Abstract
Objective
Since a quantitative polymerase chain reaction (qPCR) assay targeting the E1 region of HPV genome is cost-effective/simple to perform, we evaluated the agreement between the Roche Diagnostics Linear Array (RDLA) genotyping test and qPCR-based E1 assay to detect HR-HPV genotypes that are included or not included in HPV vaccines and compared their accuracy to detect CIN 2+.
Methods
Study population included 257 African American (AA) and 266 Caucasian American (CA) diagnosed with intraepithelial neoplasia (CIN) grades ≤CIN 1 or ≥CIN 2 (CIN 2+) and tested for HPV by the RDLA and E1 assay. The concordance was determined using Gwet’s AC1. The calculated positive predictive value (PPV) and negative predictive value (NPV) of the two assays were used to determine their suitability to detect CIN lesions.
Results
Overall, the E1 assay showed substantial agreement with the RDLA assay to detect any HR-HPV genotype and the agreement was higher in women diagnosed with CIN 2+ than ≤CIN 1. The concordance was largely higher in Cas than in Aas. The NPV and PPV values to detect CIN lesions were similar between the two assays.
Conclusion
Utilization of the HPV E1 assay as a tool for CC screening could be a cost-effective approach that applies to both vaccinated and unvaccinated populations.
Keywords: HPV, Roche Linear Diagnostic Assay, E1 qPCR assay, cervical intraepithelial neoplasia, CIN
Introduction
Cervical cancer (CC) is the fourth most common cancer in women worldwide with an estimated 570,000 new cases, accounting for 6.9% of all new cases of cancer diagnosed in 2018.1,2 Approximately, 85% of CC cases occur in the developing world3 and in such countries, CC mortality is 18-fold greater than in resource rich developed countries.4 CC rates are expected to increase disproportionately in the coming decades because of the projected increase in the population, especially in developing countries.5 Infection with carcinogenic or high-risk human papillomaviruses (HR-HPVs), which are sexually transmitted, is the main causative factor for the development of CC as more than 90% of CCs are associated with HR-HPV infections, independent of other risk factors.6–9 Studies that used improved HPV testing procedures have established HPV as a causative agent for developing higher grades of cervical intraepithelial neoplasia (CIN 2+) that are precursor lesions for developing CC.10 Prophylactic vaccines against HR-HPVs are effective in preventing CIN lesions associated with HPV genotypes included in those vaccines if given prior to exposure to those genotypes. However, vaccinated females could subsequently be infected with at least six HR-HPV genotypes and eight probable HR-HPV genotypes that are not included in those vaccines and develop CIN lesions due to those HPVs. Therefore, identification of women infected with HR-HPVs or with HR-HPV associated CIN lesions is of critical importance for preventing CC in both vaccinated and non-vaccinated populations.
Currently, there are five FDA approved HPV tests; Hybrid Capture® 2 assay based on DNA-Probe-Hybrid immunoassay technique, Cervista®HPV HR and Cervista™ HPV 16/18 based on DNA-probe technology, Cobas® 4800 HPV test based on PCR, APTIMA® HPV assay based on RNA capture and amplification of HPV RNA and BD Onclarity assay based on PCR.11 The target regions for these tests are L1, E6 or E7 genes of the HPV and the sensitivity of these assays for the detection of CIN 2 or CIN 3 ranged from 55% to 100% based on several studies.12–15 Peer-reviewed published data support >90% sensitivity for CIN 3 lesions for other PCR-based tests such as Roche Diagnostics Linear Array (RDLA) validated against the FDA approved Hybrid Capture® 2 (HC2) assay used by large epidemiological studies funded by the National Cancer Institute.16 Even though these HPV tests have demonstrated the potential for successful HPV screening in developed countries, the technical, financial, and logistic requirements of these tests are beyond the capacity of health programs as a screening tool in most low and middle income developing countries where CC represents the largest health burden. Therefore, providing a sustainable low-cost and simple to perform cervical screening test may contribute to earlier detection and a decrease in CC-related deaths in those countries. We believe that these limitations could be overcome by using a quantitative polymerase chain reaction (qPCR) assay targeting the E1 region of HPV genome which is highly conserved in a large number of HR-HPV genotypes.17 The E1 consensus region is approximately 150–200 bp in length and is much more amenable to qPCR.18 Further, since HPV requires the E1 protein for replication as it recognizes the origin of replication,17,19 E1 base testing is likely to reflect the active HPV status in a given sample.
Based on this background, the purpose of the study was to compare the qPCR assay based HPV test results with the RDLA test that targets the E1 region of 13 HR-HPV genotypes, namely 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68 by A) determining the degree of agreement between the E1 assay and the RDLA assay for detecting cervical samples infected with any of those HR-HPV genotypes or HR-HPV genotypes included or not included in the quadrivalent HPV vaccine (qHPV) (ie, HPV 16/18), samples tested negative for HR-HPV genotypes included in the qHPV but tested positive for other HR-HPV genotypes, tested positive for HR-HPV genotypes included or not included in the 9-valent HPV vaccine (9VHPV) (16, 18, 31, 33, 45, 52, and 58) and tested negative for HR-HPV genotypes included in the 9VHPV but tested positive for other HR-HPV genotypes in 523 women, and stratified by race and by CIN status (<CIN 1, CIN 1, CIN 2, CIN 3) B) compare the positive predictive value (PPV) and the negative predictive value (NPV) for the detection of histologically confirmed CIN 2 or CIN 3 lesions associated with any HR-HPV or with specific HR-HPVs that are included or not included in the vaccines, irrespective of race and stratified by race.
