Abstract
We determined biovars and serotypes of 154 isolates of Cryptococcus neoformans from clinical and environmental sources from different areas of Spain. All clinical isolates belonged to C. neoformans var. neoformans. Serotypes showed an irregular distribution. C. neoformans var. gattii serotype B was isolated from necropsy specimens from goats with pulmonary disease.
Cryptococcus neoformans is an encapsulated yeast that is responsible for life-threatening infections, particularly in immunocompromised patients (7, 21). C. neoformans exists in two varieties, C. neoformans var. neoformans and C. neoformans var. gattii (4, 32). These two varieties of C. neoformans (4, 32) are easily differentiated by their growth in l-canavanine-glycine-bromothymol blue agar (CGB) (19, 20) and agar with d-proline and d-tryptophan (9, 26). Based on the antigenic determinants of the polysaccharide capsule, serotypes A, D, and AD of C. neoformans var. neoformans and serotypes B and C of C. neoformans var. gattii have been identified (16). Differences between the two varieties with regard to pathogenicity and geographical distribution have been described. C. neoformans var. neoformans is responsible for most cases of cryptococcosis in immunocompromised patients, and C. neoformans var. gattii has been associated with infections in subjects with a normal immunologic status (10, 30). Cryptococcus neoformans var. neoformans has a worldwide distribution (3), whereas C. neoformans var. gattii has been reported to be restricted to tropical and subtropical areas (10, 11, 18, 29). Some autochthonous isolates have been obtained from temperate areas in Mexico and South America (2, 5, 14, 22) and Europe (1, 24). In Spain, the distribution of varieties and serotypes of C. neoformans isolates has not been previously assessed. We studied the distribution of biovars and serotypes of clinical and environmental isolates of C. neoformans from different geographical areas of Spain. Our findings were compared with the serotype distribution of C. neoformans isolates from several Latin American countries.
Rabbit polyclonal antibodies against each serotype, which were absorbed with the other serotypes, were prepared in our laboratory for serotyping the strains. A total of 154 C. neoformans strains isolated from different areas of Spain from 1988 through to 1997 were studied. Of these, 115 strains were from clinical human isolates; of these, only one strain was identified per patient, another 6 strains were isolated from goats with chronic pneumonia, and 33 isolates were obtained from environmental sources (soil and pigeon droppings). In 95% of the cases, samples were obtained from human immunodeficiency virus (HIV)-infected patients. All of them were intravenous drug users, except for one hemophilic patient and one homosexual patient. The non-HIV-infected patients included recipients of organ transplantation as well as patients with hematological malignancies or solid tumors. A total of 110 strains from Central and South American countries were obtained from colleagues. They originated from Buenos Aires, Argentina (44 strains), Brazil (8 strains from Porto Alegre, 8 strains from Curitiba, and 28 strains from Rio de Janeiro), and Cuba (22 strains from Havana). All of the isolates were of clinical origin except one animal isolate from Cuba. C. neoformans serotype A strains ATCC 90112 and RV56164, serotype B strain RV 20185, serotype C strains RV45978 and NCPF 3170, and serotype D strain RV68038 were used as controls.
Strains were grown on niger seed (Guizotia abyssinica) agar medium and Pal’s medium with sunflower seeds (Helianthus annuus) (31). In both media, the production of brown-pigmented colonies due to the activity of phenoloxidase was observed. Positive urease tests, auxanograms for sugars, and sensitivity to cycloheximide (Auxocolor; Sanofi, Pasteur, Paris, France) were all characteristic of the C. neoformans isolates. The biovariety study was performed by culturing the isolates in CGB medium and noting their ability to use glycine as a unique source of carbon (19, 32). Disks impregnated with d-proline and d-tryptophan on carbon base agar were used for the study of these d-amino acids as unique sources of nitrogen (9, 26). The change of color of the CGB medium or growth of the strains in the presence of d-proline and d-tryptophan was assessed after 48 to 72 h of incubation at 25°C. Serotyping was performed by a slide agglutination procedure using polyclonal antibodies obtained from the immunization of male New Zealand White rabbits with heat-killed cells from the reference strains of serotypes A, B, C, and D. The method proposed by Evans (12, 13) with modifications was used for rabbit immunization. The rabbits were inoculated with daily doses of C. neoformans for 3 weeks, with progressively higher concentrations (106, 107, and 108 CFU/ml) given each week, after jugular catheter insertion into the superior vena cava. Blood samples for determining the serum titers were obtained through the same catheter. One prophylactic dose of cloxacillin was given intramuscularly before surgery. Sera were absorbed with heterologous cells at 37°C for 2 h and left overnight at 4°C. After centrifugation, the supernatants were tested for the presence of antibodies by the slide agglutination test. The Crypto Check agglutination test (Iatron Labs Inc., Tokyo, Japan) (16, 17) was applied to 108 isolates for quality control of the procedure.
