Abstract
Monoclonal antibody treatment of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection has been widely implemented. Effects of treatment on the endogenous primary humoral response to the virus are unknown. A retrospective cohort study performed at a Veterans Health Administration medical center compared serologic responses of treated and untreated COVID-19 patients at high risk for severe outcomes. Three anti-viral spike protein IgG monoclonal treatments were used during the study period, 1) bamlanivimab, 2) casirivimab with imdevimab, and 3) bamlanivimab with etesevimab. Data were analyzed at acute (0–9 days), seroconversion (10–19 days), and maximum antibody (20–39 days) stages. SARS-Cov-2 infection induced a dynamic primary humoral response with anti-spike IgM and anti-nucleocapsid IgG seroconversion occurring after 9 days with maximum serologic indices achieved by 20–39 days. All monoclonal antibody treatments suppressed the endogenous anti-spike IgM response by 85–90% with minor effect on the anti-nucleocapsid response. Thus, passive immunization therapy may cause immunologic interference.
Keywords: SARS-CoV-2, COVID-19, Monoclonal antibody, Humoral immunity, Passive immunization
Abbreviations: ACE2, angiotensin converting enzyme 2; anti-N, anti-SARS-CoV-2 nucleocapsid protein antibodies; anti-S, anti-SARS-CoV-2 spike protein antibodies; BAM, bamlanivimab; CAS, casirivimab; BMI, body mass index; CDC, Center for Disease Control; COVID-19, Coronavirus Disease 2019; eGFR, estimated glomerular filtration rate; ETE, etesevimab; EUA, emergency use authorization; FDA, Food and Drug Administration; IMD, imdevimab; mAb, monoclonal antibody;; VHA, Veterans Health Administration
1. Introduction
The urgency surrounding the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral pandemic invoked intensive development of novel therapies to prevent or ameliorate disease. Some received Emergency Use Authorization (EUA) by the Food and Drug Administration (FDA) while the full evaluation of efficacy and safety is ongoing. Among these are neutralizing IgG monoclonal antibody (mAb) preparations for passive immunization. These antibodies are directed against SARS-CoV-2 viral surface spike (S) protein, blocking binding to host cells via the angiotensin converting enzyme 2 (ACE2) receptor [1,2]. Single agent bamlanivimab (BAM), as well as therapeutic combination preparations, casirivimab (CAS) plus imdevimab (IMD) and bamlanivimab plus etesevimab (ETE), have been employed along with other emerging formulations. When administered to patients in the early stage of infection, these agents reportedly reduce Coronavirus Disease 2019 (COVID-19)-related hospitalization and emergency room visits in patients at high risk for disease progression compared to placebo [[3], [4], [5]], but there is limited information regarding effects on endogenous immunity. Experimental animal studies suggest that the endogenous anti-viral humoral response is abrogated by passive immunization with IgG antibodies [6], but effects in humans have yet to be fully investigated. To address this issue, we undertook a retrospective cohort study in a Veterans Affairs healthcare system where these agents were regularly administered.
2. Materials and methods
2.1. Study design and data source
A retrospective cohort study approach was used for this investigation. The VA Ann Arbor Healthcare system electronic medical record was queried for a list of all unique patients with both a positive SARS-CoV-2 PCR test and SARS-CoV-2 serologic data recorded during the study period of March 2020 to May 2021. Utilizing this list, a manual chart review was performed to identify any patient who had received mAb therapy, forming the treatment group, and those who did not receive mAb therapy, forming the untreated group. Patients who were vaccinated, had evidence of primary SARS-CoV-2 infection outside the study period, were previously treated with anti-SARS-CoV-2 mAb, or were undergoing either B or T cell immunosuppressive therapies were excluded.
Data collected included age, gender, symptom onset date, medical comorbidities, administered COVID-19-specific therapies, disease outcomes, as well as date and results of SARS-CoV-2 RT-PCR along with available corresponding cycle threshold (Ct) values, SARS-CoV-2 antigen, and SARS-CoV-2 serologic tests. Clinical outcomes measures included COVID-19 related emergency room visits, hospitalizations, and deaths. The study received full review by the local institutional review board (IRB) and was approved with waiver of informed consent.
2.2. Patient selection and characteristics
All patients presented to the VA Ann Arbor Healthcare System for diagnosis, treatment, and monitoring. The overall population was 90% male and represented both outpatient and hospitalized individuals with an average age of 66 (range 23–94). Study patients had to have mild to moderate COVID-19 symptoms within 10 days of symptom onset, with high-risk comorbidities for progression to severe COVID-19.
Of 150 mAb treated patients, 64 had sufficient charted information to assess clinical outcomes; 40 of which had simultaneous diagnostic SARS-CoV-2 RT-PCR and antigen testing at presentation, and 38 had concurrent serologic testing. Of over 200 untreated patients, 34 had sufficient charted clinical, diagnostic, and concurrent serologic data to be compared with the treated group. Cohort group demographic and clinical data are summarized in Table 1 .
Table 1.
