Abstract
Two of the 25 Bartonella isolates recovered during a prevalence study of Bartonella henselae bacteremia in domestic cats from the greater San Francisco Bay region were found to differ phenotypically and genotypically from all prior B. henselae isolates. These isolates, C-29 and C-30, which were recovered from the blood of two pet cats belonging to the same household, grew on chocolate agar as pinpoint colonies following 14 days of incubation at 35°C in a candle jar but failed to grow on heart infusion agar supplemented with 5% rabbit blood. Additional phenotypic characteristics distinguished the isolates C-29 and C-30 from other feline B. henselae isolates. The restriction patterns obtained for C-29 and C-30 by citrate synthase PCR-restriction fragment length polymorphism (RFLP) analysis as well as by genomic RFLP could not be distinguished from each other but were distinctly different from that of the B. henselae type strain. In reciprocal reactions, DNAs from strains C-29 and C-30 were 97 to 100% related under optimal and stringent DNA reassociation conditions, with 0 to 0.5% divergence within related sequences. Labeled DNA from the type strain of B. henselae was 61 to 65% related to unlabeled DNAs from strains C-29 and C-30 in 55°C reactions, with 5.0 to 5.5% divergence within the related sequences, and 31 to 41% related in stringent, 70°C reactions. In reciprocal reactions, labeled DNAs from strains C-29 and C-30 were 68 to 92% related to those of the B. henselae type strain and other B. henselae strains, with 5 to 7% divergence. The 16S rRNA gene sequence of strain C-29 was 99.54% homologous to that of the type strain of B. henselae. On the basis of these findings, the two isolates C-29 and C-30 are designated a new species of Bartonella, for which we propose the name Bartonella koehlerae. The type strain of Bartonella koehlerae is strain C-29 (ATCC 700693).
Bartonella species cause a wide spectrum of human diseases. The principal causative agent of cat scratch disease in immunocompetent people is Bartonella henselae (9, 22). This bacterium is associated with bacillary angiomatosis and other clinical syndromes in immunocompromised humans (15, 21, 23, 27, 28, 32). Isolation of B. henselae from the blood of naturally infected cats, as well as the demonstration that cats remain highly bacteremic for several months, implicates cats as the major reservoir for this bacterium; the prevalence of B. henselae bacteremia in cats tested in northern California was 40% (6, 14). The prevalence of B. henselae antibodies in cats in the United States varies according to geographic region and ranges from 3.7% for Alaska to 54.6% for the southeastern states (12). The prevalence of another species, B. clarridgeiae, in a European urban stray cat population recently was demonstrated to be 16% (11), greater than that in the United States. The transmission of B. henselae from cats to humans usually occurs via scratches, and transmission from cat to cat readily occurs via the cat flea, Ctenocephalides felis (7). In the initial study identifying the prevalence of B. henselae bacteremia in domestic cats from the greater San Francisco Bay region, isolates were obtained from 41% (25 of 61) of the cats tested (14). Two of the 25 feline Bartonella isolates recovered during that study were phenotypically and genotypically characterized and found to be genetically closely related to, but distinct from, the type strain of B. henselae and, in addition, to differ phenotypically and genotypically from all other B. henselae isolates and all reported Bartonella species. These two feline isolates, C-29 and C-30, are proposed as a new Bartonella species, Bartonella koehlerae.
MATERIALS AND METHODS
Strains.
B. henselae type strain ATCC 49882 was provided by Russell Regnery, Viral and Rickettsial Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Ga. Bartonella quintana type strain ATCC VR-358 was obtained from the American Type Culture Collection (ATCC). The two Bartonella strains C-29 and C-30 were isolated from the blood of two cats belonging to the same household (14). Bartonella strains compared to C-29 by DNA hybridization analysis included B. quintana type strain ATCC VR-358, B. henselae ATCC 49882, B. henselae 88-712 (32), B. henselae G6486, B. henselae G6529, B. henselae G8378, B. henselae G5691, B. elizabethae ATCC 49927T (8), B. vinsonii ATCC VR-152T, B. vinsonii subsp. berkhoffii ATCC 51672 (17), B. clarridgeiae ATCC 51734T, B. bacilliformis ATCC 35685T (4), B. grahamii NCTC 12860T (2), and B. doshiae NCTC 12862T (2).
Primary isolation of C-29 and C-30.
Blood was obtained from two cats during a prevalence study of B. henselae in domestic cats in the greater San Francisco Bay area of northern California (14). Blood (1.5 ml) was collected after thorough cleansing of the skin with 70% alcohol, using an aseptic technique similar to that used for humans, by jugular or saphenous venipuncture of each cat. Blood was transferred into a pediatric lysis-centrifugation tube (Wampole Laboratories, Cranbury, N.J.) and centrifuged, and the pellet was resuspended in inoculation medium (15). The resuspended blood pellet was plated onto BBL heart infusion agar (Becton Dickinson, Cockeysville, Md.) containing 5% fresh defibrinated rabbit blood, as well as onto chocolate agar. The chocolate agar (19) was prepared with hemoglobin powder and GC agar base (both from Acumedia Manufacturers, Inc., Baltimore, Md.) and was supplemented with 10 ml of IsoVitalex (Becton Dickinson). The plates were incubated at 35°C in a candle extinction jar for 3 weeks. Cultures were examined every 5 days for bacterial growth.
Biochemical tests.
