Abstract
Diabetes is a systemic disease, and its progression involves multiple organ dysfunction. However, the exact mechanisms underlying pathological progression remain unclear. Small extracellular vesicles (sEVs) mediate physiological and pathological signaling communication between organs and have been shown to have important regulatory roles in diabetes and its complications in recent years. In particular, the majority of studies in the diabetes-related research field have focused on the noncoding RNAs carried by sEVs. Researchers found that noncoding RNA sorting into sEVs is not random but selective. Both tissue origin differences and environmental variations affect the cargo of sEVs. In addition, the function of sEVs differs according to the tissue they derive from; for example, sEVs derived from adipose tissue regulate insulin sensitivity in the periphery, while sEVs derived from bone marrow promote β-cell regeneration. Therefore, understanding the roles of sEVs from different tissues is important for elucidating their molecular mechanisms and is necessary for the application of sEVs as therapeutic agents for diabetes treatment in the future. In this review, we summarized current studies on the mechanisms of noncoding RNA sorting into sEVs, as well as the research progress on the effects of sEVs from different tissue origins and noncoding RNAs in diabetes and diabetic complications. The knowledge of noncoding RNAs in sEVs will help us better understand the role of sEVs in the diabetes progression.
Keywords: Extracellular vesicles, Diabetes, Noncoding RNA, Exosomes
1. Introduction
Diabetes is a chronic disease that is characterized by high blood glucose levels, which are due to absolute (type 1 diabetes, T1D) or relative insulin deficiency (type 2 diabetes, T2D). Compared with T1D, which is due to genetics and autoimmunity, the incidence of T2D is rising rapidly because of sedentary lifestyles and the increasing prevalence of obesity. Moreover, although glucose can be controlled, the incidence of diabetic complications is still high in patients [1]. Thus, exploring the pathological mechanism of diabetes and its complications is important. In recent years, extracellular vesicles (EVs) have attracted much attention. The ability of EVs to mediate cell–cell communication and crosstalk between organs has attracted a great interest in exploring the biological function and therapeutic potential of EVs [2]. Experimental evidence has demonstrated that EVs secreted by multiple cells and tissues carry various proteins, nuclei, and lipids to nearby or distant cells/tissues, promoting or protecting against diabetes and its complications [[3], [4], [5]]. The majority of studies on EVs in diabetes have focused on the noncoding RNA cargo. The noncoding RNA contained in small EVs (sEVs) has been shown to have a regulatory role in diabetic pathology in different ways, such as by regulating β-cell survival, insulin secretion, and insulin sensitivity signal-related proteins (such as PI3K/AKT, PPARα, and PPARγ) in target organs and protecting against diabetes-related complications [[6], [7], [8]]. We reviewed the pathological functions of noncoding RNAs in EVs in diabetes in 2019 [9]. Since then, the field of noncoding RNAs in diabetes and the related complications has expanded rapidly.
In this review, we will summarize EV biology and the mechanisms of noncoding RNA packaging into EVs. In particular, we focus on sEVs derived from different organs and tissues and discuss the possible role of noncoding RNAs in sEVs as both mediators and biomarkers of diabetes and its complications with an emphasis on studies published from 2019 forward, highlighting the potential use of EVs as therapeutic candidates for diabetes treatment.
2. EV biogenesis, biomarkers, and nomenclature
EVs are small vesicles derived from almost all kinds of cell types. Based on the size, EVs can be classified as large (200–1000 nm) and small (50–150 nm). Based on the biogenesis pathway, EVs can be classified as microvesicles or exosomes. Microvesicles (or microparticles) are shed directly from the surface of the plasma membrane via outward budding. The term “exosome” specifically refers to EVs that arise from the endosomal pathway. The exosome biogenesis process is initiated by an intraluminal vesicle (ILV) formation by the inward budding of the plasma membrane and subsequent multivesicular body (MVB) formation by a second invagination. Then, MVBs are either degraded by fusing with lysosomes or release exosomes by fusing with the plasma membrane [10]. Endosomal sorting complexes required for transport (ESCRT)–dependent pathways and ESCRT-independent pathways mediate exosome biogenesis [10,11].
Currently, discriminating EVs by biogenesis pathways is unsatisfactory. Some protein markers that are widely used, such as CD63 and Hsp90, are expressed on all types of EVs. CD81 and TSG101, which are widely used exosome markers, are not exclusively expressed in exosomes but are also expressed in some microvesicles [12]. Thus, protein marker detection identifies only the majority of EVs. In addition, both microvesicles and exosomes are <150 nm in size, and some EVs that are larger than 150 nm originate via the endosome pathway. Given that current EV purification methods or protein markers are unable to isolate endosome-derived EVs, according to MVSC 2018 [12], the nomenclature for EVs is recommended based on the size but not by the biogenesis pathway. In this case, as almost all studies we examined did not verify EV biogenesis, we referred to sEVs as <150 nm in the references but ignored the representation of “exosomes.”
3. Noncoding RNA sorting into EVs
The functions of EVs are largely determined by their cargoes. Proteins, lipids, and nucleic acids can all be carried in EVs, and RNAs can be encapsulated in EVs and transferred to recipient cells while retaining their function [13,14]. Interestingly, some RNAs are particularly enriched in EVs compared to their donor cells [15], suggesting that RNAs are not randomly packaged into EVs. Noncoding RNAs mainly include microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs). Despite their inability to encode proteins, these RNAs regulate multiple biological functions by recruiting epigenetic modifier proteins, controlling mRNA decay and translation, and sequestering transcription factors [16]. To date, knowledge about how each noncoding RNA is sorted into EVs is unclear. However, with biochemical, biological, and computational approaches, the mechanisms of the intracellular events responsible for the packaging of noncoding RNAs in EVs include 1) RNA-binding protein (hnRNP family, YBX1, HuR, and Ago2)-dependent pathways and 2) other protein/lipid raft-dependent pathways.
3.1. HnRNPA2B1
RNA-binding proteins (RBPs) are proteins that bind RNA through one or multiple globular RNA-binding domains (RBDs) to regulate the function of the bound RNA. Hundreds of proteins have been discovered to be RBPs [17]. Heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1), which belongs to the hnRNP family, has been proven to be a key player that regulates the noncoding RNA in EVs. On the one hand, hnRNPA2B1 regulates the EV cargo by guiding miRNAs by interacting with the GGAG motif at the 3′-end of the miRNA. For example, hnRNPA2B1 directs the loading of miRNAs (miR-575, miR-451, miR-125a, miR-198, miR-601, and miR-887) into EVs through the recognition of 3′-GGAG motifs [18]. hnRNPA2B1 selectively binds to miR-17 and miR-93 by recognizing AGG/UAG motifs and sorts them into EVs [19]. On the other hand, hnRNPA2B1 also regulates the EV cargo by inhibiting miRNA sorting. hnRNPA2B1 inhibits miR-503 sorting into sEVs in endothelial cells, whereas interrupting the hnRNPA2B1-miR503 interaction with epirubicin increases the miR-503 export to sEVs [20]. In addition to miRNA sorting, hnRNPA2B1 participates in lncRNA sorting into EVs. hnRNPA2B1 interacts with H19, mediates H19 packaging into EVs, and promotes gefitinib resistance in non-small cell lung cancer (NSCLC) cell lines [21]. hnRNPA2B1 is crucial for lncRNA-AFAP-AS1 sorting into EVs and promotes trastuzumab resistance in breast cancer cell lines [22]. These two studies did not verify the exact binding site of hnRNPA2B1 and lncRNA, but a study about the role of sEV lncRNA-LNMAT2 in lymphatic metastasis in a VEGF-C-independent pathway in bladder cancer cells showed that hnRNPA2B1 binds to LNMAT2 by a particular structure, which was located in the 1930–1960 nt region of LNMAT2 and formed a stem–loop structure [23], suggesting a different way of guiding lncRNA into sEVs compared with miRNA.
3.2. HnRNPC1/hnRNPQ
Other hnRNP family members, such as hnRNPC1 and hnRNPQ, also regulate miRNA and lncRNA sorting into EVs. hnRNPC1 binds to miR-30d and colocalizes with EVs in human endometrial epithelial cells (hEECs) [24], whereas hnRNPQ binds to miR-3470a, miR-194-2-3p, miR365-2-5p, miR-6981–5p, and miR-690, driving these RNAs to EVs in hepatic cells. In this study, the 3′-GGCU motif was identified as the binding motif for hnRNPQ and miRNA. Moreover, hnRNPA2B1 with a 3′-GGAG motif-binding capacity did not overlap with hnRNPQ, indicating sequence-specific EV sorting among these two hnRNPs [25].