Patient group characteristics.
| Treatment Groups |
||||
|---|---|---|---|---|
| Untreated | Bamlanivimab | Casirivimab + Imdevimab | Bamlanivimab + Etesevimab | |
| Demographics | ||||
| Mean Age (years) | 69.3 | 64.9 | 66.5 | 68.2 |
| Age Range | 44–93 | 45–80 | 48–86 | 43–86 |
| Group number | n = 34 | n = 24 | n = 27 | n = 13 |
| Male/Female | 32/2 | 19/5 | 25/2 | 12/1 |
| Mean BMI | 30.2 | 33.2 | 33.6 | 33.3 |
| BMI Range | 15.2–49.8 | 23.0–45.4 | 24.3–43.9 | 25.5–49.4 |
| Risk factors (incidence/total) | ||||
| Obesity (BMI > 30) | 16/34 | 15/24 | 19/27 | 10/13 |
| Diabetes | 11/34 | 10/24 | 11/27 | 9/13 |
| Hypertension | 22/34 | 18/24 | 21/27 | 6/13 |
| Kidney disease (eGFR <60) | 12/34 | 4/24 | 23/27 | 13/13 |
| Cardiovascular | 14/34 | 9/24 | 5/27 | 1/13 |
| Pulmonary | 14/34 | 6/24 | 3/27 | 3/13 |
| Cancer | 5/34 | 1/24 | 2/27 | 1/13 |
| Immunosuppressive condition | 6/34 | 5/24 | 2/27 | 1/13 |
Abbreviations: BMI, body mass index; eGFR, estimated glomerular filtration rate.
2.3. Monoclonal antibody therapies
Three EUA anti-SARS-CoV-2 spike protein-specific IgG monoclonal antibody treatments were administered as single intravenous infusions from December 2020 through April 2021 at recommended doses: 700 mg bamlanivimab (BAM) as a single agent, or the combination treatments of 1200 mg casirivimab plus 1200 mg imdevimab (CAS + IMD), or 700 mg bamlanivimab plus 1400 mg etesevimab (BAM + ETE). Treatment inclusion criteria and dosages were based on published CDC and FDA EUA guidance active during the time of use [[7], [8], [9]]. Pharmacologic details of each mAb are provided in Supplementary Table 1.
2.4. Polymerase chain reaction (RT-PCR) assays
Two FDA EUA vendor supplied rapid RT-PCR assays were employed, Xpert® Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA) and BioFire® Respiratory 2.1 Panel (bioMerieux. Marcy-l'Étoile, France) [10,11]. These are rapid assays with reported lower limits of detection of 250 and 500 viral gene copies/ml, respectively. The Xpert® Xpress test targets sequences of the nucleocapsid, envelope and RNA-dependent RNA polymerase genes. The BioFire assay targets 3 non-overlapping sequences in the viral ORF1ab and ORF8 genes. The Xpert® Xpress platform provides cycle threshold (Ct) values for the confirmatory N gene which represent a rough estimate of viral load. Both assays underwent local performance validations.
2.5. Antigen assay
Viral antigen was detected in nasal turbinate swabs using the FDA EUA BD Veritor™ System, a chromatographic digital immunoassay for the direct qualitative detection of SARS-CoV-2 nucleocapsid (N) antigens during the acute phase of infection [12].
2.6. Serologic assays
Endogenously elicited anti-spike (S) IgM and anti-nucleocapsid (N) IgG antibodies were detected in serum specimens using an FDA EUA chemiluminescent assay on an automated immunoanalyzer (Abbott Laboratories, Chicago, IL). Assay specificities and sensitivities are as follows: anti-N IgG, 100 and 99%; Anti-S IgM, 95.0 and 99.6% [13]. The numeric output is an index ratio of positive to background signal (S/Co). Negative index cutoffs were IgM (<1.0) and IgG (<1.4). Results are plotted on different scales due to lower background levels of IgM compared to IgG. These assays show high correlation to traditional enzyme-linked immunoassays [14]. Since the assays are specific for anti-S IgM and anti-N IgG, the infused recombinant anti-S IgG will not cause interference, allowing assessment of the endogenous response.
2.7. Statistical analysis
Kruskal-Wallis test with Dunn's correction for multiple comparisons was used to compare serologic and PCR numerical results of treated and untreated groups. Fisher's exact test was applied to assess differences between untreated and treated groups for positive and negative categorical parameters. Values of p < 0.05 were considered statistically significant. Calculations were performed using GraphPad Prism version 8.0.0 (GraphPad Software, San Diego, California USA).
