Abstract
Background
During the past 2 years, studies on patients with SARS-CoV-2 infection have revealed rare inborn errors of immunity (IEIs) in type interferon (IFN) pathways underlying critical COVID-19 pneumonia. This has provided insights into pathophysiological mechanisms and immune signaling circuits regulating antiviral responses to SARS-CoV-2 and governing the susceptibility to and outcome of SARS-CoV-2 infection in humans.
Objectives
In this review, the current knowledge on IEIs underlying critical COVID-19 is presented, and the clinical implications of these findings for individualized prophylaxis and treatment are outlined.
Sources
The review is based on a broad literature search, including primarily studies on whole-exome sequencing, and to a lesser extent genome-wide association studies, of patients with critical COVID-19, as well as retrospective descriptive studies of the SARS-CoV-2 disease course in individuals with known IEIs.
Content
The review describes the discovery of monogenic IEI in 9 genetic loci related to the production or responses to type I IFN in patients with critical COVID-19 pneumonia and the surprising finding of phenocopies of these, represented by neutralizing autoantibodies to type IFN in a significant proportion of patients with critical pneumonia, particularly in elderly men, and further enriched in patients with lethal disease course. Moreover insights gained from studies on SARS-CoV-2 infection, disease course, and outcome in patients with known IEI is presented. Finally, some hypotheses for a possible genetic basis of autoimmune, inflammatory, and long-term complications of SARS-CoV-2 infection are presented and discussed.
Implications
Uncovering IEIs underlying critical COVID-19 or other severe SARS-CoV-2 disease manifestations provides valuable insights into the basic principles of antiviral immune responses and pathophysiology related to SARS-CoV-2 infection. Such knowledge has important clinical implications for identification of susceptible individuals and for diagnosis, prophylaxis, and treatment of patients to reduce disease burden and improve preparedness against viral pandemics with known or emerging viruses in the future.
Keywords: COVID-19, Inborn error of immunity, Innate immunity, Interferon, MIS-C, SARS-CoV-2
Introduction: SARS-CoV-2 disease manifestations and host immune sensing of the virus
In late 2019, an increasing number of cases of acute respiratory distress syndrome were reported in China, followed in March 2020 by the WHO declaring the outbreak a pandemic and attributing COVID-19 to a novel coronavirus named SARS-CoV-2 [1]. This viral pandemic has spread to all continents of the globe, is highly transmissible, and has led to millions of cases and deaths, particularly in vulnerable individuals with old age or medical co-morbidities, such as pulmonary disease, hypertension, diabetes, obesity, or secondary immunosuppression by medications or malignant disease [2]. Although severe/critical COVID-19 pneumonia mostly affects these groups of patients, it has become apparent that otherwise supposedly healthy individuals in rare cases may experience a severe, sometimes fulminant, disease course. Moreover, in the course of the pandemic, unexpected new disease manifestations have emerged, including postinfectious multisystem inflammatory disease in children and adults (MIS-C and MIS-A9) [3] and more subtle long-term neurocognitive, pulmonary, and musculoskeletal sequels named long COVID or post-acute COVID-19 syndrome [4].
