ammonia was quantified with enzyme assays (Fig. 2E). Ammonia was likely the nitrogen source provided to other species because it was present at substantial concentrations in all supernatants. Acetate was present only in the Psy6C06 supernatant at a concentration 10.9-fold higher than the cumulative molar concentrations of all other detected metabolites. Hence, acetate was presumably the major carbon source in Psy6C06 cocultures that fueled growth of AltA3R04 (Fig. 2, D and C). Collectively, these results demonstrate that nondegrading community members are selectively supported by a combination of freely available chitin monomers, oligomers, and metabolic by-products secreted during the growth of degraders.
We suspected that exploiters also secrete metabolites that less specialized species consume and wondered whether the chitin degradation products influence these secretion profiles. To test this hypothesis, we cultivated each exploiter on colloidal chitin digested with extracellular enzymes obtained from each of the five degraders. After 36 hours, culture supernatants were analyzed with the discovery metabolomics method flow injection analysis quadrupole time-of-flight MS (FIA-QTOF-MS) (37). Overall, we detected 2521 ions, of which 216 could be annotated on the basis of accurate mass. Principal components analysis using the 216 annotated ions showed a clear clustering of the secretion profiles based on the oligomer substrate (Fig. 2F). For example, degraders Vib1A01 and VibI3M07 produced similar sets of n1-n4 GlcNAc oligomers (Fig. 2A). The secretion profiles of exploiters that used these substrates also showed no clear separation, suggesting that Vib1A01 and VibI3M07 exert similar effects on exploiters via their similar chitinases. Alternatively, degraders Psy6C06 and Vib6D03 produced primarily chitobiose and GlcNAc, respectively, and secretion profiles of exploiters grown on these digests clustered distinctly. These results demonstrate that the different chitin degradation products play a major role in determining which metabolite exploiters secrete into a shared resource pool.
Identification of strain-specific metabolic secretion and uptake profiles
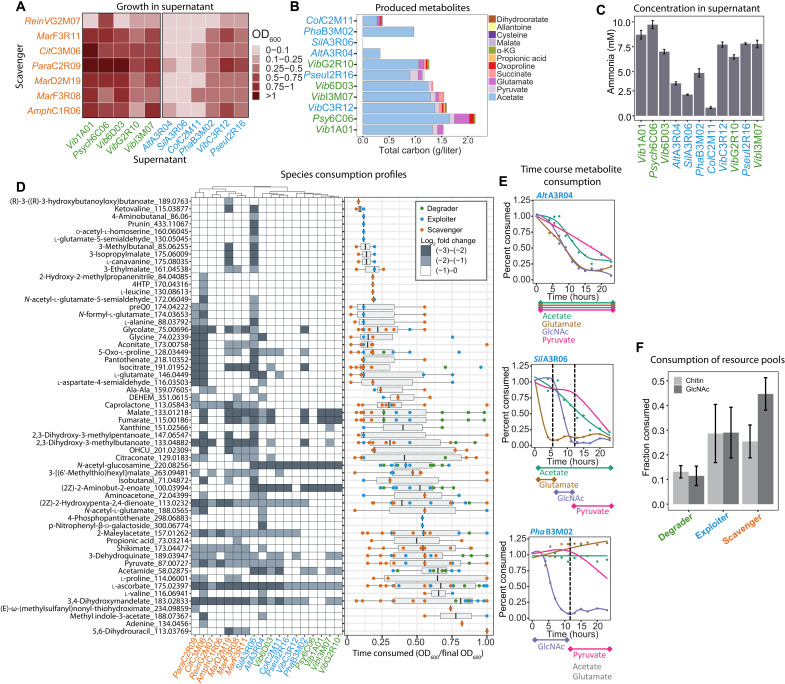
We designed two experiments to identify which of the many secreted metabolites could potentially serve as primary nutrient sources for scavengers. First, all scavengers were cultivated on cell-free supernatants collected from each of the degraders and exploiters after growth on GlcNAc that was used by all species. All supernatants except SilA3R06 supported the growth of at least two but typically more scavengers [5.27 ± 2.4 with optical density at 600 nm (OD600) > 0.1; Fig. 3A], demonstrating the secretion of large quantities of metabolites by most species. Measuring 24 absolute metabolite concentrations by targeted LC-MS and enzymatic assays confirmed acetate and ammonia as the major carbon and nitrogen sources, respectively (Fig. 3, A and C, and dataset S1), akin to the results obtained with the Psy6C06 chitin supernatant. Yet, additional metabolic niches are manifested by a variety of organic acids, amino acids, and nucleosides that made up as much 31% of the total secreted carbon pool, as in the case of ColC2M11. The second experiment was designed to test how these additional metabolites are partitioned among all community members. For this purpose, supernatants of degraders grown on chitin and degraders and exploiters grown on GlcNAc were pooled into a single mixture containing all secreted metabolites. All 18 strains were inoculated in this supernatant and grew individually to densities between 0.15 and 0.7 OD600 units, corresponding to four to six doublings (fig. S5). To identify consumed metabolites that support this growth, we determined relative changes by FIA-QTOF-MS and searched for metabolites that were (i) produced (log2 fold change of ion intensity > 1) by degraders or exploiters after growth to late exponential phase in at least one of the cultures and (ii) consumed (log2 < −0.5) by at least one species in samples taken over the course of growth in the pooled supernatant (Fig. 3D and dataset S2). Each species had a distinct metabolite consumption profile and a unique ordered hierarchy of which metabolites are consumed first (Fig. 3D). This was exemplified by acetate, GlcNAc, pyruvate, and glutamate (Fig. 3E and fig. S6). For example, AltA3R04 cometabolized all four substrates, while SilA3R06 and PhaB3M02 metabolized them with specific ordered preferences or not at all. These results suggest that species may be specialized to consume unique sets of resources with a distinct hierarchy of preferences, thus enhancing the potential for many species to coexist in environments with fluctuating nutrient availability.