3. Results
3.1. Dynamics of SARS-CoV-2 test parameters
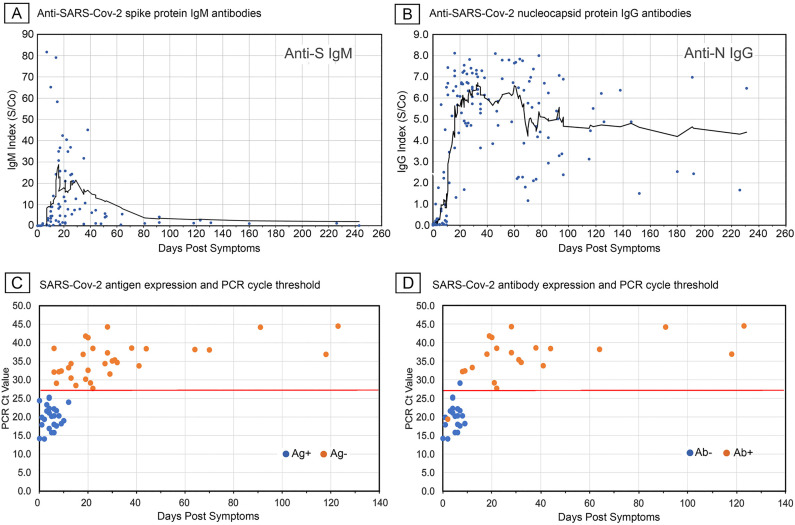
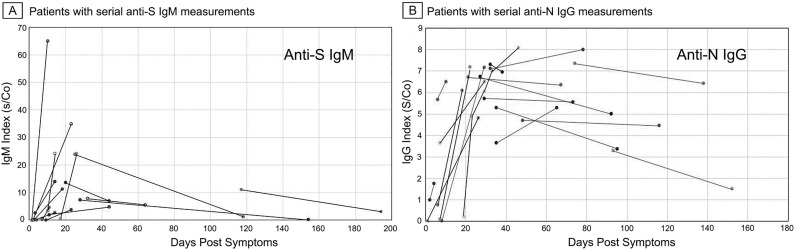
We initially established the relationship of test parameters to disease time course among unvaccinated patients not treated with mAb and without known immunosuppressive conditions (Fig. 1 ). Of the sampled charts, 85 anti-S IgM and 162 anti-N IgG results could be plotted against the time of reported symptom onset. The lesser number of IgM values was due to the later introduction of this test at our healthcare system. Fig. 1A and B show scatterplots with overlaid 10 day moving average spanning a period of about 240 days after symptom onset. Both anti-S IgM and anti-N IgG responses showed that most subjects seroconverted after day 10 post-symptom onset and reached maximum average indices by 20–40 days. While the IgM and IgG responses rose largely in parallel, the average IgM index declined after 40 days whereas IgG levels persisted. These patterns were likewise apparent when individual patients with available serial measurements were plotted (Fig. 2 ). The results were consistent with those reported by others [[15], [16], [17]] who likewise noted day 10 post symptoms as the median point of seroconversion. Moreover, the time course of index values matched that reported using quantitative anti-S IgM and anti-N assays [18,19] indicating that the chemiluminescence indices used in this study were suitable for semiquantitative measurement of antibody levels.
Fig. 1.
SARS-CoV-2 elicited anti-spike IgM and anti-nucleocapsid IgG antibody responses following symptom onset related to viral antigen detection and viral load. A and B, scatterplots with solid lines showing 10 day moving average of antibody index values. C and D, scatterplots with solid line indicating RT-PCR cycle threshold of 27.5 (Cepheid platform) above which virtually all patients were antigen negative and most seropositive.
Fig. 2.
Patients with serial anti-S IgM and anti-N IgG measurements. The kinetic pattern of anti-SARS-Cov-2 anti-S IgM and anti-N IgG antibody responses observed for the whole population was also apparent among patients with two or more serial measurements (A, anti-S IgM; B anti-N IgG, lines connect serial antibody index values for individual patients).
We also examined the relationship of RT-PCR cycle thresholds (Ct) in nasopharyngeal samples to detection of SARS-CoV-2 N-antigen in nasal turbinate specimens. Fifty-eight patients with simultaneous RT-PCR and antigen tests and reported time of symptom onset were identified (Fig. 1C). Of these, 41 also had concurrent serologic studies (Fig. 1D). As shown, antigen detection was largely restricted to 0–10 days after symptom onset when viral loads were high, as indicated by RT-PCR Ct values below 27.5. Moreover, antigen detection highly correlated with the absence of anti-SARS-CoV-2 antibodies. Thus, a negative antigen test was associated with seropositivity and higher Ct values. For this study, symptomatic subjects were considered in the acute stage of disease when presenting with a positive antigen test or low Ct value in combination with negative serology.
Based on these findings three intervals were selected to group test results for analysis, acute seronegative (0–9 days), seroconversion (10–19 days), and maximum antibody index (20–39 days) stages.