3.3. YBX1/HuR/Ago2
Other RBPs have been shown to participate in the noncoding RNA entry into EVs. YBX1 is an RNA-binding protein that recognizes the Y-box motif (5′-CTGATTGGCCAA), the single-stranded motif GGGG, or the single/double-stranded motifs CACC and CATC. A study showed that miR-223 required YBX1 sorting into EVs in HEK293T cells [26]. Then, the same group found that YBX1 was required for the sorting of various kinds of miRNAs into sEVs, including tRNAs, Y RNAs, and vault RNAs [27]. Under hypoxia/reoxygenation conditions, miR-133 is specifically sorted into EPC-derived exosomes via YBX-1 in human endothelial progenitor cells (EPCs) [28]. However, under starvation conditions, HuR (also known as ELVAL1) reversibly binds to miR-122 by ubiquitination in hepatoma cells [29], and in gastric cancer cells, HuR binds to miR-1246 by the AUUUU motif and transports it into sEVs [30]. In addition, miR-150 and miR-142 are selectively exported to EVs by binding to Ago2 in prostate cancer cells [31].
3.4. Other proteins/lipid rafts
In addition to RBPs, other proteins or lipids have been reported to participate in miRNA sorting. Most recently, EWI-2 was found to be enriched in EVs and regulate the sorting of multiple miRNAs into EVs [32]. EWI-2 is a member of the Ig protein subfamily that functions as a TGFβ signal inhibitor and regulates cancer cell proliferation and metastasis [33]. EWI-2 is localized on both plasma membranes and the nuclear envelope, and the EWI-2 knockout enforces miR-3934–5p packaging into sEVs and targets EGFR signaling [32]. Interestingly, the cytoplasmic tail of EWI-2 actively engages with the cell membrane via phosphatidylinositol phosphate (PIP) binding and palmitoylation [34], suggesting that membrane lipids could mediate RNA sorting. Lipidomic analysis showed that compared to that of the origin cells, sEV lipid composition is normally enriched in cholesterol, sphingomyelin, and glycosphingolipids (ceramide, for example) [35,36]. This composition is similar to that of membrane raft domains. Membrane rafts regulate protein sorting [37] and the biogenesis [38] of EVs. A recent work reported direct interactions between lipid rafts and miRNAs, and the authors identified four raft motifs that appear frequently on EV-associated miRNAs [39]; later, the same group confirmed that miRNAs containing raft motifs (UUGU, UCCC, CUCC, and CCCU) and some EXO motifs (CCCU and UCCU) had higher affinities for raft liposomes than aptamers without these motifs, suggesting direct RNA-sEV interactions [40]. A study reported that inhibiting the activity of nSMase2, an enzyme that synthesizes ceramide, with the chemical inhibitor GW4869 resulted in a reduced miRNA content in EVs [41]. However, GW4869 concurrently reduced the EV marker CD63, suggesting that ceramide affects EV biogenesis in addition to selectively sorting miRNAs. Table 1 summarizes the proteins and mechanisms of noncoding RNA sorting into EVs.
Table 1.
Proteins and mechanisms related to noncoding RNA sorting into EVs.
| RNA-binding Proteins | Relevant miRNA | Relevant lncRNA | EV motifs | Sorting control mechanism | EVs derived | Reference |
|---|---|---|---|---|---|---|
| HnRNPA2B1 | miR-575, miR-451, miR-125a, miR-198, miR-601, and miR-887 | – | 3′-GGAG | Bind to miRNA and promote it sorting into EVs | Primary T lymphocytes leukemic Jurkat T-cell line | 18 |
| HnRNPA2B1 | miR-17 and miR-93 | – | 3′-GGAG | Lung epithelial cells | 19 | |
| HnRNPA2B1 | miR-503 | – | NV | Bind to miR-503 and inhibit it sorting into EVs | Endothelial cells | 20 |
| HnRNPA2B1 | – | H19 | NV | Bind to miRNA and promote it sorting into EVs | Non-small cell lung cancer (NSCLC) cells | 21 |
| HnRNPA2B1 | – | AFAP-AS1 | NV | Breast cancer cell lines | 22 | |
| HnRNPA2B1 | LNMAT2 | 1930–1960 nt region | Form a stem–loop structure | Bladder cancer cells | 23 | |
| HnRNPC1 | miR-30d | – | NV | Bind to miRNA and promote it sorting into EVs | Human endometrial epithelial cells (hEECs) | 24 |
| hnRNPQ | miR-3470a, miR-194-2-3p, miR-365-2-5p, miR-6981–5p, and miR-690 | – | 3′-GGCU | Hepatic cells | 25 | |
| YBX1 | miR-223 | – | NV | HEK293T | 26 | |
| YBX1 | miR-133 | – | NV | Human endothelial progenitor cell | 28 | |
| Ago2 | miR-150 and miR-142–3p | – | NV | HEK293 | 31 | |
| HuR | miR-122 | – | NV | Hepatoma cells | 29 | |
| HuR | miR-1246 | – | 3′-AUUU | Gastric cancer cells | 30 | |
| EWI-2 | miR-3934–5p | – | NV | Prostate cancer cells | 32 |
NV: not verify.
Studies on the selection mechanisms of the noncoding RNA into sEVs are mostly conducted under normal conditions in tumor cells and immune cells, and only a few experiments have investigated hepatic cells and epithelial cells. Under high glucose conditions, the sEV packaging of miR-126 and miR-26a into coronary artery endothelial cells is reduced [42], whereas exercise elevates the level of the miR-126 expression in sEVs of endothelial progenitor cells [43], indicating possible mechanistic differences between sorting into sEVs dependent on cell metabolic status. In addition, epigenetic modifications of hnRNPA2B1, such as SUMOylation and O-GlcNAcylation, are essential for hnRNPA2B1 sorting into sEVs [18,19]. Both SUMOylation and O-GlcNAcylation on various protein substrates have been implicated in many diseases, including diabetes and its complications. For example, SUMOylation acts as a brake on glucose-stimulated insulin secretion (GSIS) [44], and O-GlcNAcylation is necessary for the maintenance of α-cell mass and regulation of glucagon secretion [45]. Therefore, SUMOylation and O-GlcNAcylation in diabetic states might contribute to noncoding RNA sorting into sEVs. In addition, the function of hnRNPA2B1 in transporting RNA to sEV is dependent on its ability to interact with cytoskeletal components [18]. Interestingly, insulin secretion and insulin-regulated GLUT4 transport also depend on their effects on cytoskeletal remodeling [46,47]. The intermediate function of the cytoskeletal structure between diabetic conditions and sEV noncoding RNA sorting is worthy of study.
4. Blood- and urine-derived sEV noncoding RNAs in diabetes
4.1. Plasma and urine sEV origins
The sEV function is largely determined by the cargo; normally, the noncoding RNA profile in EVs is dependent on the cell type they derived from or conditions to which the cells are exposed. We reviewed recent experimental studies, and the most studied diabetes-related noncoding RNAs in EVs are in circulating sEVs/urinary sEVs. Adipose tissue constitutes a major source of circulating sEV miRNAs because the fat-specific knockout of the miRNA-processing enzyme Dicer (ADicerKO) reduces serum sEV miRNA dramatically compared to total miRNA reduction [48]. A computational method to compare the distinct tissue-cellular origins from plasma sEVs (including mRNA, circRNA, and lncRNA) showed that only 0.2% of human plasma sEVs were derived from tissues, and 99.8% were generated from hemopoietic cells in the plasma. Furthermore, in this study, the authors found that plasma sEVs predominantly originated from platelets (approximately 51%), followed by lymphocytes (approximately 40%) and monocytes (approximately 3.5%). Concerning the distribution of tissue constituents, adipose tissue predominated (82–91%), followed by muscle, lung, and liver tissues (8–10%) [49]. The samples used in this study were from the plasma of patients, healthy individuals, and patients with liver disease. The researchers also found an increased enrichment in the liver fraction in samples with liver disease compared with healthy individuals, suggesting that the heterogeneity of circulating EV origins is affected by disease status. Moreover, a recent report of genetic source tracking in human urinary sEVs showed that urinary sEVs mostly express genes for bladder tissues. After analyzing the differentially expressed genes in bladder cancer, kidney cancer, and control samples, the authors showed that a certain number of differentially expressed genes in urinary sEVs are from immune cells, suggesting the role of immune activity in a urinary system-related bladder cancer progression [50]. Therefore, understanding the function of sEV noncoding RNAs of different origins deepens our insights into the mechanisms of diabetes.
4.2. Plasma and urine sEV noncoding RNAs as potential biomarkers for diabetes
In recent years, miRNAs have been investigated as potential biomarkers for the prediction of diabetes; plasma miR-146, miR-192, and miR-193b and urine miR-377 and miR-144 were all shown to be upregulated or downregulated in patients with T2D compared with healthy controls [[51], [52], [53]]. However, the miRNA expression was not consistent between studies. For example, higher miR-150 and miR-30a-5p and lower miR-15a and miR-375 levels were found in subjects with T2D than in healthy subjects, with an area under the curve (AUC) of 0.714–0.793 when clinical variables were combined with the miRNAs [54]. However, another study showed that miR-375 was 3-fold higher in subjects with prediabetes and 5.9-fold higher in subjects with T2D than in controls. In this study, miR-375 discriminated patients with prediabetes from healthy subjects with an AUC of 0.76 and patients with T2D from healthy subjects with an AUC of 0.77 [55]. Another example is miR-21, which is upregulated in peripheral blood mononuclear cells of patients with T1D compared to healthy controls [56] and is increased in patients with T2D with an AUC of 0.830 in discriminating diabetic retinopathy [57]. However, another study showed that circulating miR-21 levels were significantly decreased in the diabetic cardiomyopathy (DCM) group compared to the non-DCM group. The AUC was 0.939 when miR-21 was combined with the duration of diabetes, HbA1c%, and lipid profiles [58]. Therefore, there is no sufficient evidence from human studies to establish any miRNA as a new biomarker for T1D or T2D.