For the initial biochemical tests, standard methods were used; Gram staining and biochemical testing of the two isolates were performed as described previously (20). The inocula used for biochemical evaluations were obtained by growing both isolates, as well as the type strains B. henselae ATCC 49882 and B. quintana ATCC VR-358, on chocolate agar for 7 days at 35°C in a candle extinction jar. Preformed-enzyme tests were performed by using MicroScan Rapid Anaerobe Panels (31) (Baxter Diagnostics, Deerfield, Ill.) in accordance with the manufacturer’s instructions regarding preparation, incubation, and biochemical interpretation. The reactions and identification codes obtained for the cat isolates C-29 and C-30 were compared to those of the B. henselae ATCC 49882 and B. quintana ATCC VR-358 type strains.
PCR amplification and sequence analysis of the 16S rDNA.
Bacterial DNA was extracted from both Bartonella isolates by a standard protocol (33). The extracted DNA was then used as a template for amplification of 1,414 nucleotides of the gene encoding the 16S rRNA (16S rDNA). The 16S rDNA was amplified by using synthetic oligonucleotide primers (University of California, San Francisco, Biomolecular Resource Center) adapted from the primers POmod and PC3mod (for the 5′ half), P3mod and PC5 (for the 3′ half) (34), and rD1 (30). In addition, two oligonucleotide primers corresponding to p24E and p12B (23) were utilized. The following restriction endonuclease sites were included in the oligonucleotide primers and then used to clone the PCR products into pUC18: for POmod and P3mod, SalI; for PC3mod and PC5, HindIII; for rD1 and p12B, BamHI; and for p24E, EcoRI. We synthesized three internal 16S primers (5′-GGTTGCCCCCATTGTCC-3′, 5′-CATTCAGTTGGGCACTC-3′, and 5′-CCTTCCTCTCGGCTTAT-3′) and primer BA-9-E (5′-GGAATTCCGGAGATGGATGAGCCC-3′), the latter being located approximately 180 bp from the 5′ end of the 16S rDNA, to complete sequencing of both strands of this rDNA. All PCR runs included negative controls (containing no DNA template) and positive controls (with Bartonella DNA).
The products of DNA amplification were digested with the appropriate restriction endonucleases, ligated into the vector pUC18 (Pharmacia, Piscataway, N.J.), and used to transform Escherichia coli. The sequences of the double-stranded DNAs of recombinants were obtained by the dideoxy sequencing method (26), using a Taq Dye Primer Cycle Sequencing kit (Applied Biosystems, Inc., Foster City, Calif.), followed by analysis on an automated sequencer (Applied Biosystems). Both DNA strands of a minimum of two clones containing overlapping fragments of the 1,414 nucleotides of the 16S rDNA were sequenced. Discrepancies were resolved when necessary by sequencing additional clones. The 16S sequence from strain C-29 was aligned with the 16S sequences, retrieved from GenBank, of 10 other strains, including Bartonella species, Agrobacterium tumefaciens, and, as the outgroup, Ehrlichia canis, using the Wisconsin Sequence Analysis Package program PILEUP (Wisconsin Package version 9.0; Genetics Computer Group, Madison, Wis.). The multiple-sequence alignment was edited to remove the 5′ and 3′ hypervariable regions (10). The resulting alignment was 1,330 bases long and corresponded to bases 106 to 1457 of E. coli (5). The edited alignment was used as input to the software program PHYLIP (version 3.5; J. Felsenstein, University of Washington, Seattle) to derive a phylogenetic dendrogram via the nucleotide substitution model of Jukes and Cantor (13) and the neighbor-joining method of Saitou and Nei (25). For determination of tree robustness, the multiple-sequence alignment was used as input to the PHYLIP SEQBOOT program and a majority-rule strict consensus tree was generated by the CONSENSE program. The plot file from PHYLIP 3.5 was reformatted for the Hewlett-Packard Laserjet 4 printer by using the PRINTGL software (Ravitz Software, Inc., Lexington, Ky.).
Citrate synthase PCR-RFLP typing.
For PCR-restriction fragment length polymorphism (RFLP) analysis, primers (21) were employed to amplify a fragment of approximately 400 nucleotides from the citrate synthase gene of Bartonella species, with the following cycling program being used: 94°C for 1 min, 47°C for 1 min, and 72°C for 1 min for a total of 35 cycles. Chromosomal DNAs from B. henselae ATCC 49882 and B. quintana ATCC VR-358 type strains were used as positive controls for the PCR. Each set of reactions also included a negative control (a tube containing all of the reagents but no template DNA) and a positive control (a tube with Bartonella DNA). Undigested amplicons and amplicons digested with the enzymes HhaI, MseI, and TaqI were separated electrophoretically on an 8% acrylamide gel, and the resulting patterns compared with the RFLP patterns obtained for the B. henselae ATCC 49882 and B. quintana ATCC VR-358 type strains (21).
Genomic RFLP typing.
Genomic DNA also was subjected to RFLP analysis. The 13-day-old cultures of C-29 and C-30 were recovered by flooding the agar plates with sterile filtered phosphate-buffered saline and scraping their surfaces with a cell scraper. DNA extraction was performed by the standard cetyltrimethylammonium bromide (Sigma, St. Louis, Mo.) technique (33). NheI-digested genomic DNA fragments were separated electrophoretically on a 0.7% agarose gel, and the resulting pattern was compared with the fragment patterns of similarly digested genomic DNAs from the B. henselae ATCC 49882 and B. quintana ATCC VR-358 type strains.
