Abstract
Single-neuron actions are the basis of brain function, as clinical sequelae, neuronal dysfunction or failure for most of the central nervous system (CNS) diseases and injuries can be identified via tracing single-neurons. The bulk analysis methods tend to miscue critical information by assessing the population-averaged outcomes. However, its primary requisite in neuroscience to analyze single-neurons and to understand dynamic interplay of neurons and their environment. Microfluidic systems enable precise control over nano-to femto-liter volumes via adjusting device geometry, surface characteristics, and flow-dynamics, thus facilitating a well-defined micro-environment with spatio-temporal control for single-neuron analysis. The microfluidic platform not only offers a comprehensive landscape to study brain cell diversity at the level of transcriptome, genome, and/or epigenome of individual cells but also has a substantial role in deciphering complex dynamics of brain development and brain-related disorders. In this review, we highlight recent advances of microfluidic devices for single-neuron analysis, i.e., single-neuron trapping, single-neuron dynamics, single-neuron proteomics, single-neuron transcriptomics, drug delivery at the single-neuron level, single axon guidance, and single-neuron differentiation. Moreover, we also emphasize limitations and future challenges of single-neuron analysis by focusing on key performances of throughput and multiparametric activity analysis on microfluidic platforms.
Keywords: Single neuron analysis, Microfluidic devices, Microelectrode array, Single cell analysis, Single neuron dynamics, Omics, Single axon guidance
Graphical abstract
1. Introduction
As per the greatly debated ‘sparse-coding’ hypothesis of neural networks, only a few neurons need to fire to generate a response [1]. It has become of utmost importance to decipher single neurons. Single-cell analysis elucidates precise biological phenomena occurring at cellular physiological and morphological levels [[2], [3], [4], [5], [6]]. Particularly, for neuroscience meticulous research on single neurons as well as spatially isolated neurons provide better neuronal network dynamics functionally, chemically and at molecular level [7,8]. The studies establish the pathway for personalized medicine for neurological disorders, including epilepsy, Parkinson's disease, Alzheimer's disease, and other cognitive and motor disorders [9]. Owing to the direct effect on society, neuropathological models for disease progression and treatments reflect a great interest in biomedical research. Particularly, in vitro microfluidic devices have brought significant therapeutics or neuronal modeling advancements due to the spatiotemporal cellular microenvironment control.
In the last two decades, owing to the combinatory approach of technical development at micro and nanotechnologies with basic sciences, led to the Lab on a chip or micro total analysis systems. The confinement of cells inside the micro chambers comparable to its size has given prominent advantages to microfluidics in manipulating and analyzing single cells. The chips provide controlled, specifically designed morphological microenvironments for highly precise, automated, high-throughput, or parallelized manipulation with a high level of integration and low consumption of reagents [4,5,10]. Advanced imaging technologies have significantly increased the functionality in assessing cell proliferation, growth, gene expression, and cellular signaling pathways inside various unique microfluidic cell trapping and culture systems [11,12].
Neuroscience requires categorical multilevel analysis for a complete understanding of the systems. (a) Molecular or sub-cellular level; action potential signal and propagation, ion channels, modulators, genes along with its products, ligands and receptors, pathogens and toxins, chemical cues, lipid membranes, synapses, axons, and growth cones. Research at the sub-system level interpret the events of neurons stimulation and action potential recordings. (b) Cellular level; includes the analyses of glia, neurons, and their relationships. (c) The organs and tissue level; brains, ganglia, and spinal cord. (d) Animal models, such as snails and mice. Generally, while carrying out these studies at the bulk level there is a high chance of missing out on the cellular events occurring at the single cell level as the cell population analyses blur cellular distinctions. These missed events can be linked to the detection of rare events indicating certain disorders and diseases; thus, the contribution of an individual cell is ignored in a population. Though till now, the animal models have been successful in being the “gold standard” in neuroscience as animals mimic the cellular mechanisms of a disease, yet the tests carried on animals tend to be slow, low-throughput, complex, expensive, arguably unethical and without single neuron interrogation. Also, the use of animal models other than human sometimes results in failure for drug efficacy in humans, thus arising the need for the development of more relevant human in-vitro models for drug testing [13].
The microfluidics or Lab on-chip technology could engineer such in vitro brain models for high-throughput screening, better reproducibility, cost-effectiveness, easy operation, and effective pharmacological assessments. Therefore, deciphering the competing signals and measurements at the single neuron events associated with brain disorders and diseases becomes equally important as bulk studies [14].
The current reviews in the relevant field generally focus on highlighting the contribution of a single neuron to brain function [[15], [16], [17]] or particular tools for neuro-modulations (mostly electric field, magnetic field or optics, etc.) [[18], [19], [20]]. Our earlier publication has also highlighted the significance of single neuron in various aspects of neuroscience [8]. Based on the literature survey, none of the recent review papers covers the subjects as we proposed here. However, there is only one review paper by Wang et al. [21] which reviewed advances in microfluidic-based applications in neurobiology till the year 2009. Since then, there have been hues development of microfluidics devices in the last decade, and it has enabled to study and analyze different aspects of single neurons, ranging from single neuron isolation/trapping to the revelation of single neuron dynamics, single neuron proteomics, and genomics. Moreover, the single-cell/single neuron analysis concept came in frontier research from the last decade. However, the above-mentioned review did not cover the different aspects of single neuron analysis.
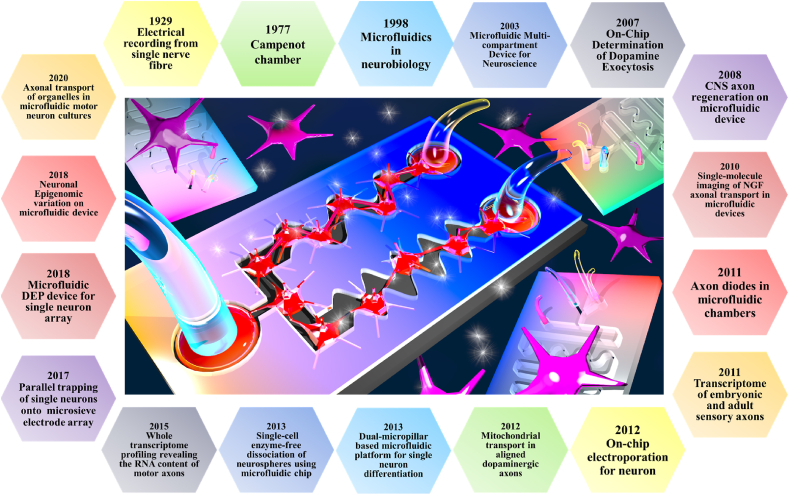
Therefore, here we aim to review all the microfluidic platforms facilitating single neuron analysis, including single neuron trapping, single neuron dynamics, single neuron proteomics, single neuron transcriptomics, drug delivery at the single neuron level, single axon guidance, and single neuron differentiation on microfluidic platforms. The single neuron differentiation lays the foundation for developmental studies in neuroscience, while microscale neuron cultures deciphering neuronal polarity help investigate axon guidance and the precise role of dendritic processes. The review finally critically analyses the advantages, drawbacks, and future challenges of single neuron analysis using microfluidic platforms. A summary of single neuron key events performed on microfluidic devices has been provided in Fig. 1.
Fig. 1.
Timeline for single neuron events performed microfluidic devices.
2. Microfluidic single neuron isolation
To explore the brain at single neuron level, it is a prerequisite to simultaneously develop reliable and high throughput isolation of single neurons along with proper quality assessment. This has been achieved via single neuron dissociation, single neuron sorting and single neuron trapping based on microfluidic platforms.
The microfluidics based dissociation of single neurons from neurospheres overcomes the innate hindrance in protease and collagenolytic activities during enzymatic dissociation of neurons by alternative synthetic enzymes. These chambers also maintain the high viability of dissociated single cells and retain their growth and differentiation features. The individual cells successively regrow neurospheres and shown to be differentiated into the three central neural lineages [22]. The optimization of microfluidic device design parameters (geometry and dimensions of the micro-pillars) and operational parameters (flow rate) improves the dissociation results [23]. As microfluidic-device-based methods are devoid of enzymatic reagents, it eradicates the contamination risk from exogenous substances.
Further, the single cell sorting refers to the sorting of a heterogeneous population of cells based on extracellular and intracellular characteristics. Firstly, inertial microfluidics was employed to dissociate single neurons from the neurosphere. The focusing on single cells at the inner wall and clusters been at the middle of the microchannels help in handling the sensitivity of single neurons towards shear stress and thus facilitating neuron sorting [24].
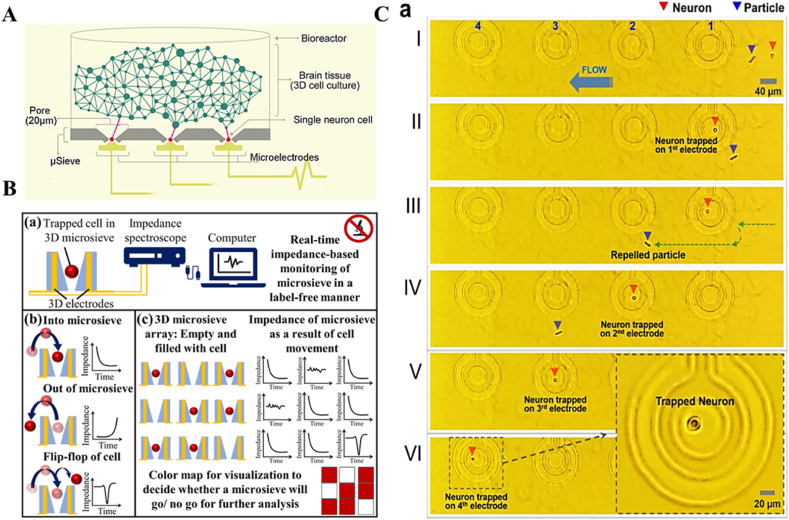
Next method for single neuron isolation known as trapping, defines contacting of single neurons to a surface during capture due to either hydrodynamic methods or diaelectrophoresis or mostly in combination owing to electrogenic nature neurons. Several single neuron trapping microfluidic devices were developed based on hydraulic pressure pump [[25], [26], [27]] and capillary action [28]. The trapping devices are often modified with functionalized electrodes for monitoring the dynamics of single neurons precisely [26]. The major section of single neuron trapping methodologies relies on the microsieve-based electrode arrays due to the direct digitalization of signal readouts at a single neuron level. Only electrical readouts happen to be completely label-free during such high parallelization. In microsieves, the 3D micropores functionalization was done with an electrode similar in the design to multichannel systems, multielectrode arrays (MEA) readout electronics of the brain at a single neuron level. The organized positioning of single neurons while single cell trapping was achieved via passive pumping and capillary phenomena, eradicating the active pumping, decreasing footprint, and also cell damage [29]. Following the laser ablation, the replica molding protocol replicated the optically transparent microsieve structures for single cell trapping, as displayed in Fig. 2A [14]. For the silicon-based platform, MEA-coupled electrodes, while coupling to contact lines of MEA, also easily integrate into the sidewalls of 3D micropores in the device. In the case of the polymeric substrate, the process is not so straightforward. These platforms can be used for dynamic optical readouts by exploiting Ca2+ imaging in SH-SY5Y cell cultures. Further, another polymeric microsieve structure coupled with a disposable add-on was developed for a reusable 3D electrode array. This helped in the real-time analysis of cell placement and distribution [30]. The schematic depicting the analysis set-up and the working principle of the device is shown in Fig. 2B.
Fig. 2.
The images show the single neuron trapping mechanisms on various microfluidic platforms. A The brain-on-a-chip concept schematic depicting a modified microsieve electrode array (μSEA) with pairing of single neurons to a single electrode, beneath is the 3-D brain tissue construct cultured atop the μSEA within a bioreactor [14]. B Analysis set-up (a) and working principles of the 3D microsieve array (b). (c) A Colour map can showing the presence of cells on the distinct microsieve pore via impedance characteristics [30]. C Images recorded for single neuron trapping on ring shaped trapping device arrays. (a) Incoming neuron (I) enters the 1st trap. (II) The neuron gets immobilized in the 1st trap electrode overcoming fluid flow. (III) After the neuron gets trapped, a repelled particle keeps moving forward with the flowing media. (IV) The 2nd trap captures the repelled neuron. (V and VI) Further neurons get trapped inside the 3rd and the 4th ring trap consecutively [31].
Another aspect of single neuron trapping involves the dielectrophoresis (DEP) technique. Under the non-uniform electric field, a polarizable particle gets trapped due to an electrokinetic phenomenon. Further using DEP, the cells are trapped, aligned, and patterned without any supplementary factors (light or magnetic field). For single neuron culture and monitoring, a completely transparent microfluidic DEP device was designed. The device comprises of MEAs fabricated by using indium-tin-oxide (ITO) and a microfluidic chip made up of polydimethylsiloxane (PDMS) (Fig. 2C). The mounting of the device inside an incubated microscope decreases the contamination during cell culture. The array of ring-shaped electrodes is arranged in a row trap and releases the target neuron sequentially. Consequently, successful monitoring of the electrophysiological parameters of single-neuronal cells was performed over time [31].
Another method for electrophoresis based single neuron separation three-layered microfluidic chip was fabricated to study dynamics in the secretion of neurotransmitters from PC12 cells [32]. The chip features nanoliter sized chamber for the perfusion of a cell controlled by pneumatic pressure valves. It also consists of a microfluidic channel for electrophoretic separation and a nanoelectrospray emitter for ionization in mass spectrophotometry (MS) detection.