Sensing and activation of the immune system by SARS-CoV-2
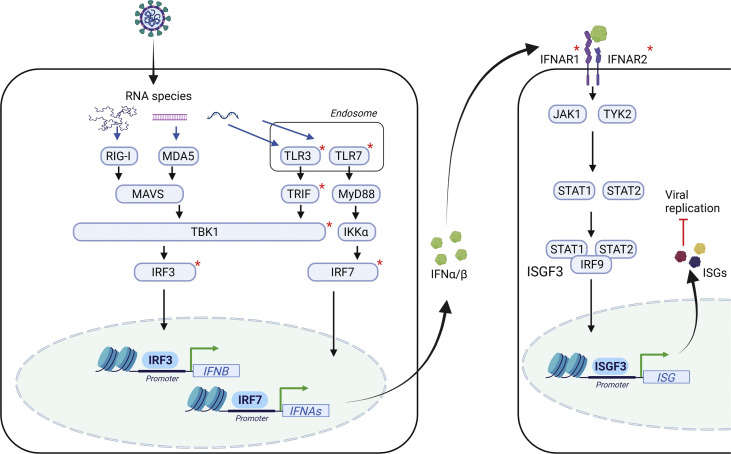
The newly emerged SARS-CoV-2 is a β-coronavirus and related to the previously identified SARS-1, the etiological source of Mediterranean respiratory syndrome. SARS-CoV-2 is a single-stranded (ss) RNA virus that infects cells through the ACE2 surface receptor and is dependent on the cellular protease TMPRRS2 to establish productive infection [5]. The innate host immune system senses the viral RNA genome, ssRNA, or double-stranded (ds) RNA replication intermediates through membrane-bound endosomal TLR3 and TLR7 or by the cytosolic RNA sensors RIG-I and MDA (Fig. 1 ). Recent data suggest that relevant leukocytes and immune cells subsets (in particular respiratory epithelial cells and plasmacytoid dendritic cells) mainly sense viral dsRNA through TLR3 and TLR7, respectively [6]. These events trigger major signalling pathways governed by and dependent on the transcription factors IRF3 and IRF7 to induce the production of type I IFN, as well as NF-κB–dependent induction of proinflammatory cytokines. Type I IFN acts in an autocrine and paracrine manner by binding to cognate receptors composed of IFNAR1 and IFNAR2 and signal through the JAK-STAT pathway to induce an antiviral program through expression of numerous IFN-stimulated genes. Like most other viruses, SARS-CoV-2 possesses mechanisms to subvert or evade these antiviral responses via targeting and antagonizing cellular IFN-inducing pathways by viral proteins [6,7].
Fig. 1.
Major cellular pathways in innate viral sensing and interferon induction in response to SARS-CoV-2 infection. Early after viral infection, viral nucleic acids in the form of single-stranded (ss)RNA or double-stranded (ds)RNA of the viral genome or replication intermediates are recognized by cytosolic or membrane-bound pattern recognition receptors (PRRs). These include the endosomal Toll-like receptors (TLR)3 and TLR7 recognizing dsRNA and ssRNA, respectively. In the cytosol, RIG-I and MDA recognize 5′triphosphorylated RNA species. These events trigger major signalling pathways, most notably governed by and dependent on the transcription factors NF-κB and interferon (IFN) regulatory factors (IRFs), inducing the production of type I IFN and proinflammatory cytokines. Type I IFN acts in an autocrine and paracrine manner by binding to cognate receptors composed of IFNAR1 and IFNAR2 and signal through the JAK-STAT pathway to induce an antiviral program through expression of numerous IFN-stimulated genes. Molecules with known genetic defects conferring susceptibility to SARS-CoV-2 infection as described in the main text are marked with a red asterisk. The figure was generated in Biorender.
Monogenic inborn errors of immunity and phenocopies in SARS-CoV-2 infection and critical COVID-19
Inborn errors of immunity in genes governing type I IFN responses in critical COVID-19 pneumonia
The clinical observation of extensive interindividual variation in disease presentation of SARS-CoV-2 infection has led to efforts to define and understand the human genetic and immunological basis of susceptibility to this virus. The COVID Human Genetic Effort (http://www.covidhge.com) consortium was the first to report major novel insights into human genetic susceptibility to severe COVID-19 [8]: It identified 23 patients with critical COVID-19 pneumonia and inborn errors of immunity (IEI) at eight genetic loci that govern TLR3-dependent type I IFN induction, amplification or response to IFN, implicating defects in TLR3, UNC93B, TRIF, TBK1, IRF3, IRF7, and IFNAR1/2 [9] (Fig. 1). Although rare, these patients with IEI showed that type I IFN immunity is indispensable for the control of SARS-CoV-2 infection and that defects in these circuits predispose individuals to critical pneumonia, particularly in the young and middle-aged, among whom an estimated 3%–5% of cases may be attributed to these genetic defects. The genetic landscape has been expanded by two independent reports of X-linked TLR7 deficiency in males with severe COVID-19, accounting for 1% of critical COVID-19 in males under 60 years of age [10,11]. Importantly, the study of TLR7 deficiency demonstrated that this defect was due to insufficient IFN production from pDCs (expressing high levels of TLR7 and IRF7), and formally proving the longstanding concept of the essential role of pDCs in type I IFN production in humans [10]. Thus, during critical SARS-CoV-2 infection, pDCs are responsible for TLR7-driven type I IFN production, whereas TLR3-dependent pathways govern mucosal type I IFN production by respiratory epithelial cells [12].