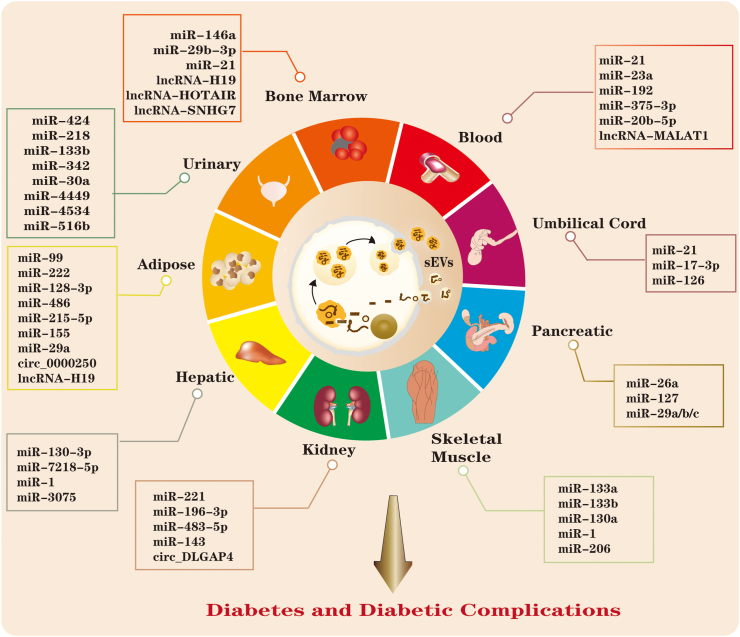
sEV miRNAs and lncRNAs have unique profiles in patients with diabetes compared with healthy controls. For example, urinary sEVs containing miR-424 and miR-218 [59] are increased, and serum sEV miR-21–5p [6] is increased in children with T1D compared with healthy controls. In addition, plasma sEV miR-23a and miR-192 are elevated in patients with T2D compared with controls, with AUCs of 0.828 and 0.717 [60,61]. Furthermore, lncRNA-MALAT1 in serum EVs from patients with T2D is reduced significantly, and this reduction is associated with total cholesterol and HDL-c levels, which are risk factors for T2D [62]. Although multiple sEV noncoding RNAs have been discovered to be differentially expressed in body fluids, the changes in sEV miRNAs/lncRNAs as causes or consequences for diabetes are unclear. Additionally, evidence has shown that sEV miRNAs or lncRNAs have different expression profiles in patients with diabetic complications compared with healthy controls or patients with no complications, making them potential biomarkers for predicting diabetic complications. For instance, the urinary sEV cargoes miR-133b, miR-342, and miR-30a were expressed at significantly elevated levels with AUCs of 0.867, 0.910, and 0.897, respectively, to discriminate diabetic nephropathy (DN) patients from healthy controls [63]. sEV miR-4449 was significantly higher in DN patients than in diabetic patients without nephropathy [64]. miR-21–5p was upregulated, and miR-30 was downregulated in DN patients and patients with chronic kidney disease but no diabetes, with an AUC of 0.813 when combined with age, sex, and HDL-C [65], indicating the biomarker potential of these two miRNAs in renal dysfunction. Later, another study showed that 7 urinary sEV miRNAs were elevated in DN patients compared to diabetic patients without nephropathy, but after correlation with proteinuria levels, only the expression of miR-4534 and miR-516b-5p showed promise as biomarkers for the progression of DN, with an AUC of 0.786 for discrimination of T2D and DN patients [66] (Figure 1, Figure 2).
Figure 1.
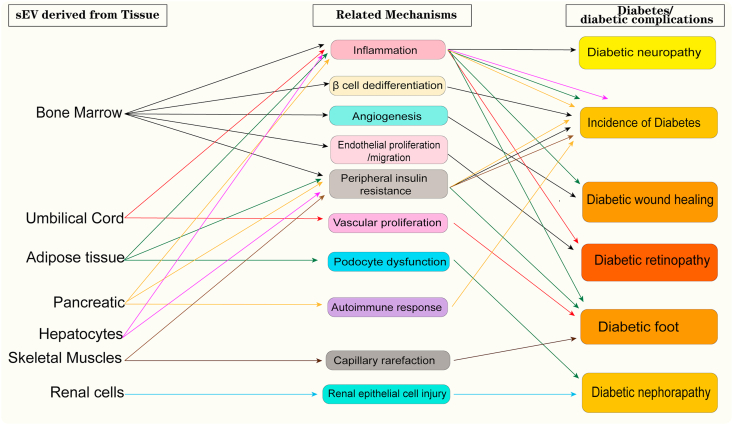
Noncoding RNAs in sEVs derived from different tissues/cells mediate the progression of diabetes and its complications. sEV noncoding RNAs derived from plasma/urinary tissue have mainly been explored as biomarkers for diabetes and its complications. sEV noncoding RNAs derived from adipose/pancreatic/skeletal muscle/liver/kidney tissue have been proven to have important roles in insulin secretion and sensitivity regulation, whereas BM-MSCs and umbilical cord-derived sEV noncoding RNAs are promising agents for diabetic therapy.
Figure 2.
An overall summary of the antidiabetic effects of sEVs at the individual tissue level.
5. Bone marrow cell-derived sEV noncoding RNAs in diabetes
Mesenchymal stem cells (MSCs) are stem cells that can differentiate into osteoblasts, adipocytes, chondroblasts, and even neuron-like cells. The high self-renewal potential, multilineage differentiation potential, and immunomodulatory properties [67] make these cells attractive for disease therapy. Bone marrow is the original source of MSCs and is still frequently used in research on MSC-derived sEVs. The majority of studies obtained bone marrow MSCs (BM-MSCs) by using a colony-forming unit-fibroblast approach, in which raw unpurified bone marrow is directly seeded into plates or flasks. To verify the phenotype of MSCs, the positive expression of biomarkers was detected on their surface, such as CD73, CD90, and CD105, and negative biomarkers, such as CD34, CD11b, CD14, CD19, CD45, and CD79a, were also examined [68].
BM-MSCs initiate pancreatic regeneration [69], indicating their promise in diabetes treatment. However, using MSCs as a therapeutic tool is associated with poor cell survival and weak biological activity because of the complex internal environment. In addition, the stemness of MSCs may cause unexpected side effects [70]. Thus, studies have been looking for strategies to enhance MSC effects but with low risk. Surprisingly, experimental studies showed that BM-MSC-derived sEVs had better effects on the gene expression (insulin, Pdx1, Smad2, Smad3, and TGFβ) than treatment with BM-MSCs in T1D rats [71]. BM-MSC-derived sEVs alleviated T2D by reversing peripheral insulin resistance in a rat model [72]. The noncoding RNAs carried by sEVs play important roles in the beneficial effects of BM-MSCs. A study exploring the protective effects of BM-MSC sEVs against diabetic peripheral neuropathy in diabetic db/db mice showed that MSC-derived sEVs did not significantly affect blood glucose levels or lipid levels but suppressed the inflammatory response in nerve tissues [73]. Later, the same group engineered BM-MSC sEVs that overexpressed miR-146a, and the results showed that engineered BM-MSC sEVs accelerated the improvements in diabetic peripheral neuropathy compared with BM-MSC-sEV treatment alone [74]. BM-MSC-derived sEV miR-146a was also proven to protect against high glucose-induced β-cell dysfunction by reversing β-cell dedifferentiation [72]. In addition, BM-MSC-derived sEV miR-21 attenuated erectile dysfunction in diabetic rats [75], suggesting that the miRNA cargoes in MSCs highly contribute to their protective effects against diabetes and diabetic complications. Moreover, lncRNAs carried by BM-MSC-derived sEVs have a beneficial effect on diabetic complications. The overexpression of HOTAIR in BM-MSC-produced sEVs enhanced angiogenesis and wound healing in a diabetic db/db mouse model [76], and BM-MSC-derived sEV lncRNA SNHG7 inhibited high glucose-induced proliferation and migration of retinal endothelial cells, which is a critical step in diabetic retinopathy development [77]. The effects of the MSC-sEV cargo also depend on cell status and the culture environment. BM-MSCs from aged mice had increased sEV miR-29b-3p release and induced insulin resistance by targeting SIRT1. Conversely, BM-MSCs specifically overexpressing miR-29b-3p in young mice induced insulin resistance [78] (Figure 1, Figure 2).