Fig. 3. Metabolite consumption and production profiles among community members.
(A) Growth of scavengers at 36 hours on cell-free supernatants obtained from degraders and exploiters after growth on GlcNAc. (B) Metabolite concentration in cultures of degraders or exploiters after growth on 20 mM GlcNAc. α-KG, α-ketoglutarate. (C) Ammonia concentration in cultures of degraders or exploiters after growth on GlcNAc. (D) Consumption of crossfed metabolites in the pooled supernatant. (Left) FIA-QTOF-MS measurements of metabolites that are consumed by degraders, exploiters, or scavengers (ion intensity log2 < −0.5). (Right) Time course measurements of metabolite consumption. Points represent the fraction of the final OD600 at which metabolite consumption is detected for each species. (E) Relative change of acetate, pyruvate, GlcNAc, and glutamate during growth in the pooled supernatant by three exploiters. Curves are fit using local polynomial regression. (F) Fraction of available metabolites contained in chitin or GlcNAc-derived resource pools that is consumed by each functional guild. All experiments were performed in triplicate. Error bars represent SD. DEHEM, 4-deoxy-α-l-erythro-hex-4-enopyranuronate-β-d-mannuronate; OHCU, 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline. Strain names in green are degraders, blue are exploiters, and orange are scavengers.
Given the sequential colonization of degraders, exploiters, and scavengers (Fig. 1C), we wanted to test whether unique metabolic preferences of guilds and species drive this succession. For this purpose, we approximated the availability of resource pools at early time points by determining which metabolites are produced by degraders after individual growth on colloidal chitin, because chitin degradation initiates community succession. Conversely, we approximated the resource pools that degraders and exploiters produce at later time points by determining metabolites they secreted after growth on GlcNAc. We then calculated the fraction of metabolites that each species consumed from the early or late resource pools (Fig. 3F). Of the 32 metabolites present in the chitin-derived resource pool, degraders, exploiters, and scavengers consumed on average 13, 28, and 25% of the available metabolites, respectively. Scavengers greatly increased their consumption to 45% of the 54 metabolites in the GlcNAc-derived resource pool, while degrader and exploiter consumption did not change much. Thus, metabolites produced at later time points in community succession are consumed preferentially by scavengers, explaining how this functional guild is specialized to thrive on resources that become available at later time points in community succession (Fig. 1C).
Inferring metabolite exchange networks that explain coculture outcomes
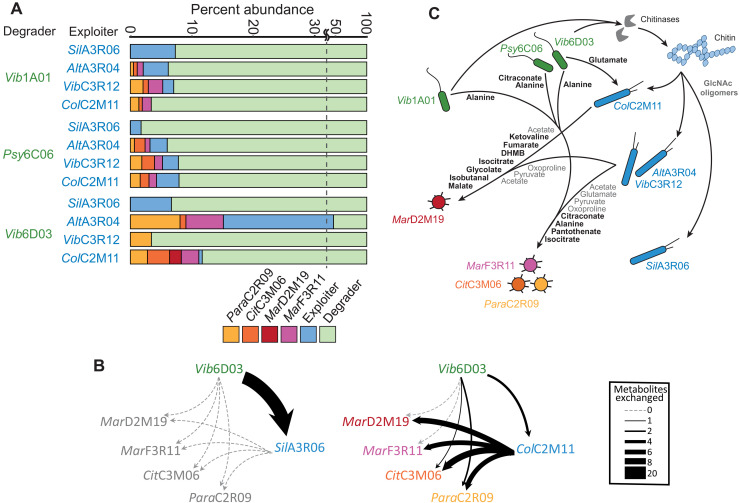
In light of the unique secretion and uptake profiles of each species, we explored how these traits may contribute to the outcome of communities in coculture. Cocultures were seeded with one of three degraders and one of four exploiters with an additional set of the same four scavengers to create 12 six-member communities (Fig. 4A). The three degraders extracellularly produced different sets of chitin degradation products (Fig. 2A), which appeared to influence the prevalence of exploiters in several cases. For example, exploiter AltA3R04 used GlcNAc but not chitobiose (Fig. 2B) and consistently coexisted well with degrader Vib6D03 (Fig. 4A) that produced GlcNAc as a major degradation product (Fig. 2A). However, degrader-exploiter coexistence was not entirely explained by their dependence on GlcNAc oligomers. For instance, exploiter ColC2M11 did not grow in coculture with degrader Vib1A01 (Fig. 4A), despite its ability to grow on chitobiose and GlcNAc formed by Vib1A01(Fig. 2A and fig. S3). This is likely the result of poor substrate affinity for GlcNAc oligomers and suggests that additional secreted metabolites may fuel ColC2M11.
Fig. 4. Outcomes of chitin-degrading cocultures and hypothetical metabolic exchange networks.
(A) Each coculture was seeded with six species: one degrader (green), one exploiter (blue), and each of the four scavengers ParaC2R09 (yellow), CitC3M06 (orange), MarD2M19 (red), and MarF3R11 (purple). Cultures were grown for 5 weeks with four serial dilutions on colloidal chitin, and relative abundance is shown for the final culture. Experiments were performed in triplicate. (B) Two of 12 hypothetical metabolite exchange networks that illustrate the number of metabolites exchanged between species that coexisted in cocultures outcomes from (A). Edge width represents the number of exchanged metabolites. Exchange networks based on the remaining 10 coculture outcomes are contained in dataset S5. (C) A graphical summary of metabolite exchanges that likely contribute to the coculture outcomes. Bolded metabolites are all inferred causal metabolites that may contribute to the growth of individual species. Gray metabolites represent broadly exchanged growth supporting substrates that are secreted by degraders or exploiters grown on GlcNAc and consumed by scavengers. All remaining secreted metabolites and cross-feeding interactions that are not included in this illustration are contained in datasets S1, S3, and S5. DHMB, 2,3-dihydroxy-2-methylbutanoate.