Within the genes so far demonstrated to confer susceptibility to critical COVID-19, there is a striking lack of genetic defects within adaptive immunity. This may suggest that viral restriction and control to avoid severe acute infection is particularly exerted by early innate antiviral pathways, whereas humoral and cellular defects play a more modest role, if any, in analogy to the situation in influenza. On a theoretical basis, genetic defects in adaptive immunity, if they exist, may be found in patients with prolonged infection or among those who develop critical COVID-19 despite vaccination, so-called breakthrough cases.
Neutralizing auto-antibodies to type I IFNs underlying critical COVID-19 pneumonia
The identification of genetic defects in antiviral type I IFN circuits in critical COVID-19 was accompanied by the almost simultaneous discovery of pre-existing neutralizing auto-antibodies against type I IFNs (most notably against IFNa and IFNo) as a phenocopy of the type I IFN-related IEI [13]. Although the presence of auto-antibodies to type I IFN were previously reported in some patients receiving IFN therapy, as well as in systemic lupus erythematosus, myasthenia gravis, thymoma, autoimmune polyendocrine syndrome-1, incontinentia pigmenti, and others, the impact on infection susceptibility was previously largely unexplored [14]. However, although the discovery of auto-antibodies to type I IFNs in COVID-19 patients was novel and unexpected, such phenocopies of genetically well-defined IEI exerted by the existence of specific auto-antibodies to these same mediators has been described previously in the case of auto-antibodies against IFNg in mycobacterial infection, against IL17 in chronic mucocutaneous candidiasis, and against IL6 in hyper-IgE syndrome [8]. In subsequent studies, neutralizing auto-antibodies against type I IFNs were confirmed in independent cohorts in over 10% of patients with severe COVID-19 [15]. Moreover, neutralizing auto-antibodies to physiologically relevant levels of type I have been demonstrated in an even larger fraction of patients with COVID-19, leading to the conclusion that neutralizing auto-antibodies to type I IFNs may be involved in the disease course in more than 20% of critical COVID-19 cases in patients over the age of 80 years as well as in fatal COVID-19 across all ages [16]. Intriguingly, auto-antibodies to type I IFN have subsequently been reported to underlie critical COVID-19 in a significant fraction of patients with autoimmune polyendocrine syndrome-1, and such autoantibodies have also been associated with the development of adverse reactions to the yellow fever live attenuated vaccine [14,17].
Collectively, these studies on the human genetics of severe COVID-19 demonstrated the importance of type I IFN in antiviral resistance against SARS-CoV-2 in humans and suggested that IEI related to type I IFNs, either genetically determined or through phenocopies, represent a major risk factor for the development of critical COVID-19. It remains to be determined whether the existence of auto-antibodies against type I IFN is fully or partially genetically determined. Moreover, although neutralizing type I IFN autoantibodies predated SARS-CoV-2 infection, in agreement with defective early antiviral defences, it remains incompletely explored whether levels of autoantibodies may be enhanced in some individuals in the course of acute infection.
The search for human genetics underlying auto-immune postinfectious manifestations and viral resistance
Auto-inflammatory and autoimmune drivers of pathogenesis in acute infection and MIS-C/A
As we begin to gain major insights into the pathogenesis of critical COVID-19 pneumonia and other SARS-CoV-2 disease manifestations, an appreciation of the significant role of inflammation, auto-immunity, and immunopathology in determining clinical outcome is unavoidable. For example, the acute inflammation or ‘viral sepsis’ observed in critical COVID-19 pneumonia has major similarities to known conditions, such as macrophage activation syndrome and haemophagocytic lymphohistiocytosis, whereas MIS-C/A bears a striking resemblance to Kawasaki disease, the latter also affecting children and being characterized by overt inflammation, fever, and endothelial/heart and gastrointestinal tract involvement [6,18]. In this context, it is interesting and of potential relevance that both macrophage activation syndrome/haemophagocytic lymphohistiocytosis and Kawasaki disease have a more or less well-established genetic basis and can be triggered by viral infection, although the specific role of an infectious trigger remains incompletely resolved in the case of Kawasaki disease [6]. These observations and data strongly suggest that genes beyond those related to innate type I IFN-mediated antiviral control may be disease causing or disease modifying.