6. Umbilical cord-derived sEV noncoding RNAs in diabetes
The human umbilical cord is another major source of MSCs. Umbilical cord-MSC (huMSC)-derived sEVs were more prominent in tissue damage repair than sEVs from other cells [79]. The noncoding RNA cargo confers protective effects. It has been reported that sEVs derived from huMSCs protect β-cells from hypoxia-induced apoptosis, and transfection of the sEV cargo miR-21 abrogated β-cell apoptosis by targeting p38 MAPK signaling [13]. Another study confirmed the protective effects of huMSC-derived sEV miR-21 on vessel proliferation and angiogenesis in the diabetic foot [80]. In addition, huMSC-derived sEV miRNAs were proven to protect against diabetic retinopathy by targeting key regulators, such as sEV miR-17–3p targeting STAT1 [81] and sEV miR-126 downregulating the HMGB1 signaling pathway [82], both of which are important proteins that mediate inflammation during the progression of diabetic retinopathy (Figure 1, Figure 2).
7. Adipose tissue-derived sEV noncoding RNAs in diabetes
Given that obesity is a high-risk factor for T2D incidence, researchers have been looking for links between adipose tissue and diabetes/diabetic complications. Adipose tissue consists of white adipose tissue (WAT), which is a major tissue type that functions as energy storage, and brown adipose tissue (BAT), which represents 1–2% of fat, generates heat under cold stress and contains a small fraction of stromal and immune cells, such as macrophages [83]. Studies using sEVs derived from adipose tissues are mostly focused on WAT from visceral fat tissue or subcutaneous fat tissue. Preadipocytes or adipose-derived stem cells (ADSCs) can be isolated from adipose tissue with collagenase I digestion and cultured in DMEM with 10–15% FBS [84,85]. To induce differentiation, adipogenic medium was used in some experiments to obtain an adipocyte phenotype. For ADSC identification, some studies verified biomarkers such as CD29, CD44, and CD105 and used a differentiation medium to verify multilineage differentiation [86].
Adipose tissue-derived miRNAs are the major circulating sEV miRNAs. sEV-derived miRNAs in plasma samples from obese and nonobese women showed miRNA patterns associated with body mass index (BMI) and homeostatic model assessment of insulin resistance (HOMA-IR) [87]. DicerKO mice exhibit insulin resistance with markedly impaired glucose tolerance test (GTT) results. GTT results were significantly improved in DicerKO mice that received BAT transplantation but not WAT transplantation. Furthermore, sEV miR-99 was shown to target FGF21 mRNA. Interestingly, only BAT-derived sEVs reduced hepatic FGF21 levels, indicating that BAT-derived sEVs may more efficiently target the liver than WAT-derived sEVs [48]. Another study confirmed that gonadal WAT-derived serum sEV miR-222 promoted insulin resistance in the liver and skeletal muscle of HFD-fed obese mice by suppressing IRS-1 expression [88]. Studies of sEVs from ADSCs focus on therapy for diabetic complications. A circRNA (circ_0000250) that is overexpressed in sEVs derived from ADSCs enhanced the beneficial effect of promoting wound healing in diabetes by absorbing miR-128–3p and upregulating SIRT1 [89]. Podocytes are highly specialized cells in the kidney glomerulus, which provide a barrier for plasma proteins. Podocyte dysfunction is considered an important pathology for kidney disease. A study showed that the ADSC-sEV cargo miR-486 ameliorated DN symptoms by inhibiting the Smad1/mTOR signaling pathway in podocytes [90]. Additionally, ADSC-sEVs mediated the shuttling of miR-215–5p to podocytes, thereby protecting against HG-induced podocyte loss [91].
During the progression of obesity and insulin resistance, the phenotypic switch of macrophages from M1 to M2, which is a proinflammatory phenotype, is an important factor that promotes insulin resistance. Preadipocyte-derived sEVs have been proven to be associated with insulin resistance development by intercommunicating with macrophages [92]. sEVs from visceral adipose tissue are taken up by blood monocytes and promote the M1 transition to induce insulin resistance [93]. In addition, sEV miRNAs derived from adipose tissue macrophages have shown that adipose macrophages secrete miR-155-containing sEVs, which regulate insulin activity by targeting PPARγ in adipocytes, the liver, and the muscle [94]. After this study, obesity-induced adipose macrophage-secreted sEVs containing miR-29a were transferred into adipocytes, myocytes, and hepatocytes and caused insulin resistance by targeting PPAR-delta [95], which is a protein that reduces inflammation and obesity. In addition, subcutaneous fat tissue-MSCs release sEVs containing lncRNA-H19 and H19 to prevent apoptosis and inflammation in fibroblasts by impairing miR-152-3p-mediated PTEN inhibition, leading to the stimulation of wound healing in a diabetic foot ulcer [96] (Figure 1, Figure 2).
8. Pancreatic tissue-derived sEV noncoding RNAs in diabetes
The loss or failure of pancreatic β-cells is the main pathological characteristic of T1D. Recently, a report demonstrated that the injection of engineered sEVs derived from human pancreas-derived MSCs increased islet numbers and β-cell mass, elevated circulating insulin, and improved glycemic control in T1D rats [97], suggesting that pancreatic cell-derived sEVs are promising in regenerative medicine. Mechanistic studies showed that the activation of the innate or adaptive immune system leads to the irreversible destruction of islet cells. sEVs secreted by human T1D pancreatic β-cells carry autoantigens, including GAD65, IA-2, and proinsulin, which are taken up by and activate dendritic cells, thereby initiating autoimmune responses in T1D [98] and indicating the key role of pancreatic β-cell sEVs in immune responses via autocrine effects. An miRNA array analysis of EVs (size 100–300 nm) derived from human pancreatic β-cells showed that approximately 208 miRNAs were found within EVs, and 8 miRNAs were specifically concentrated in EVs [99]. Another analysis of noncoding RNAs in sEVs (size 50–100 nm) derived from human islets with or without proinflammatory cytokine stress showed that a total of 31 lncRNAs, 19 miRNAs, 25 piRNAs, 8 snoRNAs, and 20 tRNAs were differentially expressed [100]. These sEVs can be taken up by islet endothelial cells and induce proangiogenic and antiapoptotic phenotypes. Experimental studies showed that the transfer of sEV miRNAs from stressed β-cells was responsible for apoptosis in neighboring β-cells [101]. Additionally, the mouse pancreatic cell line Min6 secreted sEVs containing miR-127 to promote endothelial cell migration and tube formation [102]. Furthermore, β-cells can secrete factors that affect insulin sensitivity. Primary pancreatic islet-derived sEVs containing miR-26a preserve β-cell function through paracrine effects and can be taken up by liver tissue and adipose tissue but not the heart or brain, thus enhancing peripheral insulin sensitivity in mice [103]. Another study showed that primary pancreatic islets secrete sEV miR-29a/b/c in response to high levels of free fatty acids (FFAs), and sEV miR-29a/b/c targets insulin signaling in the liver and blunts hepatic insulin sensitivity [104]. Therefore, pancreatic tissue-derived sEVs with the noncoding RNA cargo are important regulators of β-cell survival and peripheral insulin sensitivity (Figure 1, Figure 2).
9. Hepatocyte-derived sEV noncoding RNAs in diabetes
Liver is the main target organ for insulin resistance and exert important effects on systemic metabolism. The noncoding RNA regulatory crosstalk via sEVs between the liver and other organs is not fully understood. It has been reported that liver-derived sEVs containing miR-130–3p could be taken up by adipocytes and improve glucose tolerance by suppressing the PHLPP2/AKT/GLUT4 signaling pathway [105]. In high-fat diet-induced obese mice, hepatocellular sEVs miR-7218–5p promoted proliferation but not insulin secretion in the β-cell line MIN6 [106]. sEVs derived from hepatocytes from high-fat mice delivered miR-1 to promote endothelial inflammation and facilitate atherogenesis [8]. Interestingly, a recent study showed that in mice with early onset obesity, hepatocyte-derived sEVs containing miR-3075 enhanced insulin sensitivity, whereas in mice with chronic obesity, hepatocyte-derived sEVs promoted insulin resistance by inducing macrophage inflammation [107], suggesting that the effects of sEVs secreted by hepatocytes are determined by cell conditions (Figure 1, Figure 2).
10. Skeletal muscle-derived sEV noncoding RNAs in diabetes
Exercise is known to improve insulin sensitivity and benefit many other organs, including the liver, pancreas, and adipose tissue, but the exact mechanisms remain unclear. It has been reported that endurance exercise increases circulatory sEV secretion [108]. Skeletal muscle tissues secrete sEVs containing miR-1, miR-133a, miR-133b, and miR-206. However, these miRNAs are not evenly sorted into sEVs but are affected by muscle type. In one study, sEVs in the extensor digitorum longus, soleus muscles, tibialis anterior (TA), gastrocnemius, and quadriceps muscles were evaluated. The results showed that TA-derived sEVs contained higher miR-1, miR-133a, and miR-133b per muscle weight than those from the other four muscles [109]. Furthermore, exercise induced the expression of skeletal muscle-derived sEV miR-133a and miR-133b, which account for glucose tolerance improvements and insulin sensitivity increases by targeting hepatic Foxo1 [110], which is a key transcription factor for gluconeogenesis in the liver. This finding indicates that the regulatory role of miRNA cargoes in sEVs derived from skeletal muscle is one of the beneficial mechanisms that affect hepatic function. Moreover, skeletal muscle capillary rarefaction is a pathological factor in vasodilatory response dysfunction in diabetes, which contributes to a diabetic foot ulcer, and exercise supports skeletal muscle angiogenesis [111]. A study showed that exercise induced an increase in skeletal muscle-derived sEV miR-130a, which regulated endothelial cell functions via reactive oxygen species-induced NF-κB signaling and contributed to angiogenesis [112] (Figure 1, Figure 2).