Advancements in technologies led to the development of individual neuron selection for on-chip electroporator devices coupled with micrometer-sized gold mushroom-shaped microelectrode array. The microfluidic chip integrates two functions – cell electroporation and dielectrophoretic viable cell sorting [[33], [34], [35], [36]]. Apart from single neuron isolation the microfluidic channels allow the soma and axonal compartmentalization for axonal regeneration and analysis [37]. The microfluidic device incorporated with the asymmetrical-channel-based biochip enabled single-cell-resolution study. The laser cell deposition system nullifies the prior impact of cytophilic/phobic surface patterns on axon growth, thus providing systematic investigation of polarized neuron growth, synaptogenesis and development [[38], [39], [40]]. The microchannels also facilitated unidirectional axon growth between “Source” and “Target” hippocampal neural sub-networks. The neurons were cultured in PDMS microfluidic chip, aligned with a 60-microelectrode array [39]. All the microfluidic based methods for single neuron isolation are summarized in Table 1 with their details.
Table 1.
Comparison between the various single neuron isolation techniques based on microfluidics.
| Method | Principle | Modules | Advantages | Disadvantages | Capture ratio/Single cell yield | Cell viability | Reported application for single neuron analysis | Ref. |
|---|---|---|---|---|---|---|---|---|
| Laser cell capture | Laser capture micro-dissection | Gravity-assisted Microdissection; Laser pressure catapult; Laser induced forward transfer | Nullifies the effects of cytophilic/phobic surface | Slow capture rate | 1 cell/90s | 85% | Guidance axon regeneration | [[17], [18], [19]] |
| Microchannels | Mechanical pressure | Micropillars; Asymmetric channels; Layered microfluidics | Avoids protease and collagenolytic activities, No immune response | High flow rate causing cell damage | 91–95% | (80–85%) | Axon growth | [22] |
| Microsieve device | Capillary pumping, Hydrodynamic flow | Passive pumping; Microsieve electrode array, Polymer replica moulded | Label-free, Digital readout of signals at the single cell level, High parallelization, High spatial control | Labor-intensive in absence of electrodes | 80% | 90% over 7 days | Calcium imaging | [14,26,30] |
| Inertial microfluidics | Inertial lift force (FL) and a curvature-induced Dean's drag force (FD) | Differential inertial focusing; Selective inertial focusing | Low shear stress, Low flow rates | Inability to predict outcome, limited ability to deal with concentrated cellular samples | 97% | >90% | Profiling neuro-chemistry occurring in selected neurons | [24] |
| Di-electophoresis | Di-electrophoretic force | Non-transparent electrodes; transparent indium-tin-oxide (ITO) based | Non-invasive, Label-free and non-destructive, Improved homeostatic conditions reduced, contamination risk | Confound live-cell imaging | 100 | 99% at 5 days | Electrophysiological recording and neurological studies. | [31] |
| Droplet microfluidics | Shear forces | T-junction; Flow focusing; Co-flow | Maintains cellular heterogeneity, High-throughput | Low success rate of single-cell droplet isolation | 100-690,000 | 80% at 3 day | Transciptome sequencing | [86] |
3. Single neuron dynamics
The “dynamic single-cell analysis (DSCA)” is referred to real-time monitoring of the different biochemical process in a cell unit [41] helping us to understand the pathological and physiological processes at the cellular level. The most widely used techniques for cell analysis include flow cytometry, automated microscopy, and laser scanning cytometry. However, these techniques lack real-time monitoring of the cell kinetics [42]. To solve the limitations, microfluidic techniques are used to monitor individual cells in real-time as fluidic control has been considered to be a standard for dynamic manipulation for a stimulation pattern even much before the popularity for genetic or synthetic methods. Here, dynamic environmental variables can be controlled and improved for precise and high-throughput analysis of single-cell. Variations in the intracellular components and corresponding reaction of a cell to the environmental stimulus or stress can be analyzed using the microfluidic technique. Thus, we intend to review in this section regarding microfluidics platforms that has become a new era and an effective tool for dynamic analysis in case of single neuron.
3.1. Neuronal communication via action potential propagation in microfluidic platforms
Electrical signaling is known to be primary mode of neuronal communication, where action potentials (APs) are used to encode and transmit information. Thus, to understand neuronal circuits, electrophysiological readings are important. The AP amplitude are very low and impractical to discriminate, therefore making AP propagation difficult to analyze in neuronal cultures. The neuronal processes localization and improved signal-to-noise ratio can be achieved using devices combining microelectrodes and microfluidics (μEF devices) for various neuroscience applications. Such a μEF device having temperature control modulation was used to illustrate neural activity in dorsal root ganglion neurons [43]. The APs of an isolated axon was detected using MEA platform with PDMS microtunnels. This device permitted selective (axonal signals) and relative large (up to 200 μV) electrical signals recordings, including propagation speed and direction of the APs [44,45]. Neuron-on-a-chip was fabricated for axon isolation without dendrite or soma. Optical stimulation method such as femtosecond laser and blue light emitting diode at 780 nm, was used to generate neuronal signals and measure APs [46]. Also, a biomimetic artificial neuron using microfluidic platform was fabricated to resolve the problem of biocompatibility. The microfluidic chambers here represented the environment of inter and intracellular connected via Quake valves. The channels mimic the plasma membrane for the exchange of various chemical species. The membrane potential difference between the two environments (i.e., inter and intracellular) are measured using integrated electrodes [47]. Another on-a-chip system where microfluidic platform is integrated with matrix of electrodes was used to record the intracellular dynamics along with the electrical activity in axonal presynaptic projections and neuronal postsynaptic targets [48]. Lopes et al. assessed neuronal communication and axonal signal propagation along with a user-friendly computational tool, μSpikeHunter on integrated microfluidic setup. The setup records the spike propagation with high signal-to-noise ratio over the course of several weeks for which the computational tool provides detailed quantification of various communicating properties such as spiking time, conduction failure, propagation velocity, and coding mechanism [49,50].
3.2. Dynamic monitoring of stimulated release from individual neurons
Biochemical and chemical messengers that are released from single-cells, plays an important role in biological process, are also a crucial way to communicate between cells. In neuroscience, the real-time dynamic monitoring of these stimulated release of biomolecules from single neuron is significantly important to understand the mechanism and functions of neuron-to-neuron or neuron-glia communications. In different brain regions, over 200 neurotransmitters are secreted to the response of environmental, pharmacological, and various internal effects. Inappropriate release or imbalance of these chemicals can create mental illness. Thus, studying these secreted transmitters at an individual cell level can provide us with in-depth knowledge about intracellular communication and help us better understand the functionality of the neuronal cells. Different catecholamines (such as dopamine, noradrenaline, and adrenaline) are key signaling neurotransmitters in the CNS. Deformity in the exocytosis of these catecholamines leads to neurological disorders such as Alzheimer's and Parkinson's disease. Thus, Huang et al. fabricated a microfluidic device for the location, transport, and quantal secretion monitoring of single cells [25]. Here, single rat pheochromocytoma (PC12) cell was transported to a microchamber using a hydraulic pressure pump. The micromanipulator controlled positioning of the carbon fiber microelectrode 1 mm from the surface of the cell. Microelectrodes were used to detect the quantum secretion of dopamine from individual PC12 cell amperometricaly. The microfluidic device enabled manipulation of single-cell by injecting stimulants, and monitoring quantal secretion from the cell, therefore offering an appropriate platform to investigate physiological and pathological phenomena at an individual cellular level [[51], [52], [53]]. Similarly, the mercaptopropionic acid (MPA)-modified gold electrodes microchip were used to determine and monitor the release of dopamine from single PC12 cell [26]. This microchip was also used to detect diacetate released from PC12 cell using K+ stimulation. Later, a fully automated microchip platform was developed for the detection of exocytosis from single cell positioned on an aperture near a MPA modified electrode. This microchip was used for detection of calcium dependent catecholamine secretion from PC12 cell. The device can detect about 10,000 dopamine molecules in single release [27].
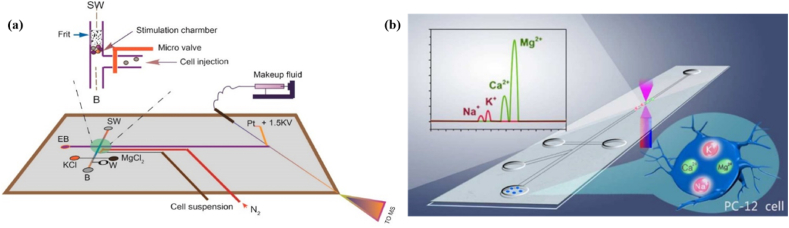
Multilayered microchip device were fabricated by Li et al. with a three-layered microfluidic chip to study dynamics in the secretion of neurotransmitters from PC12 cells [32]. The chip features nanoliter sized chamber for the perfusion of a cell controlled by pneumatic pressure valves. It also consists of a microfluidic channel for electrophoretic separation and a nanoelectrospray emitter for ionization in mass spectrophotometry (MS) detection (Fig. 3a). Thus, the microfluidic platform was deployed as an electrophoresis-mass spectrometric method (MCE-MS) which simultaneously analyzes and quantifies neurotransmitters including serotonin (5-HT), dopamine (DA), glutamic acid (Glu), and aspartic acid (Asp) without need for enrichment or labeling. Two different secretion patterns were observed-one for the amino acids (Asp and Glu) neurotransmitters and another for monoamine (DA and 5-HT). Secretion dynamics for the monoamine neurotransmitters were found to be notably different. This proposed microfluidic platform has shown helpful in monitoring cellular secretions in biological studies.
Fig. 3.
(a) Schematic representation of the microfluidic platform. The cell perfusion is carried out in the nanolitre-sized chamber as depicted in the inset [32]. (b) Schematic representation of the parallel analysis of Mg2+, Ca2+, K+, and Na+ in PC-12 single cell using a microfluidic device. The metal ions are first recognized by the fluorescent probes (Mag-Fluo-4 AM, Fluo-3 AM, and cBDP) in PC-12 cells. Further, in the five-reservoir microchip, the cells were introduced, succeeding one after the other. Later with the application of a strong electric field, the cells were lysed. Next, using this microchip electrophoresis, the ions from the cells were segregated in the microchannel. Finally, a microfluidic device (MFD) was used to detect and analyze multiple fluorescence signals from the ions [54].
Different intracellular metal ions have been shown to have a functional role in the CNS. The deficiency or surplus of essential metal ions could lead to neurodegenerative diseases. Therefore, for diagnostics and monitoring of these diseases, detection of different neuronal metal ions is crucial. Especially, single-neuron analysis for various metal ions helps to comprehend cellular differentiation and heterogeneity along with the relationship between individual cells and metal ions. Li et al. developed a sensitive, convenient and consistent technique to instantaneously detect and measure many metal ions (Na+, K+, Ca2+, and Mg2+) in the individual PC12 cells using microfluidic systems shown in Fig. 3b [54]. They also studied the variations in the metal ions using Aβ25-35 treated PC12 cells, a model for Alzheimer's disease. The study provided understandings of the synergistic function of these metal ions in controlling the neurological diseases at the level of single neurons.
Nitric oxide (NO) is one of the main inflammatory signalling molecules released from microglia in pathological conditions leading to DNA/protein damage followed by induced neuronal apoptosis. Thus to target the NO production via inhibiting inducible nitric oxide synthase (iNOS), a high-throughput microfluidic device was fabricated. The intracellular NO was fluorogenically observed in SIM-A9 microglial cells after lipopolysaccharides (LPS) and interferon gamma (IFN-γ) treatment either in presence or absence of an iNOS inhibitor. Therefore 1.6-fold rise was reported in NO production among untreated and stimulated cells, while it decreased about 0.5 times between stimulated and inhibited cells [55].
3.3. Monitoring neuronal synapses in microfluidic platforms
Microfluidic devices have been helpful for short-distance and long-distance cell-cell communication studies. Short distance communications refer to neuron communications via electrical events called action potentials and chemical neurotransmitters; while long distance communication involve systemic release of signals and transmission of information in absence of direct cell-cell contact; also known as synapses. Synaptic connections help neurons to cover more considerable distances traveling through various extracellular matrices (ECM) and process information transmission. Thus, microfluidics makes an ideal technique for the study of ordered neuron connectivity. Directed neuronal networks can be created by culturing single neuron with the help of low-melting agar etching method. 50 nm thick ITO layer is coated on the cover-slip just below the agar. The initial axon growth will help individual neurons to connect through opened microchannels to form simple networks [56]. Therefore, compartmentalized microfluidic devices for axonal guidance and growth are mainly based on microgrooves. They provide high level of controlled connectivity for neuronal networks at single cell level. The single neuron axonal guidance microfluidic platforms are briefly discussed in next section. Moreover, microfluidic devices are also used for studying neuronal interaction with glial cell or any other targeted cell. Park et al. fabricated a microfluidic device to culture and compartmentalize hippocampal and other CNS neuron [57]. The device consisted of two chambers able to load independently and also were connected with a microchannel array. Defined chemical environments was created inside the chambers using different hydrostatic pressures. This allows the interaction between the neuronal populations using their axonal projections. Later, a neuron-glial co-culture platform was devised to study CNS-glial interaction. For example, myelination process in neuron-oligodendrocyte signaling [[58], [59], [60]]. Taylor et al. fabricated microfluidic local perfusion platform to control presynaptic, synaptic and postsynaptic regions in vitro [61]. More than 100 parallel rows are connected to separate neuron that directs the formation of synapses. The chamber was also used to explore the synapse-to-nucleus signaling. Changes in calcium was also measured at dendrites and somata along with the local perfusion of glutamate.