3.2. Effect of anti-SARS-CoV-2 monoclonal therapy on endogenous SARS-CoV-2 antibody response
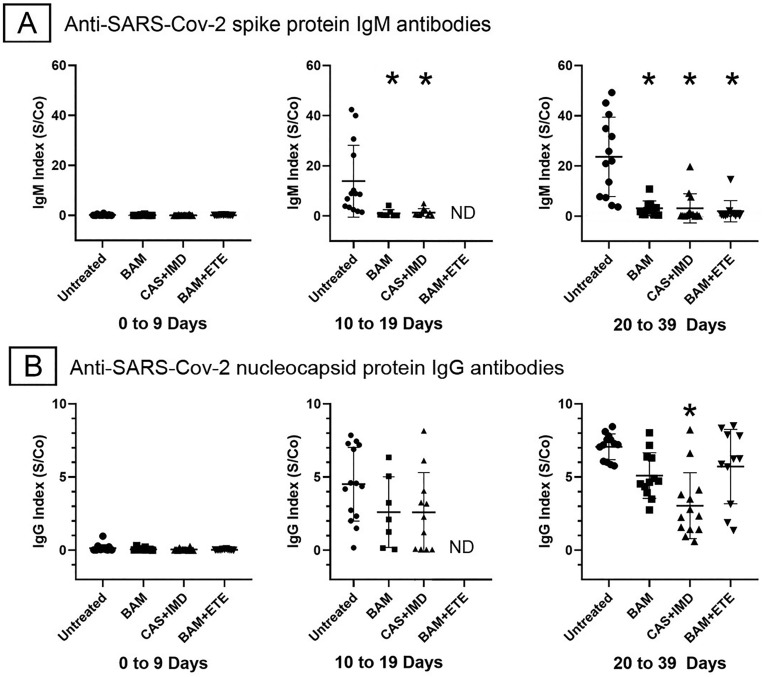
Of the mAb treated patients, 38 had serologic testing at presentation with 18 and 36 with follow up testing at 10–19 and 20–39 days after symptom onset, respectively. Of the untreated group, 15 patients had serologic testing at baseline with 14 and 13 patients tested at 10–19 and 20–39-days post symptom onset, respectively. Fig. 3A and B show anti-S IgM and anti-N IgG serologic indices among untreated and mAb treated groups at the designated stages of disease. In the acute stage (0–9 days), all subjects were seronegative. At the seroconversion stage (10–19 days) anti-S IgM levels were reduced by >90% in BAM and CAS + IMD treated subjects compared to the untreated group, anti-N IgG levels trended lower but did not reach statistical significance. Insufficient data was available for statistical analysis among BAM-ETE treated patients at this stage. In the maximum antibody stage (20–39 days), anti-S IgM levels remained reduced by 85–90% in all mAb treated groups. Anti-N IgG levels were affected to a lesser degree, with significant reductions of 50% only in the CAS + IMD treated group. A separate group of 11 patients was found to be seropositive at the time of mAb treatment. Of these, 9 were also negative for SARS-CoV-2 antigen. They were excluded from the analysis as they did not meet criteria for having been treated at the acute stage. However, follow up serology was available for 6 of the patients treated with BAM or BAM + ETE. Treatment did not significantly reduce the endogenous IgM anti-S or IgG anti-N humoral immune response in these subjects, suggesting that the suppressive effect of mAb on the primary antibody response required administration during the seronegative acute stage (Supplementary Fig. 1).
Fig. 3.
Effect of anti-SARS-CoV-2 monoclonal antibody treatments on elicited IgM anti-spike and IgG anti-nucleocapsid antibodies. Points show antibody indices with means and standard deviations of each group. Asterisks indicate p < 0.05 for comparison of untreated to treated groups, Kruskal–Wallis test with Dunn's correction for multiple comparisons. ND, not determined.
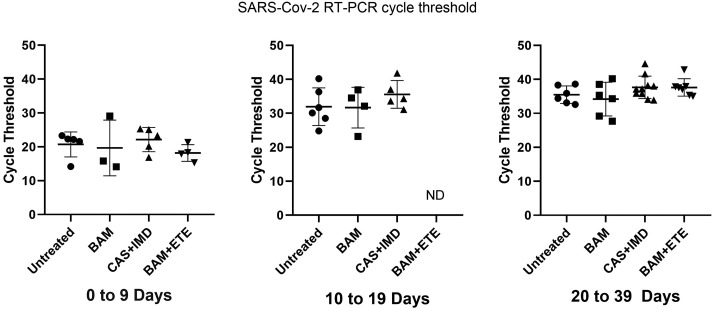
3.3. Effect of anti-SARS-CoV-2 monoclonal therapy on SARS-CoV-2 viral clearance
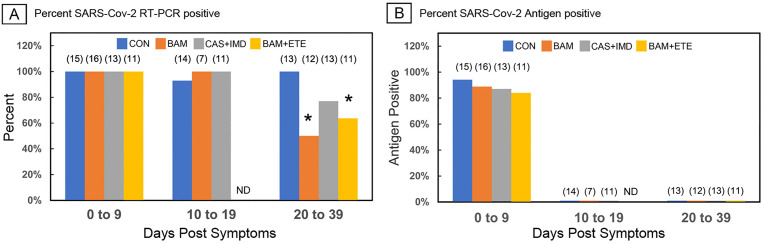
To determine if the observed reduction of the primary humoral response was due to rapid viral clearance, SARS-CoV-2 RT-PCR positivity, viral antigen and Ct values were analyzed. As shown in Fig. 4A and B, at the acute stage (0–9 days) all patients were RT-PCR positive and 80–90% were antigen positive. At 10–19 days virtually all subjects remained RT-PCR positive but had become antigen negative. At 20–39 days, all patients remained antigen negative, but differences were observed in the percentage of patients remaining RT-PCR positive. Untreated patients remained RT-PCR positive, while 50% and 40% of BAM and BAM + ETE treated patients were RT-PCR negative, respectively. Twenty percent of CAS + IMD treated subjects also became PCR negative, but this did not reach statistical significance. Fig. 5 shows the available Ct values recorded for those individuals remaining RT-PCR positive at the stage intervals. In the acute stage (0–9 days) Ct values averaged approximately 20 cycles with no significant differences among groups, indicating comparable viral loads at presentation. At 10–19 days and 20–39 days Ct values increased among all groups consistent with declining viral loads, but again with no significant statistical difference between treated and untreated groups. Thus, while mAb did promote viral clearance among some patients, there were no profound differences in viral load during the acute and seroconversion stages when antigen is first presented to the immune system.
Fig. 4.