11. Renal cell-derived sEV noncoding RNAs in diabetes
DN is a chronic loss of kidney function that occurs in patients with diabetes. Glomerular destruction is the main pathological mechanism and includes tubular cell changes, podocyte dysfunction, and mesangial cell expansion. A study showed that tubular cell-derived sEVs are reduced after high glucose (HG) stimulation, indicating that sEVs might have a regulatory role in DN [113]. Indeed, HG-induced glomerular endothelial cell-derived sEVs can be internalized by podocytes and trigger podocyte dysfunction [114]. Moreover, HG-induced podocyte-derived sEVs induce proximal tubular epithelial cell (TEC) injury. Noncoding RNAs in sEVs mediate these interactions. MiR-221 is encapsulated in sEVs derived from high glucose-induced podocytes and mediates epithelial cell injury through Wnt/β-catenin signaling [115]. LPS-induced renal TECs release sEVs with increased miR-19b-3p, and these sEVs are internalized by macrophages, leading to M1 phenotype polarization by targeting NF-κB/SOCS-1 and resulting in tubulointerstitial inflammation [116]. Another study reported that the loss of noncoding RNAs in sEVs may worsen DN. A study showed that TEC-sEVs carrying miR-483–5p target MAPK1 and TIMP2 to inhibit renal fibrosis, whereas under HG conditions, TEC-derived sEVs were eliminated by hnRNPA1-induced sEV release, resulting in DN development [117]. Similarly, HG increases the secretion of circRNA-DLGAP4 from sEVs derived from mesangial cells, circDLGAP4 functions as a miR-143 sponge, and the loss of sEV circDLGAP4 increases miR-143, thereby promoting diabetic kidney disease progression by repressing ERBB3/NF-kB/MMP-2 expression in mesangial cells [118]. In addition, a recent noncoding RNA profile in sEVs derived from human TECs with or without HG stimulation demonstrated that a total of 169 lncRNAs, 3 circRNAs, and 152 miRNAs were differentially expressed in sEVs secreted by HG-challenged TEC cells compared with controls, and these noncoding RNAs mainly participate in the TNF signaling pathway, NF-κB signaling pathway, and fatty acid metabolism according to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses [119] (Figure 1, Figure 2).
12. Limitation and challenges
To date, MSCs are the most commonly used sEV sources in experimental studies in vivo. Isolated MSC identity was determined by using CD70, CD90, and CD105 as positive markers and CD34 as a negative marker [120]. However, none of the three positive markers could be a specific marker for in vivo MSC detection. CD34 is shown to be positive in many cell types when freshly isolated but loses expression in the culture medium in vitro; thus, the origins of MSCs are questionable, leading to the unreliability and instability of the obtained sEVs. On the other hand, poor reproducibility and underpowered approaches slow the development of sEVs or other EVs as reliable biomarkers or therapeutic entities. Additionally, all the results showing that sEV miRNAs/lncRNAs have therapeutic effects on diabetes and diabetic complications are conducted in vitro or in rodent models, and whether they have the same clinical effect is unknown. Broadly, the metabolic and pathological impacts of sEVs and their cargoes remain an area that needs further exploration.
13. Conclusion
sEVs derived from multiple tissues are being investigated as potential candidates for diabetes diagnosis and therapy. In addition, noncoding RNAs within sEVs largely mediate their biological and pathological effects. However, in addition to obstacles, such as safety, mass production, and quality control, discriminating the functions of sEVs from different tissues is important for future production, which will lead us to choose the most suitable sEV origin for further investigation. Moreover, understanding the exact mechanisms of noncoding RNA sorting into sEVs and their functional role in diabetes could aid sEV-related diabetic therapeutic agent development.
Funding
This work was supported by Qingdao Postdoctoral Application and Basic Research Program and National Natural Science Foundation of China (81700704, 81970253), Major Natural Science Projects of Shandong Province (ZR2019ZD28), and the Natural Science Foundation of Shandong Province (ZR2021MC189).
Acknowledgments
W Chang, M Li, S Miao, W Yu, and L Song collected all of the data, and W Chang and J Wang drafted and wrote the manuscript. All authors have read and approved the final version of the manuscript.
Conflict of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Stolar M. Glycemic control and complications in type 2 diabetes mellitus. Americas Journal of Medicine. 2010;123(3 Suppl):S3–S11. doi: 10.1016/j.amjmed.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Chung I.M., Rajakumar G., Venkidasamy B., Subramanian U., Thiruvengadam M. Exosomes: current use and future applications. Clinica Chimica Acta. 2020;500:226–232. doi: 10.1016/j.cca.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Mathew B., Ravindran S., Liu X., Torres L., Chennakesavalu M., Huang C.C., et al. Mesenchymal stem cell-derived extracellular vesicles and retinal ischemia-reperfusion. Biomaterials. 2019;197:146–160. doi: 10.1016/j.biomaterials.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei F., Wang A., Wang Q., Han W., Rong R., Wang L., et al. Plasma endothelial cells-derived extracellular vesicles promote wound healing in diabetes through YAP and the PI3K/Akt/mTOR pathway. Aging. 2020;12(12):12002–12018. doi: 10.18632/aging.103366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gan L., Xie D., Liu J., Bond Lau W., Christopher T.A., Lopez B., et al. Small extracellular microvesicles mediated pathological communications between dysfunctional adipocytes and cardiomyocytes as a novel mechanism exacerbating ischemia/reperfusion injury in diabetic mice. Circulation. 2020;141(12):968–983. doi: 10.1161/CIRCULATIONAHA.119.042640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakhter A.J., Pratt R.E., Moore R.E., Doucette K.K., Maier B.F., DiMeglio L.A., et al. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia. 2018;61(5):1124–1134. doi: 10.1007/s00125-018-4559-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prattichizzo F., Matacchione G., Giuliani A., Sabbatinelli J., Olivieri F., de Candia P., et al. Extracellular vesicle-shuttled miRNAs: a critical appraisal of their potential as nano-diagnostics and nano-therapeutics in type 2 diabetes mellitus and its cardiovascular complications. Theranostics. 2021;11(3):1031–1045. doi: 10.7150/thno.51605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang F., Chen Q., Wang W., Ling Y., Yan Y., Xia P. Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. Journal of Hepatology. 2020;72(1):156–166. doi: 10.1016/j.jhep.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Chang W., Wang J. Exosomes and their noncoding RNA cargo are emerging as new modulators for diabetes mellitus. Cells. 2019;8(8) doi: 10.3390/cells8080853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;(6478):367. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juan T., Furthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Seminars in Cell & Developmental Biology. 2018;74:66–77. doi: 10.1016/j.semcdb.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 12.Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Chen J., Cheng Y., Fu Y., Zhao H., Tang M., et al. Mesenchymal stem cell-derived exosomes protect beta cells against hypoxia-induced apoptosis via miR-21 by alleviating ER stress and inhibiting p38 MAPK phosphorylation. Stem Cell Research & Therapy. 2020;11(1):97. doi: 10.1186/s13287-020-01610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O., et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 15.Kim K.M., Abdelmohsen K., Mustapic M., Kapogiannis D., Gorospe M. RNA in extracellular vesicles. Wiley Interdisciplinary Reviews: RNA. 2017;8(4) doi: 10.1002/wrna.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Hentze M.W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nature Reviews Molecular Cell Biology. 2018;19(5):327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 18.Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N., et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H., Li C., Zhang Y., Zhang D., Otterbein L.E., Jin Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. Journal of Experimental Medicine. 2019;216(9):2202–2220. doi: 10.1084/jem.20182313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Boza J., Boeckx A., Lion M., Dequiedt F., Struman I. hnRNPA2B1 inhibits the exosomal export of miR-503 in endothelial cells. Cellular and Molecular Life Sciences. 2020;77(21):4413–4428. doi: 10.1007/s00018-019-03425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei Y., Guo W., Chen B., Chen L., Gong J., Li W. Tumorreleased lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncology Reports. 2018;40(6):3438–3446. doi: 10.3892/or.2018.6762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Han M., Gu Y., Lu P., Li J., Cao H., Li X., et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Molecular Cancer. 2020;19(1):26. doi: 10.1186/s12943-020-1145-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Chen C., Luo Y., He W., Zhao Y., Kong Y., Liu H., et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. Journal of Clinical Investigation. 2020;130(1):404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balaguer N., Moreno I., Herrero M., Gonzalez M., Simon C., Vilella F. Heterogeneous nuclear ribonucleoprotein C1 may control miR-30d levels in endometrial exosomes affecting early embryo implantation. Molecular Human Reproduction. 2018;24(8):411–425. doi: 10.1093/molehr/gay026. [DOI] [PubMed] [Google Scholar]