Similarly, Coquinco et al. fabricated an advanced microfluidic device with three-compartment to create three different isolated environments [62]. Thus, the target neuron was placed at the center compartment, while the competing neurons were located on either side compartment. Shi et al. developed a microfluidic device where neurons and glia could co-cultured, which permitted reversible separation [63]. The communications between astrocytes and neurons and their reactions towards glutamate stimulation were studied by Gao et al. [64] using a microfluidic device. The combination of polyethylene glycol surface patterning was used precisely for cell placement and neurite guidance. Moreover, the two chambers are separated by the fabricated microgrooves [58], and genetically encoded calcium indicators were used to monitor neuron-astrocyte interactions. This technological combination has allowed analyzing real-time signaling between astrocytes and neurons. The combination of actuators with microfluidics platforms can be used for fine control of neuron-to-neuron connectivity [[64], [65], [66]]. This has provided increased interest in the study of formation of synapses in vitro. Also, a study shows microfluidic platform used to reconstruct the cortico-hippocampal neuronal network, where individual axon is easily identifiable passing through the synaptic chamber [67]. The device was used for the study of brain derived neurotrophic factor axonal transport depending upon glucocorticoid treatment. Thus, Microfluidic technology has greatly helped the neuroscience field and will continue to offer exceptional communication and neuron synapse studies.
4. Microfluidic single axon guidance and analysis
Neurons are highly polarized cells with a 1000 times difference in length of their two morphological and functional parts, i.e., axons and cell bodies. The protein-synthesizing machinery, i.e., ribosome devoid of such long axons, mediates the diffusion of protein produced in the soma for cell growth and maintenance. Thus, neurons are highly dependent on active axon transport for the materials required at synaptic terminals, including neurotransmitter receptors, synaptic proteins, lipids, ion channels and mitochondria etc. On the contrary, the target cell-derived neurotrophic signals (e.g., neuron growth factor NGF) are transported to cell bodies via retrograde axon transport from the distal-axon sites. Thus, both the parts of the neuron cell play a crucial yet dependent role. Any ambiguities or disruption in axonal transport might result in pathological conditions and neurodegenerative diseases [68].
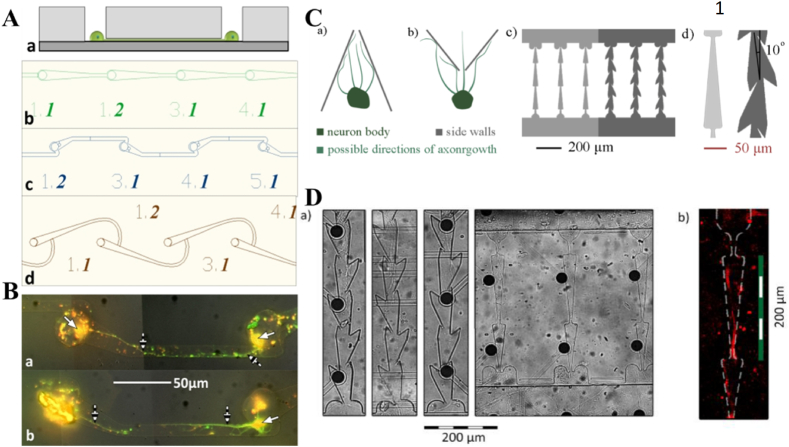
Microfluidic channels allow the soma and axonal compartmentalization for isolation studies. It facilitated the imaging of pseudo rabies virus (PRV)-Bartha viral protein dynamics in live infected neurons, separately in axon and soma [37]. Aguayo and colleagues [69] demonstrated that a suitable environment favors increasing the length of adult CNS injured neuron processes. The microfluidic device incorporated with the asymmetrical-channel-based biochip enabled single-cell-resolution study. It helped to assess the effect of channel geometry and glial cells over polarized axonal growth in the formation of distinct neuron circuits [38]. To nullify the prior impact of cytophilic/phobic surface patterns on axon growth, the single cells, including neuron-glia pairs, were placed into particular micro-wells of the microfluidic device with the aid of a laser cell deposition system. As an outcome, the “snag” channels (showed in Fig. 4A) caused axonal growth in the expected direction 4:1 over opposite direction (Fig. 4B). As displayed in Fig. 6B, axons tend to extend past sharp turns [38]. For further understanding of the neuronal network for communication and connectivity, four different types of microchannels by Malishev et al. as shown in Fig. 4C&D were designed for modeling axon “focusing,” and a “bottleneck” for backward growing axons inside “Straight” type, and “Zig-zag” type microchannel designs [39].
Fig. 4.
Single axon guidance on microfluidic devices: A depicting (a) the elastomeric membrane concept where the neurons are confined in the microwells, with neurites are directed through the short microtunnels (b) Directed (c) Snag and (d) Hook microstructure design. B Fluorescence microscopy indicating abundance of MAP2 (red) and neurofilaments (green) respectively in dendrites and axons post 72 h of deposition inside snag microstructures, and overlapping of the two stains is shown in yellow. White arrows point out neuron cell bodies while striped arrows point towards axons. (a)A complete and (b) an incomplete circuit between two neurons [38]. C Some more designs proposed for microchannels modeling a) axon “focusing,” b) a “bottleneck” for backward growing axons inside either c) “Straight” type (left) and “Zig-zag” type (right), or d) triangular segments' design, blue bar – 200 μm. D Axon growth microscopy a) bright field and b) immunofluorescence (Tau) images of axons in the proposed microchannels.
Fig. 6.
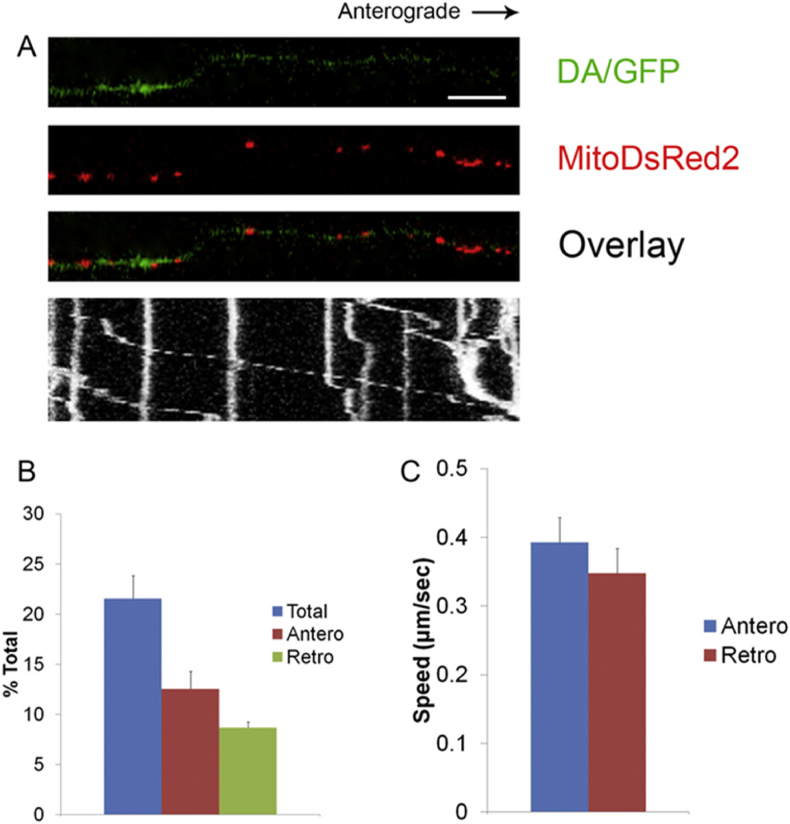
Axonal mitochondrial transport measurement inside microchannels. (A) Kymograph of mitoDsRed2 labeled DA mitochondrial normal movement in microchannels. Scale bar 10 μm. (B) Bar graph showing the percentage of moving mitochondria from 12 axons over 3 independent experiments. (C) The mitochondrial movement speed in both anterograde and retrograde directions (n = 60 mitochondria) [74].
The microchannels facilitated unidirectional axon growth between “Source” and “Target” hippocampal neural sub-networks. The neurons were cultured in PDMS microfluidic chip, aligned with a 60-microelectrode array. The “Straight” microchannels were optimum for unidirectional synaptic spike propagation during spontaneous and stimulus-evoked bursts. A minimum length of 400 μm was required for the microchannel to deliver unidirectional coupling. Such MEA coupled microfluidic chips decipher network dynamics providing realistic morphology, network-wide synaptic plasticity, and intercellular interactions among different cell types, a basic factor governing higher cognitive functions of the brain [39].
Jeong et al. developed a device, which included microgrooves (10 μm wide, 3 μm in height, and 150–900 μm in length) to isolate axons from cell bodies by capillary action [28]. The fluidic isolation of the somal and axonal compartment provides a highly tunable platform resembling neurodegeneration and CNS neuron injury [57]. Moreover, the isolation of axons renders the ability to provide localized biomolecular treatment along with physical guidance. The axon length quantification process reveals the influence of local components of ECM and brain-derived neurotropic factors. The chondroitin sulfate proteoglycan causes axon retraction during axons treatment, while stomata treatment leads to axonal growth; therefore, these microchips are highly preferred for axon growth and regeneration studies [70]. Compartmentalization and axons isolation happens to be ideal for the investigation of subcellular processes.
Owing to the smaller diameter of axons (approx. 0.2 μm diameter), the neurons could be patched mostly via somas and dendritic spines to study prevailingly counterparts employing the patch-clamp technique. Moreover, due to the physical limitation of proximity, many patch-clamp electrodes could be used while restraining the cell number or the total points onto a single cell to be patched instantaneously. But the axonal isolation using microfluidic chips overcomes the limitations mentioned above and enables long-term recordings, up to several weeks or months. These axonal isolation devices employed various isolation and axon study methods, i.e., substrate patterning with conducting polymeric materials to chemical attractants, physical barriers in terms of fluidic isolation, or controlled chemical gradient system [71].
The creation of tapered ‘axon diodes channels’ oriented the network in one direction. The combination of extracellular electrodes aligned with tapered microchannels facilitated the electrophysiological recording of both soma and axons located outside and inside the channels, respectively. Further, such kind of microfluidic isolation channels combined with microelectrode array, capable of recording and stimulation, helped in clear understanding of axonal signal behavior [72]. Lewandowska et al. fabricated a two-chamber device, separated via >30 channels, supported the long-term recording of single spike with high spatial resolution from isolated axons with signal amplitudes from 100 μV to 2 mV. Hence, such studies help to make the axonal action potential reshaping evident, and the modulations support real inferences for the release of neurotransmitters. The device involves around 10–50 electrodes for each channel to record the propagating signals along the axons. It also becomes feasible to readout single action potentials propagating along individual axons with multiple branches with high spatial resolution while considering spontaneous action potentials with characteristic shapes propagating from somas along axons in between the two compartments. Interfacing of somas and axons of a single cell with electrode arrays facilitates distinguishing soma-axon pairs via stimulating the electrode array. The single cell analysis on microfluidic channels also showed clearly the origin of complex spike shapes generated from a linear superposition of multiple axonal signals instead of signal distortion posed by the channels [71].
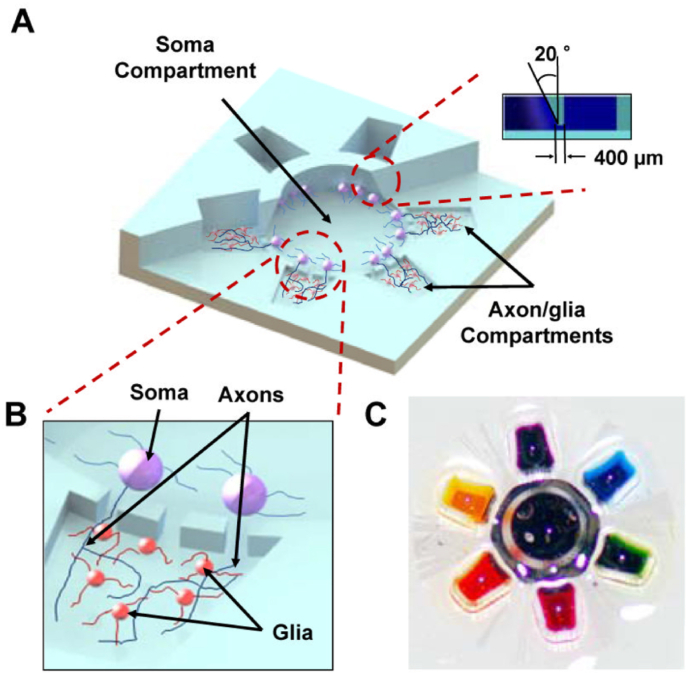
A laser-based cell-micro patterning system created a single cell analysis platform with different compartments for axon isolation with polarized neuron growth to systematically investigate synaptogenesis, development, and role [40]. The localized biomolecular treatments also helped in the understanding of CNS axon biology and glia-axon interactions. The multi-compartmentalized CNS neuron-glia microfluidic co-culture platform administers localized drug and biomolecule administration in parallel, in multiple co-culture high throughput settings prepared to evaluate axon-glia interactions. Axons developing from the soma could be physically and fluidically isolated in the six satellite axon/glia compartments facilitating localized therapy. Park et al. employed similar microfluidic chamber (shown in Fig. 5) to treat isolated axons with chondroitin sulfate proteoglycan (CSPG, concentration range 0–25 μg/ml), assessing the effect of parallel localized biomolecular treatment on axonal biology and axon-glia interactions [59].
Fig. 5.
(A) 3D graphic showing a multi-compartment neuron-glia co-culture microsystem. It facilitates multiple localized axon treatments in parallel (Inset contains the cross-sectional projection for truncated cone-shaped soma compartment). (B) Schematic shows the axon-soma isolation for localized axon-glia interaction study. (C) Digital photograph for the neuron-glia co-culture platform with seven different colour dyes for visualization [59].
