Abstract
Red light (RL) can improve egg production in Jinghai Yellow hens. Circular RNAs (circRNAs) are novel, non-coding RNAs, but the molecular mechanism underlying circRNA function during follicular development in hens under monochromatic light has not been established. Herein, we compared expression profiles of granulosa cells (GCs) from small yellow follicles (SYFs) from hens under RL and white light (WL). A total of 2,468 circRNAs were identified, of which 22 were differentially expressed (DE) in the RL and WL groups. DE circRNA host genes were enriched in ovarian steroidogenesis, and MAPK and PI3K-Akt signaling pathways. Furthermore, DE circRNA_0320 and circRNA_0185 interacted with miR-143-3p, which targets the follicle-stimulating hormone receptor and is essential for GC differentiation and follicle development. These findings will facilitate further analysis of the molecular mechanism leading to GC development in hens raised under monochromatic light, which could lead to increased egg production.
Key words: circRNA, chicken, monochromatic light, granulosa cell, small yellow follicle
INTRODUCTION
Follicle development is closely related to egg production in hens (Johnson et al., 2015). The reproductive strategy for avian species that produce clutches of eggs is dependent on the maintenance of a small cohort of viable, undifferentiated follicles (Johnson, 2015). In hens one small yellow follicle (SYF), 6 to 8 mm in diameter, is selected daily from a pool of follicles of similar size to become a preovulatory follicle (Hernandez and Bahr, 2003; Woods and Johnson, 2005). Furthermore, the proliferation and differentiation of granulosa cells (GCs) are closely related to follicular development (Jin et al., 2006). Orisaka et al. (2009) demonstrated that follicular growth is tightly regulated by intraovarian oocyte-granulosa-theca cell interactions. GCs help supply nutrients during oocyte growth through gap junction associations (Larsen et al., 1979; Buccione et al., 1990). Shen et al. (1993) reported that yolk particles passing between GCs are important for the growth of chicken oocytes (Schneider, 2009).
SYFs contain significantly more follicle-stimulating hormone receptors (FSHRs) than other follicles (You et al., 1996). FSHRs are among the earliest markers of the most recently selected follicle in the granulosa layer of the hen ovary (Woods and Johnson, 2005); however, the granulosa layer within pre-hierarchical follicles remains undifferentiated and steroidogenically inactive (Johnson and Lee, 2016). Tilly et al. (1991) demonstrated that the steroidogenic competency of GCs occurs during the transition of follicles from 6–8 mm to 9–12 mm. Differentiated GCs derived from preovulatory follicles are capable of producing progesterone, particularly in response to luteinizing hormone (LH), which eventually results in the LH surge that initiates ovulation (Robinson and Etches, 1986; Johnson, 2012). Therefore, follicle selection is associated with an increase in the cholesterol side-chain cleavage enzyme (P450scc), together with progesterone secretion in the GCs from SYFs (Jin et al., 2006). Therefore, the final differentiation of the GC layer commences at the time of follicle selection (Johnson and Lee, 2016).
Circular (circ) RNAs are novel non-coding (nc) RNAs that are ubiquitously expressed in eukaryotic cells during post-transcriptional processes (Shao and Chen, 2016). CircRNAs are unusually stable RNAs produced by circularization of exons, albeit via a poorly characterized mechanism (Ashwal-Fluss et al., 2014). Memczak et al. (2013) detected thousands of highly-expressed, stable circRNAs that are often tissue- and/or developmental stage-specific. Endogenous circRNA molecules are efficient miRNA sponges, adding to a growing repertoire of known regulatory functions in gene expression (Hansen et al., 2013; Salzman et al., 2013). In addition, endogenous circRNA molecules modulate pre-mRNA alternative splicing and have a protein-coding capacity (Wang et al., 2017). Wu et al. (2020) reported that circRNA host genes in GCs of duck preovulatory follicles are enriched in vital pathways related to oocyte development and cell proliferation. Shen et al. (2019) identified the key gene, RalGPS2, which is produced in three isoforms of circRNAs in the GCs of chicken follicles. Artificial illumination is widely used in poultry, and previous studies have reported that 300-day-old hens laid 90.61 eggs under red light (RL), but 87.44 eggs under white light (WL). Thus, RL significantly improves egg production by Jinghai Yellow hens, but the molecular mechanism underlying circRNA function during follicular development is not known.
In the present study, Jinghai Yellow chickens under monochromatic light were used to perform RNA sequencing (RNA-seq) and identify circRNAs in GCs of SYFs.
MATERIALS AND METHODS
Ethics Approval
This study was reviewed and approved by the Institutional Animal Care and Use Committee of the Department of Animal Science and Technology at Yangzhou University (Yangzhou city, China). All chicken procedures were performed according to the 2008 Standards for the Administration of Experimental Practices (Jiangsu, China).