Methods
Study Population
We randomly selected a subset of women (n = 523, 257 African American [AA], 266 Caucasian American [CA]) from 2295 women enrolled by two National Cancer Institute (NCI) funded R01 grants (R01CA105448 and R01CA102489) in order to obtain a representative sample comparable to that of the parent study with regard to the distribution of HPV genotypes and CIN status. All women were tested by the cytology-based standard Pap test in clinics of the Health Departments in the Jefferson County and surrounding counties in Alabama and were diagnosed with varying severity of abnormal cervical cells. All women were referred to the University of Alabama at Birmingham (UAB) for further examination by colposcopy and directed biopsies. Women were 19–50 years old, non-pregnant at the time of study enrollment, had no history of CC or other cancer of the lower genital tract and no history of hysterectomy or destructive therapy of the cervix. The distribution of CIN diagnosis of the population was the following: 167 women were diagnosed with CIN 2+ (cases, including CIN 2 [n = 93], CIN 3 [n = 74]) and 363 women were diagnosed with ≤ CIN 1 (non-cases, including normal cervical epithelium [n = 16], HPV cytopathic effect [HCE, n = 30], reactive nuclear enlargement [RNE, n = 37] or CIN 1 [n = 273]). All study participants provided informed consent and the study protocols complied with the Helsinki Declaration. The study was approved by the University of Alabama at Birmingham Institutional Review Board.
At the UAB colposcopy clinic, prior to obtaining a biopsy specimen, exfoliated cervical cells were collected with a cervical brush and immediately rinsed in 10 mL of PBS. These samples were kept cold in a refrigerator and transported on ice within 2 hours of collection to the laboratory of the principal investigator (PI). In the laboratory, cervical cell samples were centrifuged, and the resulting pellets were stored at −80°C until HPV detection assays were performed using the RDLA test and a qPCR-based HPV test that targeted the E1 region of any one of the 13 HR-HPV genotypes.
Detection of HR-HPV
DNA Extraction
Exfoliated cervical cells were resuspended in 200 mL of 1x PBS (Phosphate Buffer Saline). DNA was extracted using the Qiagen Min Elute Kit Media kit (Qiagen, Inc.) following the manufacturer’s instructions.
HPV Detection by the RDLA
The RDLA was performed according to the manufacturer’s instructions by a research associate trained by personnel from the Roche Diagnostics. Briefly, target DNA was amplified by PCR using the PGMY09/11 L1 consensus primer system. The human beta-globin DNA served as an internal control for adequate sample cellularity for DNA extraction. Detection and HPV genotyping were achieved using a reverse line-blot method, and this test included probes for 37 anogenital HPV genotypes [6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73 (MM9), 81, 82 (MM4), 83 (MM7), 84 (MM8), IS39, and CP6108]. Those 37 genotypes included 13 HR-HPV genotypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68), 7 probable HR-HPV genotypes (HPV 26, 53, 66, 67, 70, 73 and 82) and 17 LR-HPV genotypes. The cost of this assay, depending on the volume of tests performed could be as high as > $75.
HPV Detection by the qPCR E1 Assay
The DNA was diluted to 5ng/µl and 2µl of the diluted DNA was used for the E1 qPCR assay. The qPCR assay used primers that targeted a 100 nucleotide fragment across a region of E1 that has a high degree of homology to high-risk HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68. The forward primers and the reverse primers targeting the E1 region were CCTATAGTACATTTAAAAGGTG and CNTGTCCAATGCCAGGTAGATG, respectively,20 both at a concentration of 2 µM. Single reactions were run on the Roche Light Cycler 480 using Light Cycler 480 SYBR Green I Master with the following parameters: 10µl reactions (5µl SYBR green master mix, 2 µl of 2µm Primer mix [forward and reverse] and 1µl water) were heated at 95°C for 15 minutes to activate the enzyme. This was followed by 40 cycles of 94°C for 60 seconds and 60°C for 90 seconds with signal acquisition. Beta-globin DNA was included in each run of a plate with the E1 target. The threshold cut point of the Ct value obtained for the detection of HR-HPVs was determined by estimating the area under the curve of the receiver operating curve (ROC) considering RDLA as the standard HPV test. All values above and below the optimal threshold cut point value indicative of being HR-HPV positive with the RDLA were considered as negative or positive for HR-HPVs, respectively, using the E1 assay. The cost of this assay (<$10) varies less with the number of assays performed.
Statistical Analysis
Descriptive statistics were used to describe the study population. We categorized women as positive for any HR-HPV genotype, HR-HPV genotypes included in the qHPV, HR-HPVs included in the 9VHPV, negative for HR-HPVs included the qHPV but positive for all other HR-HPVs, negative for HR-HPVs in the 9VHPV but positive for other HR-HPVs included in the RDLA assay. The number and percentage of women positive for the E1 assay in each group in the entire population and stratified by race, lesion status and by race and by lesion status were computed. The crude concordance rate and the Gwet’s agreement coefficient (AC1) with 95% confidence intervals (CIs) were calculated to estimate the concordance between the E1 and the RDLA tests for the detection of HPV genotype groups stated above. The following classification documented by the Cohen’s Kappa coefficient was used to interpret the degree of agreement of AC1 as <0 no agreement; 0.01–0.2 slight agreement; 0.21–0.40 fair agreement; 0.41–0.60 moderate agreement; 0.61 to 0.80 substantial agreement and 0.8–1.0 almost perfect agreement.21 We also compared the PPV and NPV for the detection of CIN 2 and CIN 3 by HR-HPVs detected by the RDLA and the E1 assay in the entire population and stratified by race. The interpretation of PPV and NPV was based on a preset categorization scheme (≥90 high, ≥80-<90 moderately high, ≥70-<80 moderate, <70 relatively low).