Further, organelle tracking in individual MN axons at microfluidic platform have helped to investigate the mechanism of MN health, affecting the neuron growth, development, and persistence in space and time, ratio of organelles undergoing retrograde and anterograde movement along with transport velocity distribution [73]. The transportation of mitochondria and acidic organelles such as lysosomes, endosomes, trans-Golgi apparatus, and certain secretory vesicles in HB9:: GFP ventral spinal cord explant axons revealed information flow, organelle transport, and material transfer along the axons from soma/synapse and vice versa [73]. Lu et al. fabricated a PDMS microfluidic device comprising two large open culture chambers attached through a parallel microchannel array facilitating the transport study of mitochondria along single dopaminergic axons from mid-brain cultures. As displayed in Fig. 6, the fluidic separation of axons and soma permitted the organelle tracking. Mitochondria were tracked under a confocal microscope using either mitochondrial-targeted DsRed2 lentiviral vector or the mitochondria-specific dye Mitotracker Deep Red. The mitochondrial and acidic organelle transport defects majorly contribute to most neurodegenerative diseases, for example, Parkinson's disease. The technique can also help in better understanding of various neuroscience aspects intracellularly, such as vesicular transport and microtubule fragmentation, causing the axon degeneration onset [74].
Further, advancements were made to the microfluidic devices with programmable capability; high-power LED array integration to carbon black (CB)-PDMS compartmentalized device. It optically stimulated subcellular parts of neurons optogenetically and simultaneously in thousands of neurons to overcome the drawbacks of techniques including subcellular channel-rhodopsin-assisted circuit mapping, two-photon activation approach, and laser-pritzer approach based on fiber optic. The LED gives the feasibility of temporal control over neural stimulation. The pharmacological stimulation on the compartmentalized microfluidic chamber can provide extensive spatial control, yet misses temporal control on neural activity with no selective cell stimulation. The pharmacological treatment in the axonal chamber cannot restrict its effect on Oligodendrocyte precursor cells while aimed only to stimulate axons [75].
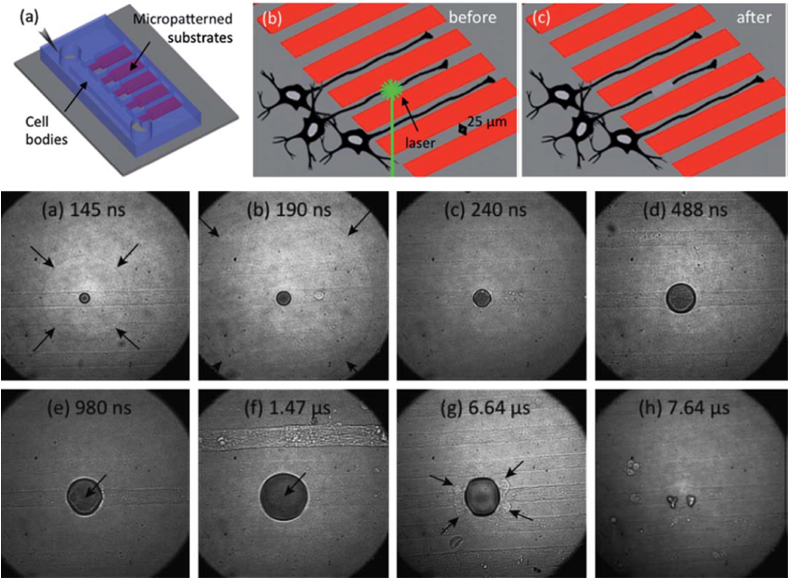
Reproducible and precise laser-based irradiated axonal injury models integrated with microfluidic platforms can be used to examine axon injury and regeneration in-vitro dynamics and manipulating the CNS neurons' spatial organization in-vivo [76,77]. The 180 ps duration of laser pulses created precise regions of axon-injury in-vitro for checking (dieback of proximal neurite segment, and anterograde degeneration involving the distal neuritic segment) the initial dieback response along with CNS neurons’ growth subsequently under incubation of cell culture media with ethylene glycol tetra-acetic acid (EGTA). EGTA chelates extracellular calcium. This microfluidic-based strip assay demonstrated the complementary use of micro-patterned cell culture and laser irradiation, as shown in Fig. 7 [76].
Fig. 7.
Axotomy of 25 mm axon strips with one 180 ps pulse of laser. Axotomy was done only for the lower strip. Images were captured post 1 min of laser pulse exposure: (a) partial and (b) complete transections, via Ep¼400 nJ and 800 nJ, respectively. (d) Time-resolved image series of laser axotomy after one 180 ps of 0.5 mJ laser microbeam. Grey scale images show Shock wave propagation (arrows, a,b), expansion of cavitation bubble (a–f), collapse (g–h), during axonal injury. Axon are getting severed (arrows, e, f) and deformed (arrows, g) [76].
The alternating surface pattern of aggrecan and poly l-lysine was used to control the polarized neurite outgrowth toward particular regions to facilitate anterograde and retrograde directions as the directionality function the growing neurite bundles [77]. The integrated microfluidic devices with laser microbeam are independent of various systemic and homeostatic conditions; it also helps to maintain the extracellular environment.
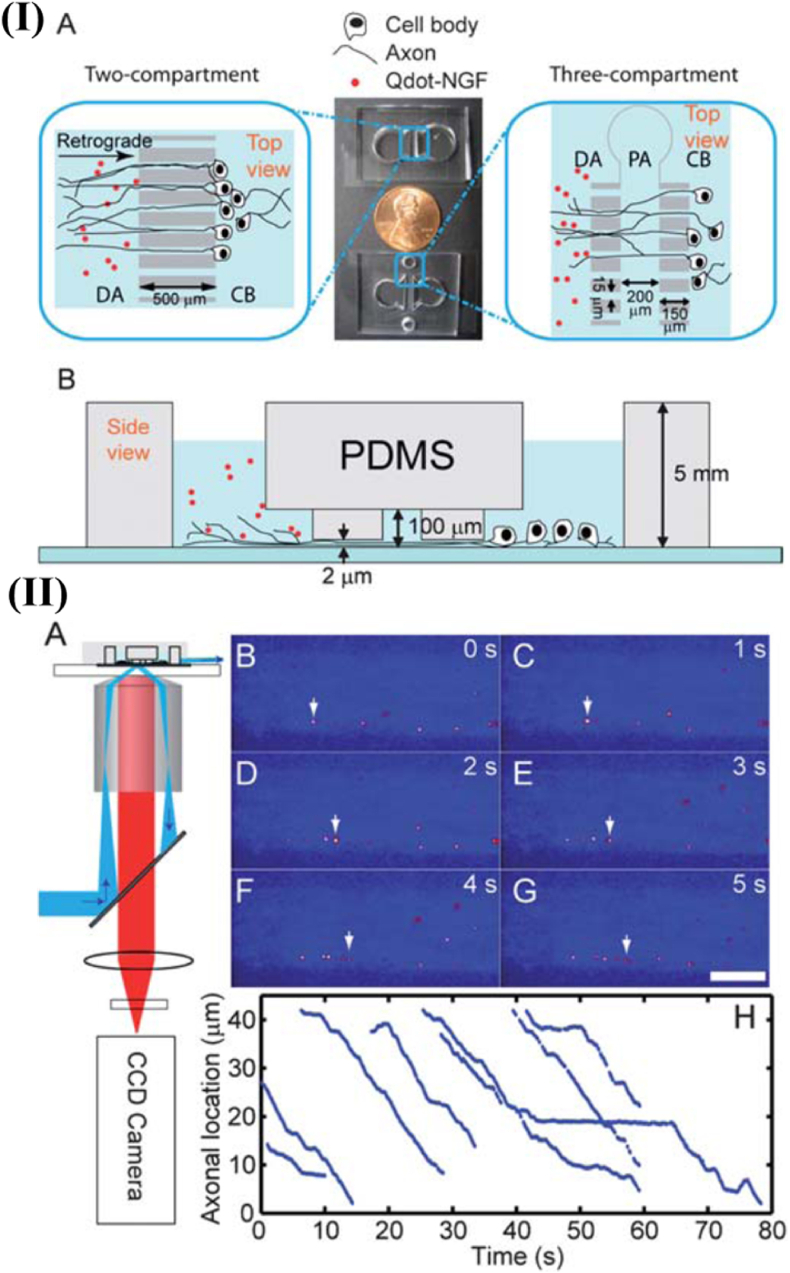
After discovering that most endosomes contain a single NGF molecule, single-molecule imaging is a critical factor in analyzing NGF studies [78]. Though compartmentalized Campenot chambers could be employed to treat axonal termini with NGF in a much-localized manner, it filters out the background fluorescence of molecules that got diffused into the surrounding solutions and axon surface spatially. But this background fluorescence gets multiplied due to vacuum grease. The grease makes the system susceptible to mechanical disturbances during transportation, offering challenges to single-molecule detection [66]. Thus, to cut down the penetration time across the distal axon compartment through the grease barriers in the earlier systems, a three-compartment microfluidic chamber was designed for NGF tracking in dorsal root ganglion (DRG) neuronal culture, as shown in Fig. 8. The retrograde movement of endosomes containing quantum dot labeled NGF was monitored along the axon to soma displaying a characteristic stop-and-go moving pattern. Even for the same axon, the individual endosome transport behavior is dependent on the movement speed and the pause duration [68].
Fig. 8.
Microfluidic devices for Single-molecule imaging of Qdot-NGF axonal transport in DRG neuron culture. (I) (A) Top view of the two- and three-compartment microfluidic devices: cell-body (CB), distal-axon (DA), and proximal-axon (PA) compartment. (B) The side view of the three-compartment device. (II) Single-molecule imaging of Qdot-NGF axonal transport with a pseudo-TIRF microscope. (A) Illustration of a pseudo-TIRF microscope. (B–G) Retrograde transport of Qdot-NGF labeled endosomes is depicted in axons (bright yellow dots, false color). White arrow points the location of the specific Qdot-NGF at different time frames in same field-of-view. Scale bar 10 mm. (H) Illustrative trajectories of 8 endosomes traveling in same axon, showing a typical stop-and-go moving pattern [68].
5. Microfluidics-based single-neuron omics
“Omics” spanning genomics, transcriptomics, proteomics and epigenomics in neuroscience refers to the tools and analysis methods allowing neuroscientists to understand the genes, proteins and molecular level variations in their preferred cell types in nervous system. This section reviews the proteomics, transcriptomics and epigenomics analysis to study the complex process occurring in different neuronal regions using microfluidic devices in the axon compartment. The reason to conduct omics analysis of the axon compartment is due to the various types of in vitro, in vivo, and ex vivo models available for the nervous system. Mature axons consist of complex and continuously changing transcriptomics, elucidating the various features of the axonal function. This requires determination and translation of the mRNAs mediated different functions that have originated obstacles in the field of axonal biology [79,80]. Using microarray studies, the axonal transcriptome assessment spotted important axonal mRNAs and illustrated their dynamic and complex nature [81,82].
Transciptome is major determinant for functioning of any individual cell. Thus, single cell transcriptome sequencing, namely scRNA-seq, analyses gene expression across the transcriptome at the resolution of single cell, it helps to identify biological differences existing in individual cells. Though significant improvements have been done in mRNA-seq analysis of individual cells separated from neurosphere. But it remains still crucial to prepare single neuron for profiling in cell culture platform. Microfluidic provides the fast and scalable approach to characterize complex brain tissues at single neuron level, under diverse conditions and environment. Thus, Gole et al. developed the microwell displacement amplification system (MIDAS) to allow for simultaneous polymerase cloning of individual neurons in thousands of nanoliter reactors. The microfluidic based system was used to detect copy number variations in human adult single neurons without much sequencing experimentation [83]. Further, single cell RNA sequencing of 466 cells at whole transcriptome level deciphered the cellular complexity of human adult and fetal brain. It was found that fetal human cortical neurons show variation in expression of genes in comparison to adult neurons, where the gradient expression of genes establishes a transition from replicating from quiescent fetal neuronal populations. The study revealed that only a class of adult neurons shows major histocompatibility complex type I (MHC-I) gene expression, while it was absent in fetal neurons [84]. Drop-Seq, a sequencing method based on the droplet microfluidics facilitated parallel mRNA expression profiling in thousands of single cells via encapsulation of individual cells inside nanolitre droplets [85]. Macosko et al. compartmentalized mouse retina cells into these microfluidic droplets for RNA sequencing. A molecular barcoding was introduced to overcome the issue of retaining a molecular memory of the cell-of-origin of each isolated mRNA. Then, Drop-Seq screened 44,808 cell profiles along cell cycles to retrieve 39 different populations. The results demonstrated use of microfluidics for large-scale single-cell analysis showing biological information of complex brain tissues and cell populations [86]. Another, droplet based single neuron transcriptome analysis method named scGESTALT, combining the lineage recording capabilities of genome editing of synthetic target arrays for lineage tracing (GESTALT) with cell-type identification by single-cell RNA sequencing, was developed. Sequencing of ∼60,000 transcriptomes from the juvenile zebrafish brain identified >100 cell types and marker genes to reveal lineage relationships, gene expression cascades, and differentiation trajectories during brain development [87,88]. In another attempt, Drop-seq performed profiling of RNA expression across 690,000 single cells collected from 9 regions of adult mouse brain. As an outcome, 565 transcriptionally distinct cell populations were identified. Cross-region analysis has shown characteristics brain organization, consisting of gene-expression module to produce axons and pre-synaptic constituents, voltage-gated ion channel co-deployment schemes, vascular cell functionalities and glutamatergic neuron specialization over cortical regions. Also, rare spiny projection neurons were discovered by systematic neuronal classifications of basal ganglia nuclei and the striatum [89].