Chicken Rearing and Sample Preparation
Eighteen generations of Jinghai Yellow chickens were raised at Jiangsu Jinghai Poultry Industry Group Co., Ltd. (Nantong, Jiangsu, China). After transfer to the laying house, hens (16 wk of age) were caged individually and provided with water ad libitum, but with restricted access to food (Supplementary file 1). The room temperature was 15 to 25°C. A total of 300 hens were divided into 2 groups and exposed to a red LED (660 nm) or a white LED (400−760 nm) for 16 h each day. There were 5 subgroups per group, with WL serving as the control group. Battery cages were oriented vertically, and the second floor was used to rear hens. The light intensity was 15.20 ± 1.05 lux, as measured with a TES-1336A light meter (TES Electrical Electronic Crop., Taipei, China). The experimental period was 118 d. Based on the pedigree record, 6 half-sibhens with an average body weight at 300 days of age were selected. Hens were properly anesthetized with isoflurane, decapitated, and SYFs (6–8 mm) were collected and gently washed in cold phosphate-buffered saline (PBS; Gibco, Grand Island, NY). SYFs were carefully isolated using a scalpel based on follicle diameter, then the follicles were immediately inverted over a suitable dish containing aqueous medium and follicular contents. The membrane and granular layers were separated, and granular layers were collected (Gilbert et al., 1977), flash-frozen in liquid nitrogen, and stored at −80°C until analysis.
RNA-seq Preparation and Sequencing
TRIzol reagent (Invitrogen, Carlsbad, CA) was used to extract total RNA from GCs of SYFs. The quality of total RNA was assessed by 1% agarose gel electrophoresis. A spectrophotometer (wavelengths of 260 and 280 nm) was used to measure the concentration, purity, and mass of the extracted RNA. Samples with an RNA integrity number >9.0 and an OD 260/280 value of ∼2.0 were used. Five micrograms of RNA from each sample were used to construct the circRNA library. Ribosomal RNA was removed, the library was built, and high-throughput RNA-seq was performed by Shanghai OE Biotech Co., Ltd. (Shanghai, China; Yu et al., 2021).
Identification of Differentially-Expressed circRNAs
Bioinformatic methods were performed for quality control; the Q20, Q30, and GC content were calculated. Clean reads were aligned to the reference genome by Bowtie2 and HISAT2 software (Langmead and Salzberg, 2012). CircRNAs were identified by find_circ and CIRI2 (Hansen et al., 2016; Gao et al., 2017), and the level of circRNA expression was measured by mapped backsplicing junction reads per million mapped reads. The DESeq package, fold-changes of the same circRNA in 2 groups ≥2, and a P-value ≤ 0.05 were used to distinguish differentially expressed (DE) circRNAs.
Bioinformatics Analysis
The potential functions of DE circRNA host genes and predicted target genes were analyzed. The clusterProfiler package within R software was used for GO and KEGG pathway annotation (Yu et al., 2012). Interactions of miRNAs and circRNAs were analyzed by miRanda with a score of ≥140 as a cut-off. TargetScan and miRDB were used to predict target genes of miRNAs (Wang, 2008), and Cytoscape software was applied to describe the miRNA-circRNA network (Wang et al., 2016).
RT-qPCR Validation
Total RNA was extracted from GCs of SYFs using a mirVana RNA isolation kit (ABI, Austin, TX) according to the manufacturer's specifications. Quantification was performed with a two-step reaction. Each reverse transcription reaction consisted of 0.5 μg of RNA, 2 μL of 5 × TransScript All-in-One SuperMix for qPCR, and 0.5 μL of gDNA Remover; the total volume was 10 μL. Reactions were performed using a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA) for 15 min at 42°C and 5 s at 85°C. The 10 μL reverse transcription reaction mix was then diluted 10-fold in nuclease-free water and held at −20°C. PCR products for cDNA and genomic DNA were analyzed by agarose gel electrophoresis, and the backsplicing sites of circRNAs were confirmed by Sanger sequencing at Sango Biotech Co. Ltd. (Shanghai, China). Primer sequences were designed according to previous studies (Shen et al., 2019; Wu et al., 2020), and primers for circRNAs and mRNAs are listed in Supplementary file 2. RT-qPCR was performed using a LightCycler 480 II real-time PCR Instrument (Roche, Switzerland) using a 10 μL reaction mixture containing 1 μL of cDNA, 5 μL of 2 × PerfectStart Green qPCR SuperMix, 0.2 μL of forward primer, 0.2 μL of reverse primer, and 3.6 μL of nuclease-free water. Reactions were incubated in a 384-well optical plate (Roche) at 94°C for 30 s, followed by 45 cycles at 94°C for 5 s, and 60°C for 30 s. Each sample was run in triplicate for analysis. Using ACTB as a reference, relative levels of gene and lncRNA expression were quantified using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Statistical Analysis
Data analyses were performed with SPSS 13.0 software (SPSS Inc., Chicago, IL). Results are expressed as the mean ± standard error. The statistical significance of differences between groups was evaluated by independent-samples t tests and a P < 0.05 was considered statistically significant.