Results
The study population consisted of 49% of AAs and 51% of CAs. Of the 523 women, 402 (76%) women were positive for any HR-HPV using the RDLA test; 50% of those tested were AAs and 50% were CAs. Forty six percent of those tested positive for HR-HPV (n = 190) were positive for HPV 16 or 18 (23% of them were positive for HPV 16 or 18 only and 24% positive for HPV 16 or 18 and one or more of other HR-HPVs). Eighty one percent of those tested positive for HR-HPVs (n = 327) were positive for HR-HPVs included in the 9VHPV (55% for any one of the HR-HPVs in the 9VHPV and 26% for HR-HPVs in the 9VHPV and one or more of the other HR-HPVs). The cut point derived by the ROC curve for the E1 Ct value was 32.375. Samples with E1 Ct values <32.375 and ≥32.375 were considered positive or negative, respectively, for any HR-HPV or specific HR-HPV genotypes.
Table 1 shows the crude concordance and Gwet’s AC1 agreement between the RDLA and E1 assay for the detection of any HR-HPV or HR-HPVs included/not included in vaccines, irrespective of race and by race. Based on the Gwet’s AC1 agreement, the E1 assay showed substantial agreement with the RDLA assay for detecting any HR-HPV and HPV 16/18 genotypes. The agreement was moderate rather than substantial for detecting other specific HR-HPV groups except for some HPV genotype groups in CAs.
Table 1.
The Concordance Between the RDLA and E1 Assay for the Detection of Vaccine and Non-Vaccine HPV Types, Irrespective of Race and by Race
| HPV Groups | Race | Number of Study Participants | HR-HPV Status (RDLA/E1) | Crude Concordance | Gwet’s AC1 Agreement | |||
|---|---|---|---|---|---|---|---|---|
| +/+ | -/- | +/- | -/+ | |||||
| Any HR-HPV | ALL | 523 | 316 | 98 | 86 | 23 | 0.79 (0.76–0.83) | 0.65 (0.58–0.71) |
| AA | 257 | 152 | 42 | 49 | 14 | 0.76 (0.70–0.81) | 0.59 (0.48–0.69) | |
| CA | 266 | 164 | 56 | 37 | 9 | 0.83 (0.78–0.87) | 0.70 (0.62–0.79) | |
| HPV 16 or 18 positive | ALL | 212 | 74 | 98 | 17 | 23 | 0.81 (0.76–0.86) | 0.63 (0.52–0.73) |
| AA | 87 | 23 | 42 | 8 | 14 | 0.75 (0.65–0.84) | 0.52 (0.33–0.70) | |
| CA | 125 | 51 | 56 | 9 | 9 | 0.86 (0.79–0.92) | 0.71 (0.59–0.84) | |
| HPV 16 or 18 negative | ALL | 333 | 149 | 98 | 63 | 23 | 0.74 (0.70–0.79) | 0.50 (0.40–0.59) |
| AA | 179 | 87 | 42 | 36 | 14 | 0.72 (0.65–0.79) | 0.48 (0.34–0.61) | |
| CA | 154 | 62 | 56 | 27 | 9 | 0.77 (0.70–0.83) | 0.53 (0.40–0.67) | |
| Positive for 9 valent HPV types | ALL | 341 | 167 | 98 | 53 | 23 | 0.78 (0.73–0.82) | 0.57 (0.48–0.66) |
| AA | 158 | 71 | 42 | 31 | 14 | 0.72 (0.64–0.79) | 0.45 (0.31–0.59) | |
| CA | 183 | 96 | 56 | 22 | 9 | 0.83 (0.78–0.89) | 0.68 (0.57–0.78) | |
| Negative for 9 valent HPV types | ALL | 196 | 56 | 98 | 19 | 23 | 0.79 (0.73–0.64) | 0.59 (0.48–0.71) |
| AA | 95 | 33 | 42 | 6 | 14 | 0.79 (0.71–0.87) | 0.58 (0.42–0.75) | |
| CA | 101 | 23 | 56 | 13 | 9 | 0.78 (0.70–0.86) | 0.61 (0.45–0.77) | |
Notes: Degree of agreement categorization <0-no agreement, 0.01–0.2 slight agreement, 0.21–0.40 fair agreement, 0.41–0.60 moderate agreement, 0.61 to 0.80 substantial agreement and 0.8–1.0 almost perfect agreement.
Abbreviations: RDLA, Roche Diagnostics Linear Array; HR-HPV, high-risk human papillomavirus; AA, African American; CA, Caucasian American.
Table 2 shows the concordance between RDLA and E1 assay for detection of any HR-HPVs or HR-HPVs included/not included in vaccines stratified by the severity of histological diagnosis of CIN 2 or 3 (cases) and <CIN 1 and CIN 1 (non-cases). The agreement between the E1 and RDLA assay for detecting any HR-HPV was substantial to almost perfect in women diagnosed with CIN 2 or 3 for all HPV genotype groups, while it was moderate among those diagnosed with either <CIN 1 or CIN 1.