For a more comprehensive and temporal view, we inferred coculture-specific hypothetical nutrient exchange networks by leveraging the sequential production and consumption data. Specifically, we compiled metabolite secretion and uptake data of individual species (dataset S3) to form pools where each metabolite was assumed to either be present or absent (dataset S4). The first metabolite pool includes only degrader-derived metabolites, i.e., those produced during chitin degradation that then become available to the exploiter that next colonized the community. The second pool consists of exploiter-derived metabolites upon growth on the first metabolite pool. The exploiter-derived metabolite pool is defined as the degrader-derived pool minus metabolites consumed by the exploiter plus those secreted by the exploiter. A hypothetical metabolite exchange occurred between degrader and exploiter if a metabolite in the degrader-derived pool was produced by the degrader and consumed by the exploiter. Metabolite exchanges with scavengers were inferred from metabolites present in the exploiter-derived pool (i.e., produced by degrader or exploiter) and consumed by a scavenger. This resulted in 12 hypothetical exchange networks (dataset S5) where nodes represent a species that survived in coculture and edges represent exchanged metabolites. Figure 4B depicts a simplified illustration of two networks where the edge width is the number of exchanged metabolites between each species. These networks identify and quantify the number of metabolites exchanged between pairs of species, providing a basis to infer which interactions may contribute to coculture outcomes.
To assess how exploiters influenced the flow of metabolites from degraders to scavengers, we evaluated how exploiters may potentially compete with scavengers for degrader-derived metabolites by identifying the percentage of degrader-derived metabolites that are consumed by exploiters in the exchange networks (table S2). No scavengers coexisted with SilA3R06 in any coculture (Fig. 4A), and on average, SilA3R06 consumed 78.3% of all degrader-derived metabolites, compared to at most 33.3% by any of the other exploiters. This suggests that SilA3R06 competes with scavengers for degrader-derived metabolites to a much higher degree than any other exploiter. Moreover, to evaluate whether SilA3R06 itself contributed metabolites to scavengers that may support their growth, we calculated the number of metabolites produced by each exploiter that scavengers may consume (table S3 and Fig. 4B), revealing that SilA3R06 did not secrete any such metabolite. On the contrary, our networks predict that, in six cases, exploiters secreted a larger variety metabolites than they consumed, suggesting that exploiters have the ability to expand the number of available metabolic niches to support a more diverse population. This is exemplified with ColC2M11 in coculture with Vib6D03 (Fig. 4B), which only consumed GlcNAc, chitobiose, and glutamate, yet it engaged in 27 hypothetical exchanges with scavengers that are mediated by 10 metabolites. Subsequently, this is the only coculture in which growth of all scavengers was observed. Collectively, these data illustrate how exploiters, after their abundance in the community is influenced by degraders (Fig. 2D), have downstream influence on community diversity by changing the resources available to scavengers.
Last, we aimed to identify metabolites that accounted for the growth of individual species in coculture (Fig. 4A). To this end, we combined the metabolic uptake/secretion profiles of individual species (dataset S3) with the six-member coculture outcomes (Fig. 4A) and their respective metabolite pools (dataset S4). Given that many metabolite abundances have only relative and not absolute quantities, we used a Boolean approach that takes into consideration only the presence or absence of each metabolite. This approach inferred a potential causal relationship between a metabolite and a given species when three conditions were true: (i) the metabolite was shown to be consumed by the species in its uptake profile, (ii) the metabolite was present in the inferred metabolite pool available to the species in all of the cocultures where it showed growth, and (iii) the metabolite was absent in the inferred metabolite pool available to the species in all cocultures where it did not grow. With this approach, we were able to identify potential causal metabolites, primarily organic acids, amino acids, and one vitamin, for five species (table S4). In the case of ColC2M11, which grew in the presence of two degraders but not in the presence of Vib1A01, glutamate was the only metabolite that could potentially explain the growth phenotype. In another case, the scavengers ParaC2R09, CitC3M06, and MarF3R11 coexisted together in 8 of the 12 cocultures (Fig. 4A), suggesting that there may be an overlap in metabolites that contribute to their growth. Our approach identified four potential causal metabolites that overlapped between two or three of these scavengers: isocitrate, alanine, pantothenate, and the isoleucine precursor, citraconate (table S4). In a more specific example, scavenger MarD2M19 only grew in coculture with exploiter ColC2M11 and degrader Vib6D03. We inferred seven metabolites in the exploiter-derived resource pool of this coculture that may underlie the growth of MarD2M19. These include malate, fumarate, and glycolate, which are only secreted by ColC2M11 in comparison to the other three exploiters. Figure 4C is a graphical summary of metabolite exchanges that we speculate contribute to the coculture outcomes based on our findings. Included are all the inferred causal metabolites (table S4) and metabolites that were likely to be broadly exchanged between many species. The latter includes metabolites that were commonly produced by degraders and exploiters after growth on GlcNAc (Fig. 3B), which were shown to be consumed by scavengers (Fig. 3D), such as glutamate, pyruvate, oxoproline, and the growth supporting substrate acetate (Fig. 3B). Collectively, these metabolite exchanges illustrate how specific metabolites, in addition to broadly exchanged metabolites, may contribute to the presence of individual species.
DISCUSSION
Resolving microbial community function to the level of contributing species is hampered by the difficulty in monitoring how nutrients are dispersed between species and how individual species can affect the broader community through metabolic interaction networks. Here, we demonstrate an approach for how community interaction networks can be learned from the characterization of individual species and how specialized biopolymer degraders scaffold these networks to shape the population. Degraders secrete chitinases that produce different oligomer profiles that influence the abundance of exploiters, which thrive primarily on oligomers that they cannot generate themselves. Moreover, the composition of oligomer profiles influences which metabolite exploiters secrete into a shared resource pool that is available to the entire population. Hence, degraders initiate a hierarchical cascade of nutrient flow into a population.