On a hypothetical basis, such susceptibility genes could be involved in immune dysregulation, autoinflammation, or autoimmunity, therefore theoretically involving gain of function or loss of inhibition of molecules and pathways involved in the regulation of cytokine and TLR signalling cascades, particularly those related to IL1 and IL6 biology [19]. Regarding MIS-C/A, disease-causing genetic defects may be partly related to tissue type, given that SARS-CoV-2 superantigen-mediated TCR Vb21.3 polyclonal T cell expansion and activation seem to play a major role in the pathogenesis [20]. In a small study on the genetics and immunology of patients with MIS-A, several gene variants related to Kawasaki disease, viral replication, and cell stress were identified, although the functional impact of these variants remains to be explored [21]. At present, evidence on genetic components underlying MIS-C/A remain incompletely clarified but should be part of future focus; this may teach us important lessons, not only about MIS-C/A but also about the closely related Kawasaki disease, for which the genetics, pathogenesis, and possible infectious trigger(s) have remained enigmatic for decades [3].
Finally, another even less well-understood, albeit relatively clinically prevalent, effect of SARS-CoV-2 infection is long COVID/post-acute COVID-19 syndrome, defined as symptoms persisting more than 12 weeks after acute infection and consisting of fatigue, dyspnoea, myalgia, and neurocognitive disturbances [4]. Several hypotheses have been presented to explain this medical entity, including metabolic and mitochondrial disturbances, persistence of virus, auto-immunity, and others, but objective biochemical, immunological, and functional evidence of such pathologies has been difficult to establish, and accordingly no genetic aetiology or associations have so far been provided [4].
Resistance to SARS-CoV-2 infection or disease
On the opposite side of the spectrum of genetic susceptibility to critical disease or auto-immune manifestations is the theoretical existence of protective variants or polymorphisms that confer genetically determined resistance to infection. Accumulating evidence suggest that some individuals avoid SARS-CoV-2 infection despite repeated intense exposure to SARS-CoV-2 from family members or other close contacts and that some patients do not become very ill despite several comorbidities. This might suggest rare resistance genes or polymorphisms protecting against SARS-CoV-2 infection and/or severe outcomes of infection. Such resistance genes may interfere with viral cell entry, replication, immune activation, or viral shedding. For example, genetic polymorphisms in the viral receptor ACE2 or the cellular protease TMPRSS2 that would interfere with SARS-CoV-2 cell entry may be envisaged to offer some level of resistance to infection. However, solid evidence for a marked effect of such variants in interfering with SARS-CoV-2 entry or the COVID-19 disease course has not yet been provided.
Epidemiological genetic associations with critical COVID-19 obtained from genome wide association studies
Despite several genetic associations between single nucleotide polymorphisms and critical COVID-19 described in some cohorts, these have not been uniformly reproducible in other cohorts, and therefore few conclusions have been reached so far concerning major genetic risk factors influencing disease severity in COVID-19. One of the largest, a meta-analysis of 13 cohorts from seven studies, found that patients hospitalized for COVID-19 were significantly more likely to belong to blood group A and less likely to belong to blood group O than controls, with pooled ORs of 1.23 and 0.77, respectively [22]. Moreover, four chromosomal regions associated with severe COVID-19 relative to the general population have been revealed through genome wide association studies (GWAS). The first region encompasses a gene cluster on chromosome 3 (3p21.31) encoding six genes, with an OR between 1.6 and 2.1 for heterozygosity for the susceptibility haplotype [23]. Furthermore, two independent GWAS studies comparing critically ill COVID-19 patients with the rest of the population identified three regions, two of which are involved in antiviral immunity [24]. The first is a region on chr12q24.13, including a cluster of OAS1, OAS2, and OAS3 genes; the second region, on chr21q22.1, includes IFNAR2; and the third is CCR2. Collectively, population-based, genetic epidemiological studies have yielded only modest ORs that do not explain the full extent of interindividual variability in infection beyond known comorbidities.
Lessons to be learned from studying the disease course of SARS-CoV-2 infection in patients with known IEIs
As clinical data and experience on COVID-19 disease presentation/course and severity in individuals with known PIDs are emerging, this should provide insights into how various genetic defects predispose to SARS-CoV-2 susceptibility and risk of severe disease. A few studies have addressed this issue; however, the time span is limited, and data are influenced by the age and socioeconomic factors of the patient cohort, increased awareness and self-isolation among IEI patients, and, not least, the introduction of effective vaccines against SARS-CoV-2/COVID-19.