Table 2.
The Concordance Between the RDLA and E1 Assay for Detection of Vaccine and Non-Vaccine HPV Types by Case Status
| HPV Groups | Case Status | Number of Study Participants | HR-HPV Results (RDLA/E1) | Crude Concordance | Gwet’s AC1 Agreement | |||
|---|---|---|---|---|---|---|---|---|
| +/+ | -/- | +/- | -/+ | |||||
| Any HR-HPV | <CIN 1 | 83 | 32 | 28 | 18 | 5 | 0.72 (0.63–0.82) | 0.45 (0.25–0.64) |
| CIN 1 | 273 | 144 | 65 | 50 | 14 | 0.76 (0.71–0.80) | 0.54 (0.45–0.63) | |
| CIN 2 | 93 | 80 | 4 | 7 | 2 | 0.90 (0.84–0.96) | 0.88 (0.80–0.96) | |
| CIN 3 | 74 | 60 | 1 | 11 | 2 | 0.82 (0.74–0.91) | 0.79 (0.66–0.91) | |
| HPV 16 or 18 positive | <CIN 1 | 46 | 9 | 28 | 4 | 5 | 0.80 (0.69–0.92) | 0.67 (0.44–0.89) |
| CIN 1 | 119 | 32 | 65 | 8 | 14 | 0.82 (0.75–0.89) | 0.66 (0.52–0.78) | |
| CIN 2 | 21 | 13 | 4 | 2 | 2 | 0.81 (0.63–0.99) | 0.68 (0.34–1.0) | |
| CIN 3 | 26 | 20 | 1 | 3 | 2 | 0.81 (0.65–97) | 0.75 (0.51–0.99) | |
| HPV 16 or 18 negative | < CIN 1 | 63 | 17 | 28 | 13 | 5 | 0.71 (0.60–0.83) | 0.45 (0.22–0.68) |
| CIN 1 | 189 | 70 | 65 | 40 | 14 | 0.71 (0.65–0.78) | 0.43 (0.30–0.56) | |
| CIN 2 | 53 | 42 | 4 | 5 | 2 | 0.87 (0.78–0.96) | 0.83 (0.69–0.96) | |
| CIN 3 | 28 | 20 | 1 | 5 | 2 | 0.75 (0.58–0.92) | 0.66 (0.38–0.94) | |
| Positive for 9 valent HPV types | < CIN 1 | 62 | 19 | 28 | 10 | 5 | 0.76 (0.65–0.87) | 0.53 (0.31–0.74) |
| CIN 1 | 184 | 76 | 65 | 29 | 14 | 0.77 (0.71–0.83) | 0.53 (0.41–0.66)) | |
| CIN 2 | 48 | 37 | 4 | 5 | 2 | 0.85 (0.75–0.96) | 0.80 (0.65–0.96) | |
| CIN 3 | 47 | 35 | 1 | 9 | 2 | 0.77 (0.64–0.89) | 0.69 (0.49–0.89) | |
| Negative for 9 valent HPV types | < CIN1 | 42 | 4 | 28 | 5 | 5 | 0.76 (0.63–0.90) | 0.64 (0.40–0.88) |
| CIN 1 | 118 | 28 | 65 | 11 | 14 | 0.79 (0.71–0.86) | 0.61 (0.47–0.76) | |
| CIN 2 | 24 | 16 | 4 | 2 | 2 | 0.83 (0.68–0.99) | 0.73 (0.45–1.0) | |
| CIN 3 | 12 | 8 | 1 | 1 | 2 | 0.75 (0.48–1.0) | 0.63 (0.13–1.0) | |
Notes: Degree of agreement categorization <0-no agreement, 0.01–0.2 slight agreement, 0.21–0.40 fair agreement, 0.41–0.60 moderate agreement, 0.61 to 0.80 substantial agreement and 0.8–1.0 almost perfect agreement.
Abbreviations: RDLA, Roche Diagnostics Linear Array; HR-HPV, high-risk human papillomavirus; CIN 1, 2, 3, cervical intraepithelial neoplasia grades.
Table 3 shows the concordance by lesion status and by race. Because of the smaller numbers of lesions, we computed the concordance after combining <CIN 1 and CIN 1 (≤CIN 1) and CIN 2 and CIN 3 (CIN 2+). The results showed that concordance was stronger (substantial to almost perfect) in CAs than AAs except for lesions not associated with HPV genotypes included in 9V HPV.
Table 3.
