Abstract
In order to investigate the seroprevalence of human herpesvirus 8 (HHV-8) infection in central and southern Italy, sera from human immunodeficiency virus (HIV)-seronegative subjects, with and without Kaposi’s sarcoma (KS), were analyzed by immunofluorescence assay, using BC-3, a cell line latently infected with HHV-8. High titers of antibody against HHV-8 lytic and latent antigens were detected in all 50 KS patients studied, while in 50 HIV-seronegative subjects without KS, 32 (64%) were found positive for HHV-8 antibodies. Titers in the sera of these patients were lower than those for KS patients. This data suggests that HHV-8 infection is not restricted to KS patients and that the prevalence of HHV-8 infection in the general population may be correlated with differing rates of prevalence of KS in different parts of the world. In view of these findings, possible nonsexual transmission routes were evaluated. Nested PCR was used to test for the presence of HHV-8 DNA in saliva, urine, and tonsillar swabs from KS and non-KS patients. In KS patients, 14 out of 32 tonsillar swabs (43.7%), 11 out of 24 saliva samples (45.8%), and just 2 out of 24 urine samples (8.3%) tested positive for HHV-8 DNA. In the control group, on the contrary, none of the 20 saliva and 20 urine specimens was positive for HHV-8 DNA; only 1 out of 22 tonsillar swabs gave a positive result. This data supports the hypothesis that HHV-8 infects the general population in a latent form. The reactivation of viral infection may result in salivary shedding of HHV-8, contributing to viral spread by nonsexual transmission routes.
Kaposi’s sarcoma (KS) is a multifocal vascular tumor, with an uneven geographic distribution. Four types have been described: classic, iatrogenic, endemic, and AIDS-associated KS. The etiology of KS is still unknown. Recently, a new herpesvirus, named KS-associated herpesvirus or human herpesvirus 8 (HHV-8) (8, 18), has been identified in virtually all KS lesions, from both human immunodeficiency virus (HIV)-seronegative and HIV-seropositive subjects, suggesting that this could be the infectious, sexually transmitted cofactor involved in KS pathogenesis (1, 5, 6, 9, 11, 14, 25).
Molecular and epidemiological studies have suggested that HHV-8 could be widespread throughout the human population, especially in geographic areas where KS is prevalent (2, 7, 10, 21, 22). High rates of HHV-8 seroprevalence have recently been reported in Africa and Italy (13, 17).
Interestingly, central and southern Italy are areas where a relatively high prevalence of classic KS has been observed (12, 20). Preliminary data suggests that rates of HHV-8 infection could in fact be higher than previously believed (7, 29, 30). This supports the hypothesis that the prevalence of KS is correlated with the prevalence of HHV-8 infection in a population.
In order to evaluate the prevalence of HHV-8 infection in the populations of central and southern Italy, serum samples from HIV-seronegative patients, with and without KS, were tested for anti-HHV-8 antibodies by indirect immunofluorescence assay (IFA).
High HHV-8 seroprevalence rates, together with the detection of antibodies in young people in areas where KS is endemic (23, 26), suggest that multiple modes of transmission are possible; as for other herpesviruses, horizontal transmission, possibly via nongenital fluids, may in fact play an important role in the spread of HHV-8 (2, 3, 4, 10, 15, 17). In order to verify this hypothesis we tested for the presence of HHV-8 DNA in blood, saliva, tonsillar swabs, and urine from KS and non-KS patients.
MATERIALS AND METHODS
Patients and specimens.
Fifty KS patients (mean age, 65.8 years) attending our Department of Dermatology were enrolled in the study. Thirty-nine of the patients (30 males and 9 females) were suffering from classic KS, and 11 were suffering from iatrogenic KS (8 males and 3 females). In all cases KS diagnosis was confirmed by routine histologic examination. Enzyme immunoassay and immunoblot analysis were used to screen all patients for antibodies to HIV types 1 and 2.
Serum samples for antibody detection were obtained from each of the KS patients; 32 tonsillar swabs, 23 saliva specimens, and 24 urine specimens were also collected.