An open question regarding the exchange of nutrients in a community is the extent to which it is mediated by broad and nonspecific cross-feeding networks (6) or by species-specific interactions with an ordered structure (7, 38, 39). Our results provide evidence that the exchange of nutrients has an ordered structure that allows individual species to shape the flow of metabolites and, subsequently, that nonspecific cross-feeding networks do not capture the complexity of nutrient exchange. We show that a multitude of secreted metabolites provide metabolic niches to support a large population, including major growth supporting substrates such as acetate, ammonium, and glutamate whose exchange appears to be nonspecific between most species. However, our results further demonstrate that scavengers, which can grow on neither chitin nor its degradation products, preferentially consume metabolites that are formed at later points in community succession, demonstrating how degraders scaffold downstream colonization of nonspecialized species. With our approach of integrating individual species consumption and secretion data, we were able to infer metabolic exchange networks that hypothesize specific metabolites that support growth of different species.
Collectively, these results demonstrate how a hierarchical structure is formed within microbial communities by radiating cross-feeding networks. In principle, this cross-feeding structure can be generalized to the multitude of communities that require specialized biopolymer degraders, including those involved in human health (1), biogeochemical transformations (2), and biotechnology (40). Although hierarchical structures may be convoluted in more complex environments, we outline a concept for generating hypotheses on cross-feeding networks when constructing smaller consortia that recapitulate key aspect of the entire community.
MATERIALS AND METHODS
Bacterial species and chemicals
All chemicals were purchased from Sigma-Aldrich unless otherwise noted. GlcNAc oligonucleotide standards were obtained from Omicron Biochemicals. Medium used are either Marine Broth 2216 (Thermo Fisher Scientific, Difco, no. 279100) or MBL minimal medium supplemented with a carbon source. MBL is mixed from several stock components: 4× concentrated seawater salts (NaCl, 80 g/liter; MgCl2*6H2O, 12 g/liter; CaCl2*2H2O, 0.6 g/liter; KCl, 2 g/liter), 1000× concentrated trace minerals (FeSO4*7H2O, 2.1 g/liter; H3BO3, 30 mg/liter; MnCl2*4H2O, 100 mg/liter; CoCl2*6H2O, 190 mg/liter; NiCl2*6H2O, 24 mg/liter; CuCl2*2H2O, 2 mg/liter; ZnSO4*7H2O, 144 mg/liter; Na2MoO4*2H2O, 36 mg/liter; NaVO3, 25 mg/liter; NaWO4*2H2O, 25 mg/liter; Na2SeO3*5H2O, 6 mg/liter), 1000× concentrated vitamins (riboflavin, 100 mg/liter; d-biotin, 30 mg/liter; thiamine hydrochloride, 100 mg/liter; l-ascorbic acid, 100 mg/liter; Ca d-pantothenate, 100 mg/liter; folate, 100 mg/liter; nicotinate, 100 mg/liter; 4-aminobenzoic acid, 100 mg/liter; pyridoxine HCl, 100 mg/liter; lipoic acid, 100 mg/liter; nicotinamide adenine dinucleotide (NAD), 100 mg/liter; thiamin pyrophosphate, 100 mg/liter; cyanocobalamin, 10 mg/liter), 1 mM phosphate dibasic, 1 mM sodium sulfate,50 mM Hepes (pH 8.2), and a carbon source. All stocks are diluted accordingly, and the volume is adjusted to reach 1× concentrations of all components.
Species were received from the laboratory of O.X.C. from Massachusetts Institute of Technology (table S1). All species were previously collected from polymer degrading communities of natural seawater communities from Canoe Beach, Nahant, MA, USA; 42°25′11.5″ N, 70°54′26.0″ W (5, 26). Species were stored in −80°C in glycerol stocks and streaked onto Marine Broth 2216 plates containing 1.5% agar (BD, no. 214010) before use.
Cross-feeding with pooled supernatant
Single colonies were transferred into liquid Marine Broth 2216 and grown overnight at 220 rpm at 27°C. Cells were centrifuged and washed in MBL containing no carbon source two times. This was then used to inoculate the respective culture to a final OD600 of 0.01. All supernatants were harvested from species grown in 125 ml of shake flasks containing 20 ml of the respective medium and were shaken at 100 rpm at 27°C. Supernatants were collected by centrifugation and further filtered using a 0.2 μM polyethersulfone (PES) membrane Stericup filter unit (Millipore). Supernatants were stored at −20°C until further use. Pooled supernatants were prepared by pooling equal volumes of each supernatant. This includes supernatants collected from each degrader grown on chitin and each degrader and each exploiter grown on GlcNAc. The pooled supernatant was diluted 25% with fresh MBL containing no carbon source before inoculation. Species were inoculated into 1.8 ml of the pooled supernatant contained in 96 deep well plates with a 4-mm glass bead. Plates were shaken at 220 rpm at 27°C. Samples were taken every 2 to 4 hours after inoculation, and cells were removed by centrifugation. The supernatant was stored at −20°C before measurements.
Preparation of colloidal chitin
Chitin powder (10 g; Sigma-Aldrich, C7170) was added to 100 ml of phosphoric acid (85 weight %) and incubated at 4°C for 48 hours. Roughly 500 ml of fresh H2O was added to this mixture and shaken until the chitin precipitated. This was then filtered using vacuum filter paper (MACHEREY-NAGEL, MN615), and fresh water was used to wash the filtrate three times. Chitin precipitate was then added into 3-kDa dialysis tubing cellulose membrane (Sigma-Aldrich) and placed in water to remove residual monomers and oligomers. Fresh water was replaced daily three times. Colloidal chitin was then collected, adjusted to pH 7 with NaOH, and homogenized using a Bosch SilentMixx Pro blender. The colloidal chitin was autoclaved before use.