Concordance Between RDLA and E1 Assay for Detection of Vaccine and Non-Vaccine HPV Types by Case Status and Race
| HPV Groups | Race | Case Status | Number of Study Participants | HR-HPV Results (RDLA/E1) | Crude Concordance | Gwet’s AC1 Agreement | |||
|---|---|---|---|---|---|---|---|---|---|
| +/+ | -/- | +/- | -/+ | ||||||
| Any HR-HPV | AA | ≤ CIN 1 | 180 | 89 | 42 | 37 | 12 | 0.73 (0.66–0.79) | 0.49 (0.36–0.62) |
| CA | ≤ CIN 1 | 176 | 87 | 51 | 31 | 7 | 0.78 (0.72–0.85) | 0.59 (0.46–0.71) | |
| AA | CIN 2+ | 77 | 63 | 0 | 12 | 2 | 0.82 (0.73–0.91) | 0.78 (0.66–0.91) | |
| CA | CIN 2+ | 90 | 77 | 5 | 6 | 2 | 0.91 (0.85–0.97) | 0.89 (0.81–0.97) | |
| Positive for HPV 16 or 18 | AA | ≤ CIN 1 | 72 | 14 | 42 | 4 | 12 | 0.78 (0.68–0.88) | 0.61 (0.43–0.80) |
| CA | ≤ CIN 1 | 93 | 27 | 51 | 8 | 7 | 0.84 (0.76–0.91) | 0.70 (0.55–0.85) | |
| AA | CIN 2+ | 15 | 9 | 0 | 4 | 2 | 0.60 (0.33–0.87) | 0.41 (−0.13–0.95) | |
| CA | CIN 2+ | 32 | 24 | 5 | 1 | 2 | 0.91 (0.80–1.0) | 0.86 (0.70–1.0) | |
| HPV 16 or 18 negative | AA | ≤ CIN 1 | 134 | 50 | 42 | 30 | 12 | 0.69 (0.61–0.77) | 0.38 (0.22–0.53) |
| CA | ≤ CIN 1 | 118 | 37 | 51 | 23 | 7 | 0.74 (0.65–0.83) | 0.48 (0.29–0.66) | |
| AA | CIN 2+ | 45 | 37 | 0 | 6 | 2 | 0.82 (0.71–0.94) | 0.79 (0.63–0.95) | |
| CA | CIN 2+ | 36 | 25 | 5 | 4 | 2 | 0.83 (0.71–0.96) | 0.75 (0.53–0.96) | |
| Positive for HR-HPV types included in 9VHPV vaccines | AA | ≤ CIN 1 | 113 | 39 | 42 | 20 | 12 | 0.72 (0.63–0.80) | 0.43 (0.27–0.60) |
| CA | ≤ CIN 1 | 133 | 56 | 51 | 19 | 7 | 0.75 (0.67–0.83) | 0.50 (0.34–0.66) | |
| AA | CIN 2+ | 45 | 32 | 0 | 11 | 2 | 0.71 (0.58–0.85) | 0.62 (0.39–0.85) | |
| CA | CIN 2+ | 50 | 40 | 5 | 3 | 2 | 0.90 (0.82–0.99) | 0.87 (0.74–0.99) | |
| Negative for HR-HPV types included in 9V HPV vaccines | AA | ≤ CIN 1 | 80 | 20 | 42 | 6 | 12 | 0.78 (0.68–0.87) | 0.58 (0.40–0.77) |
| CA | ≤ CIN 1 | 80 | 12 | 51 | 10 | 7 | 0.79 (0.70–0.88) | 0.66 (0.49–0.82) | |
| AA | CIN 2+ | 15 | 13 | 0 | 0 | 2 | 0.87 (0.68–1.0) | 0.85 (0.60–1.0) | |
| CA | CIN 2+ | 21 | 11 | 5 | 3 | 2 | 0.76 (0.57–0.96) | 0.56 (0.18–0.95) | |
Notes: Degree of agreement categorization <0-no agreement, 0.01–0.2 slight agreement, 0.21–0.40 fair agreement, 0.41–0.60 moderate agreement, 0.61 to 0.80 substantial agreement and 0.8–1.0 almost perfect agreement.
Abbreviations: RDLA, Roche Diagnostics Linear Array; HR-HPV, high-risk human papillomavirus; AA, African American; CA, Caucasian American; CIN 1, 2, 3, cervical intraepithelial neoplasia grades.
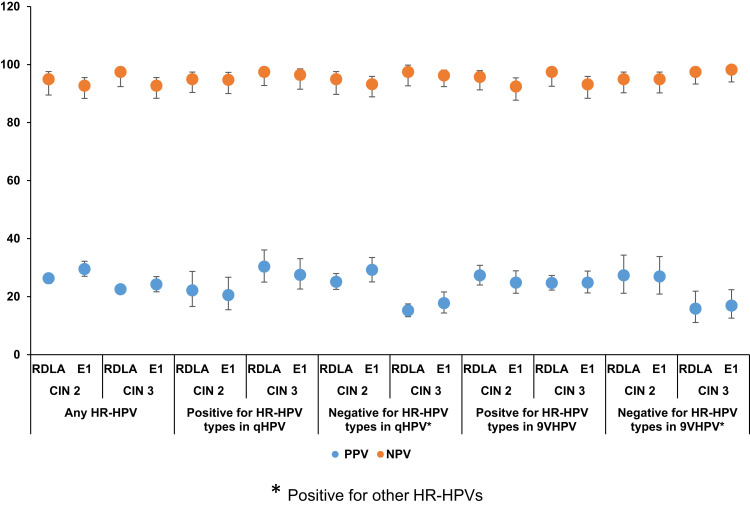
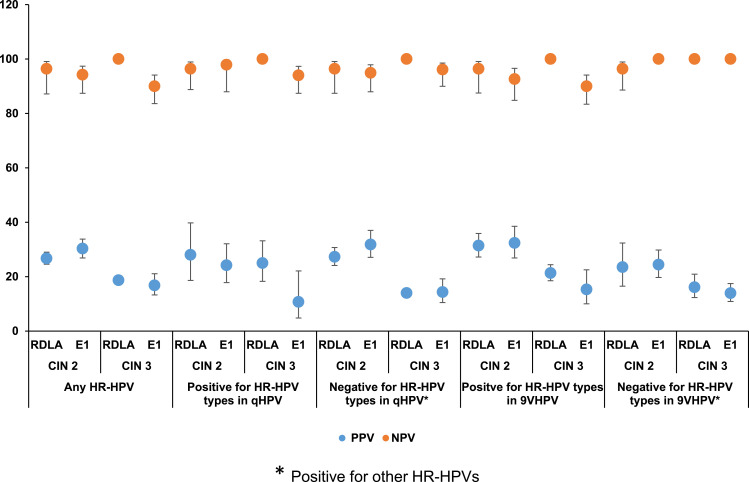
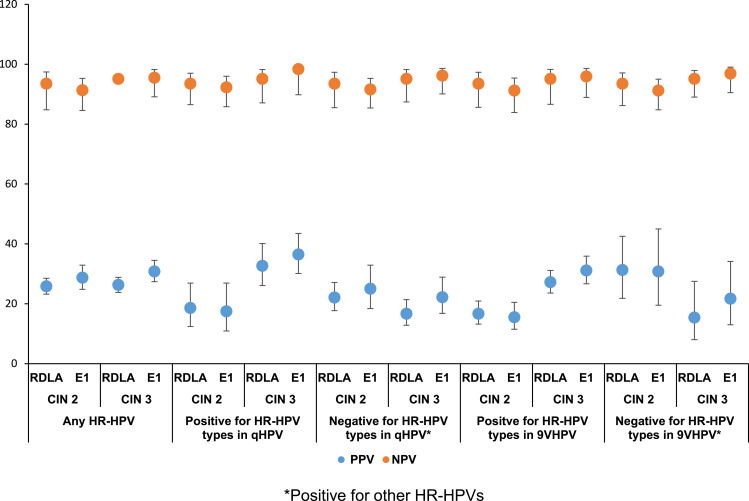
Figure 1 shows the comparison of the PPV and NPV of the RDLA and E1 assay results for the detection of histologically confirmed CIN 2 and CIN 3 lesions associated with any HR-HPV or with specific HR-HPVs included or not included in the vaccines, irrespective of the race. With regard to excluding the possibility of having underlying CIN lesions, the E1 assay, similar to the RDLA assay showed ≥90% NPV for CIN 2 or 3 lesions associated with any HR-HPV or with specific HR-HPVs included or not included in the vaccines. The PPV for detecting CIN 2 and CIN 3 lesions associated with any HR-HPV or specific HPV genotypes was also similar between the RDLA and the E1 assay, ie, all values below 40%. Figures 2 and 3 show the comparison of the PPV and NPV of the RDLA and E1 assay results for the detection of histologically confirmed CIN 2 and CIN 3 lesions with any HR-HPV or with specific HR-HPVs included or not included in the vaccines in AAs and CAs, respectively. In both races, similar to the RDLA assay, E1 assay had ≥90% NPV value for excluding the possibility of CIN 2 or 3 lesions associated with any HR-HPV or with specific HR-HPVs included or not included in the vaccines. The PPV for detection of CIN 2 or CIN 3 was also similar between the E1 and RDLA assay for both races, ie, all values below 40%.