DNA hybridization methods.
C-29 and C-30 cells were grown on chocolate agar at 35°C in an atmosphere of 5% CO2 for 7 days, scraped from the agar surface, and washed twice with sterile 0.1 M NaCl. The methods used for DNA extraction and purification as well as the hydroxyapatite method have been described previously (3). DNA was labeled enzymatically in vitro with [α-32P]dCTP by use of a nick translation reagent kit (GIBCO BRL, Gaithersburg, Md.) according to the manufacturer’s instructions. Divergence of related sequences was estimated to be approximately 1% for each degree decrease in thermal stability of a heterologous reassociated DNA duplex compared with that of the homologous reassociated DNA duplex (3). Divergence was calculated to the nearest 0.5%.
Nucleotide sequence accession numbers.
The sequence for the C-29 16S rDNA has been assigned accession no. AF076237 in GenBank.
RESULTS
Recovery of Bartonella isolates C-29 and C-30.
The two asymptomatic cats, C-29 and C-30, were 5 and 6 months old, respectively, lived indoors and outdoors on a farm, and were infested with cat fleas (C. felis). The isolates obtained from cats C-29 and C-30 were visible on chocolate agar following incubation at 35°C in a candle jar for 14 days, much longer than for all other B. henselae feline isolates (usually 3 to 5 days).
Phenotypic characteristics.
Both strains grew poorly and appeared as minute pinpoint colonies after 14 days of incubation on chocolate agar. In contrast to B. henselae, which grows preferentially on heart infusion agar supplemented with 5% rabbit blood rather than on chocolate agar, C-29 and C-30 grew preferentially on chocolate agar and usually failed to produce any growth on heart infusion agar supplemented with 5% rabbit blood. On subsequent passage, we also observed a much longer doubling time for C-29 and C-30 than for other B. henselae isolates. Gram staining revealed tiny, short, straight or slightly curved gram-negative bacilli.
Tests for production of catalase and oxidase were negative. Using a MicroScan Rapid Anaerobe Panel to test for preformed enzymes, our isolates C-29 and C-30 were negative for l-proline-β-naphthylamide (PRO) as well as for l-lysine-β-naphthylamide (acid) (LYA). In contrast, B. henselae ATCC 49882 and B. quintana ATCC VR-358 were positive for PRO. B. henselae ATCC 49882 was positive and B. quintana ATCC VR-358 was negative for LYA. In summary, B. henselae and the new species differed with regard to the preformed enzymes PRO and LYA whereas B. quintana and the new species differed only with regard to the preformed enzyme PRO. No other differences between our two strains and the reference strains were observed, resulting in the code 10073240 for C-29 and C-30. The code obtained for B. henselae ATCC 49882 was 10077640, and the one obtained for B. quintana ATCC VR-358 was 10073640. The codes for our reference strains were consistent with the ones previously reported for B. henselae and B. quintana (31). The biochemical characteristics of our isolates C-29 and C-30 were most similar to, but distinct from, those of the B. henselae type strain.
Citrate synthase PCR-RFLP analysis.
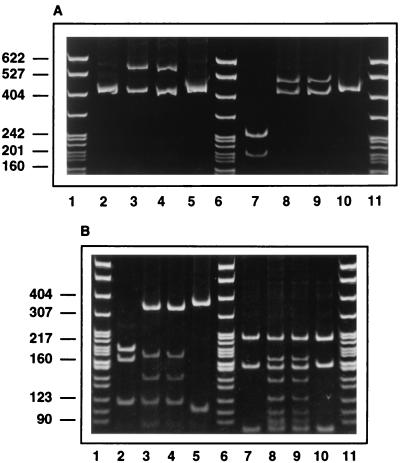
Citrate synthase PCR-RFLP analysis was used to compare isolates C-29 and C-30. In contrast to the B. henselae and B. quintana type strains, two PCR products were observed for C-29 and C-30. The smaller amplicon was the same size as the single amplicons of the B. henselae and B. quintana type strains (approximately 400 bp), but the second one was larger (approximately 550 bp) and was not seen for B. henselae and B. quintana (Fig. 1). The patterns produced by digestion of the amplicons of both C-29 and C-30 were identical, but the sizes and number of digestion products differed from those of the B. henselae and B. quintana type strains (Fig. 1).
FIG. 1.
Citrate synthase gene PCR-RFLP analysis of DNA from feline isolates C-29 and C-30, electrophoresed on 8% acrylamide gels and stained with ethidium bromide. (A) Undigested (lanes 2 to 5) or HhaI-digested (lanes 7 to 10) PCR-amplified citrate synthase gene fragments. (B) TaqI-digested (lanes 2 to 5) or MseI-digested (lanes 7 to 10) PCR-amplified citrate synthase gene fragments. Lanes 1, 6, and 11, size standards (in base pairs); lanes 2 and 7, B. henselae ATCC 49882; lanes 3 and 8, feline isolate C-29; lanes 4 and 9, feline isolate C-30; lanes 5 and 10, B. quintana ATCC VR-358.
Genomic RFLP analysis.