Briefly, the first study published included 94 patients, more than half of whom had antibody deficiencies and only 15% combined and 3% innate immunodeficiency; the morbidity and mortality were not striking, at around 10% of these [25]. Another study, including 121 individuals, concluded that the specific IEI was not a factor predicting severity, and the identified predictors, such as bronchiectasis and cardiopathy, were similar to those established in the general population [26]. An Israeli study found that the COVID-19 pandemic did not have a major impact on individuals with IEI and even suggested that in some cases a lack of strong inflammatory immune responses may be paradoxically a protective measure against the development of severe disease and sequelae [27]. Finally, a Danish study of patients with common variable immunodeficiency of mixed genetic background also reported only mild disease/low morbidity [28]. Collectively, larger cohorts of (unvaccinated) individuals are needed in order to ascertain associations between genetically well-defined IEIs and risk of severe COVID-19. In particular these studies should focus on patients predicted to experience increased vulnerability to SARS-CoV-2, such as innate deficiencies in type I IFN induction and signalling, T cell defects, or combined immunodeficiencies. A recent review of the characteristics of SARS-CoV-2 infection in 648 patients with different IEI found that combined immunodeficiencies, immune dysregulation disorders, and innate immune defects impairing type I IFN responses were associated with severe disease course in some patients [29]. However, for most patients, the underlying IEI did not represent an independent risk factor for severe COVID-19—on the contrary, some IEI might be protective due to impaired inflammation and consequently less severe immunopathology.
Clinical therapeutic implications and concluding remarks
The discoveries of IEI affecting type I IFN circuits and phenocopies thereof in severe COVID-19 have improved our understanding of fundamental antiviral disease mechanisms in humans. Moreover, these insights may have important clinical implications for the management of patients. Single-patient proof-of-principle case reports and, not least, larger clinical trials are now needed to determine how this new knowledge can be translated into clinical medicine to improve the management of the SARS-CoV-2 infection at the individual and population level. The therapeutic implications to be further explored include identification of susceptible individuals for accelerated vaccination schedules, early (even presymptomatic) antiviral treatment, or intensified treatment during acute disease. More specifically, current treatment options include treatment of vulnerable patients (due to genetic IEI or autoantibodies) with monoclonal antibodies against the SARS-CoV-2 spike protein, administration of antivirals, or possibly type I IFN nasally or systemically. Indeed, proof of principle for the potential benefit of IFN therapy among particularly vulnerable COVID-19 patients has been provided by reports of IFNa2 treatment of patients with TLR3-and IRF3-deficiencies [30] as well as IFNb treatment of patients with IFN auto-antibodies and incontinentia pigmenti [31]. Although the precise role of type I IFN treatment in COVID-19 infection remains to be determined, a recent study demonstrated impaired antiviral type I IFN immunity in the nasal mucosa in patients with such auto-antibodies, suggesting a beneficial effect of IFN therapy [32]. In the case of neutralizing auto-antibodies, these may be detected by a rapid point-of-care test and be removed by plasma exchange before progression into critical COVID-19 pneumonia with a high fatality rate.
As previously documented over the past decades during the unravelling of the genetic basis of viral infectious diseases, the discoveries of IEIs in patients with various clinical SARS-CoV-2 infection phenotypes may teach us important lessons on SARS-CoV-2 infection pathogenesis, virus–host interactions, and correlates of protective immunity. Many questions remain unanswered in relation to the human genetics of different SARS-CoV-2 disease manifestations, including the development of breakthrough infection, post-infectious autoimmune disorders such as MIS-C/A, susceptibility to long COVID-19, post-vaccine myocarditis, and possible viral resistance. There are undoubtedly additional IEI underlying these disease manifestations that remain to be identified. Therefore, future efforts should focus on patient cohorts with different SARS-CoV-2 disease manifestations, with the goal of addressing unresolved issues related to the molecular genetics, immunological pathways/circuits, and pathophysiology involved in the SARS-CoV-2 disease course and outcome in patients. Hopefully, this approach will continue to provide further insights into the essential role of human genetics in governing antiviral immune responses in humans and how this may be translated into clinical medicine.
Transparency declaration
The author has no conflicts of interest to declare. No financial support was received for writing of this review. THM is supported by the Novo Nordisk Foundation (NNF21OC0067157; NNF20OC0064890) and The Independent Research Fund Denmark (0134-00006B).