Assay for acetate quantification
The following master mix was used for assays: 100 mM tris (pH 7.4), 5 mM MgCl2, 1 mM phosphoenolpyruvate (PEP), 1 mM adenosine triphosphate, pyruvate kinase (3 U/ml), and acetate kinase (3 U/ml). The assay was started by adding 20 μl of sample to 200 μl of master mix. The assay was monitored by measuring an absorbance at OD230, and concentrations were determined by comparing the rate of PEP consumption over time compared to acetate standards.
Assay for ammonia quantification
The following master mix was used for assays: 100 mM tris (pH 8.3), 7 mM α-ketoglutarate, 0.25 mM reduced form of nicotinamide adenine dinucleotide phosphate (NADPH), and glutamate dehydrogenase (0.1 U/ml). The assay was started by adding 20 μl of sample to 200 μl of master mix. Consumption of NADPH was measured over time by measuring an absorbance at OD340, and concentrations were determined by comparing slopes to an analytical standard of ammonium chloride.
Concentration of extracellular chitinases and enzyme digests
Degraders were inoculated into 50 ml of colloidal chitin MBL (2 g/liter) after growing in Marine Broth 2216 as a preculture. Cultures were grown until mid-late log phase (OD600 ~ 1), and all remaining chitin and cells were removed by centrifugation at 4°C. The supernatant was filtered using a 0.2 μM PES Stericup Filter unit (Millipore). Enzymes were concentrated from the supernatant 50-fold using a 10-kDa centrifugation protein concentration filter (Amicon Ultra, Millipore). Following this, the enzyme was added to fresh MBL containing chitin (5 g/liter) and sampled every 24 hours to measure for degradation products. The resulting digest was further sterile-filtered and diluted by a factor of 2.5 and used to support growth of exploiters. Protein concentration was determined using the Bradford reagent.
Chitin coculture experiments
Individual colonies were inoculated into Marine Broth 2216 from solid medium and grown overnight at 220 rpm at 27°C. Cells were then washed twice in MBL with no carbon and further inoculated into MBL with chitin (2 g/liter) at 1 × 106 colony-forming units (CFU)/ml, except for degraders, which were inoculated at 5 × 106 CFU/ml for quantitative polymerase chain reaction (qPCR) experiments and 2 × 107 CFU/ml for 16S coculture experiments. All coculture experiments used 96 deep well plates containing 1.8 ml of medium and a 4-mm glass bead. Samples for quantifying species abundance were taken of the planktonic phase. Removal of colloidal chitin from samples was done by centrifuging the cultures at 500 relative centrifugal force for 30 s, and the top later of culture from each well was sampled. Samples were stored at −20°C. Cell abundance was measured with qPCR or 16S sequencing.
Genomic DNA purification
Genomic DNA purification was done using the DNAdvance Kit (Beckman Coulter, A48705) following the manufacturer’s protocol with the modification that the input for the kit is 100 μl of Lysis Master Mix combined with 100 μl of bacterial culture that is diluted to an OD600 of less than 0.2.
Cell absolute quantification with qPCR
qPCR was performed using the GoTaq qPCR Master Mix (Promega) following the manufacturer’s protocol. qPCR reactions were prepared with a final volume of 15 μl using prepared genomic DNA as a template. Samples were measured and data were analyzed using a QuantStudio 3 Real-Time PCR System with the ΔΔCt method. Absolute quantification of species was obtained by comparing Ct values with those obtained from standard cultures that have known CFU/ml values. PCR primers were designed to have an efficiency of 90 to 105%, and sequences are listed in table S5. Primers were tested for specificity to the bacterial species used in each coculture.
16S sequencing
Measuring relative abundance of species in coculture was performed with amplicon 16S sequencing using the MetaFast protocol (Fasteris, Switzerland) including proprietary MetaFast barcoded primers. Amplicons were prepared by amplifying purified genomic DNA samples with a unique pair of MetaFast barcoded primers with Kapa HiFi DNA polymerase. Further library preparation was performed by Fasteris, and samples were sequenced with an Illumina MiSeq platform.
Paired, quality-filtered reads obtained from 16S sequencing were processed using dada2 version 1.16 to determine ESVs. Except where noted, the default parameters were used (41). Briefly, sequences were trimmed to remove adapters and further trimmed from positions 25 to 175, yielding 150–base pair sequences with a quality score > 38. The maximum expected error rate was set to maxEE(2,5) to ensure stringent trimming and better differentiate reads belonging to species with very similar sequences. After learning error rates, samples were pooled to define variants. Species were matched to ESVs using a custom BLAST built from the full-length 16S genes of species used in the experiment (42). The taxonomy of the ESVs was further confirmed by taxonomic identification using DECIPHER. The SILVA SSU r138 database from 2019 was used for DECIPHER analysis (43).
Genome annotation
The species used for this analysis were isolated and sequenced as part of two experiments previously performed in the laboratory of O.X.C. for studying the succession of natural microbial communities from seawater on polysaccharide particles (5, 26). For each genome, protein-coding genes were predicted and translated using Prodigal v2.6.3 (44). Predicted protein sequences were compared to a custom database of profile hidden Markov models (HMMs) of proteins involved in growth on chitin using the hmmsearch function of HMMER v3.3 with default parameters (45). Publicly available HMMs were downloaded from the Pfam v33.1 (46) or TIGRFAM v15.0 databases (see dataset S6 for accession numbers) (47). Custom HMMs were made by identifying experimentally verified proteins of interest (48, 49), finding their homologs in the UniProtKB/Swiss-Prot v2020_06 database (50), creating a seed alignment using MAFFT v7 with default parameters (51, 52), and building the profile HMMs using the hmmbuild function of HMMER with default parameters (see dataset S6 for details on each custom HMM). The isolate strain’s proteins were annotated on the basis of the hmmsearch results; if the protein length was at least 100 amino acids, then the independent E value was less than 1 × 10−9, and the domain score was greater than 30. Only the annotation with the highest score was used for each protein sequence. Gene copy numbers were calculated for each genome by tallying the number of annotations made for each protein group. Genome sequences for each organism can be found using National Center for Biotechnology Information (NCBI) accession numbers listed in table S1.