RESULTS
Identification of circRNAs in GCs of SYFs
Six circRNA libraries were built from the RL (n = 3) and WL (n = 3) groups. An Illumina Hiseq 4000 instrument was used to sequence these samples, and the obtained reads were mapped to the chicken reference genome (Gallus-gallus-5.0/galGal5). After removing low-quality sequences and adapters, 55.35 Gb of clean reads was obtained with an average GC content of 48.2%. CIRI and circBase were used to identify circRNAs (Glažar et al., 2014; Gao et al., 2015). A total of 2,468 circRNAs were identified in regenerating GCs from SYFs in the RL and WL groups. Nearly 85% of circRNAs had a predicted splice length <2,000 bp, 46.64% of circRNAs were >500 bp, 26.26% were 500 to 1,000 bp, and the average length was 2,564 bp (Figure 1A). The identified circRNAs in GCs were distributed across almost all chromosomes, with the fewest number on chromosome (chr) 32 and the highest number on chr1 (Figure 1B). According to the genomic loci, circRNAs were grouped into exon, intron, and intergenic regions. Most (2,180) circRNAs originated from sense genic_exonic regions (Figure 1C).
Figure 1.
Profiling of circRNAs in GCs from SYFs of chickens under monochromatic light. (A) Splice length of circRNAs. (B) Chromosomes of circRNAs. (C) Source of circRNAs. Abbreviations: circRNAs, circular RNAs; GCs, granulosa cells; SYFs, small yellow follicles.
Analysis of DE circRNAs
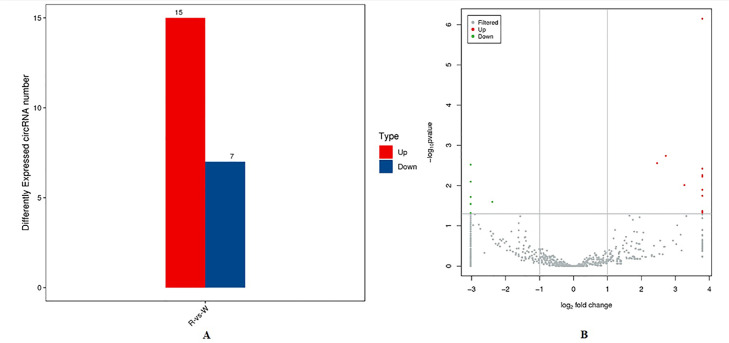
The levels of circRNA expression of GC libraries from SYFs were compared between the RL and WL groups; 22 DE circRNAs were identified (P < 0.05). Of the 22 DE circRNAs, 15 were upregulated and 7 were downregulated (Figure 2A; Supplementary file 3). Furthermore, spliced reads per million (RPM) was used to measure the level of circRNA expression (Li et al., 2017) and volcano plots of DE circRNAs were plotted (Figure 2B).
Figure 2.
Features of DE circRNAs. (A) Number of DE circRNAs. (B) Volcano plot of DE circRNAs. Abbreviations: circRNAs, circular RNAs.
GO and KEGG Analyses of Host Genes Related to DE circRNAs
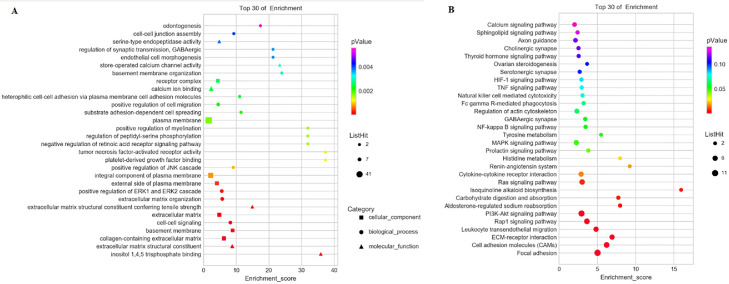
The biological functions of host genes related to DE circRNAs were further analyzed. GO enrichment analysis was performed on DE circRNAs involved in the positive regulation of cell migration, the ERK1and ERK2 cascade, cell-cell signaling, and inositol 1,4,5-trisphosphate binding (Figure 3A; Supplementary file 4). KEGG pathway enrichment analysis of DE circRNAs identified ovarian steroidogenesis, as well as MAPK, prolactin, PI3K-Akt, and thyroid hormone signaling pathways (Figure 3B; Supplementary file 5).
Figure 3.
GO and KEGG analyses of host genes of DE circRNAs. (A) The top 10 GO enrichment terms in BP, CC, and MF categories for host genes related to DE circRNAs. (B) The top 20 KEGG enrichment terms of host genes related to DE circRNAs. Abbreviations: circRNAs, circular RNAs.