- 25.Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling MicroRNA sorting. Cell Reports. 2016;17(3):799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 26.Shurtleff M.J., Temoche-Diaz M.M., Karfilis K.V., Ri S., Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5 doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shurtleff M.J., Yao J., Qin Y., Nottingham R.M., Temoche-Diaz M.M., Schekman R., et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proceedings of the National Academy of Sciences of the U S A. 2017;114(43):E8987–E8995. doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin F., Zeng Z., Song Y., Li L., Wu Z., Zhang X., et al. YBX-1 mediated sorting of miR-133 into hypoxia/reoxygenation-induced EPC-derived exosomes to increase fibroblast angiogenesis and MEndoT. Stem Cell Research & Therapy. 2019;10(1):263. doi: 10.1186/s13287-019-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee K., Ghoshal B., Ghosh S., Chakrabarty Y., Shwetha S., Das S., et al. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Reports. 2016;17(8):1184–1203. doi: 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y., Wang Z., Zhu X., Chen L., Ma Y., Wang J., et al. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. International Journal of Clinical Oncology. 2020;25(1):89–99. doi: 10.1007/s10147-019-01532-9. [DOI] [PubMed] [Google Scholar]
- 31.Guduric-Fuchs J., O’Connor A., Camp B., O’Neill C.L., Medina R.J., Simpson D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu C., Zhang Q., Wang A., Yang S., Jiang Y., Bai L., et al. EWI-2 controls nucleocytoplasmic shuttling of EGFR signaling molecules and miRNA sorting in exosomes to inhibit prostate cancer cell metastasis. Mol Oncol. 2021;15(5):1543–1565. doi: 10.1002/1878-0261.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H.X., Sharma C., Knoblich K., Granter S.R., Hemler M.E. EWI-2 negatively regulates TGF-beta signaling leading to altered melanoma growth and metastasis. Cell Research. 2015;25(3):370–385. doi: 10.1038/cr.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corken M., Porter J. Is vitamin B(6) deficiency an under-recognized risk in patients receiving haemodialysis? A systematic review: 2000-2010. Nephrology. 2011;16(7):619–625. doi: 10.1111/j.1440-1797.2011.01479.x. [DOI] [PubMed] [Google Scholar]
- 35.Skotland T., Sagini K., Sandvig K., Llorente A. An emerging focus on lipids in extracellular vesicles. Advanced Drug Delivery Reviews. 2020;159:308–321. doi: 10.1016/j.addr.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Chang W., Xiao D., Fang X., Wang J. Phospholipids in small extracellular vesicles: emerging regulators of neurodegenerative diseases and cancer. Cytotherapy. 2022;24(2):93–100. doi: 10.1016/j.jcyt.2021.09.013. [DOI] [PubMed] [Google Scholar]
- 37.de Gassart A., Geminard C., Fevrier B., Raposo G., Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102(13):4336–4344. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 38.Skryabin G.O., Komelkov A.V., Savelyeva E.E., Tchevkina E.M. Lipid rafts in exosome biogenesis. Biochemistry. 2020;85(2):177–191. doi: 10.1134/S0006297920020054. [DOI] [PubMed] [Google Scholar]
- 39.Janas T., Janas P., Sapon K., Janas T. Binding of RNA aptamers to membrane lipid rafts: implications for exosomal miRNAs transfer from cancer to immune cells. International Journal of Molecular Sciences. 2020;21(22) doi: 10.3390/ijms21228503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manka R., Janas P., Sapon K., Janas T., Janas T. Role of RNA motifs in RNA interaction with membrane lipid rafts: implications for therapeutic applications of exosomal RNAs. International Journal of Molecular Sciences. 2021;22(17) doi: 10.3390/ijms22179416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. Journal of Biological Chemistry. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jansen F., Wang H., Przybilla D., Franklin B.S., Dolf A., Pfeifer P., et al. Vascular endothelial microparticles-incorporated microRNAs are altered in patients with diabetes mellitus. Cardiovascular Diabetology. 2016;15:49. doi: 10.1186/s12933-016-0367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma C., Wang J., Liu H., Chen Y., Ma X., Chen S., et al. Moderate exercise enhances endothelial progenitor cell exosomes release and function. Medicine & Science in Sports & Exercise. 2018;50(10):2024–2032. doi: 10.1249/MSS.0000000000001672. [DOI] [PubMed] [Google Scholar]
- 44.Davey J.S., Carmichael R.E., Craig T.J. Protein SUMOylation regulates insulin secretion at multiple stages. Scientific Reports. 2019;9(1):2895. doi: 10.1038/s41598-019-39681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Essawy A., Jo S., Beetch M., Lockridge A., Gustafson E., Alejandro E.U. O-linked N-acetylglucosamine transferase (OGT) regulates pancreatic alpha-cell function in mice. Journal of Biological Chemistry. 2021;296:100297. doi: 10.1016/j.jbc.2021.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khayat Z.A., Tong P., Yaworsky K., Bloch R.J., Klip A. Insulin-induced actin filament remodeling colocalizes actin with phosphatidylinositol 3-kinase and GLUT4 in L6 myotubes. Journal of Cell Science. 2000;113 Pt 2:279–290. doi: 10.1242/jcs.113.2.279. [DOI] [PubMed] [Google Scholar]
- 47.Arous C., Halban P.A. The skeleton in the closet: actin cytoskeletal remodeling in beta-cell function. American Journal of Physiology. Endocrinology and Metabolism. 2015;309(7):E611–E620. doi: 10.1152/ajpendo.00268.2015. [DOI] [PubMed] [Google Scholar]
- 48.Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., He X., Li Q., Lai H., Zhang H., Hu Z., et al. EV-origin: enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Computational and Structural Biotechnology Journal. 2020;18:2851–2859. doi: 10.1016/j.csbj.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu Q., Cheng L., Deng C., Huang L., Li J., Wang Y., et al. The genetic source tracking of human urinary exosomes. Proceedings of the National Academy of Sciences of the U S A. 2021;118(43) doi: 10.1073/pnas.2108876118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mensa E., Giuliani A., Matacchione G., Gurau F., Bonfigli A.R., Romagnoli F., et al. Circulating miR-146a in healthy aging and type 2 diabetes: age- and gender-specific trajectories. Mechanism of Ageing and Development. 2019;180:1–10. doi: 10.1016/j.mad.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Parrizas M., Brugnara L., Esteban Y., Gonzalez-Franquesa A., Canivell S., Murillo S., et al. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. The Journal of Cinical Endocrinology and Metabolism. 2015;100(3):E407–E415. doi: 10.1210/jc.2014-2574. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y., Xiao L., Li J., Kanwar Y.S., Liu F., Sun L. Urine miRNAs: potential biomarkers for monitoring progression of early stages of diabetic nephropathy. Medical Hypotheses. 2013;81(2):274–278. doi: 10.1016/j.mehy.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jimenez-Lucena R., Camargo A., Alcala-Diaz J.F., Romero-Baldonado C., Luque R.M., van Ommen B., et al. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: from the CORDIOPREV study. Experimental & Molecular Medicine. 2018;50(12):1–12. doi: 10.1038/s12276-018-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Muhtaresh H.A., Al-Kafaji G. Evaluation of two-diabetes related microRNAs suitability as earlier blood biomarkers for detecting prediabetes and type 2 diabetes mellitus. Journal of Clinical Medicine. 2018;7(2) doi: 10.3390/jcm7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mostahfezian M., Azhir Z., Dehghanian F., Hojati Z. Expression pattern of microRNAs, miR-21, miR-155 and miR-338 in patients with type 1 diabetes. Archives of Medical Research. 2019;50(3):79–85. doi: 10.1016/j.arcmed.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Jiang Q., Lyu X.M., Yuan Y., Wang L. Plasma miR-21 expression: an indicator for the severity of Type 2 diabetes with diabetic retinopathy. Bioscience Reports. 2017;37(2) doi: 10.1042/BSR20160589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao L., Huang X., Xu M., Qin Z., Zhang F., Hua F., et al. Value of circulating miRNA-21 in the diagnosis of subclinical diabetic cardiomyopathy. Molecular and Cellular Endocrinology. 2020;518:110944. doi: 10.1016/j.mce.2020.110944. [DOI] [PubMed] [Google Scholar]