Effect of anti-SARS-CoV-2 monoclonal antibody treatments on viral detection parameters. A, Bars show percent RT-PCR positive patients by Cepheid Xpert® Xpress or BioFire® platforms, asterisks indicate p < 0.05, Fisher exact test. B, Bars show percent of patients with positive nasal turbinate viral antigen test. Numbers in parentheses show group number. CON, untreated. ND, not determined.
Fig. 5.
Effect of anti-SARS-CoV-2 monoclonal antibody treatments on RT-PCR cycle thresholds. Points show cycle thresholds with means and standard deviations of RT-PCR positive patients in each group. Testing performed on Cepheid platform. ND, not determined. Significant differences were not identified among groups.
3.4. Effect of anti-SARS-CoV-2 monoclonal therapy on COVID-19 disease course and outcomes
Clinical trials show mAb treatment reduces emergency room visits and hospitalizations when administered during the early symptomatic stage of disease [[3], [4], [5]]. To determine if similar benefit occurred among our treatment cohort, disease outcomes were assessed. As shown in Table 2 , both emergency room visits, and hospitalizations were lower among all mAb treated patients at 5.9% and 2.9%, respectively, compared to 20.6% and 70.5% for the untreated group. The rates among all mAb treated patients were comparable to those previously reported [3]. A multi-facility study of VA medical centers reported admission rates among all veterans presenting with COVID-19 at about 20% with high-risk patients comprising most admissions [20]. Hence, the high rate of admissions among the untreated study group was not unexpected as this was a retrospective study specifically biased to patients at highest risk for admission. As recently reported, COVID-19 admission rates among high-risk patients with one or more comorbidities adjusted for age, sex, and race/ethnicity may range from 2.5 to 5 times the overall community rate [21]. While not the central aim of this study, the findings further support the clinical benefit of mAb treatment and indicated efficacious use at our facility.
Table 2.
Effect of monoclonal antibody treatment on clinical outcomes.
| Treatment Groups |
|||||
|---|---|---|---|---|---|
| Untreated | Bamlanivimab | Casirivimab + Imdevimab | Bamlanivimab + Etesevimab | All mAb treated | |
| Clinical course | |||||
| ED visit for COVID-19 | 7/34 | 2/24 | 2/27 | 0/13 | 4/64 |
| 20.6% | 8.3% | 6.5% | 0.0% | 5.9% | |
| Hospitalized for COVID-19 | 24/34 | 0/24 | 2/27 | 0/13 | 2/64 |
| 70.5% | 0.0% | 6.5% | 0.0% | 2.9% | |
| Deaths due to COVID-19 | 1/34 | 0/24 | 0/27 | 0/13 | 0/64 |
| 2.9% | 0.0% | 0.0% | 0.0% | 0.0% | |
| Other COVID-19 therapiesa | |||||
| Corticosteroids | 17/34 | 0/24 | 1/27 | 0/13 | 1/64 |
| Remdesivir | 16/34 | 0/24 | 1/27 | 0/13 | 1/64 |
Administered during hospitalizations.
4. Discussion
Passive immunization with mAb is an important therapy for the treatment of high-risk individuals presenting early with mild to moderate symptomatic COVID-19. However, a nuanced understanding of the effects on both the virus and host are needed to optimally employ these agents. Little is known about the effects of monoclonal antibody administration on host endogenous immunity. Animal studies have demonstrated that passive immunization inhibits the host primary anti-viral antibody response [6], but this has not been fully assessed in humans [4]. The intensive use of anti-viral spike protein (S) mAb therapy at our medical center during the SARS-CoV-2 pandemic offered an opportunity to approach this question through retrospective cohort analysis. Here we demonstrate that anti-SARS-CoV-2 mAb administration suppresses the primary endogenous humoral immune response.
A major technical obstacle to the analysis of the humoral response in passively immunized subjects is that it is difficult to distinguish between endogenously induced and administered antibodies. To circumvent this, we examined the host anti-S IgM and anti-nucleocapsid (N) IgG antibody response measured by a laboratory-based chemiluminescence assay which does not detect the therapeutically administered anti-spike (S) IgG mAb. We initially established the post symptom onset induction time course of these antibodies which closely matched that reported in non-veteran care settings [[15], [16], [17]]. Thus, despite being predominantly male high-risk patients, our veteran cohort showed similar serologic kinetics to the general population.