All isolates were urease positive and also gave a positive reaction for phenoloxidase in the niger seed and Pal’s media. There was a complete agreement among the three methods used for determining the biovariety, and the biovars correlate with the serotypes. The strains of C. neoformans var. gattii grew, producing a color change in CGB medium, and all assimilated d-proline and d-tryptophan with enhanced growth around the disk. The serotype distribution of the Spanish isolates of C. neoformans according to their source is shown in Table 1. Most of the human and environmental isolates belonged to serotype A; only those isolated from goats were serotype B. Complete agreement in results was found with the 108 isolates tested with the Crypto Check agglutination method. The geographical origin and serotype distribution of the clinical Spanish isolates are shown in Table 2. Distribution of clinical serotypes around the country was uneven (Fig. 1). Complete clinical data were obtained in 73% of the Spanish cases. Only three of them were HIV negative, and all belonged to serotype A. In the HIV-positive patients, serotype A was predominant (59.3%) but a high number of serotype D (28.4%) and serotype AD (12.3%) isolates were also found. No C serotype isolates were observed. Serotypes of the clinical isolates of C. neoformans from Brazil, Cuba, and Argentina are shown in Table 3. C. neoformans var. gattii was not found among the clinical isolates from Argentina and Cuba. One strain of C. neoformans var. gattii, however, was obtained from a necropsied cheetah (Acinonyx jubatus) from the National Zoo of Havana. In contrast, 18 isolates of C. neoformans var. gattii were identified among the Brazilian strains, 14 of them having been isolated from HIV-negative patients.
TABLE 1.
Distribution of the serotypes of 154 C. neoformans isolates from Spain with respect to their sources
| Source | Total no. of strains | No. (%) of strains of indicated serotype
|
||||
|---|---|---|---|---|---|---|
| A | D | AD | B | C | ||
| Humans | 115 | 71 (62) | 33 (29) | 11 (9) | 0 | 0 |
| Animals | 6 | 0 | 0 | 0 | 6 (100) | 0 |
| Environment | 33 | 26 (79) | 5 (15) | 2 (16) | 0 | 0 |
TABLE 2.
Geographical origin and serotype distribution of C. neoformans among Spanish clinical isolates
| Geographic region | Total no. of strains | No. (%) of strains of indicated serotype
|
||||
|---|---|---|---|---|---|---|
| A | D | AD | B | C | ||
| Barcelona | 35 | 27 (77) | 6 (17) | 2 (6) | 0 | 0 |
| Valencia | 11 | 0 | 11 (100) | 0 | 0 | 0 |
| Mallorca | 21 | 21 (100) | 0 | 0 | 0 | 0 |
| Sevilla | 20 | 12 (60) | 4 (20) | 4 (20) | 0 | 0 |
| Madrid | 17 | 6 (35) | 9 (53) | 2 (12) | 0 | 0 |
| Salamanca | 1 | 0 | 1 (100) | 0 | 0 | 0 |
| Pais Vasco | 9 | 4 (45) | 2 (22) | 3 (33) | 0 | 0 |
| Santander | 1 | 1 (100) | 0 | 0 | 0 | 0 |
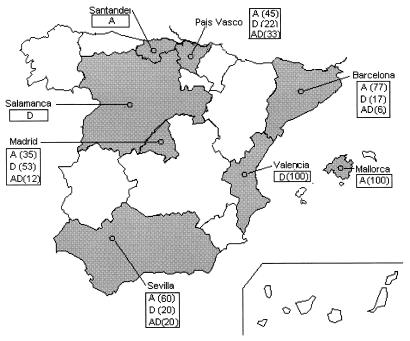
FIG. 1.