A control group was created by including 70 patients affected by dermatological diseases other than KS and healthy subjects. All subjects came from the same geographic areas as the KS patients.
Serum samples for antibody detection were collected from 50 subjects, 35 males and 15 females aged 30 to 87 years (mean age, 65.5 years). From 20 control subjects, 16 males and 4 females (mean age, 60.8 years), tonsillar swabs, saliva samples, and urine samples were collected and analyzed for HHV-8 DNA.
Informed consent was obtained from all patients.
IFA.
An IFA was developed using the HHV-8-positive and Epstein-Barr virus (EBV)-negative B-cell line BC-3 (American Type Culture Collection, Manassas, Va.). The EBV producer cell line P3HR-1 (American Type Culture Collection) and the HHV-8- and EBV-negative cell line Ramos (American Type Culture Collection) were used as controls. Cells were grown in RPMI 1640 medium (Eurobio, Les Ulis, France), supplemented with 10% heat-inactivated fetal calf serum, 200 mM glutamine, and antibiotics. By using BC-3 and P3HR-1 cells, the lytic replicative cycle of the viruses (HHV-8- and EBV, respectively) was induced by incubating 106 cells/ml with 20 ng of the phorbol ester 12-O-tetradecanoyl phorbol-13-acetate (TPA) (Sigma, St. Louis, Mo.) per ml for 72 h (24, 27). Uninduced and TPA-induced BC-3 and TPA-induced P3HR-1 and Ramos cells were collected, washed in phosphate-buffered saline (PBS) (pH 7.4), spotted onto welled slides (inner diameter, 5 mm; 10 wells/slide; bioMérieux sa, Marcy l’Etoile, France), air dried in a laminar-flow hood, and fixed in cold acetone for 10 min. Cells (105/well) were incubated for 30 min at 37°C with 20 μl of twofold dilutions of human sera, beginning at 1:20.
After incubation, slides were washed three times in PBS for 10 min each time. The slides were then incubated for 30 min at 37°C with a prestandardized dilution of Kallestad fluorescein-conjugated goat F(ab′)2 fragment anti-human immunoglobulin G (Sanofi Diagnostics Pasteur, Chaska, Minn.) and a 1:20,000 dilution of Evans blue (Sigma). They were then washed three times in PBS for 10 min each time and once in distilled water for 1 min, air dried, and mounted with mounting fluid (Light Diagnostics, Temecula, Calif.). Slides were examined under a fluorescence microscope. Specific reactivity at a dilution of 1:40 or more with uninduced and TPA-induced BC-3 cells was considered positive for HHV-8 antigens.
Sample preparation and DNA extraction.
Tonsillar swabs were immediately stored in 2 ml of sterile PBS and then vigorously vortexed for 2 min to dislodge the cells from the swabs, before removing the latter. The suspensions were then centrifuged and the cell pellets were used for DNA detection. Aliquots of 300 to 500 μl of saliva and urine specimens were stored at −80°C until analysis. Nucleic acids from tonsillar swabs, saliva samples, and urine samples were extracted by Gene-Fizz solution (Eurobio) directly into PCR tubes before amplification reactions, as recommended by the supplier.
PCR detection of HHV-8 DNA.
All samples were assayed for detection of HHV-8 DNA by nested PCR. The KS330233 primer set (8) and the P1 and P2 primers (19), corresponding to positions 711 to 728 and 1412 to 1430 of the published 1,853-bp KS330 Bam region, were used in this study. Nested PCR was performed by using P1 and P2 as outer primers and KS330233 sequences as inner primers. The outer PCR protocol was as follows: 98°C for 20 s followed by 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s. The inner PCR was run for a further 25 cycles with the following thermal profile: 94°C for 20 s, 60°C for 20 s, and 72°C for 20 s.
Each PCR mixture, containing 40 pmol of each primer, 200 μM concentrations of deoxynucleoside triphosphates (Boehringer GmbH, Mannheim, Germany), 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.0), 50 mM KCl, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer, Cetus, Norwalk, Conn.), in a final volume of 100 μl, was performed in a GeneAmp PCR system (model 9600; Perkin-Elmer, Cetus).