We also related SARS-CoV-2 RT-PCR and antigen detection assays to serologic findings. This not only validated the assays for comparative analysis but allowed the definition of interval stages of the disease based on laboratory studies. The acute stage could be defined by negative serology with a low RT-PCR Ct value, positive nasal antigen test, or combination of the two. Optimal benefit of mAb treatment requires administration in the early post symptom stage of infection [4]. As subjective symptom onset reporting may be inaccurate, our findings would endorse including baseline serology along with other laboratory diagnostic tests to confirm acute stage disease. Using this analytic framework, we demonstrated that passive immunization of COVID-19 patients with anti-S monoclonal IgG preparations profoundly suppressed the induction of the endogenous anti-S IgM response and to a lesser extent the anti-N IgG response. The suppressive effect of passive immunization on the induction of endogenous humoral immunity is well-described in mouse models [[22], [23], [24]] and the concept is employed empirically in humans to prevent Rh hemolytic disease in newborns through peripartum administration of anti-RhD immunoglobulin to block induction of anti-Rh antibodies in Rh-incompatible mothers [25]. The mechanism of suppression has been controversial, and it was thought that infused antibody simply caused clearance of the foreign antigen preventing access to immune responding cells. However, animal studies point to antigen-specific blockade, such that antibody can “mask” a specific epitope on a single cell or molecule without affecting the response to other antigens [22,24,26,27]. Blockade or steric interference by passive antibody can potentially inhibit viral antigen binding to cognate B cell receptors during early B cell activation [28]. Early-stage interference would be consistent with our observation that mAb did not suppress the response after seroconversion (Supplementary Fig. 1). Intuitively, reduction of viral load in the early stage of infection might be expected to result in a reduced antibody response. Indeed, anti-SARS-CoV-2 mAb preparations reportedly can reduce viral load [4,29], but the effect is modest, observed at 7 days after symptoms and with only combination therapy. Therefore, it is unclear if this is sufficient antigen reduction to affect the endogenous antibody response. One study comparing COVID-19 patients with mild and severe disease showed no difference in anti-viral IgM and IgG levels or their kinetics of onset despite 2–3 log lower viral loads in mild disease [30]. In models of influenza infection, the critical period of B cell activation is within 3–7 days after maximum viral load [28]. While a rough measure of viral load, we observed similar RT-PCR Ct values among treated and untreated patients during the early and seroconversion stages suggesting comparable viral load during this critical period. Still, we cannot rule out a hybrid model as clearance and blockade mechanisms are not mutually exclusive. Since mAb treatments were directed at the S-protein, the profound suppression of the anti-S IgM response may primarily reflect antigen masking. There was a lesser effect on the anti-N response but with a trend to lower indices which reached statistical significance in only the CAS-IMD group at 20–39 days. The anti-N response could be more resistant to suppression due to a greater immunogenicity of this protein [31], but a partial effect would be consistent with accelerated viral clearance as suggested by the greater number PCR negative patients in the treated groups at 20–39 days. By this period, PCR Ct values in nasopharyngeal samples of those subjects with persisting PCR positivity were approaching 40 cycles in all groups which suggested very low viral loads in the upper airways. Since only Ct values were available for the upper respiratory tract, we cannot rule out the possibility of greater differences in viral loads in other locations. Viral loads persist longer in the lower respiratory tract of those with more severe disease as would be the case among untreated subjects [32].
Our study agrees with the recent report of Zhang et al. [33]. That study examined the endogenous anti-SARS-CoV-2 antibody response in BAM and BAM + ETE treated subjects by measuring antibody titers against portions of the spike protein not recognized by the mAb. They demonstrated significantly reduced titers as well as impaired viral neutralization from 15 to 85 days after mAb treatment with lesser effect on the anti-nucleocapsid response. However, only BAM and BAM + ETE preparations were examined without separation of IgM and IgG responses. Reduced viral load was suggested as a likely etiology but baseline viral loads of treated and control subjects were not compared. Our findings extend these by showing a profound effect on the IgM anti-S response by CAM + IMD as well as BAM and BAM + ETE. Our results also suggest that specific antigen-blockade during early antigen presentation must be considered as a potential major interference mechanism during passive immunization.
A serious implication of our study is the potential effect on long-term immunity. A recent mouse study showed that passive antibody suppressed not only the IgM response but also the IgG and memory B cell response [22]. The reduced antibody titers reported at 85 days after mAb treatment by Zhang et al. [33] suggests this might be the case. Still, this has yet to be demonstrated in humans and would apply only to the humoral response. Our study did not assess effects on CD8 T killer and T helper cell anti-SARS-CoV-2 responses which may be unaffected by mAb treatment. Carefully designed prospective studies would be needed to clarify these points. Nevertheless, our study supports prudent management with follow up vaccination of mAb treated patients after clearance of infused antibodies.
In summary, this study provides evidence that virus-targeted passive immunization with engineered monoclonal antibodies during early-stage infection inhibits the host endogenous primary antibody response. This effect of passive immunization has been observed in animal models and our study suggests it likely applies to humans. While potentially beneficial, the full effects of anti-pathogen monoclonal antibody therapies must be understood to guide effective follow-up vaccination timing.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgements
Indirect support for this project was provided by the Department of Veterans Affairs as part of its ongoing mission to improve and advance medical care. We wish to likewise recognize Chelsea McIntyre, Cheryl Rollins and Lurielyn Fernandez for their expert technical efforts in validating the serologic assays used for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clim.2022.108959.
Appendix A. Supplementary data
Supplementary Table and Figure
References
- 1.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z., Huang Q., Xu L., Tang J., Tian Q., Yao W., Hu L., Yan X., Zhou X., Wu Y., Deng K., Zhang Z., Qian Z., Chen Y., Ye L. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. (Epub 2020 April 22. PMC7167496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. (Epub 2020 May 27) [DOI] [PubMed] [Google Scholar]
- 3.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., Shawa I., Adams A.C., Van Naarden J., Custer K.L., Shen L., Durante M., Oakley G., Schade A.E., Sabo J., Patel D.R., Klekotka P., Skovronsky D.M. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. (Epub 2020 October 29. PMC7646625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. (Epub 2021 April 19. PMC8054133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., Perry C., Pan C., Hosain R., Mahmood A., Davis J.D., Turner K.C., Hooper A.T., Hamilton J.D., Baum A., Kyratsous C.A., Kim Y., Cook A., Kampman W., Kohli A., Sachdeva Y., Graber X., Kowal B., DiCioccio T., Stahl N., Lipsich L., Braunstein N., Herman G., Yancopoulos G.D. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. (Epub 2020 December 18. PMC7781102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowe J.E., Jr., Firestone C.Y., Murphy B.R. Passively acquired antibodies suppress humoral but not cell-mediated immunity in mice immunized with live attenuated respiratory syncytial virus vaccines. J. Immunol. 2001;167:3910–3918. doi: 10.4049/jimmunol.167.7.3910. (Epub 2001 September 21) [DOI] [PubMed] [Google Scholar]
- 7.https://www.fda.gov/media/143603/download, Fact sheet for health care providers emergency use authorization (EUA) of bamlanivimab, 2020.