Distribution of C. neoformans clinical serotypes in Spain (results shown in percentages). Only single cultures from Santander and Salamanca were evaluated.
TABLE 3.
Distribution of C. neoformans serotypes in human and animal isolates from a West Indian and two South American countries
| Country | Total no. of strains | No. (%) of strains of indicated serotype
|
||||
|---|---|---|---|---|---|---|
| A | D | AD | B | C | ||
| Brazil | 44 | 26 (59) | 0 | 0 | 18 (41) | 0 |
| Cuba | 22 | 21 (95) | 0 | 0 | 1a (5) | 0 |
| Argentina | 44 | 39 (88) | 3 (7) | 2 (5) | 0 | 0 |
Isolate of animal origin.
This was the first epidemiological survey performed in Spain on the prevalence and geographical distribution of the biovarieties and serotypes of C. neoformans. As in other European countries in which epidemiologic studies of C. neoformans have been carried out, C. neoformans var. neoformans is the cause of cryptococcosis in HIV-infected patients and other immunocompromised hosts. Until now, in Spain, no isolates of C. neoformans var. gattii have been observed in humans, but this variety has been isolated from goats with cryptococcosis suffering from severe pulmonary disease (1). This finding suggests that the autochthonous distribution of serotype B of C. neoformans is not limited to tropical and subtropical areas but also includes areas with temperate climates, such as Spain, Portugal, and Italy (23, 24). There seems to be a relationship between the distribution of five species of Eucalyptus, including E. blakelyi, E. camaldulensis, E. gomphocephala, E. rudis, and E. tereticornis (11, 27, 28) and C. neoformans var. gattii isolates. Most of these Eucalyptus species are found in some Spanish geographical locations, but no isolates of C. neoformans var. gattii have been previously identified (6). In Europe, there have been a few reports in which autochthonous human isolates of C. neoformans var. gattii have been described (18, 25). The three methods used for the differentiation of the two varieties were accurate, and the results were in agreement. All isolates of the C. neoformans var. gattii assimilated d-proline and d-tryptophan and grew in CGB medium; therefore, any one of these methods could have been used. The assimilation test for d-amino acids is easy to perform and less expensive than growth in CGB medium.
Serotype A is prevalent in clinical specimens in Spain, but serotypes D (29%) and AD (9%) are frequent and predominant in some areas (Fig. 1). This pattern also occurs with the environmental isolates, with serotypes D and AD frequently occurring. These data are consistent with the relative prevalence of serotype D in some areas of France and Italy (8, 15, 33). We found an irregular distribution of serotype D among clinical isolates, with the highest prevalence of serotype D occurring in Valencia (eastern Spain) and Madrid (central Spain). No serotype C isolates were found either in Spain or Europe. Data from American isolates also showed a higher prevalence of serotype A than serotype D in Brazil and Cuba. The exception to this pattern was for isolates from Argentina, possibly because Buenos Aires has a temperate climate similar to that of Spain. We would like to draw attention to the first isolation of C. neoformans var. gattii in a cheetah from the National Zoo in Havana (Cuba). This animal is another mammalian victim of cryptococcosis.
Currently, studies using molecular biology techniques are in progress to establish the genomic DNA patterns of C. neoformans and thus obtain a better understanding of the epidemiology of cryptococcosis in Spain.
Acknowledgments
This work was supported by grant 96/1991-01 from the Fondo de Investigaciones sanitarias (FIS) of the Spanish Ministry of Health.
We thank all of our colleagues who provided C. neoformans strains. We are grateful to Libero Ajello for constructive criticisms and critical review and to Marta Pulido for editing the manuscript and for editorial assistance.