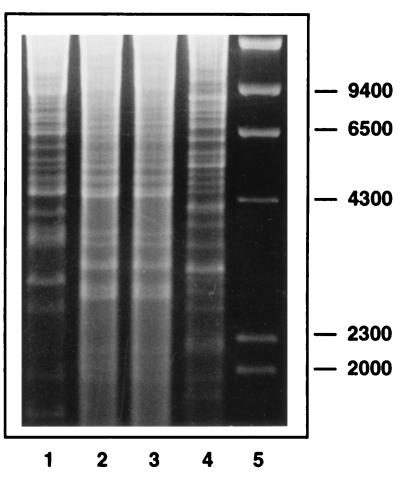
Genomic RFLP analysis was used to compare isolates C-29 and C-30 with the B. henselae and B. quintana type strains. The patterns observed for the C-29 and C-30 isolates when the enzyme NheI was used were identical to each other but were distinctly different from those of the B. henselae and B. quintana type strains (Fig. 2).
FIG. 2.
Genomic RFLP analysis of feline Bartonella isolates C-29 and C-30. Genomic DNA was extracted and digested with NheI, and the restriction fragments were electrophoresed on a 0.7% agarose gel and then stained with ethidium bromide. Lane 1, B. henselae ATCC 49882; lanes 2 and 3, feline blood isolates C-29 and C-30, respectively; lane 4, B. quintana ATCC VR-358; lane 5, size standards (in base pairs).
16S rDNA sequence analysis.
The 16S rDNA sequences of cat isolates C-29 and C-30 were identical. The C-29 sequence was compared with the 16S rDNA sequences of 10 other Bartonella species or subspecies, obtained from the GenBank database. A total of 1,414 nucleotide positions, corresponding to E. coli sequence positions 23 to 1490 (5), were determined for both cat isolates. The 16S rDNA sequence obtained for C-29 was most similar to that of the B. henselae type strain. Comparing an overlapping region of 1,414 nucleotides of the C-29 and the B. henselae type strain 16S rDNAs, six differences were noted: at position 267, C-29 had a substitution of a cytosine residue for the thymine residue of the B. henselae type strain; at positions 860 and 861, there was an inversion of a guanosine and a cytosine residue; at position 940, there was an insertion of a guanosine residue; at position 1136, there was substitution of a cytosine residue for a thymine residue; and at position 1137, as well as at position 1455, there was a substitution of an adenine residue for a guanosine residue.
The 16S rDNA sequence of strain C-29 was most similar to that of B. henselae (99.54% similarity). The levels of similarity determined between the C-29 16S rDNA sequence and those of other Bartonella species (Table 1) were as follows: for B. doshiae, 99.39%; for B. vinsonii, 99.39%; for B. vinsonii subsp. berkhoffii, 99.16%; for B. taylorii, 99.09%; and for B. grahamii, 99.02%. The C-29 16S rDNA sequence showed less similarity to the sequences of B. quintana (98.86%), B. elizabethae (98.78%), B. bacilliformis (98.33%), and B. clarridgeiae (98.10%). The percentages of similarity between a number of other Bartonella species were identical to or greater than the 99.54% similarity observed between B. henselae and C-29 (e.g., 99.54% between B. vinsonii and B. taylorii, as well as between B. grahamii and B. taylorii, and even higher percentages of similarity between B. grahamii and B. vinsonii [99.62%], B. taylorii and B. doshiae [99.7%], and B. vinsonii and B. doshiae [99.85%]). C-29 was 94.89% similar to Brucella abortus and 93.99% similar to A. tumefaciens.
TABLE 1.
16S rDNA similarity values
| Organism | % Similarity
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C-29 | B. hen-selae | B. eliza-bethae | B. dosh-iae | B. bacilli-formis | B. clar-ridgeiae | B. quin-tana | B. vin-sonii | B. vinsonii subsp. berkhoffii | B. tay-lorii | B. gra-hamii | Brucella abortus | A. tume-faciens | E. canis | |
| C-29 | 100.00 | |||||||||||||
| B. henselae | 99.54 | 100.00 | ||||||||||||
| B. elizabethae | 98.78 | 98.86 | 100.00 | |||||||||||
| B. doshiae | 99.39 | 99.39 | 99.24 | 100.00 | ||||||||||
| B. bacilliformis | 98.33 | 98.17 | 98.25 | 98.33 | 100.00 | |||||||||
| B. clarridgeiae | 98.10 | 98.10 | 98.25 | 98.41 | 97.49 | 100.00 | ||||||||
| B. quintana | 98.86 | 98.63 | 98.41 | 98.71 | 97.73 | 97.79 | 100.00 | |||||||
| B. vinsonii | 99.39 | 99.39 | 99.24 | 99.85 | 98.33 | 98.25 | 98.55 | 100.00 | ||||||
| B. vinsonii subsp. berkhoffii | 99.16 | 99.16 | 99.31 | 99.62 | 98.17 | 98.41 | 98.71 | 99.62 | 100.00 | |||||
| B. taylorii | 99.09 | 99.09 | 99.02 | 99.70 | 98.25 | 98.25 | 98.79 | 99.54 | 99.39 | 100.00 | ||||
| B. grahamii | 99.02 | 99.16 | 99.39 | 99.62 | 98.17 | 98.25 | 98.55 | 99.62 | 99.54 | 99.54 | 100.00 | |||
| Brucella abortus | 94.89 | 94.89 | 95.58 | 95.50 | 94.74 | 95.20 | 94.74 | 95.50 | 95.58 | 95.50 | 95.50 | 100.00 | ||
| A. tumefaciens | 93.99 | 93.69 | 93.99 | 93.99 | 93.61 | 93.84 | 93.46 | 94.14 | 94.08 | 93.84 | 93.99 | 93.60 | 100.00 | |
| E. canis | 82.84 | 82.68 | 82.68 | 82.68 | 82.76 | 82.46 | 82.76 | 82.76 | 82.70 | 82.61 | 82.53 | 82.34 | 82.24 | 100.00 |
The phylogenetic dendrogram generated by the comparison of C-29 with 10 named Bartonella species and subspecies and B. abortus is shown in Fig. 3. In this figure, as well as in a dendrogram including 10 unnamed Bartonella strains, six brucellae, A. tumefaciens, and E. canis (data not shown), the bartonellae formed a monophyletic group, with B. clarridgeiae and B. bacilliformis exhibiting the highest degrees of divergence from the group. When the majority-rule consensus tree, derived from 100 trees constructed from the same input file, was used, C-29 was grouped with B. henselae in 86% of the trees; in the remaining 14 trees, C-29 was grouped with other bartonellae, but none of the groupings occurred more than 4% of the time.