Further, the recent breakthroughs in individual-cell proteomics have helped to differentiate various cellular subpopulations using profiling of proteins and its process. However, single neuron proteomic profile remains unknown. This leads to the assessment of mRNA levels to conclude amount of protein from single-cells. Single cell proteome could bring distinctive information regarding brain biological processes. This indeed can help in finding the signaling events in specific brain cell along with spatial identification of the brain regions, where laser-based microdissection was employed to capture a part of brain showing analysis with cell-type distinction. A chemical cytometry-based on microfluidics capillary electrophoresis technology was used for single-cell chemical proteomics (SCCP) to recognize proteins present on the individual primary neuron membrane. A functional single-neuron was isolated from a mouse and injected into a capillary for generating capillary electrophoresis-laser-induced fluorescence to study receptor protein, labeled by activity-based probes [90]. The electropherogram results conclude the heterogeneous expression of GB1 probably owing to the presence of two important G-protein (GB1) variant splices (GB1a and GB1b, in CNS) or precise post-translational alterations. The antagonist for GABAB receptors (G-protein coupled receptors for gamma-aminobutyric acid) CGP64213, decreased the electrophoretic peak, indicating the selective activity-based trimodular probe (ABP) labeling onto GB1 ligand-binding site. Similarly, microprobe capillary electrophoresis mass spectrometry was used for single neuron selection from substantia nigra, where 160 proteins were identified from dopaminergic neuron [91]. Few proteins such as tho complex subunit 2 (Thoc), pro low-density lipoprotein receptor related protein (Lrp1), and zinc finger and scan domain containing protein 2 (Zscan2), were favorably present in dopaminergic neurons.
Though till recent past the genome sequence was considered to be the basis of organism's biology, but recently scientists have discovered another level of control namely the epigenome. The epigenome consists of chemicals that dot the DNA, deciding the level of gene expression as well as its location. This branch of “omics” is still infancy state, thus very few attempts have been made in epigenomics of neuroscience. Arguably, investigation of epigenetics at single neuron level makes more sense as intercellular variation can be studied more prominently in comparison to bulk analysis [92]. Firstly, Ma et al. demonstrated a microfluidics based chromatin immunoprecipitation (ChIP) SurfaceChIP-seq, where antibodies coated on the surface of channels allow multiplexing with ultralow-input profiling of the histone modifications present on NeuN+ (neuron) and NeuN− (glia) fractions in mouse prefrontal cortx (PFC) and cerebellum. The microfluidic based antibody coated surface avoided the use of immunomagnetic beads; therefore, making the device design simple along with several assays in-parallel. Minimum input was of 30 cells per assay with single chip throughput of 8 parallel assays. Around 3 million to 13 million unique reads per data set (with input of 30–1000 cells per assay, respectively) were achieved, with data characteristics superior than conventional analyses [93]. Another semi-automated microfluidic oscillatory washing–based ChIP followed by sequencing (MOWChIP-seq) has shown histone modifications profiling with as low as 100 cells per assay with same throughput of 8 in single run from human and mouse neural tissues. In MOWChIP-seq, the chromatin fragments are passed through a packed bed of antibody-coated beads, later strong microfluidic oscillatory motions perform washing [94]. The MOWChIP-seq technology was also used to profile acetylation of histone H3 at lysine 27 (H3K27ac) along with transcriptome from neuronal nuclei of mouse frontal cortex [95]. The results confirm that the epigenomic variations in synaptic plasticity withstand long-term antidepressant action of psychedelics, it also advise against probable substrate-overlap with genetic risks in psychiatric disorders. Later, in order to address the issue of high amount of DNA for methylomic studies, a microfluidic diffusion-based reduced representative bisulfite sequencing (MID-RRBS) was developed. It allows superior methylomic profiling in the nuclei from mouse-brain cells and sort them into NeuN+ and NeuN− sets, and to maintain methylomes particular to the cell-type even in nanogram-to- single-cell quantities. Many small molecules can be loaded or released in diffusion-based reagent exchange method without compromising on significant amount of DNA. The microfluidic platform with simple design allows simultaneous processing for multiple samples in-parallel. Though this device is unable to perform library preparation in the present form, thus library is prepared outside the microfluidic device [96].
6. Single neuron on-chip drug delivery and analysis
Microfluidic systems have also been useful for patient-specific and physiological in vitro assays, via improving the rate of therapeutic drug approval. Though a complete complexity of in vivo systems is challenging to capture, these devices help summarize and stimulate it in in vitro models. The technologies allow to regulate the quantity of fluid straight into the device to create near microenvironment at molecular and cellular levels [37]. However, multiplex and integrated in vitro microfluidic devices targeting disease therapeutics and neuron modeling need the potential to attain very high spatial resolution and localized access to a targeted cell environment [99].
Focused on-chip electroporation is one such tool for such purposes. Most of the electroporating devices are structured for suspension cultures or large population of cells. While the on-chip electroporation device generates electropores distributed over plasma membrane of an individual cell. Hai and Spira used on-chip electroporator device where micrometer-sized gold mushroom-shaped microelectrode array electroporate adhered selective neurons [33]. This device enables membrane transient in-cell recordings and repair dynamics. Similarly, a wave generator, biochip consists of microelectrode arrays along with a control system allowing the signal transfer to single microelectrode, achieved successful transfection of cos-7 cells and individual-neurons with oligonucleotides [34,35]. A microfluidic chip to integrate two functions – cell electroporation and dielectrophoretic viable cell sorting electro transfected pEGFP plasmid into range of cell types like Neuro-2a (neuroblastoma cell line), HUVEC cells (primary human umbilicalvein endothelial cell), and HSF cells (primary human skin fibroblast cell) at individual cell level [36]. In comparison to unsorted cells, the viable sorted N2a cells showed enhanced survival rate (30–80%) while maintaining the transfection rate of about 65%. Though another microfluidic device has been employed to validate electroporation with transfection efficiency of ∼95% for propidium iodide and up to 50% for plasmids. The device also facilitated on-chip differentiation of neuronal cells, followed by transfecting the post-mitotic neurons with GFP plasmid [100]. In addition to this, various miniaturized devices with integrated microneedle electrodes or even microchannels have also been used for individual-cell electroporation [8,101].
The microfluidic chip consisting of detachable and resealable layers permit multiple operational mode in time of cell culture, fluidic isolation for selected transfection and low-angle electrode recording access [102]. Such a flexible technique can bring usefulness to functional studies related to specific expression, such as optogenetic tools in presynaptic neurons. In a study by Tolomeo et al. microfluidic platform was used in high efficiency delivery of Neurogenin-2 modified m-RNA (mmRNA) for neuronal transcriptional programming in hiPSCs [103]. Also, this microfluidic devices was previously used for high efficient transfection in mmRNA-based reprogramming [[104], [105], [106]].
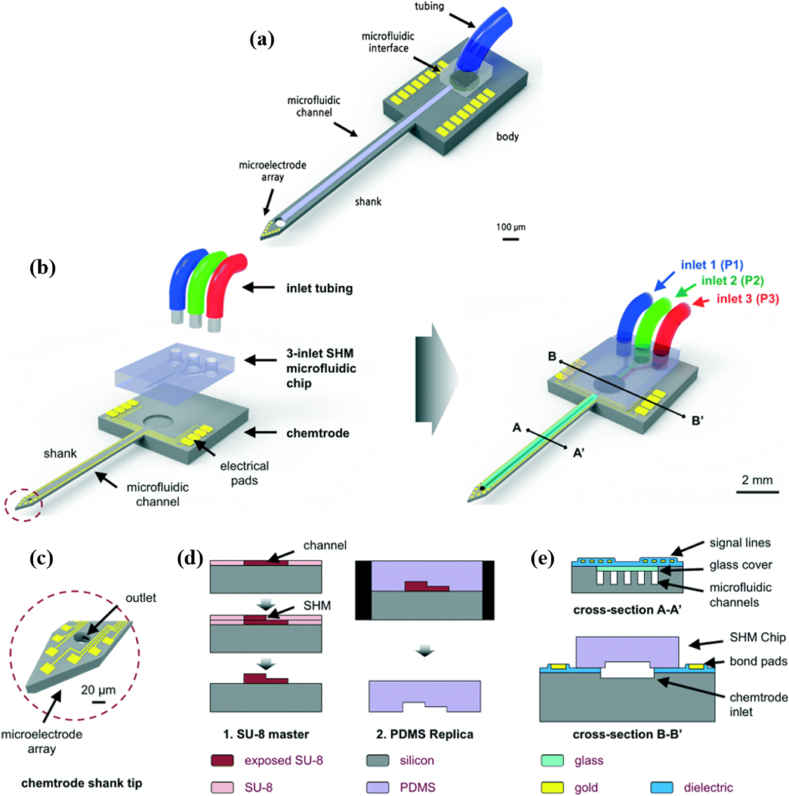
In neuroscience, multi-functional neural probes are emergent tools to facilitate simultaneous observations of a single neural spike after applying different stimulation methods such as optical, chemical, and electrical, etc. [107,108]. Thus, they are broadly used to investigate pathology, physiology, and electrogenic cell connectivity in 2D neuronal cell cultures and brain tissue. Microfluidic channels integrated with neural probes (chemtrodes) when compared to the other techniques (such as systemic administration) for direct brain infusion is the most effective way for the delivery of the drugs while in the presence of a blood-brain barrier (BBB) [[109], [110], [111], [112]]. Targeted drug delivery is enabled using chemtrodes with a precise quantity and concentration of drugs that needed to be applied on specific neurons. During drug delivery, changes in neural signals are monitored in near real-time. The essential benefit of this method is that it stimulates and records excitable cells with no mechanical damage inflicted [113]. Such tools have an important application in identifying different neuro-degenerative disorders, i.e. Parkinson's, Alzheimer's, and multiple sclerosis diseases. Lee et al. used a multi-functional neural probe integrated with MEA and microfluidic channel up to 70 mm length and cross-sectional dimension in the range of 5–30 μm to analyze the neural signals influenced via drug delivery (Fig. 9a) [114]. The group fabricated thin neural probes of 40 μm in thickness with embedding microchannels on a silicon substrate. The implementation of this multi-functional probe created a challenge because of its increased size with added functionalities. The amount of damage incurred during insertion is closely related to the size of the neural probe. Thus, it is beneficial to determine the ability of the neural probe in detection of signals. Further, multiplexing in an implant requires an external manifold that helps to achieve multi-drug delivery. Shin et al. implemented a staggered herringbone mixer (SHM) using a 3-inlet microfluidic chip as a fluidic interface; integrated with silicon chemtrode (Fig. 9b-e) [115]. The functionality of the chemtrode for multi-drug delivery was assessed using dyes in in vivo mice experiments. They showed the successful modulation of neural activities with two different drug concentrations (pilocarpine and tetrodotoxin) and cell nuclei staining near the site of injection.
Fig. 9.
(a) Schematic representation of multi-functional neural probe with drug delivery capability [114]. (b) Conceptual representation of various components (chemtrode, 3-inlet SHM chip, and tubing) of the neural probe system for multi-drug delivery. (c) Close-in schematic diagram of the chemtrode shank tip integrated with microelectrode array and an outlet. (d) PDMS mold and two layers of the SU-8 process were used for the fabrication of the SHM chip. (e) Chemtrode shank cross-section (A-A′) and the probe body (B–B′) show the SHM chip outlet, which is aligned to the inlet [115].
Another major aspect of neuropharmacology involves observation of spike-wave discharge for thorough analysis of the absence seizure (a specific type of epileptic seizure) using electroencephalogram (EEG), which mostly [116] reads average neural signals. Thus alone, EEG is incapable of pinpointing the brain circuits precisely responsible for the onset of absence seizure. Therefore, devices for deep-brain recordings, i.e. neural probes and wire bundles, are utilized to analyze the brain circuits [117]. Individual neurons provide immense information by neural spikes recording using microelectromechanical systems (MEMS) neural probes with MEAs. Moreover, with added functionalities, like MEMS neural probes integrated with drug delivery cues, it can be beneficial to manage the timings of seizure initiation and termination along with parallel recoding of every single neural spike by injecting seizure-inducing drugs for the in-depth analysis [114].
Therefore, microfluidic chips support a unique electronic interfacing to neurons enabling single neuron selection for on-chip electroporation, optogenetics, simultaneous observations of a single neural spike while applying various stimulation methods such as optical, chemical, electrical and observation of spike wave discharge to analyze brain circuits.
7. Microfluidic neural stem cell differentiation
The neural stem cells (NSCs) are multipotent, self-renewing cells competent in getting differentiated into neurons and glial cells. To facilitate differentiation, long-term in vitro culture and proliferation of NSCs is carried out either as a cluster [118] or as an adherent monolayer [119]. Thus, to induce differentiation into cells or to carry out clonal analysis, neurospheres are dissociated either mechanically or chemically or both. Neurospheres are considered to be great tool for assessing proliferation, self-renewal, and multipotency of NSCs. Neither of the dissociating methods gives the entire single-cell population. For single stem cell investigation, microfluidic chips provide more optimum microenvironment for their growth and differentiation.
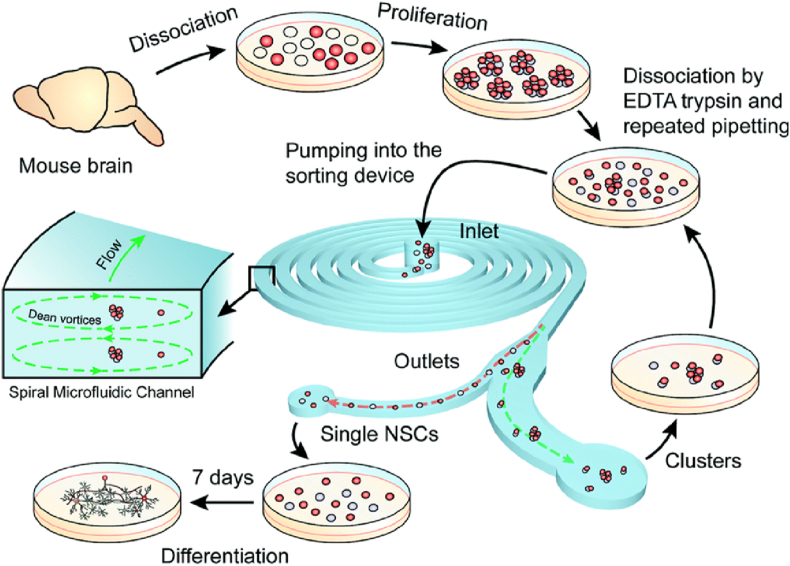
Nathamgari et al. used an inertial microfluidic device (based on variation in size) to isolate single-cells from clustered mixed populations and chemically dissociated neurospheres [24]. The authors presented mainly two engineering challenges while sorting single cells from dissociated populations – first, substantial inconsistency in the size of single cells and clusters. Secondly, primary NSCs are shear stress sensitive. Thus, the force was tuned to Dean's force to apply contradictory effects on clusters and single cells. Due to this effect, the single cells are concentrated near the inner wall, while the clusters are in the middle of the microchannel (Fig. 10). The process has shown to be applicable and helped maintain the stem cells' multipotency by the sorted cell population differentiating into astrocytes and neurons. The present method can be demonstrated equally to other sphere-forming cell lines apart from neurosphere, where a dissociated single-cell population is necessary.