Metagenomic 16S rRNA dynamics
The 16S ribosomal RNA (rRNA) sequence of each genome was determined by first amplifying it using the 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) + 1492R (5′-GGTTACCTTGTTACGACTT-3′) universal bacterial primers (19, 20), followed by Sanger sequencing. We mapped the 16S sequences of our isolates back to the metagenomic 16S trajectories (PRJNA319196 and PRJNA478695) using BLAST (42). For each isolate, 16S matches in the metagenome were found using a moving threshold approach—we searched for matches starting at 100% identity and going down to 97% identity at 1% identity steps, and if a match was found in any identity threshold, then the search was ended, and all metagnomic hits above that threshold were considered to belong to that isolate 16S. The read counts were transformed into relative frequencies, and only entries with a relative abundance of >10−3 were retained for downstream analysis. Each experiment was originally performed in triplicate, and for each isolate, we take the mean relative abundance at each time point. For each isolate, the trajectories were normalized by their highest relative abundance (so that the highest point of any trajectory is 1), and the average of these normalized trajectories is what is reported in Fig. 1C.
Untargeted metabolite profiling and analysis
Untargeted methods were carried out using FIA-QTOF-MS. All supernatants were first diluted 100-fold into water before measurements. Metabolomics was carried out with a binary LC pump (Agilent Technologies) and a MPS2 Autosampler (Gerstel) coupled to an Agilent 6520 time-of-flight mass spectrometer (Agilent Technologies). This was operated in negative mode, at 2 GHz for extended dynamic range, with a mass/charge ratio (m/z) range of 50 to 1000, as described previously (37). The mobile phase consisted of isopropanol:water (60:40, v/v) with 5 mM ammonium fluoride buffer at pH 9, and the flow rate was 150 μl/min. Raw data for all measurements were subjected to a spectral processing and alignment pipeline using MATLAB (MathWorks, Natick, MA) as described previously (37).
Overall, we detected 2521 ions, of which 216 could be annotated on the basis of accurate mass, corresponding to a maximum of 526 unique metabolites. Ions were annotated with a tolerance of 0.005 Da against a curated compound library that contains all metabolites predicted to be present in at least one of the species used in this study based on BioCyc databases. If a single m/z ion is matched with multiple isomeric or isobaric compounds within the compound library, then the compound that participates in the largest number of enzymatic reactions in pathway databases for all 18 organisms used in this work is chosen as the top annotation. Certain isomers were resolved using LC-MS methods as described below, in which case the correct annotations were manually edited.
LC-MS exometabolomic compound measurements
Extracellular metabolomics measurements were performed as described previously (53). Briefly, chromatographic separation was performed using a Poroshell 120 EC-CN 2.1 mm–by–150 mm, 2.7-μm column (Agilent Technologies) in an Agilent 1290 infinity stack set to 40°C. The method performed was isocratic, with a mobile phase consisting of 10 mM ammonia acetate (pH 5.9) and 5% acetonitrile with a flow rate of 250 μl/min and a 5-μl injection volume. An Agilent 6520 series QTOF-MS was used for measurements with an m/z range of 20 to 400 operated in negative mode. Before LC-MS measurements, samples were diluted both 20- and 100-fold in water to maximize the number of measurable compounds within the limits of quantification. Data analysis was performed using Agilent Masshunter Quantitative Analysis software. Peaks were integrated using spectral summation integration with integration windows that are defined on the basis of retention times observed from analytical standards. The Agilent software was used to determine polynomial calibration curves with a goodness of fit (R2 > 0.97).
GlcNAC oligomer quantification
GlcNAc oligomers were quantified using LC-MS with an Agilent 1100 series stack. Chromatographic separation used a Poroshell 120 HILIC-Z 2.1 mm–by–100 mm, 2.7-μm column (Agilent) kept at a temperature of 30°C and a constant flow rate of 1 ml/min. Five microliters of sample was injected after fivefold dilution in 80% acetonitrile. Mobile phase A consists of 10 mM ammonia acetate (pH 9) in water, and mobile phase B consists of 10 mM ammonia acetate (pH 9) in 90% acetonitrile. At injection, the mobile phase contains 90% B phase for 2 min, followed by a gradient to 40% B phase until 12 min and held for one more minute before being reequilibrated at the starting condition. An Agilent 6520 series QTOF-MS was used for measurements, operated in negative mode at 2 GHz for extended dynamic range, with an m/z range of 100 to 1500. Calibration curves were measured using analytical standards and used to calculate concentrations of oligomers in the sample.