- 59.Kong Q., Guo X., Guo Z., Su T. Urinary exosome miR-424 and miR-218 as biomarkers for type 1 diabetes in children. Clinical Laboratory. 2019;65(6) doi: 10.7754/Clin.Lab.2018.180921. [DOI] [PubMed] [Google Scholar]
- 60.Liu C., Gao Y., Wu J., Zou J. Exosomal miR-23a and miR-192, potential diagnostic biomarkers for type 2 diabetes. Clinical Laboratory. 2021;67(2) doi: 10.7754/Clin.Lab.2020.200612. [DOI] [PubMed] [Google Scholar]
- 61.Fu Q., Jiang H., Wang Z., Wang X., Chen H., Shen Z., et al. Injury factors alter miRNAs profiles of exosomes derived from islets and circulation. Aging. 2018;10(12):3986–3999. doi: 10.18632/aging.101689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tello-Flores V.A., Valladares-Salgado A., Ramirez-Vargas M.A., Cruz M., Del-Moral-Hernandez O., Cahua-Pablo J.A., et al. Altered levels of MALAT1 and H19 derived from serum or serum exosomes associated with type-2 diabetes. Noncoding RNA Res. 2020;5(2):71–76. doi: 10.1016/j.ncrna.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eissa S., Matboli M., Bekhet M.M. Clinical verification of a novel urinary microRNA panal: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomedicine & Pharmacotherapy. 2016;83:92–99. doi: 10.1016/j.biopha.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 64.Kim H., Bae Y.U., Jeon J.S., Noh H., Park H.K., Byun D.W. The circulating exosomal microRNAs related to albuminuria in patients with diabetic nephropathy. Journal of Translational Medicine. 2019;17(1):236. doi: 10.1186/s12967-019-1983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zang J., Maxwell A.P., Simpson D.A., McKay G.J. Differential expression of urinary exosomal MicroRNAs miR-21-5p and miR-30b-5p in individuals with diabetic kidney disease. Scientific Reports. 2019;9(1):10900. doi: 10.1038/s41598-019-47504-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y., Shen A., Guo F., Song Y., Jing N., Ding X., et al. Urinary exosomal MiRNA-4534 as a novel diagnostic biomarker for diabetic kidney disease. Frontiers in Endocrinology. 2020;11:590. doi: 10.3389/fendo.2020.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mushahary D., Spittler A., Kasper C., Weber V., Charwat V. Isolation, cultivation, and characterization of human mesenchymal stem cells. Cytometry. 2018;93(1):19–31. doi: 10.1002/cyto.a.23242. [DOI] [PubMed] [Google Scholar]
- 68.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 69.Hess D., Li L., Martin M., Sakano S., Hill D., Strutt B., et al. Bone marrow-derived stem cells initiate pancreatic regeneration. Nature Biotechnology. 2003;21(7):763–770. doi: 10.1038/nbt841. [DOI] [PubMed] [Google Scholar]
- 70.Li H., Zhu H., Ge T., Wang Z., Zhang C. Mesenchymal stem cell-based therapy for diabetes mellitus: enhancement strategies and future perspectives. Stem Cell Reviews and Reports. 2021;17(5):1552–1569. doi: 10.1007/s12015-021-10139-5. [DOI] [PubMed] [Google Scholar]
- 71.Sabry D., Marzouk S., Zakaria R., Ibrahim H.A., Samir M. The effect of exosomes derived from mesenchymal stem cells in the treatment of induced type 1 diabetes mellitus in rats. Biotechnology Letters. 2020;42(8):1597–1610. doi: 10.1007/s10529-020-02908-y. [DOI] [PubMed] [Google Scholar]
- 72.He Q., Song J., Cui C., Wang J., Hu H., Guo X., et al. Mesenchymal stem cell-derived exosomal miR-146a reverses diabetic beta-cell dedifferentiation. Stem Cell Research & Therapy. 2021;12(1):449. doi: 10.1186/s13287-021-02371-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Fan B., Li C., Szalad A., Wang L., Pan W., Zhang R., et al. Mesenchymal stromal cell-derived exosomes ameliorate peripheral neuropathy in a mouse model of diabetes. Diabetologia. 2020;63(2):431–443. doi: 10.1007/s00125-019-05043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fan B., Chopp M., Zhang Z.G., Liu X.S. Treatment of diabetic peripheral neuropathy with engineered mesenchymal stromal cell-derived exosomes enriched with microRNA-146a provide amplified therapeutic efficacy. Experimental Neurology. 2021;341:113694. doi: 10.1016/j.expneurol.2021.113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huo W., Li Y., Zhang Y., Li H. Mesenchymal stem cells-derived exosomal microRNA-21-5p downregulates PDCD4 and ameliorates erectile dysfunction in a rat model of diabetes mellitus. The FASEB Journal. 2020;34(10):13345–13360. doi: 10.1096/fj.202000102RR. [DOI] [PubMed] [Google Scholar]
- 76.Born L.J., Chang K.H., Shoureshi P., Lay F., Bengali S., Hsu A.T.W., et al. HOTAIR-loaded mesenchymal stem/stromal cell extracellular vesicles enhance angiogenesis and wound healing. Advanced Healthcare Materials. 2021 doi: 10.1002/adhm.202002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao X., Xue L.D., Di Y., Li T., Tian Y.J., Song Y. MSC-derived exosomal lncRNA SNHG7 suppresses endothelial-mesenchymal transition and tube formation in diabetic retinopathy via miR-34a-5p/XBP1 axis. Life Sciences. 2021;272:119232. doi: 10.1016/j.lfs.2021.119232. [DOI] [PubMed] [Google Scholar]
- 78.Su T., Xiao Y., Xiao Y., Guo Q., Li C., Huang Y., et al. Bone marrow mesenchymal stem cells-derived exosomal MiR-29b-3p regulates aging-associated insulin resistance. ACS Nano. 2019;13(2):2450–2462. doi: 10.1021/acsnano.8b09375. [DOI] [PubMed] [Google Scholar]
- 79.Wang Z.G., He Z.Y., Liang S., Yang Q., Cheng P., Chen A.M. Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells. Stem Cell Research & Therapy. 2020;11(1):511. doi: 10.1186/s13287-020-02032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang C., Luo W., Wang Q., Ye Y., Fan J., Lin L., et al. Human mesenchymal stem cells promote ischemic repairment and angiogenesis of diabetic foot through exosome miRNA-21-5p. Stem Cell Research. 2021;52:102235. doi: 10.1016/j.scr.2021.102235. [DOI] [PubMed] [Google Scholar]
- 81.Li W., Jin L.Y., Cui Y.B., Xie N. Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-17-3p ameliorates inflammatory reaction and antioxidant injury of mice with diabetic retinopathy via targeting STAT1. International Immunopharmacology. 2021;90:107010. doi: 10.1016/j.intimp.2020.107010. [DOI] [PubMed] [Google Scholar]
- 82.Zhang W., Wang Y., Kong Y. Exosomes derived from mesenchymal stem cells modulate miR-126 to ameliorate hyperglycemia-induced retinal inflammation via targeting HMGB1. Investigative Ophthalmology & Visual Science. 2019;60(1):294–303. doi: 10.1167/iovs.18-25617. [DOI] [PubMed] [Google Scholar]
- 83.Kahn C.R., Wang G., Lee K.Y. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. Journal of Clinical Investigation. 2019;129(10):3990–4000. doi: 10.1172/JCI129187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mahmoudifar N., Doran P.M. Mesenchymal stem cells derived from human adipose tissue. Methods in Molecular Biology. 2015;1340:53–64. doi: 10.1007/978-1-4939-2938-2_4. [DOI] [PubMed] [Google Scholar]
- 85.Palumbo P., Lombardi F., Siragusa G., Cifone M.G., Cinque B., Giuliani M. Methods of isolation, characterization and expansion of human adipose-derived stem cells (ASCs): an overview. International Journal of Molecular Sciences. 2018;19(7) doi: 10.3390/ijms19071897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Megaloikonomos P.D., Panagopoulos G.N., Bami M., Igoumenou V.G., Dimopoulos L., Milonaki A., et al. Harvesting, isolation and differentiation of rat adipose-derived stem cells. Current Pharmaceutical Biotechnology. 2018;19(1):19–29. doi: 10.2174/1389201019666180418101323. [DOI] [PubMed] [Google Scholar]
- 87.Santamaria-Martos F., Benitez I.D., Latorre J., Lluch A., Moreno-Navarrete J.M., Sabater M., et al. Comparative and functional analysis of plasma membrane-derived extracellular vesicles from obese vs. nonobese women. Clinical Nutrition. 2020;39(4):1067–1076. doi: 10.1016/j.clnu.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 88.Li D., Song H., Shuo L., Wang L., Xie P., Li W., et al. Gonadal white adipose tissue-derived exosomal MiR-222 promotes obesity-associated insulin resistance. Aging. 2020;12(22):22719–22743. doi: 10.18632/aging.103891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shi R., Jin Y., Hu W., Lian W., Cao C., Han S., et al. Exosomes derived from mmu_circ_0000250-modified adipose-derived mesenchymal stem cells promote wound healing in diabetic mice by inducing miR-128-3p/SIRT1-mediated autophagy. American Journal of Physiology - Cell Physiology. 2020;318(5):C848–C856. doi: 10.1152/ajpcell.00041.2020. [DOI] [PubMed] [Google Scholar]