Verification of DE circRNAs and Host Genes by RT-qPCR
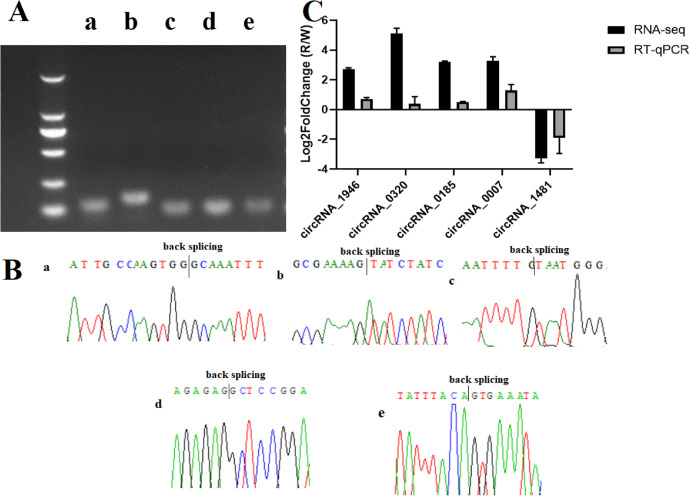
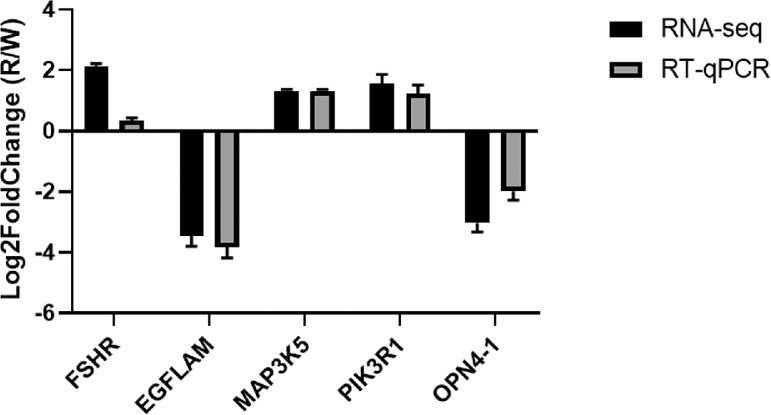
To validate the levels of DE circRNA expression and host genes to support further analysis of the molecular mechanism underlying monochromatic light on follicular development in hens, 5 circRNAs (circRNA_1946, circRNA_0320, circRNA_0185, circRNA_0007, and circRNA_1481) and 5 genes (FSHR, EGF-like fibronectin type III and laminin G domain [EGFLAM], mitogen-activated protein kinase kinase kinase 5 [MAP3K5], phosphoinositide-3-kinase regulatory subunit 1 [PIK3R1], and photopigment melanopsin-like [OPN4-1]) were analyzed by RT-qPCR, and the results were compared with the high-throughput RNA-seq results (Figures 4 and 5). The WL group served as a control group, and the pattern of expression revealed that the 2 methods gave consistent results.
Figure 4.
qRT-PCR analysis of circRNA expression. (A) Agarose gel electrophoresis of primers used to amplify circRNAs. (B) Sanger sequencing confirmation of the backsplicing junction of circRNAs. (C) RT-qPCR validation of different DE circRNAs in GCs of SYFs. a, b, c, d, and e represent circRNAs circRNA_1946, circRNA_0320, circRNA_0185, circRNA_0007, and circRNA_1481, respectively. Abbreviations: circRNAs, circular RNAs; GCs, granulosa cells
Figure 5.
qRT-PCR analysis of mRNA expression.
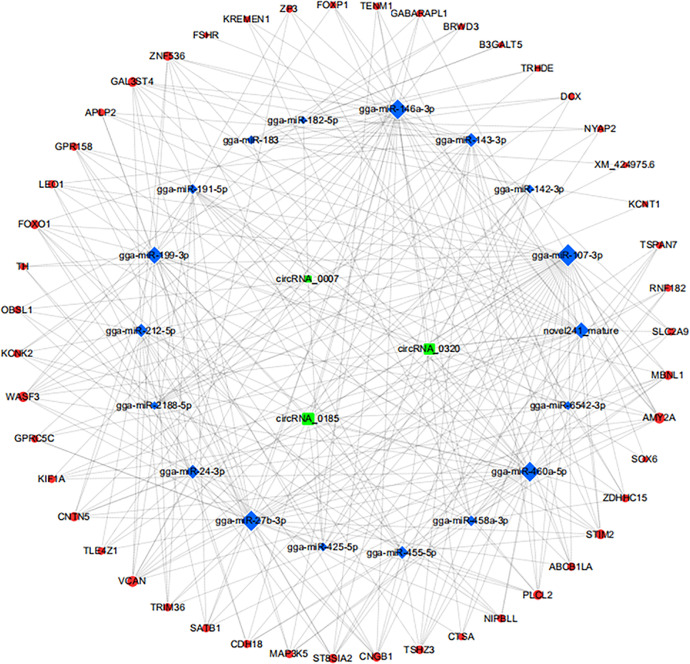
Co-expression Networks of Predicted Target miRNAs and Genes
It has been well-established that one of the main functions of circRNAs is to serve as miRNA sponges (Hansen et al., 2013). Indeed, circRNA-miRNA interactions can predict the characteristics of circRNAs (Salzman et al., 2013). Herein, 24 miRNAs and 193 mRNAs were predicted by miRBase (Wang, 2008); the results are shown in Supplementary files 6 and 7. Cytoscape software was used to construct circRNA-miRNA-mRNA co-expression networks (Wang et al., 2016). CircRNA_1805 and circRNA_0007 were shown to share common targets (miR-425-5p, circRNA_0320, and circRNA_1805) with the same 11 miRNAs, including miR-143-3p, miR-142-3p, miR-107-3p, miR-27b-3p, and miR-460a-5p. The molecular interactions between these 3 circRNAs and their miRNAs and target genes are illustrated in Figure 6. Target genes, including FSHR, EGFLAM, MAP3K5, and PIK3R1, are therefore related to chicken reproductive performance.