To assess the detection sensitivity, serial dilutions of a 233-bp PCR product cloned into pCR2.1 vector (Invitrogen, San Diego, Calif.), quantitated by UV absorption and mixed with HHV-8-negative genomic DNA (peripheral blood mononuclear cells), were amplified by a single PCR with KS330233 primers (11). The single-PCR assay allows the detection of <10 copies of the cloned HHV-8 target sequence in a background of 200,000 human cells. Tenfold dilutions of KS lesion biopsy specimens were assayed by single and nested PCRs, and a 10-fold increase of the sensitivity was obtained by using the nested-PCR protocol, as visualized by 2.5% agarose gel electrophoresis and ethidium bromide staining.
The specificity of the PCR products was confirmed by Southern blot hybridization with an internal oligonucleotide probe that was specific for HHV-8 and was digoxigenin labelled (3′-Oligonucleotide labelling kit; Boehringer GmbH) (8).
Positive samples (tonsillar swab, saliva, and urine) were also confirmed by amplification with a primer pair selected outside the region amplified by nested PCR. Primers KS-201 F and R (5′-GGTCGTGTCGTGTGATGCTTA-3′ and 5′-AAGAACTGCCGACAAGGACTG-3′), corresponding to positions 85 to 105 and 265 to 285 of the published 1,853-bp KS330 Bam region were used. To assess the integrity and suitability of the HHV-8-negative DNA samples, a 268-bp fragment of the human β-globin gene was amplified.
In order to closely monitor the occurrence of PCR false-positive results, negative controls, including reaction mixtures lacking any DNA template and HHV-8-negative human DNA, were regularly analyzed in each reaction. Negative controls were also included during both sample preparation and DNA extraction. Procedures to prevent specimen contamination and PCR carryover were rigorously observed at every step in this analysis (16).
Statistical analysis.
Differences in the frequencies of results for the study groups were analyzed by the chi-square test. P values of <0.005 were considered statistically significant.
RESULTS
Sera from KS patients and control subjects were tested by IFA with uninduced and TPA-induced BC-3 cells. All of the samples from KS patients tested positive for HHV-8; antibody titers ranged from 1:40 to 1:10,240, with a geometric mean titer of 1,414.17. Thirty-two (64%) of the 50 sera analyzed for the control group were HHV-8 seropositive; antibody titers for these sera were lower than those for KS patients, ranging from 1:40 to 1:640, with a geometric mean titer of 238.64.
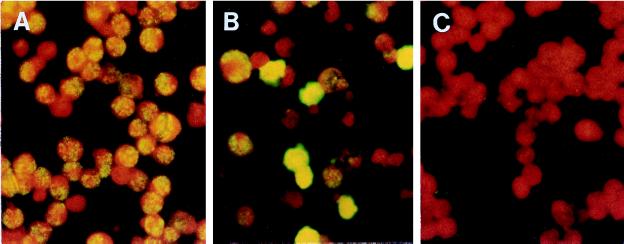
The specific staining patterns representing positive reactivity with both HHV-8 latent and lytic antigens are shown in Fig. 1A and B, respectively. Typical speckled nuclear fluorescence, indicating reactivity of antibodies to latent antigens, was seen in both latently infected and TPA-induced cells. A cytoplasmic fluorescence pattern was observed in a much higher proportion only after TPA induction to activate the lytic cycle and release viral particles.
FIG. 1.
Staining patterns of antibodies to HHV-8 latent (A) and lytic (B) antigens observed with positive sera reacting with TPA-induced BC-3 cells. (C) Negative control with only secondary fluorescein-conjugated antibody.
Antibodies to latent antigens were found in virtually all KS patients, though at lower titers than those to lytic antigens. In the control HHV-8-positive sera, typical nuclear fluorescence was observed in latently infected cells less frequently, probably due to low titers in the sera of these patients. No immunofluorescence background was observed when BC-3 cells were tested with only the secondary antibody (Fig. 1C).