Construction of hypothetical metabolite exchange networks
Metabolic exchange networks were constructed on the basis of coculture outcomes (Fig. 4A). This includes secretion profiles of degraders grown on chitin (datasets S1 and S2), GlcNAc oligomers that degraders form extracellularly (Fig. 3A), secretion profiles of exploiters after growth on the pooled supernatant (dataset S2), and metabolites consumed by exploiters and scavengers in the pooled supernatant (Fig. 3D). Combined metabolite production and consumption data used for construction of metabolic networks is contained in dataset S3. The metabolic networks for each coculture infer two metabolite pools that mimic the sequential colonization of chitin particles by degraders, exploiters, and lastly scavengers (dataset S4). To incorporate both targeted datasets, which include absolute quantification, and untargeted metabolomics datasets, which include a broad coverage of metabolites without absolute quantification, metabolites were noted to have one of two possible states: present or absent. The first metabolite pool is a degrader-derived metabolite pool consisting of metabolites produced by the degrader in the culture and represents resources available to support growth of the exploiter. The second resource pool is an exploiter-derived metabolite pool that represents metabolites available to support scavengers after the exploiter consumed or added metabolites to the first, degrader-derived metabolite pool. To construct the exploiter-derived metabolite pool, metabolites that are consumed by an exploiter are subtracted from the degrader-derived metabolite pool, and metabolites that are produced an exploiter are added to the degrader-derived metabolite pool. We next used these pools to infer metabolite exchanges between the species in coculture to construct 12 coculture specific metabolite exchange networks (dataset S5). Each species that survived in coculture is a node in these networks, and exchanged metabolites are represented by edges. To illustrate these networks in Fig. 4B, the edge width represents the number of exchanged metabolites. Metabolites exchanged between a degrader and an exploiter are metabolites present in the degrader-derived metabolite pool that are consumed by the exploiter. Metabolites exchanged between a scavenger and a degrader or exploiter are metabolites consumed by the scavenger from the exploiter-derived metabolite pool that are secreted by either the degrader or exploiter. To identify potentially causal metabolites, we used a Boolean approach where three conditions were needed to be simultaneously true to infer a causal relationship between a metabolite and a given strain: (i) the metabolite was shown to be consumed by the species in its uptake profile; (ii) the metabolite was present in the degrader-derived or exploiter-derived metabolite pools of all cocultures where the exploiter or scavenger showed growth, respectively; and (iii) the metabolite was absent in the degrader-derived or exploiter-derived metabolite pools of all cocultures where the exploiter or scavenger did not show growth, respectively. These criteria were only applicable for species that did not grow in at least one coculture. These inferred causal metabolites are in table S5.
Acknowledgments
Funding: This work was supported by a grant from the Simons Foundation (ID608247 to S.Pon. and ID542395 to O.X.C.), as part of the collaboration on Principles of Microbial Ecosystems (PriME). R.S. was supported by NSF GRFP grant no.174530.
Author contributions: S.Pon. designed and performed all experiments, analyzed data, and wrote the manuscript. R.S. performed genomic analysis of bacterial isolates. S.Pol. mapped 16S sequences of bacterial isolates to ESVs of developing chitin communities. J.S. performed 16S amplicon sequencing analysis. D.L.-T performed network analysis of chitin coculture outcomes. O.X.C. assisted in data interpretation and writing the manuscript. U.S. designed the experiments and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Metabolomics spectral files can be accessed from the MassIVE database (ftp://massive.ucsd.edu/MSV000087455/). The short read 16S rRNA amplicon sequencing data (BioSample: SAMN11023523-SAMN11023755) have been deposited in NCBI with the BioProject identifier PRJNA478695. All deposited genome sequences have been deposited in NCBI with BioProject identifier and BioSample IDs specified in table S1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S5
Other Supplementary Material for this manuscript includes the following:
Datasets S1 to S6
REFERENCES AND NOTES
- 1.Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I., The human microbiome project. Nature 449, 804–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konopka A., What is microbial community ecology. ISME J. 3, 1223–1230 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Ponomarova O., Gabrielli N., Sévin D. C., Mülleder M., Zirngibl K., Bulyha K., Andrejev S., Kafkia E., Typas A., Sauer U., Ralser M., Patil K. R., Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 5, 345–357.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baran R., Brodie E. L., Mayberry-Lewis J., Hummel E., Da Rocha U. N., Chakraborty R., Bowen B. P., Karaoz U., Cadillo-Quiroz H., Garcia-Pichel F., Northen T. R., Exometabolite niche partitioning among sympatric soil bacteria. Nat. Commun. 6, 8289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta M. S., Sliwerska E., Gore J., Polz M. F., Cordero O. X., Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7, 11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldford J. E., Lu N., Bajić D., Estrela S., Tikhonov M., Sanchez-Gorostiaga A., Segrè D., Mehta P., Sanchez A., Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasche S., Kim Y., Mars R. A. T., Machado D., Maansson M., Kafkia E., Milanese A., Zeller G., Teusink B., Nielsen J., Benes V., Neves R., Sauer U., Patil K. R., Metabolic cooperation and spatiotemporal niche partitioning in a kefir microbial community. Nat. Microbiol. 6, 196–208 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pacheco A. R., Moel M., Segrè D., Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. Commun. 10, 103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louca S., Jacques S. M. S., Pires A. P. F., Leal J. S., Srivastava D. S., Parfrey L. W., Farjalla V. F., Doebeli M., High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 1, 0015 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Cordero O. X., Ventouras L. A., DeLong E. F., Polz M. F., Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. U.S.A. 109, 20059–20064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hehemann J. H., Arevalo P., Datta M. S., Yu X., Corzett C. H., Henschel A., Preheim S. P., Timberlake S., Alm E. J., Polz M. F., Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat. Commun. 7, 12860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romine M. F., Rodionov D. A., Maezato Y., Osterman A. L., Nelson W. C., Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities. ISME J. 11, 1434–1446 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kešnerová L., Mars R. A. T., Ellegaard K. M., Troilo M., Sauer U., Engel P., Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15, e2003467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stams A. J. M., Plugge C. M., Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568–577 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Wei Z., Yang T., Friman V., Xu Y., Shen Q., Jousset A., Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6, 8413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodionov D. A., Arzamasov A. A., Khoroshkin M. S., Iablokov S. N., Leyn S. A., Peterson S. N., Novichkov P. S., Micronutrient requirements and sharing capabilities of the human gut microbiome. Front. Microbiol. 10, 1316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moens F., Verce M., De Vuyst L., Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int. J. Food Microbiol. 241, 225–236 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Gallon J. R., N2 fixation in phototrophs: Adaptation to a specialized way of life. Plant and Soil 230, 39–48 (2001). [Google Scholar]