FIG. 3.
Phylogenetic dendogram based on a neighbor-joining comparison of 1,330-nucleotide sequences of 16S rRNA genes. The scale bar at the bottom of the dendrogram represents a 1% difference. The strain designations and accession numbers for the sequences retrieved from GenBank are as follows: B. clarridgeiae CIP 104772, accession no. X97822; B. bacilliformis, accession no. M65249; B. henselae, accession no. M73229; B. quintana, accession no. U28268; B. grahamii V2, accession no. Z31349; B. elizabethae ATCC 49927, accession no. L01260; B. vinsonii subsp. berkhoffii G7464, accession no. U26258; B. taylorii M6, accession no. Z31350; B. doshiae R18, accession no. Z31351; B. vinsonii ATCC VR-152, accession no. L01259; and B. abortus 11-19, accession no. X13695.
DNA hybridization studies.
Levels of DNA relatedness were determined by hybridizing labeled DNA from the cat isolates C-29 and C-30, the type strain of B. henselae (ATCC 49882), and an additional strain of B. henselae (G6486) with unlabeled DNA from C-29, C-30, and 14 Bartonella species or subspecies (Table 2). (Not all combinations of reciprocal hybridization were performed.) The cat strains C-29 and C-30 showed average levels of relatedness of 97 and 100%, with 0.0 and 0.5% divergence, in optimal DNA reassociation reactions at 55°C. In stringent DNA reassociation reactions at 70°C, they were 98 and 100% related. Labeled DNA from the type strain of B. henselae ATCC 49882 showed levels of relatedness to the cat Bartonella isolate C-29 of 65% at 55°C, with 5.5% divergence, and 41% at 70°C, and its levels of relatedness to the cat Bartonella isolate C-30 were 61% at 55°C, with 5.0% divergence, and 31% at 70°C. Labeled DNA from B. henselae G6486 showed levels of relatedness to the cat Bartonella isolate C-29 of 79%, with 6.0% divergence at 55°C, and 61% at 70°C, and its levels of relatedness to the cat Bartonella isolate C-30 were 73%, with 5.5% divergence, at 55°C and 54% at 70°C. Using labeled C-29 DNA hybridized with unlabeled DNA from the type strain of B. henselae, C-29 showed levels of relatedness of 84% at 55°C, with 6.5% divergence, and 51% at 70°C. For C-30, the levels of relatedness to the type strain of B. henselae were 92%, with 6.0% divergence, at 55°C and 69% at 70°C. Based on DNA hybridization studies, our isolate C-29 was most closely related to B. henselae (range of levels of relatedness to the type strain and other strains, 65 to 84%, with 5 to 7% divergence, at 55°C). The levels of relatedness of C-29 and C-30 to B. quintana ATCC VR-358, B. vinsonii ATCC VR-152, and B. vinsonii subsp. berkhoffii ATCC 51672 ranged from 46 to 68% at 55°C. DNA from C-29 was least closely related (28 to 38%) to the type strains of B. grahamii, B. clarridgeiae, B. bacilliformis, and B. doshiae, as determined by DNA-DNA hybridization studies. Since no strains of B. taylorii, B. talpae, or B. peromysci are available (11a), it was not possible to include these species in the DNA hybridization studies.
TABLE 2.