Sera positive for HHV-8 failed to react when tested with HHV-8-, EBV-negative cells (Ramos) and with Hep2 cells, routinely used for detection of antinuclear autoantibodies (data not shown). HHV-8 IFA titers were not modified after absorption of the positive sera with the EBV producer P3HR-1 cells when TPA induced. In addition, there was no evidence of cross-reaction with EBV-infected cells (P3HR-1) or EBV- and HHV-8-negative B cells (Ramos).
All specimens included in the study were analyzed for HHV-8 DNA by nested PCR. HHV-8 DNA was found in 14 (43.7%) out of 32 tonsillar swabs obtained from KS patients. Viral sequences were also identified in saliva samples from 11 (45.8%) out of 24 patients with KS. In 22 patients, saliva and tonsillar swabs were collected at the same time, producing concordant results in 77.2% of cases. No significant correlation was observed between positivity in saliva and tonsillar swabs and titers of anti-HHV-8 antibody in serum; positivity did not correlate with the clinical staging of the patients. Positive urine specimens were found in just two (8.3%) of the 24 KS patients studied. Of the two positives, one was a transplant recipient who died of KS while the other was a patient with an aggressive cutaneous KS.
No viral DNA sequences in saliva and urine specimens from HIV-seronegative subjects without KS were amplified.
In the control group only 1 out of 20 tonsillar swabs analyzed was positive for HHV-8 DNA. The control patient, who had a positive tonsillar swab but whose saliva tested negative, was a 64-year-old man from Latium. When he was enrolled in the study he was suffering from dysplastic nevus; no significant risk factors or symptoms of KS have been detected for this patient.
Statistical analysis of the data shows a significant frequency of HHV-8 DNA detection in tonsillar swabs and saliva from KS patients compared with control subjects (P < 0.005).
DISCUSSION
Although molecular and serological assays have been used to define the epidemiology and transmission of HHV-8 infections, the exact prevalence rates are still uncertain. Most groups are however agreed that the prevalence of HHV-8 varies widely between different countries and geographical regions.
Different seropositivity rates for KS and non-KS subjects have been reported by various authors (13, 17, 28). One of the largest studies, performed in the United States by Lennette et al. (17), found 87 out of 91 HIV-positive KS patients to be seropositive for HHV-8. The study also reported seropositivity rates of 28% for HIV-seronegative adult women and of 20% for blood donors in general. Simpson et al. (28) failed to detect any seropositivity for HHV-8 antigens among HIV-seronegative subjects.
In the present study, HHV-8 seroprevalence was investigated in HIV-seronegative subjects by an immunofluorescence assay to detect antibodies to both latent and lytic HHV-8 antigens. In general, we found a high HHV-8 seropositivity (64%) rate for HIV-seronegative age- and sex-matched control subjects from central and southern Italy. When HIV-seronegative KS patients from the same regions were analyzed, a 100% seropositivity rate was determined.
To our knowledge, the first serological study of KS in Italy was the investigation by Gao et al., who tested 14 AIDS-related KS patients (71% of whom tested positive for HHV-8), 11 HIV-seronegative KS patients (100% of whom tested positive for HHV-8), and 107 blood donors (4% of whom tested positive for HHV-8) (13). This study was carried out with nonstimulated BCP1 cells, detecting from sera only antibodies against latent viral antigens, reported to be present at a lower prevalence both in KS and non-KS subjects (17, 29).
Numerous molecular epidemiological studies have reported differences between Italian regions in the HHV-8 DNA positivity rate in blood from non-KS subjects (5, 7, 11, 31). The work of Whitby et al. supports this finding, reporting a higher rate of seroprevalence in southern Italy than northern Italy (32). The present study confirms this result. The seropositivity rate in our control group was, however, higher than that found by previous authors. We believe this is probably due not only to the particular geographic area chosen for study, but in addition, to the age distribution of our patients.