- 19.Kramer J., Özkaya Ö., Kümmerli R., Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zelezniak A., Andrejev S., Ponomarova O., Mende D. R., Bork P., Patil K. R., Metabolic dependencies drive species cooccurrence in diverse microbial communities. Proc. Natl. Acad. Sci. U.S.A. 112, 6449–6454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venturelli O. S., Carr A. V., Fisher G., Hsu R. H., Lau R., Bowen B. P., Hromada S., Northen T., Arkin A. P., Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 14, e8157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajic D., Sanchez A., The ecology and evolution of microbial metabolic strategies. Curr. Opin. Biotechnol. 62, 123–128 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Cole J. K., Hutchison J. R., Renslow R. S., Kim Y. M., Chrisler W. B., Engelmann H. E., Dohnalkova A. C., Hu D., Metz T. O., Fredrickson J. K., Lindemann S. R., Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: Model systems for the study of autotroph-heterotroph interactions. Front. Microbiol. 5, 109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stams A. J. M., De Bok F. A. M., Plugge C. M., Van Eekert M. H. A., Dolfing J., Schraa G., Exocellular electron transfer in anaerobic microbial communities. Environ. Microbiol. 8, 371–382 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Ha P. T., Lindemann S. R., Shi L., Dohnalkova A. C., Fredrickson J. K., Madigan M. T., Beyenal H., Syntrophic anaerobic photosynthesis via direct interspecies electron transfer. Nat. Commun. 8, 1392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enke T. N., Datta M. S., Schwartzman J., Cermak N., Schmitz D., Barrere J., Pascual-García A., Cordero O. X., Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29, 1528–1535.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Flint H. J., Scott K. P., Duncan S. H., Louis P., Forano E., Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 3, 289–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayer E. A., Morag E., Lamed R., The cellulosome – a treasure- trove for biotechnology. Trends Biotechnol. 12, 379–386 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Belzer C., Chia W., Aalvink S., Chamlagain B., Piironen V., de Vos W. M., Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and Vitamin B12 production by intestinal. MBio 8, e00770-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayer B., Pelikan C., Bittner M. J., Reinthaler T., Könneke M., Herndl G. J., Proteomic response of three marine ammonia-oxidizing archaea to hydrogen peroxide and their metabolic interactions with a heterotrophic alphaproteobacterium. Am. Soc. Microbiol. 4, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris J. J., Johnson Z. I., Szul M. J., Keller M., Zinser E. R., Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLOS ONE 6, e16805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilja E. E., Johnson D. R., Segregating metabolic processes into different microbial cells accelerates the consumption of inhibitory substrates. ISME J. 10, 1568–1578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiene R. P., Linn L. J., Bruton J. A., New and important roles for DMSP in marine microbial communities. J. Sea Res. 43, 209–224 (2000). [Google Scholar]
- 34.Morono Y., Terada T., Nishizawa M., Ito M., Hillion F., Takahata N., Sano Y., Carbon and nitrogen assimilation in deep subsea fl oor microbial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 18295–18300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yesiltepe Y., Morton B. R., Cory A. B., Cardamone H. C., Hofmockel K. S., Lipton M. S., Moran J. J., Renslow R. S., Fredrickson J. K., Lindemann R., Nitrogen source governs community carbon metabolism in a model hypersaline benthic phototrophic biofilm. Msystems 5, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peter M. G., Applications and environmental aspects of chitin and chitosan. J. Macromol. Sci. A. 32, 629–640 (1995). [Google Scholar]
- 37.Fuhrer T., Heer D., Begemann B., Zamboni N., High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 83, 7074–7080 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Goyal A., Dubinkina V., Maslov S., Multiple stable states in microbial communities explained by the stable marriage problem. ISME J. 12, 2823–2834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estrela S., Vila J. C. C., Lu N., Bajić D., Rebolleda-Gómez M., Chang C.-Y., Goldford J. E., Sanchez-Gorostiaga A., Sánchez Á., Functional attractors in microbial community assembly. Cell Syst. 2021, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao J. C., Mi L., Pontrelli S., Luo S., Fuelling the future: Microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 14, 288–304 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Callahan B. J., Mcmurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.T. Madden, The BLAST sequence analysis tool, in The NCBI Handbook [Internet] (National Center for Biotechnology Information, ed. 2, 2013). [Google Scholar]
- 43.Wright E. S., Using DECIPHER v2. 0 to analyze big biological sequence data in R. R Journal 8, 352–359 (2016). [Google Scholar]
- 44.Hyatt D., Chen G., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eddy S. R., Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G. A., Sonnhammer E. L. L., Tosatto S. C. E., Paladin L., Raj S., Richardson L. J., Finn R. D., Bateman A., Pfam: The protein families database in 2021. Nucleic Acid Res. 49, D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haft D. H., Loftus B. J., Richardson D. L., Yang F., Eisen J. A., Paulsen I. T., White O., TIGRFAMs: A protein family resource for the functional identification of proteins. Nucleic Acids Res. 29, 41–43 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meibom K. L., Li X. B., Nielsen A. T., Wu C., Roseman S., Schoolnik G. K., The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101, 2524–2529 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eisenbeis S., Lohmiller S., Valdebenito M., Leicht S., Braun V., NagA-dependent uptake of N-Acetyl-Glucosamine and N-Acetyl-Chitin Oligosaccharides across the outer membrane of caulobacter crescentus. J. Bacteriol. 190, 5230–5238 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability article fast track. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madeira F., Park Y., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A. R. N., Potter S. C., Finn D., Lopez R., The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pontrelli S., Sauer U., Salt-tolerant metabolomics for exometabolomic measurements of marine bacterial isolates. Anal. Chem. 93, 7164–7171 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S5
Datasets S1 to S6