- 8.https://www.fda.gov/media/145611/download, Fact sheet for health care providers emergency use authorization (EUA) of casirivimab and imdevimab, 2020.
- 9.https://www.fda.gov/media/145802/download, Fact sheet for health care providers emergency use authorization (EUA) of bamlanivimab and etesevimab, 2021.
- 10.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V., Love N. Detection of SARS-CoV-2 by use of the Cepheid Xpert Xpress SARS-CoV-2 and Roche Cobas SARS-CoV-2 assays. J. Clin. Microbiol. 2020;58 doi: 10.1128/jcm.00772-20. e00772–00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckbo E.J., Locher K., Caza M., Li L., Lavergne V., Charles M. Evaluation of the BioFire® COVID-19 test and Respiratory Panel 2.1 for rapid identification of SARS-CoV-2 in nasopharyngeal swab samples. Diagn. Microbiol. Infect. Dis. 2021;99:115260. doi: 10.1016/j.diagmicrobio.2020.115260. (Epub 2020 December 20. PMC7654322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young S., Taylor S.N., Cammarata C.L., Varnado K.G., Roger-Dalbert C., Montano A., Griego-Fullbright C., Burgard C., Fernandez C., Eckert K., Andrews J.C., Ren H., Allen J., Ackerman R., Cooper C.K. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS antigen point-of-care test. J. Clin. Microbiol. 2020;59 doi: 10.1128/jcm.02338-20. e02338–02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance, EUA Authorized Serology Test Performance, 2021.
- 14.Mendoza R., Silver M., Zuretti A.R., Christian M., Das B., Norin A.J., Borgen P., Libien J., Bluth M.H. Correlation of automated chemiluminescent method with enzyme-linked immunosorbent assay (ELISA) antibody titers in convalescent COVID-19 plasma samples: development of rapid, cost-effective semi-quantitative diagnostic methods. J. Blood Med. 2021;12:157–164. doi: 10.2147/jbm.s296730. (2021 March 17. PMC7982562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng D.L., Goldgof G.M., Shy B.R., Levine A.G., Balcerek J., Bapat S.P., Prostko J., Rodgers M., Coller K., Pearce S., Franz S., Du L., Stone M., Pillai S.K., Sotomayor-Gonzalez A., Servellita V., Martin C.S.S., Granados A., Glasner D.R., Han L.M., Truong K., Akagi N., Nguyen D.N., Neumann N.M., Qazi D., Hsu E., Gu W., Santos Y.A., Custer B., Green V., Williamson P., Hills N.K., Lu C.M., Whitman J.D., Stramer S., Wang C., Reyes K., Hakim J.M.C., Sujishi K., Alazzeh F., Pham L., Oon C.Y., Miller S., Kurtz T., Hackett J., Jr., Simmons G., Busch M.P., Chiu C.Y. SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient blood from the San Francisco Bay area. medRxiv. 2020 doi: 10.1101/2020.05.19.20107482. (Epub 2020 June 9. PMC7273245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A., Jerome K.R., Mathias P.C., Greninger A.L. Performance characteristics of the Abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020;58 doi: 10.1128/jcm.00941-20. (Epub 2020 May 10. PMC7383515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S.Y., Lee Y.L., Lin Y.C., Lee N.Y., Liao C.H., Hung Y.P., Lu M.C., Wu J.L., Tseng W.P., Lin C.H., Chung M.Y., Kang C.M., Lee Y.F., Lee T.F., Cheng C.Y., Chen C.P., Huang C.H., Liu C.E., Cheng S.H., Ko W.C., Hsueh P.R., Chen S.C. Multicenter evaluation of two chemiluminescence and three lateral flow immunoassays for the diagnosis of COVID-19 and assessment of antibody dynamic responses to SARS-CoV-2 in Taiwan, Emerg. Microbes Infect. 2020;9:2157–2168. doi: 10.1080/22221751.2020.1825016. (PMC7580576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F., Astudillo M., Yang D., Miller T.E., Oliver E., Fischinger S., Atyeo C., Iafrate A.J., Calderwood S.B., Lauer S.A., Yu J., Li Z., Feldman J., Hauser B.M., Caradonna T.M., Branda J.A., Turbett S.E., LaRocque R.C., Mellon G., Barouch D.H., Schmidt A.G., Azman A.S., Alter G., Ryan E.T., Harris J.B., Charles R.C. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1101/2020.07.18.20155374. (Epub 2020 August 4. PMC7386524) [DOI] [Google Scholar]