In our study, control subjects came from regions in Italy where a high level of classic KS has previously been observed (12, 20, 31), that is, 14 (28%) from Sardinia, 12 (25%) from Latium, 5 (10%) each from Sicily and Campania, and the remainder from other regions in central and southern Italy. Importantly, the mean age of the control group (65.5 years) was comparable with the mean age of the KS patients (65.8 years) and was higher than the mean age for other control groups reported in the literature. We note that Olsen et al. have recently shown that HHV-8 seroprevalence increases linearly with age, rising to 71% among people aged 50 years and older in Zambia, where there is a high prevalence of endemic KS (26).
Our findings support the hypothesis that HHV-8 is prevalent in regions where there is a higher prevalence of KS (12, 20, 31) and that, when other predisposing factors are also present, this can contribute to the development of KS. The presence of anti-HHV-8 antibodies to latent and lytic antigens in all KS patients is consistent with a possible role of the virus in the pathogenesis of KS. At the moment, detection of antibodies against latent and lytic viral antigens by IFA seems to be a more specific and sensitive assay for detecting HHV-8 infection than either the use of IFA to detect latent antibodies only or the use of immunoenzymatic assay with recombinant viral protein (13, 28, 32).
In view of the high proportion of HHV-8 seropositivity found in our control group, we used viral DNA amplification to investigate the possibility that a nonsexual transmission route is involved in the spread of the infection through the general population. HHV-8 DNA was detected in saliva and tonsillar swabs from about half of the KS patients. None of the KS patients for whom these specimens were positive for HHV-8 DNA had oral KS lesions. We could therefore exclude such lesions as the origin of the HHV-8 DNA present in the saliva and tonsillar swabs. The presence of the virus with an almost equal frequency (45.8 and 43.8%) in both specimens suggests that HHV-8 replicates in tonsils, a part of the lymphoid system, and then sheds into saliva, which could contribute to HHV-8 transmission.
The reported presence of infectious HHV-8 particles in the saliva of KS patients (30) further supports the hypothesis that the HHV-8 DNA found in saliva specimens could be related to the reactivation of the lytic replicative cycle of the virus induced by genetic and immunologic factors. Our results confirm previous reports (15, 29) that KS patients have higher titers of anti-HHV-8 antibodies than non-KS patients. This is additional evidence for the reactivation of the viral infection.
In this study, the infrequent detection of HHV-8 DNA in urine, negative in all but two cases of aggressive classic KS, suggests that this route is unlikely to be involved in the spread of the virus. In our control group, saliva and urine samples were found to be negative and only one tonsillar swab showed a positive result for viral DNA. This data, together with the serological evidence for the spread of HHV-8 infection in the control group, supports the view that this virus might infect the general population in a latent form. Lower titers in serum, which are associated with a lower frequency of detection of antibody to latent antigen in non-KS patients, are consistent with this hypothesis.
The high rate of HHV-8 seroprevalence in HIV-seronegative individuals from central and southern Italy supports the hypothesis that KS occurs more frequently in populations with a higher rate of HHV-8 infection.
In conclusion, our results suggest that HHV-8 may establish infections in the general population which remain latent until various factors contribute to the reactivation of the lytic cycle of the virus. The virus may be localized in body sites and fluids involved in viral spreading via nonsexual routes and may contribute to the pathogenesis of KS.
ACKNOWLEDGMENTS
This work was partly supported by grant 97.04111.CTO4 from the Consiglio Nazionale delle Ricerche (C.N.R.) and by grant 7021304 from the Ministero dell’Università e della Ricerca Scientifica e Tecnologica (MURST), Rome, Italy.
Thanks are due to Franca Pedone and Creola Rocchetti for their expert technical assistance.