Fig. 10.
Outline of the experimental procedure for inertial microfluidic device to isolate single-cells from clustered mixed populations and chemically dissociated neurospheres. The sorted single NSCs are collected at the outlet and cultured in a low serum containing medium to facilitate differentiation [24].
The process helped in maintaining the multipotency of stem cells by the differentiating them into two nervous system lineages i.e. astrocytes and single neurons. Lin et al. developed a high-throughput single-neuron capture micro-fabricated dual-well (DW) device [120]. To increase the single cell loading efficiency, small microwells were used to trap single cell and then with the help of gravity, the cells were transferred into larger microwells for cells to grow and proliferate. The authors have used this device to demonstrate single cell proliferation, and differentiation using adult mouse brain neural stem/progenitor KT98 cells and other cancer cells. The KT98 cells were differentiated in DW device by replacing the culture medium with differentiation medium. The change in the medium resulted in division of one KT98 cell into six cells which exhibited neurite morphology specific to neuronal cells.
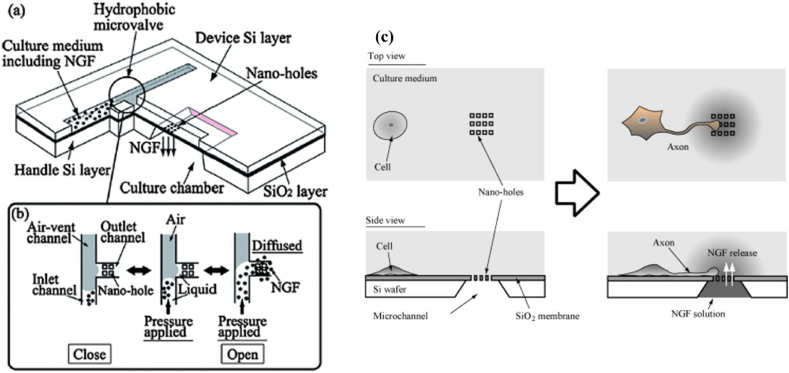
Many studies have recently proved that the stem cell differentiation fate is determined mainly by signaling molecules, growth factors, cell-to- ECM, cell-to-cell interactions, and chemical indications [[121], [122], [123], [124]]. Nakashima and Yasuda presented a microfluidic device for neural stem cell differentiation, where it consists a cell culture chamber (200 μm × 200 μm), microvalve, microchannel, and nano-hole array (500 nm in diameter) (Fig. 11) [125]. The nanohole array governed the microvalve switching and also helped to accurately control the release quantity and frequency of nerve growth factor (NGF), which successfully lead the PC12 (adrenal pheochromocytoma) cell differentiation and axon elongation. The microfluidic device construction allowed the study of differentiation and axon elongation at individual cell level overcoming the issues of bulk culture. Similarly, on-chip localized electroporation device (LEPD) demonstrated differentiation along with transfection on the neurons derived from neurospheres [100]. These single neural stem cells were plated on poly-d-Lysine coated LEPDs to differentiate into neurons using serum free medium and human recombinant epidermal growth factor (FGF). Over time, neuronal junctions were observed between neighboring cells. The neural stem cell differentiation was further confirmed using a marker for neurons named β-tubulin III. Lee et al. has explained the effect of shear-sheltering dual-micropillar arrays and hydrodynamic resistance on uniform docking of single embryonic stem cells (ES) [126]. It was observed that the applied flow rate was proportional to number of single ES cells trapped and docked within inner micropillars. While the outer micropillars were used to generate uniform shear stress and velocity. These single ES cells cultured for six days in the device and employed with N2 and ITS supplements. These individual ES cells differentiated into Tuj1-positive neurons, showing about 72% differentiated into neuronal cells. The morphology of the single ES cell-induced neuron on microfluidic platform was round. Therefore, single ES cell-derived neuronal differentiation can be easily regulated using potentially powerful tool such as microfluidic platform.
Fig. 11.
Schematics of a microfluidic device for the guidance of cell differentiation. (a) It consists of a microchannel for transport of chemicals, a nano-hole array for chemical release, a cell culture chamber, and a hydrophobic passive microvalve for regulating the chemical release. (b) The schematics of liquid switching principle for the microvalve. (c) Schematics of cell stimulation on the fabricated microfluidic device [125].
Generally, individual stem cells are differentiated on microfluidic devices with the motivation of further assessment of neuron dynamics. As Elie et al. investigated the formation of the synapse by differentiating the neurons from rat blastocyst-derived murine embryonic stem cells (ES-J1) and pheochromocytoma cells (PC12) on interdigitated microelectrode arrays [127]. In this in vitro study, ES or PC12 cells were differentiated by incubating them with forskolin and nerve growth factor (NGF) or treatment with basic fibroblast growth factor (FGF-2) and by deprivation of serum, respectively. After seven days, the neuronal cells enlarged very long and ended in pili-like contact formations on top of the ITO electrodes, representing growth cone structures. Neuron–microelectrode interfaces are advantageous for the non-invasive analysis in response to signal transmission and neuroactive drugs. Moreover, microfluidics integrated with microelectrodes is a systematic approach for studying neuron bio-sensors, neuroelectrophysiology and neuropharmacology etc.
The stem cell differentiation process is often be characterized and quantified using its intrinsic cellular electrical markers such as, cytoplasmic conductivity (σcytoplasm) and specific membrane capacitance (Cspecific membrane). However, there has been absence of such specific tools for large number of differentiated cells or stem cells. Thus Zhao et al. developed such a microfluidic platform where population of single neural stem cells go through differentiation can be enabled for high throughput quantification of σcytoplasm and Cspecific membrane without biochemical staining [128]. The initiation of neural stem cells differentiation process was experimentally quantified using biochemical markers (such as, Tubulin, GFAP, Nestin) showing loss of stemness and increase in cellular markers of neurons and glial cells. Before the differentiation process was initiated, individual neural stem cells showed large distribution difference in σcytoplasm. While, during differentiation process, a large distribution difference was observed in Cspecific membrane. Thus, different expressions of membrane proteins indicated the heterogeneity in the differentiation process.
8. Conclusions
With the advancements in neuroscience, single neuron analysis is proven to be the gold standard for understanding the basic system of neurophysiology. Tremendous breakthroughs have been achieved from these micro scale neuron cultures; ranging from deciphering neuronal polarity to investigating axon guidance and precise role of dendritic processes. The, microfluidic devices are benefiting the neuroscience at various perspectives. Considering this further advancements and sophistication are being introduced to the microfluidic devices, aiming to solve unsolved and complex queries of the neuroscience. The microfluidic devices offer unprecedented level of ease and simplicity for controlling the microenvironment chemically, physically, and fluidically for a developing neuron and also assessing the alteration imposed on cell morphology and functions. However, the efforts are the pioneer to address single neuron analysis dealing with real-life issues, i.e., neurologic and psychiatric diseases. To overcome averaging the information and take account of single neurons, it is worth focusing on microfluidic systems for pathological conditions such as traumatic brain injury, stroke, dementia, neuromuscular disease, depression, personality disorders like bipolar, schizophrenia, obsessive-compulsive disorder, or even multiple critical health conditions. Thus there lies a huge opportunity for the amalgamation of basic science and clinical interventions to overcome the most prevalent and tedious neurological issues influencing the brain and also to construct the next-gen neurotechnologies for maintaining and restoring neurologic functions. Neuroengineering and translational neuroscience are on the onset of integrating this vital tool, one-neuron-at-a-time, for the people with neurologic or psychiatric diseases.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by DBT/Wellcome Trust India Alliance Fellowship grant number IA/E/16/1/503062 awarded to Dr. Tuhin Subhra Santra.
References
- 1.Ledford H. The power of a single neuron. Nature. 2007:1–2. doi: 10.1038/news.2007.392. [DOI] [Google Scholar]
- 2.Shinde A., Illath K., Gupta P., Shinde P., Lim K.T., Nagai M., Santra T.S. vol. 10. Cells; 2021. (A Review of Single-Cell Adhesion Force Kinetics and Applications). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinde P., Mohan L., Kumar A., Dey K., Maddi A., Patananan A.N., Tseng F.-G., Chang H.-Y., Nagai M., Santra T.S. Current trends of microfluidic single-cell technologies. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tseng F.-G., Santra T.S. 2016. Essentials of Single-Cell Analysis: Concepts, Applications and Future Prospects. [DOI] [Google Scholar]
- 5.Tuhin Subhra Santra. Tseng Fan-Gang. Springer Singapore; 2020. Handbook of Single Cell Technologies. [DOI] [Google Scholar]
- 6.Tseng F., Santra T.S. MDPI AG Basel; Switzerland: 2016. Single Cell Analysis in Biotechnology and Systems Biology. [Google Scholar]
- 7.Smith I.T. The influence of a single neuron on its network. Nature. 2019;567:320–321. doi: 10.1038/d41586-019-00687-9. [DOI] [PubMed] [Google Scholar]
- 8.Gupta P., Balasubramaniam N., Chang H.Y., Tseng F.G., Santra T.S. A single-neuron: current trends and future prospects. Cells. 2020;9:1528. doi: 10.3390/cells9061528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross A., Schoendube J., Zimmermann S., Steeb M., Zengerle R., Koltay P. Technologies for single-cell isolation. Int. J. Mol. Sci. 2015 doi: 10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santra T.S., Tseng F.G. MDPI AG; 2014. Micro/nanofluidic Devices for Single Cell Analysis. [DOI] [Google Scholar]
- 11.Pal A., Glaß H., Naumann M., Kreiter N., Japtok J., Sczech R., Hermann A. High content organelle trafficking enables disease state profiling as powerful tool for disease modelling. Sci. Data. 2018;5:1–15. doi: 10.1038/sdata.2018.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H.J., Park J.W., Byun J.H., Poon W.W., Cotman C.W., Fowlkes C.C., Jeon N.L. Quantitative analysis of axonal transport by using compartmentalized and surface micropatterned culture of neurons. ACS Chem. Neurosci. 2012;3:433–438. doi: 10.1021/cn3000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perrin S. Make mouse studies work. Nature. 2014;507:423–425. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
- 14.Moonen E., Luttge R., Frimat J.P. Single cell trapping by capillary pumping using NOA81 replica moulded stencils. Microelectron. Eng. 2018;197:1–7. doi: 10.1016/j.mee.2018.04.010. [DOI] [Google Scholar]
- 15.Smith I.T. The ripple effect of a single neuron. Nature. 2019;567:320–321. doi: 10.1038/d41586-019-00687-9. http://www.nature.com/articles/d41586-019-00687-9 [DOI] [PubMed] [Google Scholar]
- 16.Cash S.S., Hochberg L.R. The emergence of single neurons in clinical neurology. Neuron. 2015;86:79–91. doi: 10.1016/j.neuron.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Economo M.N., Winnubst J., Bas E., Ferreira T.A., Chandrashekar J. Single-neuron axonal reconstruction: the search for a wiring diagram of the brain. J. Comp. Neurol. 2019;527:2190–2199. doi: 10.1002/cne.24674. [DOI] [PubMed] [Google Scholar]
- 18.Roet M., Hescham S.-A., Jahanshahi A., Rutten B.P.F., Anikeeva P.O., Temel Y. Progress in neuromodulation of the brain: a role for magnetic nanoparticles? Prog. Neurobiol. 2019;177:1–14. doi: 10.1016/j.pneurobio.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Emmenegger V., Obien M.E.J., Franke F., Hierlemann A. Technologies to study action potential propagation with a focus on HD-MEAs. Front. Cell. Neurosci. 2019;13 doi: 10.3389/fncel.2019.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S., Loke G., Fink Y., Anikeeva P. Flexible fiber-based optoelectronics for neural interfaces. Chem. Soc. Rev. 2019;48:1826–1852. doi: 10.1039/C8CS00710A. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Ren L., Li L., Liu W., Zhou J., Yu W., Tong D., Chen S. Microfluidics: a new cosset for neurobiology. Lab Chip. 2009;9:644–652. doi: 10.1039/b813495b. [DOI] [PubMed] [Google Scholar]
- 22.Lin C.H., Chang H.C., Hsu C.H. Enzyme-free dissociation of neurospheres by a microfluidic chip-based method. Methods Mol. Biol. 2016;1516:289–297. doi: 10.1007/7651_2016_348. [DOI] [PubMed] [Google Scholar]
- 23.Lin C.H., Lee D.C., Chang H.C., Chiu I.M., Hsu C.H. Single-cell enzyme-free dissociation of neurospheres using a microfluidic chip. Anal. Chem. 2013;85:11920–11928. doi: 10.1021/ac402724b. [DOI] [PubMed] [Google Scholar]