- 19.Brochot E., Demey B., Touzé A., Belouzard S., Dubuisson J., Schmit J.-L., Duverlie G., Francois C., Castelain S., Helle F. Anti-spike, anti-nucleocapsid and neutralizing antibodies in SARS-CoV-2 inpatients and asymptomatic individuals. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.584251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardemil C.V., Dahl R., Prill M.M., Cates J., Brown S., Perea A., Marconi V., Bell L., Rodriguez-Barradas M.C., Rivera-Dominguez G., Beenhouwer D., Poteshkina A., Holodniy M., Lucero-Obusan C., Balachandran N., Hall A.J., Kim L., Langley G. COVID-19-related hospitalization rates and severe outcomes among veterans from 5 veterans affairs medical centers: hospital-based surveillance study. JMIR Public Health Surveill. 2021;7 doi: 10.2196/24502. (Epub 2021 January 22. PMC7836907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko J.Y., Danielson M.L., Town M., Derado G., Greenlund K.J., Kirley P.D., Alden N.B., Yousey-Hindes K., Anderson E.J., Ryan P.A., Kim S., Lynfield R., Torres S.M., Barney G.R., Bennett N.M., Sutton M., Talbot H.K., Hill M., Hall A.J., Fry A.M., Garg S., Kim L. Risk factors for Coronavirus Disease 2019 (COVID-19)-associated hospitalization: COVID-19-associated hospitalization surveillance network and behavioral risk factor surveillance system. Clin. Infect. Dis. 2021;72:e695–e703. doi: 10.1093/cid/ciaa1419. (PMC7543371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergström J.J.E., Xu H., Heyman B. Epitope-specific suppression of IgG responses by passively administered specific IgG: evidence of epitope masking. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gadd S.J., Brogan M.P., Ashman L.K. Specificity of the passive antibody-induced suppression of the humoral immune response of mice to surface antigens on human cells. Immunology. 1985;54:223–231. (PMC1453498) [PMC free article] [PubMed] [Google Scholar]
- 24.Maier C.L., Mener A., Patel S.R., Jajosky R.P., Bennett A.L., Arthur C.M., Hendrickson J.E., Stowell S.R. Antibody-mediated immune suppression by antigen modulation is antigen-specific. Blood Adv. 2018;2:2986–3000. doi: 10.1182/bloodadvances.2018018408. (Epub 2018 November 11. PMC6234375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbaniak S.J., Greiss M.A. RhD haemolytic disease of the fetus and the newborn. Blood Rev. 2000;14:44–61. doi: 10.1054/blre.1999.0123. [DOI] [PubMed] [Google Scholar]
- 26.Xu H., Zhang L., Heyman B. IgG-mediated immune suppression in mice is epitope specific except during high epitope density conditions. Sci. Rep. 2018;8:15292. doi: 10.1038/s41598-018-33087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H., Heyman B. IgG-mediated suppression of antibody responses: hiding or snatching epitopes? Scand. J. Immunol. 2020;92 doi: 10.1111/sji.12921. (Epub 2020 July 16) [DOI] [PubMed] [Google Scholar]
- 28.Lam J.H., Baumgarth N. The multifaceted B cell response to influenza virus. J. Immunol. 2019;202:351–359. doi: 10.4049/jimmunol.1801208. (PMC6327962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., Shawa I., Kumar P., Adams A.C., Van Naarden J., Custer K.L., Durante M., Oakley G., Schade A.E., Holzer T.R., Ebert P.J., Higgs R.E., Kallewaard N.L., Sabo J., Patel D.R., Klekotka P., Shen L., Skovronsky D.M. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J., Tang X., Bai R., Liang C., Zeng L., Lin H., Yuan R., Zhou P., Huang X., Xiong Q., Peng J., Cui F., Ke B., Su J., Liu Z., Lu J., Tian J., Sun R., Ke C. The kinetics of viral load and antibodies to SARS-CoV-2. Clin. Microbiol. Infect. 2020;26 doi: 10.1016/j.cmi.2020.08.043. 1690.e1691–1690.e1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smits V.A.J., Hernández-Carralero E., Paz-Cabrera M.C., Cabrera E., Hernández-Reyes Y., Hernández-Fernaud J.R., Gillespie D.A., Salido E., Hernández-Porto M., Freire R. The Nucleocapsid protein triggers the main humoral immune response in COVID-19 patients. Biochem. Biophys. Res. Commun. 2021;543:45–49. doi: 10.1016/j.bbrc.2021.01.073. (Epub 2021 January 22. PMC7825866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen P.Z., Bobrovitz N., Premji Z.A., Koopmans M., Fisman D.N., Gu F.X. SARS-CoV-2 shedding dynamics across the respiratory tract, sex, and disease severity for adult and pediatric COVID-19. Elife. 2021;10 doi: 10.7554/eLife.70458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Poorbaugh J., Dougan M., Chen P., Gottlieb R.L., Huhn G., Beasley S., Daniels M., Trinh T. Ngoc Vy, Crisp M., Freitas J.J., Vaillancourt P., Patel D.R., Nirula A., Kallewaard N.L., Higgs R.E., Benschop R.J. Endogenous antibody responses to SARS-CoV-2 in patients with mild or moderate COVID-19 who received Bamlanivimab alone or Bamlanivimab and Etesevimab together. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.790469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table and Figure