REFERENCES
- 1.Baró T, Torres-Rodriguez J M, Hermoso de Mendoza M, Morera Y, Alía C. First identification of autochthonous Cryptococcus neoformans var. gattii isolated from goats with predominantly severe pulmonary disease in Spain. J Clin Microbiol. 1998;36:458–461. doi: 10.1128/jcm.36.2.458-461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bava A J, Negroni R. Características epidemiológicas de 105 casos de criptococosis diagnosticados en la República Argentina entre 1981–1990. Rev Inst Med Trop Sao Paulo. 1992;34:335–340. [PubMed] [Google Scholar]
- 3.Bennett J E, Kwon-Chung K J, Howard D H. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 4.Bennett J E, Kwon-Chung K J, Theodore T S. Biochemical differences between serotypes of Cryptococcus neoformans. Sabouraudia. 1978;16:167–174. [PubMed] [Google Scholar]
- 5.Bustamante B, Swinne D. Cryptococcus neoformans var. gattii isolates from two Peruvian patients. Rev Iberoam Micol. 1998;15:22–24. [PubMed] [Google Scholar]
- 6.Colom M F, Alberdi M, Meseguer I, Torres-Rodríguez J M. Aislamiento de Cryptococcus neoformans, en muestras de medio ambiente de Alicante. Rev Iberoam Micol. 1997;14:63–64. [PubMed] [Google Scholar]
- 7.Chuck S L, Sande M A. Infections with Cryptococcus neoformans in the acquired immunodeficiency syndrome. N Engl J Med. 1989;321:794–799. doi: 10.1056/NEJM198909213211205. [DOI] [PubMed] [Google Scholar]
- 8.Dromer F, Mathoulin S, Dupont B, Letenneur L, Ronin O the French Cryptococcosis Study Group. Individual and environmental factors associated with infection due to Cryptococcus neoformans serotype D. Clin Infect Dis. 1996;23:91–96. doi: 10.1093/clinids/23.1.91. [DOI] [PubMed] [Google Scholar]
- 9.Dufait R, Velho R, De Vroey C. Rapid identification of the two varieties of Cryptococcus neoformans by D-Proline. Mykosen. 1987;30:483. [PubMed] [Google Scholar]
- 10.Ellis D H. Cryptococcus neoformans var. gattii in Australia. J Clin Microbiol. 1987;25:430–431. doi: 10.1128/jcm.25.2.430-431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis D H, Pfeiffer T J. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990;28:1642–1644. doi: 10.1128/jcm.28.7.1642-1644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans E E, Kessel J F. The antigenic composition of Cryptococcus neoformans. II. Serologic studies with the capsular polysaccharide. J Immunol. 1951;67:109–114. [PubMed] [Google Scholar]
- 13.Evans E E, Sorensen L J, Walls K W. The antigenic composition of Cryptococcus neoformans. A survey of cross-reactions among strains of Cryptococcus and other antigens. J Bacteriol. 1953;66:287–293. doi: 10.1128/jb.66.3.287-293.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gezuele E, Calegari L, Sanabria D, Davel G, Civila E. Isolation in Uruguay of Cryptococcus neoformans var. gattii from a nest of the wasp Polybia occidentalis. Rev Iberoam Micol. 1993;10:5–6. [Google Scholar]
- 15.Griseo G, Gallo M. Serotyping of Cryptococcus neoformans isolates from environmental and clinical sources in extreme southern Italy (Calabria and Sicily, central Mediterranean area) Mycoses. 1997;40:95–100. doi: 10.1111/j.1439-0507.1997.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda R, Shinoda T, Fukazawa Y, Kaufman L. Antigenic characterization of Cryptococcus neoformans serotypes and its application to serotyping clinical isolates. J Clin Microbiol. 1982;16:22–29. doi: 10.1128/jcm.16.1.22-29.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabasawa K, Itagaki H, Ikeda R, Shinoda T, Kagaya K, Fukazawa Y. Evaluation of a new method for identification of Cryptococcus neoformans which uses serologic tests aided by selected biologic tests. J Clin Microbiol. 1991;29:2873–2876. doi: 10.1128/jcm.29.12.2873-2876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohl K H, Hof H, Schrettenbrunner A, Seeliger H P R, Kwon-Chung K J. Cryptococcus neoformans var. gattii in Europe. Lancet. 1985;29:1515. doi: 10.1016/s0140-6736(85)92300-1. [DOI] [PubMed] [Google Scholar]