DNA relatedness of C-29 and C-30 to other Bartonella species
| Source of unlabeled DNA | % Relatedness to labeled DNA from:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C-29
|
C-30
|
B. henselae ATCC 49882T
|
B. henselae G6486
|
|||||||||
| 55°C | Da | 70°C | 55°C | D | 70°C | 55°C | D | 70°C | 55°C | D | 70°C | |
| C-29 | 100 | 0.0 | 100 | 100 | 0.5 | 100 | 65 | 5.5 | 41 | 79 | 6.0 | 61 |
| C-30 | 97 | 0.0 | 98 | 100 | 0.0 | 100 | 61 | 5.0 | 31 | 73 | 5.5 | 54 |
| B. henselae ATCC 49882T | 84 | 6.5 | 51 | 92 | 6.0 | 69 | 100 | 0.0 | 100 | 100 | 0.5 | 100 |
| B. henselae G6486 | 78 | 6.0 | 54 | —b | — | — | 83 | 0.5 | 90 | 100 | 0.0 | 100 |
| B. henselae G6529 | 68 | 5.5 | 54 | — | — | — | 85 | 0.5 | 88 | — | — | — |
| B. henselae G8378 | 79 | 6.5 | 60 | — | — | — | 92 | 0.0 | 94 | — | — | — |
| B. henselae 88-712 | 83 | 7.0 | 62 | — | — | — | 100 | 0.5 | 100 | — | — | — |
| B. henselae G5691 | 84 | 5.0 | 48 | — | — | — | 95 | 0.5 | 95 | — | — | — |
| B. quintana ATCC VR-358T | 56 | 9.5 | 31 | 68 | 9.5 | 40 | 46 | 8.5 | 20 | 57 | 9.0 | 34 |
| B. vinsonii ATCC VR-152T | 55 | 10.5 | 29 | 67 | 10.0 | 36 | 50 | 8.0 | 26 | 63 | 8.5 | 43 |
| B. vinsonii subsp. berkhoffii ATCC 51672T | 46 | 12.0 | — | 57 | 12.0 | — | 42 | 10.5 | — | — | — | — |
| B. elizabethae ATCC 49927T | 53 | 13.0 | 21 | 63 | 12.5 | — | 46 | 11.0 | 21 | — | — | — |
| B. clarridgeiae ATCC 51734T | 31 | 12.5 | — | — | — | — | — | — | — | — | — | — |
| B. bacilliformis ATCC 35685T | 34 | 12.5 | — | 46 | 13.0 | — | 33 | 11.0 | — | — | — | — |
| B. grahamii NCTC 12860T | 28 | 11.5 | — | — | — | — | — | — | — | — | — | — |
| B. doshiae NCTC 12862T | 38 | 10.0 | — | — | — | — | — | — | — | — | — | — |
D, percent divergence at 55°C within related sequences, calculated to the nearest 0.5%.
—, not done.
DISCUSSION
The recovery of these two new, distinct Bartonella isolates from two pet cats provides further evidence of the diversity of Bartonella species that infect the domestic cat, Felis domesticus. To date, two species, B. henselae and B. clarridgeiae, have been isolated from domestic cats, and apparently B. clarridgeiae is cultivable from a higher percentage of European cats than cats in the United States (1, 11). Repetitive-element PCR on B. henselae isolates from cats also demonstrated diversity among B. henselae isolates (24), but all B. henselae isolates obtained to date have been closely related both phenotypically and genotypically. Recent data suggest that an even greater diversity of Bartonella species infect small mammals such as mice, moles, and voles in the United States and the United Kingdom (2, 18).
Different phenotypic characteristics distinguish isolates C-29 and C-30 from B. henselae feline isolates. B. henselae and B. clarridgeiae are readily isolated from feline blood cultures, and colonies are usually apparent only 3 days after plating on agar, in contrast to isolates from human blood and tissue, whose colonies do not appear until after a minimum of 8 days of incubation (16). We observed only tiny colonies after 14 days of incubation of primary cultures, and these initially were ascribed to cellular debris from lysed erythrocytes and leukocytes present after processing of the blood in lysis-centrifugation tubes. Our isolates were slow growers, appeared only as minute pinpoint colonies, and, in contrast to other B. henselae strains, produced only slightly larger colonies on subsequent passaging. Drawing on our extensive experience in isolation and cultivation of Bartonella species from cat blood, as well as from human blood and tissues, and of Bartonella type strains, C-29 apparently has the most-fastidious growth characteristics of any Bartonella species cultured to date.
Bartonella species have distinctive preferences for growth on different agar media, with B. henselae growing preferentially on heart infusion agar supplemented with 5% rabbit blood and B. quintana growing preferentially on chocolate agar (16). The C-29 and C-30 isolates were unique among our cat isolates because they did not grow on heart infusion agar supplemented with 5% rabbit blood but rather grew only on chocolate agar. They also failed to grow on Columbia or sheep blood agar. Although we found that these isolates represented 2 of the 25 Bartonella isolates (8%) recovered during documentation of B. henselae bacteremia in cats from the greater San Francisco Bay region, the prevalence of this new species remains unknown and will be difficult to quantitate because of the chocolate agar preference and extremely fastidious growth characteristics. Additionally, it is not known whether this new Bartonella species can infect humans.
The two isolates C-29 and C-30 were recovered from the blood of two kittens that had lived both outdoors and indoors on a farm with seven other cats; B. henselae was isolated from the blood of two of these seven cats, and the remaining five animals were culture negative. The relationship between infection of these kittens with this new species and their association with farm animals (one kitten lived primarily in the barn) is notable; the remainder of the 61 cats cultured in this study were urban or suburban, and those that were infected with a Bartonella species had B. henselae (14).
Although biochemical profiles cannot be used routinely and reliably for differentiation of Bartonella species because of the relatively inert nature of bartonellae, we found that C-29 and C-30 yielded a reproducibly distinct code for preformed enzymes when tested with the MicroScan Rapid Anaerobe Panel. The obtained code was similar, but not identical, to the ones reported for other Bartonella species (31). This observation may be of practical diagnostic value for laboratories attempting to distinguish among different Bartonella species.
PCR-RFLP analysis of the citrate synthase gene is commonly used to identify Bartonella isolates to the species level (21). The distinctive citrate synthase PCR-RFLP product observed for C-29 and C-30 provides additional evidence that C-29 and C-30 represent a distinct species. The pattern obtained by genomic RFLP analysis for our two cat isolates also was clearly different from those of the B. henselae and B. quintana type strains. The 16S rDNA sequence analysis confirmed that cat isolate C-29 is a member of the genus Bartonella, is most closely related to B. henselae, and is most distantly related to B. grahamii.