Figure 1.
The comparison of the PPV and NPV of the RDLA and E1 assay results for the detection of CIN 2 and CIN 3 lesions associated with any HR-HPV or with specific HR-HPVs*.
Figure 2.
The comparison of the PPV and NPV of the RDLA and E1 assay results for the detection of CIN 2 and CIN 3 lesions associated with any HR-HPV or with specific HR-HPVs* in African Americans.
Figure 3.
The comparison of the PPV and NPV of the RDLA and E1 assay results for the detection of CIN 2 and CIN 3 lesions associated with any HR-HPV or with specific HR-HPVs* in Caucasian Americans.
Discussion
For over 50 years, the Pap test has been the gold standard for CC screening because of its profound effect on lowering CC mortality in nations that have adopted screening programs based on this test.22–24 However, recent scientific evidence illustrates that HPV testing is more sensitive than the Pap test. Therefore, WHO recommends implementation of primary HPV screening in countries where Pap-based screening programs do not exist or are ineffective.25 However, most resource-limited countries are unable to implement currently FDA approved HPV testing for CC screening. This is of concern, as targeted referral of women infected with HR-HPVs for further care will play a major role in reducing the risk of developing this preventable cancer, especially in many regions of the world with high rates of CC due to limited resources to implement other preventive measures such as prophylactic HPV vaccines.26 In addition to having lower HPV vaccine availability and lower acceptance of it in developing countries, millions of women are already infected with HPVs that are highly prevalent and uncontrolled, leaving those women with no opportunity for such preventive approaches. Therapeutic HPV vaccines present far more challenges than prophylactic HPV vaccines and have not reached adequate efficacy to be licensed in clinical practice.27
In developed countries where HPV vaccines are available, those are effective in preventing CIN or CC due to HPV genotypes included in the vaccines if given prior to exposure to such genotypes.28 However, studies have reported that infection with other HR-HPV genotypes have the same risk of developing CIN 2+ or CC.29,30 We also have documented that lesions that may develop due to HR-HPV genotypes that are not included in HPV vaccines are likely to have similar malignant potential to that of lesions associated with HR-HPVs included in both qHPV bivalent and 9VHPV vaccines,31 suggesting that continuation of screening programs is essential to manage the risk CC in the post-vaccine era. To reduce the added cost of continued screening, developed countries can also benefit from a cost-effective and simple HPV assay that can be performed in any laboratory rather than in specialized laboratories. We expect that the E1 qPCR assay will fulfil this need as it is not only cost effective and simple to perform but also has the ability to identify women infected with any one of the 13 HR-HPV genotypes irrespective of whether those are included or not included in HPV vaccines and their relationship to CIN status.