Editor: L. Scudeller
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sancho-Shimizu V., Brodin P., Cobat A., Biggs C.M., Toubiana J., Lucas C.L., et al. SARS-CoV-2-related MIS-C: a key to the viral and genetic causes of Kawasaki disease? J Exp Med. 2021;218 doi: 10.1084/jem.20210446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paludan S.R., Mogensen T.H. Innate immunological pathways in COVID-19 pathogenesis. Sci Immunol. 2022;7 doi: 10.1126/sciimmunol.abm5505. [DOI] [PubMed] [Google Scholar]
- 7.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casanova J.L., Su H.C., Effort C.H.G. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181:1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casanova J.L., Abel L. Mechanisms of viral inflammation and disease in humans. Science. 2021;374:1080–1086. doi: 10.1126/science.abj7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bastard P., Orlova E., Sozaeva L., Levy R., James A., Schmitt M.M., et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med. 2021;218 doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Wijst M.G.P., Vazquez S.E., Hartoularos G.C., Bastard P., Grant T., Bueno R., et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastard P., Michailidis E., Hoffmann H.H., Chbihi M., Le Voyer T., Rosain J., et al. Auto-antibodies to type I IFNs can underlie adverse reactions to yellow fever live attenuated vaccine. J Exp Med. 2021;218 doi: 10.1084/jem.20202486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter-Timofte M.E., Jorgensen S.E., Freytag M.R., Thomsen M.M., Brinck Andersen N.S., Al-Mousawi A., et al. Deciphering the role of host genetics in susceptibility to severe COVID-19. Front Immunol. 2020;11:1606. doi: 10.3389/fimmu.2020.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreews M., Le Gouge K., Khaldi-Plassart S., Pescarmona R., Mathieu A.L., Malcus C., et al. Polyclonal expansion of TCR Vbeta 21.3(+) CD4(+) and CD8(+) T cells is a hallmark of multisystem inflammatory syndrome in children. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abh1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronit A., Jorgensen S.E., Roed C., Eriksson R., Iepsen U.W., Plovsing R.R., et al. Host genetics and antiviral immune responses in adult patients with multisystem inflammatory syndrome. Front Immunol. 2021;12:718744. doi: 10.3389/fimmu.2021.718744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golinelli D., Boetto E., Maietti E., Fantini M.P. The association between ABO blood group and SARS-CoV-2 infection: a meta-analysis. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., et al. Severe Covid GWAS Group Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 25.Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E., et al. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147:520–531. doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goudouris E.S., Pinto-Mariz F., Mendonca L.O., Aranda C.S., Guimaraes R.R., Kokron C., et al. Outcome of SARS-CoV-2 infection in 121 patients with inborn errors of immunity: a cross-sectional study. J Clin Immunol. 2021;41:1479–1489. doi: 10.1007/s10875-021-01066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcus N., Frizinsky S., Hagin D., Ovadia A., Hanna S., Farkash M., et al. Minor clinical impact of COVID-19 pandemic on patients with primary immunodeficiency in Israel. Front Immunol. 2020;11:614086. doi: 10.3389/fimmu.2020.614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drabe C.H., Hansen A.E., Rasmussen L.D., Larsen O.D., Moller A., Mogensen T.H., et al. Low morbidity in Danish patients with common variable immunodeficiency disorder infected with severe acute respiratory syndrome coronavirus 2. Infect Dis (Lond) 2021;53:953–958. doi: 10.1080/23744235.2021.1957144. [DOI] [PubMed] [Google Scholar]
- 29.Bucciol G., Tangye S.G., Meyts I. Coronavirus disease 2019 in patients with inborn errors of immunity: lessons learned. Curr Opin Pediatr. 2021;33:648–656. doi: 10.1097/MOP.0000000000001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy R., Bastard P., Lanternier F., Lecuit M., Zhang S.Y., Casanova J.L. IFN-alpha2a therapy in two patients with inborn errors of TLR3 and IRF3 infected with SARS-CoV-2. J Clin Immunol. 2021;41:26–27. doi: 10.1007/s10875-020-00933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastard P., Levy R., Henriquez S., Bodemer C., Szwebel T.A., Casanova J.L. Interferon-beta therapy in a patient with incontinentia pigmenti and autoantibodies against type i ifns infected with SARS-CoV-2. J Clin Immunol. 2021;41:931–933. doi: 10.1007/s10875-021-01023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez J., Mommert M., Mouton W., Pizzorno A., Brengel-Pesce K., Mezidi M., et al. Early nasal type I IFN immunity against SARS-CoV-2 is compromised in patients with autoantibodies against type I IFNs. J Exp Med. 2021;218 doi: 10.1084/jem.20211211. [DOI] [PMC free article] [PubMed] [Google Scholar]