Figure 6.
Network analysis of circRNA-miRNA-mRNA interactions.
DISCUSSION
Chicken ovaries are an excellent model of follicular selection because the size of follicles can be easily visualized (Hernandez and Bahr, 2003). Many of the processes of follicle growth and differentiation are initiated within the GC layer (Johnson, 2015). Furthermore, compared with the WL group, the RL group had a significant increase in egg production. Elucidating the follicular development of SYFs of hens in response to monochromatic light could benefit the modern poultry industry. CircRNAs are ubiquitous in the transcriptome of humans and mice; circRNA expression is a feature of all eukaryotes (Wang et al., 2014; Lu et al., 2015; Salzman, 2016). Most circRNA sponges are formed through backsplicing, and are enriched in miRNA-binding sites (Zhang et al., 2013; Kulcheski et al., 2016). Thus, DE circRNAs, miRNAs, and mRNAs of GCs from SYFs of Jinghai Yellow chickens were investigated under RL and WL exposure to identify the key circRNAs involved in follicular development.
A total of 2,468 circRNAs were identified in GCs from SYFs, which is greater than the number (1,800) in livers of ALV-J-resistant chickens (Zhang et al., 2017), but less than the number (4,254) in spleens of ALV-J-infected chickens (Qiu et al., 2018). This finding is possibly due to tissue-specific factors. In the present study, host genes of DE circRNAs were enriched in ovarian steroidogenesis, as well as the MAPK, prolactin, and PI3K-Akt signaling pathways. Previous studies concluded that ovarian steroidogenesis is important for GC differentiation and follicle development (Wojtysiak et al., 2011; Wang et al., 2013; Johnson and Lee, 2016). MAPK signaling prevents premature differentiation of the GC layer within pre-hierarchical follicles (Johnson and Woods, 2009), which is involved in the regulation of follicle growth. The PI3K pathway is known to be essential for regulation of cell proliferation, survival, and migration, and is also likely to play a role in regulating the activation of primordial follicles in mouse ovaries (Dupont et al., 2012). Thus, it can be concluded that DE circRNAs play an important role in GC development and follicular maturation.
Previous studies have shown that chr5:13061358|13064218 binds to miR-27b-3p and participates in follicular development of chicks (Shen et al., 2019), and aplacirc_013267 directly binds to and inhibits miR-1-13 in duck ovarian follicles (Wu et al., 2020). In the present study we found that circRNA_0320 and circRNA_0185 were more abundant in the RL group than the WL group, and a number of miRNAs were predicted to interact with these circRNAs, including miR-27b-3p, miR-143-3p, and miR-142-3p (Figure 5). Furthermore, the circRNA-miRNA-mRNA network also revealed that miR-143p-3p targets FSHR. These findings are in agreement with the findings of Zhang et al. (2019) and Du et al. (2016); specifically, miR-143 affects GC apoptosis by binding to FSHR. One of the earliest markers for differentiating GCs is an elevated expression of FSHR (Johnson and Woods, 2009); FSHR mRNA levels in the GC layer were highest within the cohort of 6 to 8 mm SYFs (You et al., 1996). The FSHR gene is associated with stimulating egg production (Xu et al., 2017). Expression of FSHR was higher in GCs from SYFs in the RL group, which also had higher egg production. Genes, including EGFLAM, MAP3K5, PIK3R1, and OPN4-1, were also identified as targets of DE circRNAs, and play an important role in follicular development. FSH promotes the accumulation of MAP3K5 (Sirotkin et al., 2008), which is a reliable marker for dominant follicles in cattle, and at d 2.5 post-ovulation there are clear differences in expression of MAP3K5 between the dominant follicle and the next largest follicle 1 d later (Pfeffer et al., 2007). In the present study RL increased the expression of FSH and MAP3K5, which may be related to follicular development. OPNs play a pivotal role in non–image-forming responses to light, including physiologic adaptations to ambient light (Wang et al., 2020). Direct photic regulation of sleep in mice is predominantly mediated by OPN4 (Lupi et al., 2008). OPN4−/− mice display a severely attenuated phase resetting in response to brief pulses of monochromatic light (Panda et al., 2002). RL decreased the expression of OPN4, which may explain why light of a relatively long wavelength weakens the circadian rhythm, resulting in persistent follicle development. PIK3R1 plays an important role in regulating ovarian activities in seasonal reproduction of yaks (Xu et al., 2020). Chen et al. (2021) concluded that PIK3R1 is upregulated in follicles with higher egg production, which is in agreement with the findings of the present study. Furthermore, PIK3R1 is also involved in signaling pathways that control GC proliferation and oocyte maturation (Zheng et al., 2012). EGFLAM is involved in cellular growth, differentiation, and proliferation (Naturil-Alfonso et al., 2017), but Vicente et al. (2013) found that expression of EGFLAM is negatively correlated with foetal growth. Herein, expression of EGFLAM levels was lower in the RL group, demonstrating a negative correlation with follicle maturation. Analysis of competing endogenous RNAs (ceRNAs) revealed that circRNAs compete with miRNAs to alter the expression of key genes involved in GC differentiation and follicle development. Further characterization of the functions of circRNAs and associated molecules related to follicular development are required to confirm the roles of ceRNAs in GC differentiation; interference studies could prove useful for this purpose.