In our study, we compared the E1 assay with the RDLA assay that has been shown to have >80% and >65% agreement with the FDA approved Cobas HPV assay and HC2 assay, respectively.32 We observed that overall, the E1 assay was in substantial agreement with the RDLA assay. We, however, noted that there were differences in the degree of agreement by the severity of CIN lesion status and by race. The reasons for those differences are unclear but could be due to factors such as differences in the HPV genotypes, viral load or the integration status of the HPVs that could vary by the severity of lesion status or by race.27,33,34 Despite those differences, the NPV to exclude the possibility of having underlying CIN lesions caused by any HPV genotype, included or not included in HPV vaccines, was highly satisfactory (≥90%) in both AA and CA women. Therefore, the utilization of the HPV E1 assay as a tool for CC screening could be a cost-effective approach that applies to both vaccinated and unvaccinated populations comprised varying races or ethnicities in developed and underdeveloped countries.
One of the main strengths that adds validity to our results is that all women in our study have histological diagnoses rather than cytological diagnoses that lead to misclassification of women for the severity of CIN lesions. Also, the RDLA used as the standard assay to compare with E1 assay results had similar performance to that of FDA approved Cobas HPV assay and HC2 assay. Further, since we had access to a larger population of women, we were able to report our results based on specific HPV genotypes that were included or not included in HPV vaccines, allowing us to report applicability of the E1 assay in both vaccinated and unvaccinated women. Since this is the first study that took this approach, however, replication of these findings in other populations is necessary to increase the scientific credibility and its wider applications in CC screening programs.
We conclude that overall, the E1 assay showed substantial agreement with the RDLA assay to detect any HR-HPV genotype group and the agreement was higher in women diagnosed with CIN 2+ than ≤ CIN 1. The NPV and PPV values to detect CIN lesions were also similar between the two assays.
Acknowledgments
This study was supported by R01 105448 and R01 102489 (National Cancer Institute) and T37-MD001448 (Minority Health Research Training grant, National Institute on Minority Health and Health Disparities). The authors thank Nuzhat Rahman Siddiqui and Michelle M Chambers for their excellent technical support.
Highlights
HPV assay targeting the E1 region is cost effective and easier to perform than other HPV assays.
Overall, the E1 assay is in substantial agreement with the RDLA assay to detect HPV.
The E1 assay has similar NPVs and PPVs to that of the RDLA assay to detect CIN.
The HPV E1 assay is applicable in both vaccinated and unvaccinated populations.
Cost-effectiveness of the E1 assay will improve CC screening rate.
Disclosure
The authors report no conflicts of interest for this work and have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8(2):e191–e203. doi: 10.1016/S2214-109X(21)00554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worldwide cancer data- Global cancer statistics for the most common cancers. Available from: https://www.wcrf.org/dietandcancer/cancer-trends/worldwide-cancer-data. Accessed December 10, 2020.
- 3.LaVigne AW, Triedman SA, Randall TC, et al. Cervical cancer in low and middle income countries: addressing barriers to radiotherapy delivery. Gynecol Oncol Rep. 2017;22:16–20. doi: 10.1016/j.gore.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012: estimated cancer incidence, mortality and prevalence worldwide in 2012 v1.0. International Agency for Research on Cancer; 2014. Available from: http://globocan.iarc.fr. Accessed July 18.
- 5.Sankarnarayanan R, Bhudhuk AM, Rajkumar R. Effective screening programmes for cervical cancer in low- and middle-income developing countries. Bull World Health Organ. 2001;79(10):954–962. [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch FX, Manos MM, Muñoz N, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) study group. J Natl Cancer Inst. 1995;87(11):796–802. doi: 10.1093/jnci/87.11.796 [DOI] [PubMed] [Google Scholar]
- 7.Franco EL, Rohan TE, Villa LL. Epidemiologic evidence and human papillomavirus infection as a necessary cause of cervical cancer. J Natl Cancer Inst. 1999;91(6):506–511. doi: 10.1093/jnci/91.6.506 [DOI] [PubMed] [Google Scholar]
- 8.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi: [DOI] [PubMed] [Google Scholar]
- 9.Stark E.Human papillomaviruses: IARC Monographs on the evaluation of carcinogenic risks to humans. IARC Monogr Eval Carcinog Risks Hum. 2007;90:1–636. [PMC free article] [PubMed] [Google Scholar]
- 10.Kovachev SM, Slavov VD. Causative relations between human papilloma virus infection and cervical intraepithelial neoplasia. Biotechnol Biotechnol Equip. 2016;30(3):558–561. doi: 10.1080/13102818.2016.1159922 [DOI] [Google Scholar]
- 11.Salazar KL, Duhon DJ, Olsen R, et al. A review of the FDA-approved molecular testing platforms for human papillomavirus. J Am Soc Cytopathol. 2019;8(5):284–292. doi: 10.1016/j.jasc.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 12.Rebolj M, Bonde J, Ejegod D, et al. A daunting challenge: human Papillomavirus assays and cytology in primary cervical screening of women below age 30 years. Eur J Cancer. 2015;51(11):1456–1466. doi: 10.1016/j.ejca.2015.04.012 [DOI] [PubMed] [Google Scholar]