- 24.Nathamgari S.S.P., Dong B., Zhou F., Kang W., Giraldo-Vela J.P., McGuire T., McNaughton R.L., Sun C., Kessler J.A., Espinosa H.D. Isolating single cells in a neurosphere assay using inertial microfluidics. Lab Chip. 2015;15:4591–4597. doi: 10.1039/c5lc00805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W.H., Cheng W., Zhang Z., Pang D.W., Wang Z.L., Cheng J.K., Cui D.F. Transport, location, and quantal release monitoring of single cells on a microfluidic device. Anal. Chem. 2004;76:483–488. doi: 10.1021/ac035026y. [DOI] [PubMed] [Google Scholar]
- 26.Spégel C., Heiskanen A., Acklid J., Wolff A., Taboryski R., Emnéus J., Ruzgas T. On-chip determination of dopamine exocytosis using mercaptopropionic acid modified microelectrodes. Electroanalysis. 2007;19:263–271. doi: 10.1002/ELAN.200603720. [DOI] [Google Scholar]
- 27.Spégel C., Heiskanen A., Pedersen S., Emnéus J., Ruzgas T., Taboryski R. Fully automated microchip system for the detection of quantal exocytosis from single and small ensembles of cells. Lab Chip. 2008;8:323–329. doi: 10.1039/B715107A. [DOI] [PubMed] [Google Scholar]
- 28.Park J.W., Vahidi B., Kim H.J., Rhee S.W., Jeon N.L. Quantitative analysis of CNS axon regeneration using a microfluidic neuron culture device. Biochip J. 2008;2:44–51. [Google Scholar]
- 29.Sci J.V., Schurink B. vol. 1. 2017. (Passive Pumping for the Parallel Trapping of Single Neurons onto a Microsieve Electrode Array). [DOI] [Google Scholar]
- 30.Demircan Yalcin Y., Luttge R. 3D-electrode integrated microsieve structure as a rapid and cost-effective single neuron detector. J. Vac. Sci. Technol. B. 2020;38 doi: 10.1116/6.0000518. [DOI] [Google Scholar]
- 31.Kim H., Lee I.K., Taylor K., Richters K., Baek D.H., Ryu J.H., Cho S.J., Jung Y.H., Park D.W., Novello J., Bong J., Suminski A.J., Dingle A.M., Blick R.H., Williams J.C., Dent E.W., Ma Z. Single-neuronal cell culture and monitoring platform using a fully transparent microfluidic DEP device. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-31576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Hu H., Zhao S., Liu Y.M. Microfluidic platform with in-chip electrophoresis coupled to mass spectrometry for monitoring neurochemical release from nerve cells. Anal. Chem. 2016;88:5338–5344. doi: 10.1021/acs.analchem.6b00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hai A., Spira M.E. On-chip electroporation , membrane repair dynamics and transient in-cell recordings by arrays of gold mushroom-shaped microelectrodes. Lab Chip. 2012;12:2865–2873. doi: 10.1039/C2LC40091J. [DOI] [PubMed] [Google Scholar]
- 34.Vassanelli S., Cellere G., Srl N. Biochip electroporator and its use in multi-site, single-cell electroporation. U.S. Pat. 2011;8:367. [Google Scholar]
- 35.Packer A.M., Roska B., Häuser M. Targeting neurons and photons for optogenetics. Nat. Neurosci. 2013;16:805–815. doi: 10.1038/nn.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Z., Li X., Zhao D., Yan H., Hu Z., Liang Z., Li Z. Flow-through cell electroporation microchip integrating dielectrophoretic viable cell sorting. Anal. Chem. 2014;86:10215–10222. doi: 10.1021/AC502294E/SUPPL_FILE/AC502294E_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- 37.Liu W.W., Goodhouse J., Jeon N.L., Enquist L.W. A microfluidic chamber for analysis of neuron-to-cell spread and axonal transport of an alpha-herpesvirus. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pirlo R.K., Sweeney A.J., Ringeisen B.R., Kindy M., Gao B.Z. Biochip/laser cell deposition system to assess polarized axonal growth from single neurons and neuron/glia pairs in microchannels with novel asymmetrical geometries. Biomicrofluidics. 2011;5 doi: 10.1063/1.3552998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malishev E., Pimashkin A., Gladkov A., Pigareva Y., Bukatin A., Kazantsev V., Mukhina I., Dubina M. Microfluidic device for unidirectional axon growth. J. Phys. Conf. Ser. 2015:12025. doi: 10.1088/1742-6596/643/1/012025. [DOI] [Google Scholar]
- 40.Huang T., Pirlo B.Z.G. Russell K., Wan Qin, Lin Yongliang, Wei Lina, Schmidt Lucas, Erdman Nick, Xi Tingfei, DeSilva Mauris N. Development of a Compartmentalized Biochip for Axonal Isolation and Neuronal-Circuit Formation at the Single-Cell Level. Neuromethods; 2015. pp. 83–104. [DOI] [Google Scholar]
- 41.Di Carlo D., Lee L.P. Dynamic single-cell analysis for quantitative biology. Anal. Chem. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Chen Z.Z., Li Q.L. Microfluidic techniques for dynamic single-cell analysis. Microchim. Acta. 2010;168:177–195. doi: 10.1007/s00604-010-0296-2. [DOI] [Google Scholar]
- 43.Pearce T.M., Wilson J.A., Oakes S.G., Chiu S.Y., Williams J.C. Integrated microelectrode array and microfluidics for temperature clamp of sensory neurons in culture. Lab Chip. 2005;5:97–101. doi: 10.1039/B407871C. [DOI] [PubMed] [Google Scholar]
- 44.Dworak B.J., Wheeler B.C. Novel MEA platform with PDMS microtunnels enables the detection of action potential propagation from isolated axons in culture. Lab Chip. 2009;9:404–410. doi: 10.1039/B806689B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howell B., Huynh B., Grill W.M., Eladly A., Del Valle J., Pan L., Alagapan S., Franca E., Brewer G.J., Wheeler B.C. Propagation of action potential activity in a predefined microtunnel neural network. J. Neural. Eng. 2011;8 doi: 10.1088/1741-2560/8/4/046031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jang J.M., Lee J., Kim H., Jeon N.L., Jung W. One-photon and two-photon stimulation of neurons in a microfluidic culture system. Lab Chip. 2016;16:1684–1690. doi: 10.1039/C6LC00065G. [DOI] [PubMed] [Google Scholar]
- 47.Levi T., Fujii T. Microfluidic neurons, a new way in neuromorphic engineering? Micromachines. 2016;7:146. doi: 10.3390/MI7080146. 7 (2016) 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moutaux E., Charlot B., Genoux A., Saudou F., Cazorla M. An integrated microfluidic/microelectrode array for the study of activity-dependent intracellular dynamics in neuronal networks. Lab Chip. 2018;18:3425–3435. doi: 10.1039/C8LC00694F. [DOI] [PubMed] [Google Scholar]
- 49.Lopes C.D.F., Mateus J.C., Aguiar P. Interfacing microfluidics with microelectrode arrays for studying neuronal communication and axonal signal propagation. JoVE (Journal Vis. Exp. 2018;2018 doi: 10.3791/58878. [DOI] [PubMed] [Google Scholar]
- 50.Heiney K., Mateus J.C., Lopes C.D.F., Neto E., Lamghari M., Aguiar P. μSpikeHunter: an advanced computational tool for the analysis of neuronal communication and action potential propagation in microfluidic platforms. Sci. Rep. 2019;91:1–14. doi: 10.1038/s41598-019-42148-3. 9 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gencoglu A., Minerick A.R. Electrochemical detection techniques in micro- and nanofluidic devices. Microfluid. Nanofluidics. 2014;17:781–807. doi: 10.1007/s10404-014-1385-z. [DOI] [Google Scholar]
- 52.Dittami G.M., Rabbitt R.D. Electrically evoking and electrochemically resolving quantal release on a microchip. Lab Chip. 2010;10:30–35. doi: 10.1039/B911763F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganguly R., Lee B., Kang S., Kim Y.S., Jeong S.-G., Kim J.S., Park S.Y., Yohei Y., Lee C.-S. Microfluidic single-cell trapping and cultivation for the analysis of host-viral interactions. Biotechnol. Bioproc. Eng. 2021;26:179–187. doi: 10.1007/s12257-020-0143-1. [DOI] [Google Scholar]
- 54.Li L., Fan Y., Li Q., Sheng R., Si H., Fang J., Tong L., Tang B. Simultaneous single-cell analysis of Na+, K+, Ca2+, and Mg2+ in neuron-like PC-12 cells in a microfluidic system. Anal. Chem. 2017;89:4559–4565. doi: 10.1021/acs.analchem.6b05045. [DOI] [PubMed] [Google Scholar]
- 55.Sibbitts J., Culbertson C.T. Measuring stimulation and inhibition of intracellular nitric oxide production in SIM-A9 microglia using microfluidic single-cell analysis. Anal. Methods. 2020;12:4665–4673. doi: 10.1039/D0AY01578D. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki I., Sugio Y., Moriguchi H., Jimbo Y., Yasuda K. Modification of a neuronal network direction using stepwise photo-thermal etching of an agarose architecture. J. Nanobiotechnol. 2004;2:1–8. doi: 10.1186/1477-3155-2-7/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park J.W., Vahidi B., Taylor A.M., Rhee S.W., Jeon N.L. Microfluidic culture platform for neuroscience research. Nat. Protoc. 2006 doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 58.Cohen M.S., Orth C.B., Kim H.J., Jeon N.L., Jaffrey S.R. Neurotrophin-mediated dendrite-to-nucleus signaling revealed by microfluidic compartmentalization of dendrites. Proc. Natl. Acad. Sci. Unit. States Am. 2011 doi: 10.1073/pnas.1012401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J., Koito H., Li J., Han A. Multi-compartment neuron-glia co-culture platform for localized CNS axon-glia interaction study. Lab Chip. 2012;12:3296–3304. doi: 10.1039/c2lc40303j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang I.H., Gary D., Malone M., Dria S., Houdayer T., Belegu V., McDonald J.W., Thakor N. Axon myelination and electrical stimulation in a microfluidic, compartmentalized cell culture platform. NeuroMolecular Med. 2012;14:112–118. doi: 10.1007/S12017-012-8170-5/FIGURES/3. [DOI] [PubMed] [Google Scholar]
- 61.Taylor A.M., Dieterich D.C., Ito H.T., Kim S.A., Schuman E.M. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 2010;66:57–68. doi: 10.1016/J.NEURON.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coquinco A., Kojic L., Wen W., Wang Y.T., Jeon N.L., Milnerwood A.J., Cynader M. A microfluidic based in vitro model of synaptic competition. Mol. Cell. Neurosci. 2014;60:43–52. doi: 10.1016/j.mcn.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Shi M., Majumdar D., Gao Y., Brewer B.M., Goodwin C.R., McLean J.A., Li D., Webb D.J. Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab Chip. 2013;13:3008–3021. doi: 10.1039/c3lc50249j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao Y., Broussard J., Haque A., Revzin A., Lin T. Functional imaging of neuron–Astrocyte interactions in a compartmentalized microfluidic device. Microsystems Nanoeng. 2016;2:1–9. doi: 10.1038/micronano.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Y., Majumdar D., Jovanovic B., Shaifer C., Lin P.C., Zijlstra A., Webb D.J., Li D. A versatile valve-enabled microfluidic cell co-culture platform and demonstration of its applications to neurobiology and cancer biology. Biomed. Microdevices. 2011;13:539–548. doi: 10.1007/S10544-011-9523-9/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brunello C.A., Jokinen V., Sakha P., Terazono H., Nomura F., Kaneko T., Lauri S.E., Franssila S., Rivera C., Yasuda K., Huttunen H.J. Microtechnologies to fuel neurobiological research with nanometer precision. J. Nanobiotechnol. 2013;11:1–8. doi: 10.1186/1477-3155-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lenoir S., Genoux A., Agasse F., Saudou F., Humbert S. Recreating mouse cortico-hippocampal neuronal circuit in microfluidic devices to study BDNF axonal transport upon glucocorticoid treatment. STAR Protoc. 2021;2:100382. doi: 10.1016/J.XPRO.2021.100382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang K., Osakada Y., Vrljic M., Chen L., Mudrakola H.V., Cui B. Single-molecule imaging of NGF axonal transport in microfluidic devices. Lab Chip. 2010;10:2566–2573. doi: 10.1039/c003385e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.David S., Aguayo A.J. Axonal elongation into peripheral nervous system “bridges” after central nervous system injury in adult rats. Science. 1981 doi: 10.1126/science.6171034. 80. [DOI] [PubMed] [Google Scholar]
- 70.Park J., Kim S., Park S.I., Choe Y., Li J., Han A. A microchip for quantitative analysis of CNS axon growth under localized biomolecular treatments. J. Neurosci. Methods. 2014;221:166–174. doi: 10.1016/j.jneumeth.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewandowska M.K., Bakkum D.J., Rompani S.B., Hierlemann A. Recording large extracellular spikes in microchannels along many axonal sites from individual neurons. PLoS One. 2015;10:1–24. doi: 10.1371/journal.pone.0118514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peyrin J.M., Deleglise B., Saias L., Vignes M., Gougis P., Magnifico S., Betuing S., Pietri M., Caboche J., Vanhoutte P., Viovy J.L., Brugg B. Axon diodes for the reconstruction of oriented neuronal networks in microfluidic chambers. Lab Chip. 2011;11:3663–3673. doi: 10.1039/c1lc20014c. [DOI] [PubMed] [Google Scholar]
- 73.Altman T., Maimon R., Ionescu A., Pery T.G., Perlson E. Axonal transport of organelles in motor neuron cultures using microfluidic chambers system. JoVE. 2020;2020 doi: 10.3791/60993. [DOI] [PubMed] [Google Scholar]