REFERENCES
- 1.Beral V, Peterman T A, Berkelman R L, Jaffe H W. Kaposi’s sarcoma among persons with AIDS: a sexually transmitted infection? Lancet. 1990;335:123–128. doi: 10.1016/0140-6736(90)90001-l. [DOI] [PubMed] [Google Scholar]
- 2.Bigoni B, Dolcetti R, de Lellis L, Carbone A, Boiocchi M, Cassai E, Di Luca D. Human herpesvirus 8 is present in the lymphoid system of healthy persons and can reactivate in the course of AIDS. J Infect Dis. 1996;173:542–549. doi: 10.1093/infdis/173.3.542. [DOI] [PubMed] [Google Scholar]
- 3.Blackbourn D J, Lennette E T, Ambroziak J, Mourich D V, Levy J A. Human herpesvirus 8 detection in nasal secretions and saliva. J Infect Dis. 1998;177:213–216. doi: 10.1086/517356. [DOI] [PubMed] [Google Scholar]
- 4.Boldogh I, Szaniszlo P, Bresnahan W A, Flaitz A M, Nichols M C, Albrecht T. Kaposi’s sarcoma herpesvirus-like DNA sequences in the saliva of individuals infected with human immunodeficiency virus. Clin Infect Dis. 1996;23:406–407. doi: 10.1093/clinids/23.2.406. [DOI] [PubMed] [Google Scholar]
- 5.Brambilla L, Boneschi V, Berti E, Corbellino M, Parravicini C. HHV8 cell-associated viraemia and clinical presentation of Mediterranean Kaposi’s sarcoma. Lancet. 1996;347:1338. doi: 10.1016/s0140-6736(96)90989-7. [DOI] [PubMed] [Google Scholar]
- 6.Buonaguro F M, Tornesello M L, Beth-Giraldo E, Hatzakis A, Mueller N, Downing R, Biryamwaho B, Sempala S D K, Giraldo G. Herpesvirus-like DNA sequences detected in endemic, classic, iatrogenic and epidemic Kaposi’s sarcoma (KS) biopsies. Int J Cancer. 1996;65:25–28. doi: 10.1002/(SICI)1097-0215(19960103)65:1<25::AID-IJC5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Cattani P, Capuano M, Lesnoni La Parola I, Guido R, Santangelo R, Cerimele F, Masini C, Nanni G, Fadda G, Cerimele D. Human herpesvirus 8 (HHV8) in Italian HIV-seronegative patients with Kaposi’s sarcoma. Arch Dermatol. 1998;134:695–699. doi: 10.1001/archderm.134.6.695. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y, Moore P S. Kaposi’s sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis. 1996;5:215–222. [PubMed] [Google Scholar]
- 10.Chang Y, Ziegler J, Wabinga H, Katangole-Mbidde E, Boshoff C, Schulz T, Whitby D, Maddalena D, Jaffe H W, Weiss R A, Moore P S the Uganda Kaposi’s Sarcoma Study Group. Kaposi’s sarcoma-associated herpesvirus and Kaposi’s sarcoma in Africa. Arch Intern Med. 1996;156:202–204. [PubMed] [Google Scholar]
- 11.Corbellino M, Poirel L, Bestetti G, Pizzuto M, Aubin J T, Capra M, Bifulco C, Berti E, Agut H, Rizzardini G, Galli M, Parravicini C. Restricted tissue distribution of extralesional Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS patients with Kaposi’s sarcoma. AIDS Res Hum Retroviruses. 1996;12:651–657. doi: 10.1089/aid.1996.12.651. [DOI] [PubMed] [Google Scholar]
- 12.Cottoni F, De Marco R, Montesu M A. Classical Kaposi’s sarcoma in North-east Sardinia: an overview from 1977 to 1991. Br J Cancer. 1996;72:1132–1133. doi: 10.1038/bjc.1996.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P. KSHV antibodies among Americans, Italians, and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 14.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–923. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 15.Koelle D M, Huang M L, Chandran B, Vieira J, Piepkorn M, Corey L. Frequent detection of Kaposi’s sarcoma-associated herpesvirus (HHV8) DNA in saliva of immunodeficiency virus infected men: clinical and immunologic correlates. J Infect Dis. 1997;176:94–102. doi: 10.1086/514045. [DOI] [PubMed] [Google Scholar]