- 13.Einstein MH, Martens MG, Garcia FA, et al. Clinical validation of the Cervista HPV HR and 16/18 genotyping tests for use in women with ASC-US cytology. Gynecol Oncol. 2010;118(2):116–122. doi: 10.1016/j.ygyno.2010.04.013 [DOI] [PubMed] [Google Scholar]
- 14.Szarewski A, Ambroisine L, Cadman L, et al. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3033–3042. doi: 10.1158/1055-9965.EPI-08-0508 [DOI] [PubMed] [Google Scholar]
- 15.Cuzick J, Cadman L, Mesher D, et al. Comparing the performance of six human papillomavirus tests in a screening population. Br J Cancer. 2013;108(4):908–913. doi: 10.1038/bjc.2013.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gravitt PE, Schiffman M, Solomon D, et al. A comparison of linear array and hybrid capture 2 for detection of carcinogenic human papillomavirus and cervical precancer in ASCUS-LSIL triage study. Cancer Epidemiol Biomarkers Prev. 2008;17(5):1248–1254. doi: 10.1158/1055-9965.EPI-07-2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergvall M, Melendy T, Archambault J. The E1 proteins. Virology. 2013;445(1–2):35–56. doi: 10.1016/j.virol.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seaman WT, Andrews E, Couch M, et al. Detection and quantitation of HPV in genital and oral tissues and fluids by real time PCR. Virol J. 2010;7:194. doi: 10.1186/1743-422X-7-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride AA. Mechanisms and strategies of papillomavirus replication. Biol Chem. 2017;398(8):919–927. doi: 10.1515/hsz-2017-0113 [DOI] [PubMed] [Google Scholar]
- 20.Nichols AC, Dhaliwal SS, Palma DA, et al. Does HPV type affect outcome in oropharyngeal cancer? J Otolaryngol Head Neck Surg. 2013;42(1):9. doi: 10.1186/1916-0216-42-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med. 2012;22:276–282. doi: 10.11613/BM.2012.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinn M, Babb P, Jones J, et al. Effect of screening on incidence of and mortality from cancer of cervix in England: evaluation based on routinely collected statistics. BMJ. 1999;318(7188):904–908. doi: 10.1136/bmj.318.7188.904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasieni P, Adams J. Effect of screening on cervical cancer mortality in England and Wales: analysis of trends with an age period cohort model. BMJ. 1999;318(7193):1244–1245. doi: 10.1136/bmj.318.7193.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Aa MA, Pukkala E, Coebergh JW, et al. Mass screening programmes and trends in cervical cancer in Finland and the Netherlands. Int J Cancer. 2008;122(8):1854–1858. doi: 10.1002/ijc.23276 [DOI] [PubMed] [Google Scholar]
- 25.Pan American Health Organization/World Health Organization/Centers for Disease Control and Prevention. Integrating HPV testing in cervical cancer screening programs: a manual for program managers; 2016. Available from: http://iris.paho.org/xmlui/handle/123456789/31393. Accessed September 27, 2017.
- 26.Garbuglia AR, Lapa D, Sias C, et al. The use of both therapeutic and prophylactic vaccines in the therapy of papillomavirus disease. Front Immunol. 2020;11:188. doi: 10.3389/fimmu.2020.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montealegre JR, Varier I, Bracamontes CG, et al. Racial/ethnic variation in the prevalence of vaccine-related human papillomavirus genotypes. Ethn Health. 2019;24(7):804–815. doi: 10.1080/13557858.2017.1373073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braaten KP, Laufer MR. Human Papillomavirus (HPV), HPV-related disease, and the HPV vaccine. Rev Obstet Gynecol. 2008;1(1):2–10. [PMC free article] [PubMed] [Google Scholar]
- 29.Uijterwaal MH, Polman NJ, Van Kemenade FJ, et al. Five-year cervical (pre) cancer risk of women screened by HPV and cytology testing. Cancer Prev Res. 2015;8(6):502–508. doi: 10.1158/1940-6207.CAPR-14-0409 [DOI] [PubMed] [Google Scholar]
- 30.Sung YE, Ki EY, Lee YS, et al. Can human papillomavirus (HPV) genotyping classify non-16/18 high-risk HPV infection by risk stratification? J Gynecol Oncol. 2016;27(6):e56. doi: 10.3802/jgo.2016.27.e56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badiga S, Chambers MM, Huh W, et al. Expression of p16INK4A in cervical precancerous lesions that is unlikely to be preventable by human papillomavirus vaccines [published correction appears in Cancer. 2017;123(16):3198]. Cancer. 2016;122(23):3615–3623. doi: 10.1002/cncr.30229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gage JC, Sadorra M, Lamere BJ, et al. Comparison of the cobas Human Papillomavirus (HPV) test with the hybrid capture 2 and linear array HPV DNA tests. J Clin Microbiol. 2012;50(1):61–65. doi: 10.1128/JCM.05989-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Risley C, Clarke MA, Geisinger KR, et al. Racial differences in HPV type 16 prevalence in women with ASCUS of the uterine cervix. Cancer Cytopathol. 2020;128(8):528–534. doi: 10.1002/cncy.22267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montealegre JR, Peckham-Gregory EC, Marquez-Do D, et al. Racial/ethnic differences in HPV 16/18 genotypes and integration status among women with a history of cytological abnormalities. Gynecol Oncol. 2018;148(2):357–362. doi: 10.1016/j.ygyno.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]