- 19.Kwon-Chung K J, Polacheck I, Bennett J E. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C) J Clin Microbiol. 1982;15:535–537. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon-Chung K J, Bennett J E. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984;120:123–130. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 21.Levitz S M. The ecology of Cryptococcus neoformans and the epidemiology of cryptococcosis. Rev Infect Dis. 1991;13:1163–1169. doi: 10.1093/clinids/13.6.1163. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Martinez R, Soto-Hernandez J L, Ostrosky-Zeichner L, Castañon-Olivares L R, Angeles-Morales V, Sotelo J. Cryptococcus neoformans var. gattii among patients with cryptococcal meningitis in Mexico. First observations. Mycopathologia. 1996;134:61–64. doi: 10.1007/BF00436865. [DOI] [PubMed] [Google Scholar]
- 23.Martins H M, Bernardo F M, Martins M L. Abstracts of the Third Meeting of the European Confederation of Medical Mycology, Lisbon, Portugal. 1996. Cryptococcus spp. associated with Eucalyptus trees in Lisboa, Portugal, abstr. 7.1. [Google Scholar]
- 24.Montagna M T, Tortorano A M, Fiore L, Viviani M A, Barbuti S. Cryptococcus neoformans var. gattii en Italy. Note I. Premier cas autochtone de mëningite à sérotype B chez un sujet VIH positif. J Mycol Med. 1997;7:90–92. [Google Scholar]
- 25.Montagna M T, Viviani M A, Pulito A, Aralla C, Tortorano A M, Fiore L, Barbuti S. Cryptococcus neoformans var. gattii in Italy. Note II. Environmental investigation related to an autochthonous clinical case in Apulia. J Mycol Med. 1997;7:93–96. [Google Scholar]
- 26.Mukamurangwa P, Raes-Wuytack C, De Vroey C. Cryptococcus neoformans var. gattii can be separated from the var. neoformans by its ability to assimilate d-tryptophan. J Med Vet Mycol. 1995;33:419–420. [PubMed] [Google Scholar]
- 27.Pfeiffer T, Ellis D. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. J Med Vet Mycol. 1992;30:407–408. [PubMed] [Google Scholar]
- 28.Pfeiffer T, Ellis D. Abstracts of the 13th Congress of the International Society for Human and Animal Mycology (ISHAM) 1997. North Adelaide, Australia: International Society for Human and Animal Mycology; 1997. Additional eucalyptus hosts for Cryptococcus neoformans var. gattii, abstr. FIII; p. 106. [PubMed] [Google Scholar]
- 29.Sorrell T C, Brownlee A G, Ruma P, Malik R, Pfeiffer T J, Ellis D H. Natural environmental sources of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1996;34:1261–1263. doi: 10.1128/jcm.34.5.1261-1263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speed B, Dunt D. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin Infect Dis. 1995;21:28–34. doi: 10.1093/clinids/21.1.28. [DOI] [PubMed] [Google Scholar]
- 31.Staib F, Seibold M, Artweiler E, Frohlich B, Weber S, Blisse A. The brown colour effect (BCE) of Cryptococcus neoformans in the diagnosis, control and epidemiology of C. neoformans infections in AIDS patients. Zentbl Bakteriol Hyg A. 1987;266:167–177. doi: 10.1016/s0176-6724(87)80030-5. [DOI] [PubMed] [Google Scholar]
- 32.Swinne D, De Vroey C. Epidemiologie de la cryptococcose. Rev Iberoam Micol. 1987;4:77–83. [Google Scholar]
- 33.Tortorano A M, Viviani M A, Rigoni A L, Cogliati M, Roverselli A, Pagano A. Prevalence of serotype D in Cryptococcus neoformans isolates from HIV-positive and HIV-negative patients in Italy. Mycoses. 1997;40:297–302. doi: 10.1111/j.1439-0507.1997.tb00235.x. [DOI] [PubMed] [Google Scholar]