CONCLUSIONS
In conclusion, compared with the WL group, 2,468 circRNAs were identified in GCs from SYFs of Jinghai Yellow chickens following RL exposure. Knowledge on these circRNAs could improve egg production. Furthermore, DE circRNA_0320 and circRNA_0185 were shown to interact with miR-143-3p that targets FSHR, an essential protein for GC differentiation. These findings will facilitate further analysis of the molecular mechanism underlying circRNAs in GCs and provide insight into ceRNA interactions in follicular development, which could help improve egg production in Jinghai Yellow chickens.
ACKNOWLEDGMENTS
This study was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, China Agricultural Research Systems (grant No. CARS-41). The funding body did not have any role in the study design, data collection, or analysis and interpretation of the data.
Availability of data and materials: The datasets generated and/or analyzed in this work are available in the NCBI Sequence Read Archive (SRA) repository under accession number SRP278036 and BioProject accession number PRJNA657681. Additional datasets supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions: Conceptualization, Y.W., J.W., and G.Z.; methodology, Y.W., Z.G., C.Z., P.W., X.L, and L.C.; validation, Y.W.; writing—original draft preparation, Y.W.; writing—review and editing, J.W.; supervision, J.W., and G.Z.; funding acquisition, J.W. All authors have read and agreed to the published version of the manuscript.
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.101734.
Appendix. Supplementary materials
REFERENCES
- Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. CircRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Buccione R., Schroeder A.C., Eppig J.J. Interactions between somatic cells and germ cells throughout mammalian oogenesis. Biol. Reprod. 1990;43:543–547. doi: 10.1095/biolreprod43.4.543. [DOI] [PubMed] [Google Scholar]
- Chen X., Sun X., Chimbaka I.M., Qin N., Xu X., Liswaniso S., Xu R.F., Gonzalez J.M. Transcriptome analysis of ovarian follicles reveals potential pivotal genes associated with increased and decreased rates of chicken egg production. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.622751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Zhang L., Li X., Pan Z., Liu H., Li Q. TGF-β signaling controls FSHR signaling-reduced ovarian granulosa cell apoptosis through the SMAD4/miR-143 axis. Cell Death Dis. 2016;7:2476. doi: 10.1038/cddis.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont J., Reverchon M., Cloix L., Froment P., Rame C. Involvement of adipokines, AMPK, PI3K and the PPAR signaling pathways in ovarian follicle development and cancer. Int. J. Dev. Biol. 2012;56:959–967. doi: 10.1387/ijdb.120134jd. [DOI] [PubMed] [Google Scholar]
- Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:1–16. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhang J., Zhao F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2017;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- Gilbert A.B., Evans A.J., Perry M.M., Davidson M.H. A method for separating the granulosa cells, the basal lamina and the theca of the preovulatory ovarian follicle of the domestic fowl (Gallus domesticus) Reproduction. 1977;50:179–181. doi: 10.1530/jrf.0.0500179. [DOI] [PubMed] [Google Scholar]
- Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Jørgen K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hansen T.B., Venø M.T., Damgaard C.K., Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A.G., Bahr J.M. Role of FSH and epidermal growth factor (EGF) in the initiation of steroidogenesis in granulosa cells associated with follicular selection in chicken ovaries. Reproduction. 2003;125:683–691. [PubMed] [Google Scholar]
- Jin Y., Zhang C., Lin X., Zeng W. Prostaglandin involvement in follicle-stimulating hormone-induced proliferation of granulosa cells from chicken prehierarchical follicles. Prostaglandins Other Lipid Mediat. 2006;81:45–54. doi: 10.1016/j.prostaglandins.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Johnson A.L., Woods D.C. Dynamics of avian ovarian follicle development: cellular mechanisms of granulosa cell differentiation. Gen. Comp. Endocrinol. 2009;163:12–17. doi: 10.1016/j.ygcen.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Johnson A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015;94:781–785. doi: 10.3382/ps/peu008. [DOI] [PubMed] [Google Scholar]