- 74.Lu X., Kim-Han J.S., O'Malley K.L., Sakiyama-Elbert S.E. A microdevice platform for visualizing mitochondrial transport in aligned dopaminergic axons. J. Neurosci. Methods. 2012;209:35–39. doi: 10.1016/j.jneumeth.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee H.U., Nag S., Blasiak A., Jin Y., Thakor N., Yang I.H. Subcellular optogenetic stimulation for activity-dependent myelination of axons in a novel microfluidic compartmentalized platform. ACS Chem. Neurosci. 2016;7:1317–1324. doi: 10.1021/acschemneuro.6b00157. [DOI] [PubMed] [Google Scholar]
- 76.Hellman A.N., Vahidi B., Kim H.J., Mismar W., Steward O., Jeon N.L., Venugopalan V. Examination of axonal injury and regeneration in micropatterned neuronal culture using pulsed laser microbeam dissection. Lab Chip. 2010;10:2083–2092. doi: 10.1039/b927153h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim H.J., Park J.W., Park J.W., Byun J.H., Vahidi B., Rhee S.W., Jeon N.L. Integrated microfluidics platforms for investigating injury and regeneration of CNS axons. Ann. Biomed. Eng. 2012;40:1268–1276. doi: 10.1007/s10439-012-0515-6. [DOI] [PubMed] [Google Scholar]
- 78.Cui B., Wu C., Chen L., Ramirez A., Bearer E.L., Li W.P., Mobley W.C., Chu S. One at a time, live tracking of NGF axonal transport using quantum dots. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deglincerti A., Jaffrey S.R. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:120079. doi: 10.1098/rsob.120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jung H., Yoon B.C., Holt C.E. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gumy L.F., Yeo G.S., Tung Y.C.L., Zivraj K.H., Willis D., Coppola G., Lam B.Y., Twiss J.L., Holt C.E., Fawcett J.W. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. Rna. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zivraj K.H., Chun Y., Tung L., Piper M., Gumy L., Fawcett J.W., Yeo G.S.H., Holt C.E. Cellular/molecular subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gole J., Gore A., Richards A., Chiu Y.-J., Fung H.-L., Bushman D., Chiang H.-I., Chun J., Lo Y.-H., Zhang K. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat. Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thorsen T., Roberts R.W., Arnold F.H., Quake S.R. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 86.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., Trombetta J.J., Weitz D.A., Sanes J.R., Shalek A.K., Regev A., McCarroll S.A. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raj B., Gagnon J.A., Schier A.F. Large-scale reconstruction of cell lineages using single-cell readout of transcriptomes and CRISPR–Cas9 barcodes by scGESTALT. Nat. Protoc. 2018;13:2685–2713. doi: 10.1038/s41596-018-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raj B., Wagner D.E., McKenna A., Pandey S., Klein A.M., Shendure J., Gagnon J.A., Schier A.F. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 2018;36:442–450. doi: 10.1038/nbt.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saunders A., Macosko E.Z., Wysoker A., Goldman M., Krienen F.M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S., Goeva A., Nemesh J., Kamitaki N., Brumbaugh S., Kulp D., McCarroll S.A. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174:1015–1030. doi: 10.1016/j.cell.2018.07.028. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu F., Zhao H., Feng X., Chen L., Chen D., Zhang Y., Nan F., Liu J., Liu B.F. Single-cell chemical proteomics with an activity-based probe: identification of low-copy membrane proteins on primary neurons. Angew. Chem. Int. Ed. 2014;53:6730–6733. doi: 10.1002/ANIE.201402363. [DOI] [PubMed] [Google Scholar]
- 91.Lombard-Banek C., Choi S.B., Nemes P. Single-cell proteomics in complex tissues using microprobe capillary electrophoresis mass spectrometry. Methods Enzymol. 2019;628:263–292. doi: 10.1016/BS.MIE.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clark S.J., Lee H.J., Smallwood S.A., Kelsey G., Reik W. Single-cell epigenomics : powerful new methods for understanding gene regulation and cell identity. Genome Biol. 2016:1–10. doi: 10.1186/s13059-016-0944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma S., Hsieh Y.P., Ma J., Lu C. Low-input and multiplexed microfluidic assay reveals epigenomic variation across cerebellum and prefrontal cortex. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aar8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu B., Hsieh Y.P., Murphy T.W., Zhang Q., Naler L.B., Lu C. 2019. MOWChIP-Seq for Low-Input and Multiplexed Profiling of Genome-wide Histone Modifications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de la Fuente Revenga M., Zhu B., Guevara C.A., Naler L.B., Saunders J.M., Zhou Z., Toneatti R., Sierra S., Wolstenholme J.T., Beardsley P.M., Huntley G.W., Lu C., González-Maeso J. Prolonged epigenomic and synaptic plasticity alterations following single exposure to a psychedelic in mice. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma S., De La Fuente Revenga M., Sun Z., Sun C., Murphy T.W., Xie H., González-Maeso J., Lu C. Cell-type-specific brain methylomes profiled via ultralow-input microfluidics. Nat. Biomed. Eng. 2018;2:183–194. doi: 10.1038/s41551-018-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pinto M., Howell R.W. Concomitant quantification of targeted drug delivery and biological response in individual cells. Biotechniques. 2007;43:64–71. doi: 10.2144/000112492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kang W., Giraldo-Vela J.P., Nathamgari S.S.P., McGuire T., McNaughton R.L., Kessler J.A., Espinosa H.D. Microfluidic device for stem cell differentiation and localized electroporation of postmitotic neurons. Lab Chip. 2014;14:4486–4495. doi: 10.1039/C4LC00721B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang L., Wang Y.C., Ershad F., Yang R., Yu C., Fan Y. Wearable devices for single-cell sensing and transfection. Trends Biotechnol. 2019;37:1175–1188. doi: 10.1016/J.TIBTECH.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sakha P., Brunello C., Heikkinen J., Jokinen V., Huttunen H.J. Asymmetric genetic manipulation and patch clamp recording of neurons in a microfluidic chip. Microfluid. Compart. Platforms Neurobiol. Res. 2015:59–81. doi: 10.1007/978-1-4939-2510-0_4. [DOI] [Google Scholar]
- 103.Tolomeo A.M., Laterza C., Grespan E., Michielin F., Canals I., Kokaia Z., Muraca M., Gagliano O., Elvassore N. NGN2 mmRNA-based transcriptional programming in microfluidic guides hiPSCs toward neural fate with multiple identities. Front. Cell. Neurosci. 2021;15:13. doi: 10.3389/FNCEL.2021.602888/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Giulitti S., Magrofuoco E., Prevedello L., Elvassore N. Optimal periodic perfusion strategy for robust long-term microfluidic cell culture. Lab Chip. 2013;13:4430–4441. doi: 10.1039/C3LC50643F. [DOI] [PubMed] [Google Scholar]
- 105.Gagliano O., Luni C., Qin W., Bertin E., Torchio E., Galvanin S., Urciuolo A., Elvassore N. Microfluidic reprogramming to pluripotency of human somatic cells. Nat. Protoc. 2019;14:722–737. doi: 10.1038/s41596-018-0108-4. [DOI] [PubMed] [Google Scholar]
- 106.Luni C., Giulitti S., Serena E., Ferrari L., Zambon A., Gagliano O., Giobbe G.G., Michielin F., Knöbel S., Bosio A., Elvassore N. High-efficiency cellular reprogramming with microfluidics. Nat. Methods. 2016;13:446–452. doi: 10.1038/nmeth.3832. [DOI] [PubMed] [Google Scholar]
- 107.Kim T.T.-I.T., McCall J.G., Jung Y.H., Huang X., Siuda E.R., Li Y., Song J., Song Y.M., Pao H.A., Kim R.-H.R.-H., Lu C., Lee S.D., Song I.-S., Shin G., Al-Hasani R., Kim S., Tan M.P., Huang Y., Omenetto F.G., Rogers J.a., Bruchas M.R. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science. 2013;340:211–216. doi: 10.1126/science.1232437. 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pongracz A., Fekete Z., Márton G., Bérces Z., Ulbert I., Fürjes P. Deep-brain silicon multielectrodes for simultaneous in vivo neural recording and drug delivery. Sensor. Actuator. B Chem. 2013;189:97–105. [Google Scholar]
- 109.Wang Y.I., Abaci H.E., Shuler M.L. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017;114:184–194. doi: 10.1002/BIT.26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vatine G.D., Barrile R., Workman M.J., Sances S., Barriga B.K., Rahnama M., Barthakur S., Kasendra M., Lucchesi C., Kerns J., Wen N. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell. 2019;24:995–1005. doi: 10.1016/j.stem.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 111.Walter F.R., Valkai S., Kincses A., Petneházi A., Czeller T., Veszelka S., Ormos P., Deli M.A., Dér A. Elsevier B.V.; 2016. A Versatile Lab-On-A-Chip Tool for Modeling Biological Barriers. [DOI] [Google Scholar]
- 112.Wang X., Hou Y., Ai X., Sun J., Xu B., Meng X., Zhang Y., Zhang S. Potential applications of microfluidics based blood brain barrier (BBB)-on-chips for in vitro drug development, Biomed. Pharma. 2020;132:110822. doi: 10.1016/j.biopha.2020.110822. [DOI] [PubMed] [Google Scholar]
- 113.Egert U., Schlosshauer B., Fennrich S., Nisch W., Fejtl M., Knott T., Müller T., Hämmerle H. A novel organotypic long-term culture of the rat hippocampus on substrate-integrated multielectrode arrays. Brain Res. Protoc. 1998;2:229–242. doi: 10.1016/s1385-299x(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 114.Lee H.J., Son Y., Kim J., Lee C.J., Yoon E.S., Cho I.J. A multichannel neural probe with embedded microfluidic channels for simultaneous in vivo neural recording and drug delivery. Lab Chip. 2015;15:1590–1597. doi: 10.1039/c4lc01321b. [DOI] [PubMed] [Google Scholar]
- 115.Shin H., Lee H.J., Chae U., Kim H., Kim J., Choi N., Woo J., Cho Y., Lee C.J., Yoon E.S., Cho I.J. Neural probes with multi-drug delivery capability. Lab Chip. 2015;15:3730–3737. doi: 10.1039/c5lc00582e. [DOI] [PubMed] [Google Scholar]
- 116.Nersesyan H., Hyder F., Rothman D.L., Blumenfeld H. Dynamic fMRI and EEG recordings during spike-wave seizures and generalized tonic-clonic seizures in WAG/rij rats. Journals.Sagepub.Com. 2004;24:589–599. doi: 10.1097/01.WCB.0000117688.98763.23. [DOI] [PubMed] [Google Scholar]
- 117.Kim D., Song I., Keum S., Lee T., Jeong M.J., Kim S.S., McEnery M.W., Shin H.S. Lack of the burst firing of thalamocortical relay neurons and resistance to absence seizures in mice lacking α1G T-type Ca2+ channels. Neuron. 2001;31:35–45. doi: 10.1016/s0896-6273(01)00343-9. [DOI] [PubMed] [Google Scholar]
- 118.Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. 80. [DOI] [PubMed] [Google Scholar]
- 119.Sun Y., Pollard S., Conti L., Toselli M., Biella G., Parkin G., Willatt L., Falk A., Cattaneo E., Smith A. Long-term tripotent differentiation capacity of human neural stem (NS) cells in adherent culture. Mol. Cell. Neurosci. 2008;38:245–258. doi: 10.1016/j.mcn.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 120.Lin C.H., Hsiao Y.H., Chang H.C., Yeh C.F., He C.K., Salm E.M., Chen C., Chiu I.M., Hsu C.H. A microfluidic dual-well device for high-throughput single-cell capture and culture. Lab Chip. 2015;15:2928–2938. doi: 10.1039/C5LC00541H. [DOI] [PubMed] [Google Scholar]
- 121.Reubinoff B.E., Pera M.F., Fong C.Y., Trounson A., Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 122.Ilkhanizadeh S., Teixeira A.I., Hermanson O. Biomaterials; 2007. Inkjet Printing of Macromolecules on Hydrogels to Steer Neural Stem Cell Differentiation. [DOI] [PubMed] [Google Scholar]
- 123.Lee D.H., Koo D.B., Ko K., Ko K., Kim S.M., Jung J.U., Ryu J.S., Jin J.W., Yang H.J., Do S.I., Jung K.Y. Effects of daunorubicin on ganglioside expression and neuronal differentiation of mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2007;362:313–318. doi: 10.1016/j.bbrc.2007.07.142. [DOI] [PubMed] [Google Scholar]
- 124.Su H., Chu T.H., Wu W. Lithium enhances proliferation and neuronal differentiation of neural progenitor cells in vitro and after transplantation into the adult rat spinal cord. Exp. Neurol. 2007;206:296–307. doi: 10.1016/j.expneurol.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 125.Nakashima Y., Yasuda T. Cell differentiation guidance using chemical stimulation controlled by a microfluidic device. Sensors Actuators A Phys. 2007;139:252–258. [Google Scholar]
- 126.Lee J.M., Kim J.E., Borana J., Chung B.H., Chung B.G. Dual-micropillar-based microfluidic platform for single embryonic stem cell-derived neuronal differentiation. Electrophoresis. 2013;34:1931–1938. doi: 10.1002/ELPS.201200578. [DOI] [PubMed] [Google Scholar]
- 127.Bieberich E., Guiseppi-Elie A. Neuronal differentiation and synapse formation of PC12 and embryonic stem cells on interdigitated microelectrode arrays:: contact structures for neuron-to-electrode signal transmission (NEST) Biosens. Bioelectron. 2004;19:923–931. doi: 10.1016/j.bios.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 128.Zhao Y., Liu Q., Sun H., Chen D., Li Z., Fan B., George J., Xue C., Cui Z., Wang J., Chen J. Electrical property characterization of neural stem cells in differentiation. PLoS One. 2016;11 doi: 10.1371/JOURNAL.PONE.0158044. [DOI] [PMC free article] [PubMed] [Google Scholar]