DNA-DNA hybridization remains the method of choice for defining a species. It has been recommended that a species be considered to consist of strains whose DNAs are 70% or more related under optimal DNA reassociation conditions and 55% or more related under stringent DNA reassociation conditions, with 5% or less divergence within related sequences (3, 29). From the results of our DNA-DNA hybridization study, we conclude that C-29 and C-30 represent the same species (average levels of relatedness to each other of 97 and 100%, with 0.0 and 0.5% divergence under optimal DNA reassociation reactions, at 55°C and of 98 and 100% at 70°C).
C-29 showed levels of relatedness to the type strain of B. henselae of 84%, with 6.5% divergence, at 55°C and 51% at 70°C, and for C-30 the levels of relatedness to the type strain of B. henselae were 92%, with 6.0% divergence, at 55°C and 69% at 70°C. Regardless of the high degree of relatedness to B. henselae determined at 55°C when using labeled DNA from C-29 and C-30, the degree of relatedness of C-29 and C-30 to B. henselae does not fulfill the species definition because the divergence is greater than 5%. The percent DNA relatedness was substantially lower in reciprocal reactions, in which labeled DNA from the type strain of B. henselae showed levels of relatedness to the cat Bartonella isolate C-29 of 65%, with 5.5% divergence, at 55°C and 41% at 70°C and to C-30 of 61%, with 5.0% divergence, at 55°C and 31% at 70°C. This type of nonreciprocity in relatedness values has been seen in strains from many genera (2a) and emphasizes the importance of performing reciprocal DNA relatedness determinations when studying strains that are close to the species definition. Nonreciprocity can result from differences in the genome sizes of the two species being compared.
In regard to divergence and relatedness under stringent conditions, our isolates C-29 and C-30 do not fulfill the strict criteria for belonging to the species B. henselae and thus represent a new species. This is even more evident when taking into consideration the relatedness and divergence in the reciprocal reactions. Also, the hybridization of labeled C-29 DNA to that of other B. henselae strains demonstrates that the species definition is not fulfilled with any of the five other B. henselae strains tested, again substantiating that C-29 represents a new species, distinct from B. henselae.
These data demonstrate that our two cat isolates represent a new species of Bartonella which is genotypically and phenotypically different from all other Bartonella isolates. The cat isolates C-29 and C-30 can be readily differentiated from the B. henselae type strain on the basis of their slower growth, smaller colony size, clear preference for growth on chocolate agar, unique code in the MicroScan Rapid Anaerobe Panel, distinct RFLP pattern for the citrate synthase gene as well as for genomic DNA, and the presence of six base pair changes in their 16S rDNA compared with that of the B. henselae type strain. C-29 thus fulfills the criteria for a new species: it is closely related to B. henselae but exhibits genetic and phenotypic divergence (29). A new species for these organisms is proposed and described below.
Description of Bartonella koehlerae sp. nov.
Bartonella koehlerae (koeh′ ler. ae. N. L. fem. adj. koehlerae), in honor of Jane E. Koehler, who was the first to isolate Bartonella species from bacillary angiomatosis lesions and whose studies of B. quintana and B. henselae isolates from human immunodeficiency virus-infected patients have contributed to an improved understanding of the molecular epidemiology, reservoirs, and vectors of Bartonella-associated disease in humans. Growth is optimal on chocolate agar, and primary colonies are observed after 14 days of incubation at 35°C in a CO2-enriched environment. The code for preformed enzymes obtained with a MicroScan Rapid Anaerobe Panel is 10073240. The new species shows patterns for citrate synthase PCR-RFLP and for genomic RFLP which are not previously described and differ from those of B. henselae. The 16S rRNA gene sequence of C-29 is distinguished from that of the B. henselae type strain by six base pair changes. The two strains thus far isolated were from healthy cats; to date, no feline or human pathogenicity has been attributed to this species. The type strain, C-29 (ATCC 700693), was recovered from the blood of a healthy kitten during a prevalence study of B. henselae in domestic cats in the greater San Francisco Bay area of northern California.
ACKNOWLEDGMENTS
We thank Carol Glaser, Barbara Lee, and the staff of the Evergreen Veterinary Clinic for their assistance with this study.
REFERENCES
- 1.Bergmans A M C, de Jong C M A, van Amerongen G, Schot C S, Schouls L M. Prevalence of Bartonella species in domestic cats in The Netherlands. J Clin Microbiol. 1997;35:2256–2261. doi: 10.1128/jcm.35.9.2256-2261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birtles R J, Harrison T G, Molyneux D H. Grahamella in small woodland mammals in the U.K.: isolation, prevalence and host specificity. Ann Trop Med Parasitol. 1994;88:317–327. doi: 10.1080/00034983.1994.11812872. [DOI] [PubMed] [Google Scholar]
- 2a.Brenner, D. J. Personal communication.