- Johnson A.L., Lee J. Granulosa cell responsiveness to follicle stimulating hormone during early growth of hen ovarian follicles. Poult. Sci. 2016;95:108–114. doi: 10.3382/ps/pev318. [DOI] [PubMed] [Google Scholar]
- Johnson P.A. Follicle selection in the avian ovary. Reprod. Domest. Anim. 2012;47:283–287. doi: 10.1111/j.1439-0531.2012.02087.x. [DOI] [PubMed] [Google Scholar]
- Johnson P.A., Stephens C.S., Giles J.R. The domestic chicken: causes and consequences of an egg a day. Poult. Sci. 2015;94:816–820. doi: 10.3382/ps/peu083. [DOI] [PubMed] [Google Scholar]
- Kulcheski F.R., Christoff A.P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie2. Nat. Methods. 2012;9:357–369. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen W.J., Tung H.N., Murray S.A., Swenson C.A. Evidence for the participation of actin microfilaments and bristle coats in the internalization of gap junction membrane. J. Cell Biol. 1979;83:576–587. doi: 10.1083/jcb.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Guo J., Chen Y., Chang C.X.C. Comprehensive CircRNA expression profile and selection of key CircRNAs during priming phase of rat liver regeneration. BMC Genomics. 2017;18:80. doi: 10.1186/s12864-016-3476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu T., Cui L., Zhou Y., Zhu C., Fan D., Gong H., Zhao Q., Zhou C.C., Zhao Y., Lu D.F., Luo J.H., Wang Y.C., Tian Q.L., Feng Q., Huang T., Han B. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupi D., Oster H., Thompson S., Foster R.G. The acute light-induction of sleep is mediated by OPN4-based photoreception. Nat. Neurosci. 2008;11:1068–1073. doi: 10.1038/nn.2179. [DOI] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Naturil-Alfonso C., Peñaranda D.S., Vicente J.S., Marco-Jiménez F. Feed restriction regime in a rabbit line selected for growth rate alters oocyte maturation manifested by alteration in MSY2 gene expression. Reprod. Domest. Anim. 2017;52:976–984. doi: 10.1111/rda.13006. [DOI] [PubMed] [Google Scholar]
- Orisaka M., Tajima K., Tsang B.K., Kotsuji F. Oocyte-granulosa-theca cell interactions during preantral follicular development. J. Ovarian Res. 2009;2:9. doi: 10.1186/1757-2215-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S., Sato T.K., Castrucci A.M., Rollag M.D., DeGrip W.J., Hogenesch J.B., Provencio I., Kay S.A. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science. 2002;298:2213–2216. doi: 10.1126/science.1076848. [DOI] [PubMed] [Google Scholar]
- Pfeffer P.L., Sisco B., Donnison M., Somers J., Smith C. Isolation of genes associated with developmental competency of bovine oocytes. Theriogenology. 2007;68:84–90. doi: 10.1016/j.theriogenology.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Qiu L., Chang G., Bi Y., Liu X., Chen G. Circular RNA and mRNA profiling reveal competing endogenous RNA networks during avian leukosis virus, subgroup J-induced tumorigenesis in chickens. PloS one. 2018;13 doi: 10.1371/journal.pone.0204931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson F.E., Etches R.J. Ovarian steroidogenesis during foillicular maturation in the domestic fowl (Gallus Domesticus) Biol. Reprod. 1986;35:1096–1105. doi: 10.1095/biolreprod35.5.1096. [DOI] [PubMed] [Google Scholar]
- Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Gene. 2013;9 doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.J. Receptor-mediated mechanisms in ovarian follicle and oocyte development. Gen. Comp. Endocrinol. 2009;163:18–23. doi: 10.1016/j.ygcen.2008.11.032. [DOI] [PubMed] [Google Scholar]
- Shao Y., Chen Y. Roles of circular RNAs in neurologic disease. Front. Mol. Neurosci. 2016;9:25. doi: 10.3389/fnmol.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Li T., Zhang G., Wu P., Chen F., Lou Q., Chen L., Yin X., Zhang T., Wang J.Y. Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genomics. 2019;20:96. doi: 10.1186/s12864-019-5462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Steyrer E., Retzek H., Sanders E.J., Schneider W.J. Chicken oocyte growth: receptor-mediated yolk deposition. Cell Tissue Res. 1993;272:459–471. doi: 10.1007/BF00318552. [DOI] [PubMed] [Google Scholar]
- Sirotkin A.V., Benco A., Tandlmajerova A., Vasicek D., Kotwica J., Darlak K., Valenzuela F. Transcription factor p53 can regulate proliferation, apoptosis and secretory activity of luteinizing porcine ovarian granulosa cell cultured with and without ghrelin and FSH. Reproduction. 2008;136:611. doi: 10.1530/REP-08-0229. [DOI] [PubMed] [Google Scholar]