- 16.Kwok R K, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 17.Lennette E T, Blackbourn D J, Levy J A. Antibodies to human herpesvirus type 8 in the general population and in Kaposi’s sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- 18.Levy J A. A new human herpesvirus: KSHV or HHV8. Lancet. 1995;34:786. doi: 10.1016/s0140-6736(95)91611-3. [DOI] [PubMed] [Google Scholar]
- 19.Lin J C, Lin S C, Mar E C, Pelleyy P E, Stamey F R, Stewart J A, Spira T J. Is Kaposi’s sarcoma-associated herpesvirus detectable in semen of HIV-infected homosexual men? Lancet. 1995;346:1601–1602. doi: 10.1016/s0140-6736(95)91931-7. [DOI] [PubMed] [Google Scholar]
- 20.Lospalluti M, Mastrolonardo M, Loconsole F, Conte A, Rantuccio F. Classical Kaposi’s sarcoma: a survey of 163 cases observed in Bari, south Italy. Dermatology. 1995;191:104–108. doi: 10.1159/000246525. [DOI] [PubMed] [Google Scholar]
- 21.Luppi M, Torelli G. The new lymphotropic herpesviruses (HHV6, HHV7, HHV8) and hepatitis C virus (HCV) in human lymphoproliferative diseases: an overview. Haematologica. 1996;81:265–281. [PubMed] [Google Scholar]
- 22.Luppi M, Barozzi P, Majorana A, Collina G, Ferrari M G, Marasca R, Morselli M, Rossi E, Ceccherini-Nelli L, Torelli G. Frequency and distribution of herpesvirus-like DNA sequences (KSHV) in different stages of classic Kaposi’s sarcoma and in normal tissue from an Italian population. Int J Cancer. 1996;66:427–431. doi: 10.1002/(SICI)1097-0215(19960516)66:4<427::AID-IJC3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Mayama S, Cuevas L E, Sheldon J, Omar O H, Smith D H, Okong P, Silvel B, Hart C A, Schulz T F. Prevalence and transmission of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in Ugandan children and adolescents. Int J Cancer. 1998;77:817–820. doi: 10.1002/(sici)1097-0215(19980911)77:6<817::aid-ijc2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 24.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma lesions from persons with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 26.Olsen S J, Chang Y, Moore P S, Biggar R J, Melbye M. Increasing Kaposi’s sarcoma-associated herpesvirus seroprevalence with age in a highly Kaposi’s sarcoma endemic region, Zambia in 1985. AIDS. 1998;12:1921–1925. doi: 10.1097/00002030-199814000-00024. [DOI] [PubMed] [Google Scholar]
- 27.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 28.Simpson G R, Schultz T F, Whitby D, Cook P M, Boshoff C, Rainbow R, Howard M R, Gao S J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V D, Weiss R A, Moore P S. Prevalence of Kaposi’s sarcoma-associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 29.Smith M S, Bloomer C, Horvat R, Goldstein E, Casparian J M, Chandran B. Detection of human herpesvirus 8 DNA in Kaposi’s sarcoma lesions and peripheral blood of human immunodeficiency virus-positive patients and correlation with serologic measurements. J Infect Dis. 1997;176:84–93. doi: 10.1086/514043. [DOI] [PubMed] [Google Scholar]
- 30.Vieira J, Huang M L, Koelle D M, Corey L. Transmissible Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi’s sarcoma. J Virol. 1997;71:7083–7087. doi: 10.1128/jvi.71.9.7083-7087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viviano E, Vitale F, Ajello F, Perna A M, Villafrate M R, Bonura F, Aricò M, Mazzola G, Romano N. Human herpesvirus type 8 DNA sequences in biological samples of HIV-positive and negative individuals in Sicily. AIDS. 1997;11:607–612. doi: 10.1097/00002030-199705000-00008. [DOI] [PubMed] [Google Scholar]
- 32.Whitby D, Luppi M, Barozzi P, Boshoff C, Weiss R A, Torelli G. Human herpesvirus 8 seroprevalence in blood donors and lymphoma patients from different regions of Italy. J Natl Cancer Inst. 1998;90:395–397. doi: 10.1093/jnci/90.5.395. [DOI] [PubMed] [Google Scholar]