- 90.Jin J., Shi Y., Gong J., Zhao L., Li Y., He Q., et al. Exosome secreted from adipose-derived stem cells attenuates diabetic nephropathy by promoting autophagy flux and inhibiting apoptosis in podocyte. Stem Cell Research & Therapy. 2019;10(1):95. doi: 10.1186/s13287-019-1177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jin J., Wang Y., Zhao L., Zou W., Tan M., He Q. Exosomal miRNA-215-5p derived from adipose-derived stem cells attenuates epithelial-mesenchymal transition of podocytes by inhibiting ZEB2. BioMed Research International. 2020;2020:2685305. doi: 10.1155/2020/2685305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kranendonk M.E., Visseren F.L., van Balkom B.W., Nolte-’t Hoen E.N., van Herwaarden J.A., de Jager W., et al. Human adipocyte extracellular vesicles in reciprocal signaling between adipocytes and macrophages. Obesity. 2014;22(5):1296–1308. doi: 10.1002/oby.20679. [DOI] [PubMed] [Google Scholar]
- 93.Deng Z.B., Poliakov A., Hardy R.W., Clements R., Liu C., Liu Y., et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes. 2009;58(11):2498–2505. doi: 10.2337/db09-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B., et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171(2):372–384 e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 95.Liu T., Sun Y.C., Cheng P., Shao H.G. Adipose tissue macrophage-derived exosomal miR-29a regulates obesity-associated insulin resistance. Biochemical and Biophysical Research Communications. 2019;515(2):352–358. doi: 10.1016/j.bbrc.2019.05.113. [DOI] [PubMed] [Google Scholar]
- 96.Li B., Luan S., Chen J., Zhou Y., Wang T., Li Z., et al. The MSC-derived exosomal lncRNA H19 promotes wound healing in diabetic foot ulcers by upregulating PTEN via MicroRNA-152-3p. Molecular Therapy - Nucleic Acids. 2020;19:814–826. doi: 10.1016/j.omtn.2019.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cooper T.T., Sherman S.E., Bell G.I., Dayarathna T., McRae D.M., Ma J., et al. Ultrafiltration and injection of islet regenerative stimuli secreted by pancreatic mesenchymal stromal cells. Stem Cells and Development. 2021;30(5):247–264. doi: 10.1089/scd.2020.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cianciaruso C., Phelps E.A., Pasquier M., Hamelin R., Demurtas D., Alibashe Ahmed M., et al. Primary human and rat beta-cells release the intracellular autoantigens GAD65, IA-2, and proinsulin in exosomes together with cytokine-induced enhancers of immunity. Diabetes. 2017;66(2):460–473. doi: 10.2337/db16-0671. [DOI] [PubMed] [Google Scholar]
- 99.Figliolini F., Cantaluppi V., De Lena M., Beltramo S., Romagnoli R., Salizzoni M., et al. Isolation, characterization and potential role in beta cell-endothelium cross-talk of extracellular vesicles released from human pancreatic islets. PLoS One. 2014;9(7):e102521. doi: 10.1371/journal.pone.0102521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krishnan P., Syed F., Jiyun Kang N., Mirmira R.G., Evans-Molina C. Profiling of RNAs from human islet-derived exosomes in a model of type 1 diabetes. International Journal of Molecular Sciences. 2019;20(23) doi: 10.3390/ijms20235903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guay C., Menoud V., Rome S., Regazzi R. Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Communication and Signaling. 2015;13:17. doi: 10.1186/s12964-015-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen Z., Jiang H., Hsu H.T., Qian L., Fu Q., Shen M., et al. MicroRNA-127 inhibits cell proliferation via targeting Kif3b in pancreatic beta cells. Aging. 2019;11(5):1342–1355. doi: 10.18632/aging.101835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu H., Du X., Xu J., Zhang Y., Tian Y., Liu G., et al. Pancreatic beta cell microRNA-26a alleviates type 2 diabetes by improving peripheral insulin sensitivity and preserving beta cell function. PLoS Biology. 2020;18(2):e3000603. doi: 10.1371/journal.pbio.3000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li J., Zhang Y., Ye Y., Li D., Liu Y., Lee E., et al. Pancreatic beta cells control glucose homeostasis via the secretion of exosomal miR-29 family. Journal of Extracellular Vesicles. 2021;10(3):e12055. doi: 10.1002/jev2.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu J., Dong T., Chen T., Sun J., Luo J., He J., et al. Hepatic exosome-derived miR-130a-3p attenuates glucose intolerance via suppressing PHLPP2 gene in adipocyte. Metabolism. 2020;103:154006. doi: 10.1016/j.metabol.2019.154006. [DOI] [PubMed] [Google Scholar]
- 106.Fu Q., Li Y., Jiang H., Shen Z., Gao R., He Y., et al. Hepatocytes derived extracellular vesicles from high-fat diet induced obese mice modulate genes expression and proliferation of islet beta cells. Biochemical and Biophysical Research Communications. 2019;516(4):1159–1166. doi: 10.1016/j.bbrc.2019.06.124. [DOI] [PubMed] [Google Scholar]
- 107.Ji Y., Luo Z., Gao H., Dos Reis F.C.G., Bandyopadhyay G., Jin Z., et al. Hepatocyte-derived exosomes from early onset obese mice promote insulin sensitivity through miR-3075. Nature Metabolism. 2021;3(9):1163–1174. doi: 10.1038/s42255-021-00444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Safdar A., Saleem A., Tarnopolsky M.A. The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nature Reviews Endocrinology. 2016;12(9):504–517. doi: 10.1038/nrendo.2016.76. [DOI] [PubMed] [Google Scholar]
- 109.Mytidou C., Koutsoulidou A., Katsioloudi A., Prokopi M., Kapnisis K., Michailidou K., et al. Muscle-derived exosomes encapsulate myomiRs and are involved in local skeletal muscle tissue communication. The FASEB Journal. 2021;35(2):e21279. doi: 10.1096/fj.201902468RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Castano C., Mirasierra M., Vallejo M., Novials A., Parrizas M. Delivery of muscle-derived exosomal miRNAs induced by HIIT improves insulin sensitivity through down-regulation of hepatic FoxO1 in mice. Proceedings of the National Academy of Sciences of the U S A. 2020;117(48):30335–30343. doi: 10.1073/pnas.2016112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olver T.D., Laughlin M.H. Endurance, interval sprint, and resistance exercise training: impact on microvascular dysfunction in type 2 diabetes. American Journal of Physiology - Heart and Circulatory Physiology. 2016;310(3):H337–H350. doi: 10.1152/ajpheart.00440.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nie Y., Sato Y., Garner R.T., Kargl C., Wang C., Kuang S., et al. Skeletal muscle-derived exosomes regulate endothelial cell functions via reactive oxygen species-activated nuclear factor-kappaB signalling. Experimental Physiology. 2019;104(8):1262–1273. doi: 10.1113/EP087396. [DOI] [PubMed] [Google Scholar]
- 113.Wen J., Ma Z., Livingston M.J., Zhang W., Yuan Y., Guo C., et al. Decreased secretion and profibrotic activity of tubular exosomes in diabetic kidney disease. American Journal of Physiology - Renal Physiology. 2020;319(4):F664–F673. doi: 10.1152/ajprenal.00292.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu X., Gao Y., Xu L., Dang W., Yan H., Zou D., et al. Exosomes from high glucose-treated glomerular endothelial cells trigger the epithelial-mesenchymal transition and dysfunction of podocytes. Scientific Reports. 2017;7(1):9371. doi: 10.1038/s41598-017-09907-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Su H., Qiao J., Hu J., Li Y., Lin J., Yu Q., et al. Podocyte-derived extracellular vesicles mediate renal proximal tubule cells dedifferentiation via microRNA-221 in diabetic nephropathy. Molecular and Cellular Endocrinology. 2020;518:111034. doi: 10.1016/j.mce.2020.111034. [DOI] [PubMed] [Google Scholar]
- 116.Lv L.L., Feng Y., Wu M., Wang B., Li Z.L., Zhong X., et al. Exosomal miRNA-19b-3p of tubular epithelial cells promotes M1 macrophage activation in kidney injury. Cell Death & Differentiation. 2020;27(1):210–226. doi: 10.1038/s41418-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu D., Liu F., Li Z., Pan S., Xie J., Zhao Z., et al. HNRNPA1-mediated exosomal sorting of miR-483-5p out of renal tubular epithelial cells promotes the progression of diabetic nephropathy-induced renal interstitial fibrosis. Cell Death & Disease. 2021;12(3):255. doi: 10.1038/s41419-021-03460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bai S., Xiong X., Tang B., Ji T., Li X., Qu X., et al. Exosomal circ_DLGAP4 promotes diabetic kidney disease progression by sponging miR-143 and targeting ERBB3/NF-kappaB/MMP-2 axis. Cell Death & Disease. 2020;11(11):1008. doi: 10.1038/s41419-020-03169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou S., Fang J., Hu M., Pan S., Liu D., Xing G., et al. Determining the influence of high glucose on exosomal lncRNAs, mRNAs, circRNAs and miRNAs derived from human renal tubular epithelial cells. Aging. 2021;13(6):8467–8480. doi: 10.18632/aging.202656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin C.S., Xin Z.C., Dai J., Lue T.F. Commonly used mesenchymal stem cell markers and tracking labels: limitations and challenges. Histology & Histopathology. 2013;28(9):1109–1116. doi: 10.14670/hh-28.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]