- Tilly J.L., Kowalski K.I., Johnson A.L. Stage of ovarian follicular development associated with the initiation of steroidogenic competence in avian granulosa cells. Biol. Reprod. 1991;44:305–314. doi: 10.1095/biolreprod44.2.305. [DOI] [PubMed] [Google Scholar]
- Vicente J.S., Saenz-de-Juano M.D., Jiménez-Trigos E., Viudes-De-Castro M.P., Penaranda D.S., Marco-Jiménez F. Rabbit morula vitrification reduces early foetal growth and increases losses throughout gestation. Cryobiology. 2013;67:321–326. doi: 10.1016/j.cryobiol.2013.09.165. [DOI] [PubMed] [Google Scholar]
- Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T., Gong Y., Liu J., Dong Y.H., Li N., Li P.F. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- Wang M., Yu F., Wu W., Zhang Y., Chang W., Ponnusamy M., Wang K., Li P.F. Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 2017;13:1497–1506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PloS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Leader A., Tsang B.K. Follicular stage-dependent regulation of apoptosis and steroidogenesis by prohibitin in rat granulosa cells. J. Ovarian Res. 2013;6:1–10. doi: 10.1186/1757-2215-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xiao H., Pi J., Zhang H., Pan A., Pu Y., Liang Z.H., Shen J., Du J.P. The circular RNA aplacirc_13267 upregulates duck granulosa cell apoptosis by the apla-miR-1-13/THBS1 signaling pathway. J. Cell Physiol. 2020;235:5750–5763. doi: 10.1002/jcp.29509. [DOI] [PubMed] [Google Scholar]
- Wang Y., Lan Y., Lu H. Opsin3 downregulation induces apoptosis of human epidermal melanocytes via mitochondrial pathway. Photochem. Photobiol. 2020;96:83. doi: 10.1111/php.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtysiak D., Okólski A., Sechman A. Structure and steroidogenic activity of the granulosa layer of F1 preovulatory ovarian follicles of the hen (Gallus domesticus) Folia. Biologica. 2011;59:59–64. doi: 10.3409/fb59_1-2.59-64. [DOI] [PubMed] [Google Scholar]
- Woods D.C., Johnson A.L. Regulation of follicle-stimulating hormone- receptor messenger RNA in hen granulosa cells relative to follicle selection. Biol. Reprod. 2005;72:643–650. doi: 10.1095/biolreprod.104.033902. [DOI] [PubMed] [Google Scholar]
- Xu S.R., Wei P., Yang Q.L., Jia G.X., Ma S.K., Yang Q.E., Jun Z., Zhang R.N. Transcriptome analysis revealed key signaling networks regulating ovarian activities in the domestic yak. Theriogenology. 2020;147:50–56. doi: 10.1016/j.theriogenology.2020.02.023. [DOI] [PubMed] [Google Scholar]
- Xu J., Gao X., Li X., Ye Q., Jebessa E., Abdalla B.A., Nie Q.H. Molecular characterization, expression profile of the FSHR gene and its association with egg production traits in muscovy duck. J. Genet. 2017;96:341–351. doi: 10.1007/s12041-017-0783-x. [DOI] [PubMed] [Google Scholar]
- You S., Bridgham J.T., Foster D.N., Johnson A.L. Characterization of a chicken follicle-stimulating hormone receptor (cFSH-R) cDNA, and expression of cFSH-R mRNA in the ovary. Biol. Reprod. 1996;55:1055–1062. doi: 10.1095/biolreprod55.5.1055. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.L, Mi C.H., Wang Q., Zou W.B., Dai G.J., Zhang T., Zhang G.X., Xie K.Z., Wang J.Y., Shi H.Q. Comprehensive analyses of circRNA expression profiles and function prediction in chicken cecums after Eimeria tenella. Infection. Front. Cell Infect. Mi. 2021;11:108. doi: 10.3389/fcimb.2021.628667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yan Y., Lei X., Li A., Zhang H., Dai Z., Li X.J., Chen W.G., Lin W.C., Chen F., Ma J.Y., Xie Q.M. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget. 2017;8:34961–34970. doi: 10.18632/oncotarget.16442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang X.O., Chen T., Xiang J.F., F.Yin Q., Xing Y.H., Zhu S.S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Chen C.Z., Xu M.Q., Zhang L.Q., Liu J.B., Gao Y., Jiang H., Yuan B., Zhang J.B. MiR-31 and miR-143 affect steroid hormone synthesis and inhibit cell apoptosis in bovine granulosa cells through FSHR. Theriogenology. 2019;123:45–53. doi: 10.1016/j.theriogenology.2018.09.020. [DOI] [PubMed] [Google Scholar]
- Zheng W., Nagaraju G., Liu Z., Liu K. Functional roles of the phosphatidylinositol 3-kinases (PI3Ks) signaling in the mammalian ovary. Mol. Cell. Endocrinol. 2012;356:24–30. doi: 10.1016/j.mce.2011.05.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.