- 3.Brenner D J, McWhorter A C, Leete Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner D J, O’Connor S P, Hollis D G, Weaver R E, Steigerwalt A G. Molecular characterization and proposal of a neotype strain for Bartonella bacilliformis. J Clin Microbiol. 1991;29:1299–1302. doi: 10.1128/jcm.29.7.1299-1302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomel B B, Abbott R C, Kasten R W, Floyd-Hawkins K A, Kass P H, Glaser C A, Pedersen N C, Koehler J E. Bartonella henselae prevalence in domestic cats in California: risk factors and association between bacteremia and antibody titers. J Clin Microbiol. 1995;33:2445–2450. doi: 10.1128/jcm.33.9.2445-2450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomel B B, Kasten R W, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, Gurfield A N, Abbott R C, Pedersen N C, Koehler J E. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O’Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolan M J, Wong M T, Regnery R L, Jorgensen J H, Garcia M, Peters J, Drehner D. Syndrome of Rochalimaea henselae adenitis suggesting cat scratch disease. Ann Intern Med. 1993;118:331–336. doi: 10.7326/0003-4819-118-5-199303010-00002. [DOI] [PubMed] [Google Scholar]
- 10.Gray M W, Samkoff D, Cedergren R J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984;12:5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heller R, Artois M, Xemar V, De Briel D, Gehin H, Jaulhac B, Monteil H, Piemont Y. Prevalence of Bartonella henselae and Bartonella clarridgeiae in stray cats. J Clin Microbiol. 1997;35:1327–1331. doi: 10.1128/jcm.35.6.1327-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Holmes, B. Personal communication.
- 12.Jameson P, Greene C, Regnery R, Dryden M, Marks A, Brown J, Cooper J, Glaus B, Greene R. Prevalence of Bartonella henselae antibodies in pet cats throughout regions of North America. J Infect Dis. 1995;172:1145–1149. doi: 10.1093/infdis/172.4.1145. [DOI] [PubMed] [Google Scholar]
- 13.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 14.Koehler J E, Glaser C A, Tappero J W. Rochalimaea henselae infection. A new zoonosis with the domestic cat as reservoir. JAMA. 1994;271:531–535. doi: 10.1001/jama.271.7.531. [DOI] [PubMed] [Google Scholar]
- 15.Koehler J E, Quinn F D, Berger T G, LeBoit P E, Tappero J W. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N Engl J Med. 1992;327:1625–1631. doi: 10.1056/NEJM199212033272303. [DOI] [PubMed] [Google Scholar]
- 16.Koehler J E, Sanchez M A, Garrido C S, Whitfeld M J, Chen F M, Berger T G, Rodriguez-Barradas M C, LeBoit P E, Tappero J W. Molecular epidemiology of Bartonella infections in patients with bacillary angiomatosis-peliosis. N Engl J Med. 1997;337:1876–1883. doi: 10.1056/NEJM199712253372603. [DOI] [PubMed] [Google Scholar]
- 17.Kordick D L, Swaminathan B, Greene C E, Wilson K H, Whitney A M, O’Connor S, Hollis D G, Matar G M, Steigerwalt A G, Malcolm G B, Hayes P S, Hadfield T L, Breitschwerdt E B, Brenner D J. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int J Syst Bacteriol. 1996;46:704–709. doi: 10.1099/00207713-46-3-704. [DOI] [PubMed] [Google Scholar]
- 18.Kosoy M Y, Regnery R L, Tzianabos T, Marston E L, Jones D C, Green D, Maupin G O, Olson J G, Childs J E. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am J Trop Med Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 19.MacFaddin J F. Isolation-cultivation-maintenance of medical bacteria. Vol. 1. Baltimore, Md: Williams and Wilkins; 1985. [Google Scholar]
- 20.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 21.Regnery R L, Anderson B E, Clarridge III J E, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regnery R L, Olson J G, Perkins B A, Bibb W. Serological response to “Rochalimaea henselae” antigen in suspected cat-scratch disease. Lancet. 1992;339:1443–1445. doi: 10.1016/0140-6736(92)92032-b. [DOI] [PubMed] [Google Scholar]
- 23.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis: an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Barradas M C, Hamill R J, Houston E D, Georghiou P R, Clarridge J E, Regnery R L, Koehler J E. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J Clin Microbiol. 1995;33:1089–1093. doi: 10.1128/jcm.33.5.1089-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slater L N, Welch D F, Hensel D, Coody D W. A newly recognized fastidious gram-negative pathogen as a cause of fever and bacteremia. N Engl J Med. 1990;323:1587–1593. doi: 10.1056/NEJM199012063232303. [DOI] [PubMed] [Google Scholar]
- 28.Tappero J W, Mohle-Boetani J, Koehler J E, Swaminathan B, Berger T G, LeBoit P E, Smith L L, Wenger J D, Pinner R W, Kemper C A, et al. The epidemiology of bacillary angiomatosis and bacillary peliosis. JAMA. 1993;269:770–775. [PubMed] [Google Scholar]
- 29.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Trüper H G. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 30.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch D F, Hensel D M, Pickett D A, San Joaquin V H, Robinson A, Slater L N. Bacteremia due to Rochalimaea henselae in a child: practical identification of isolates in the clinical laboratory. J Clin Microbiol. 1993;31:2381–2386. doi: 10.1128/jcm.31.9.2381-2386.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch D F, Pickett D A, Slater L N, Steigerwalt A G, Brenner D J. Rochalimaea henselae sp. nov., a cause of septicemia, bacillary angiomatosis, and parenchymal bacillary peliosis. J Clin Microbiol. 1992;30:275–280. doi: 10.1128/jcm.30.2.275-280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1995. pp. 2.4.1–2.4.2. [Google Scholar]
- 34.Wilson K H, Blitchington R B, Greene R C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]